Abstract
Background
Colorectal cancer is one of the most common cancers and the third leading cause of cancer death in both sexes. The disease progresses as a multistep process and is associated with genetic alterations. One of the characteristic features of cancer is telomerase activation. We sought to evaluate the differences in telomerase activity between colon cancer and adjacent normal tissue and to correlate the differences in telomerase activity between different locations with clinicopathological factors and survival.
Methods
Matched colon tumour samples and adjacent normal mucosa samples 10 cm away from the tumour were collected during colectomy. We assessed telomerase activity using real time polymerase chain reaction. Several pathological characteristics of tumours, including p53, Ki-67, p21, bcl2 and MLH1 expression were also studied.
Results
We collected samples from 49 patients. There was a significantly higher telomerase activity in colon cancer tissue than normal tissue. Adenocarcinomas of the right colon express significantly higher telomerase than left-side cancers. Colon cancers and their adjacent normal tissue had significantly more telomerase and were more positive to MLH1 than rectal cancers. The expression of p53 negatively correlated to telomerase activity and was linked to better patient survival.
Conclusion
Colon and rectal cancers seem to have different telomerase and MLH1 profiles, and this could be another factor for their different biologic and clinical behaviour and progression. These results support the idea that the large bowel cannot be considered a uniform organ, at least in the biology of cancer.
Abstract
Contexte
Le cancer colorectal est l’un des cancers les plus répandus; il arrive au troisième rang des décès attribuables au cancer chez les hommes et les femmes. La maladie progresse en plusieurs étapes et est associée à des anomalies génétiques. L’une des principales caractéristiques du cancer est l’activation de la télomérase. Nous avons voulu évaluer les différences d’activité de la télomérase entre les tissus touchés par le cancer du côlon et les tissus adjacents normaux afin d’établir une corrélation entre les différences d’activité de la télomérase selon la localisation d’une part et les facteurs clinicopathologiques et la survie d’autre part.
Méthodes
Lors de colectomies, nous avons recueilli des échantillons tissulaires jumelés de tumeur du côlon et de muqueuse adjacente normale à 10 cm du foyer tumoral. Nous avons mesuré l’activité de la télomérase à l’aide de la réaction en chaîne de la polymérase en temps réel. Plusieurs caractéristiques pathologiques des tumeurs ont aussi été analysées, y compris l’expression des gènes p53, Ki-67, p21, bcl2 et MLH1.
Résultats
Nous avons recueilli des échantillons auprès de 49 patients. Nous avons noté une activité nettement plus intense de la télomérase dans les tissus touchés par le cancer du côlon que dans les tissus normaux. Les adénocarcinomes du côlon ascendant expriment une activité de la télomérase significativement plus intense que les cancers du côlon descendant. Les cancers du côlon et les tissus adjacents normaux présentaient une activité significativement plus intense de la télomérase et étaient plus souvent positifs à l’égard du MLH1 comparativement aux cancers rectaux. L’expression du p53 a été en corrélation négative avec l’activité de la télomérase et a été associée à une meilleure survie chez les patients.
Conclusion
Les cancers du côlon et du rectum semblent avoir des profils différents au plan de la télomérase et du gène MLH1. Ce facteur pourrait entre autre expliquer leur comportement et leur progression biologiques et cliniques distincts. Ces résultats appuient l’hypothèse selon laquelle le côlon ne peut être considéré comme un organe uniforme, du moins en ce qui concerne la biologie du cancer.
Cancers of the colon and rectum have been classically viewed as the same disease entity, namely colorectal cancer (CRC), one of the most common cancers in humans and the third leading cause of cancer death. However, more than 2 decades ago, Bufill1 proposed the existence of 2 distinct categories of CRC according to the location of the tumour in the proximal or distal segments of the colon. Comparison of the risk factors of proximal and distal CRC revealed that type 2 diabetes was associated with distal CRC in men while cholelithiasis was associated with proximal colon cancer in women.2 Patients with tumours located within the proximal colon have twice the risk for metachronous CRC than patients with distal initial disease,3 and a higher incidence of advanced disease has been reported.4
Moreover, several pathological molecular differences between proximal and distal disease have been demonstrated. The mucinous histotype was more frequent in the proximal than in the distal CRCs.5 Two forms of genetic instability have been described in patients with CRC: chromosomal instability (CIN) and microsatellite instability (MSI). However, CIN has been more commonly described in the distal colon than MSI, which is more common with proximal sporadic colon cancers.5,6 Mutations in p53 are more frequently associated with distal cancers, whereas Ki-ras mutations are more frequent in proximal tumours.7 These different characteristics may indicate diverse genetic pathways of carcinogenesis and support the hypothesis of distinct mechanisms of neoplastic transformation in the proximal and distal colon, with potential implications in the therapeutic approach. As a consequence, it has been suggested that CRC should be divided into proximal colon cancer, distal colon cancer and rectal cancer.8
Malignant transformation during colon cancer development arises as a result of the accumulation of genetic alterations and epigenetic alterations in a multistep process, including the activation of oncogenes, the inactivation of tumour suppressor genes and uncontrolled mitogenic stimulation that transforms colonic epithelial cells to colon adenocarcinoma cells.9 Throughout this mutation process, telomerase activation plays a catalytic role in the tumour progression cascade. Human telomeres function as a protective structure capping both ends of the chromosome and act as an intrinsic “counting” mechanism of the aging cellular process. Thus, telomeres limit the capacity of a cell to replicate by inducing senescence as a sort of tumour-suppressing mechanism.10 Telomerase is an enzymatic ribonucleoprotein complex that acts as a reverse transcriptase and is the main positive regulator of telomere length. Telomerase activity is the most general molecular marker for the identification of human cancer and can be detected in 85%–90% of all tumours.11 Although telomerase activity has been detected in almost 95% of patients with CRC, many discrepancies exist, and telomerase activity between the different anatomic locations of CRC has not been studied adequately.
The aim of this study, therefore, was to evaluate the telomerase activity in colon and rectal cancer tissues and their corresponding adjacent normal mucosal tissues from patients with colorectal adenocarcinoma. We compared telomerase activity in tumours located in different colon locations and analyzed the association between telomerase activity and patient clinicopathological features and survival. We also studied MLH1 as an indicator of MSI, while p53 and other molecular markers were also studied and correlated with telomerase activity.
Methods
Patients
Between July 2002 and October 2004, we prospectively recruited patients with biopsy-confirmed CRC for participation in this study. The study was approved by our local ethics committee, and we obtained written informed consent from all participants.
At the time of surgery, once the tumour was resected, 4 cancer tissue samples and 4 apparently normal mucosal samples about 10 cm distal to the resection margin were sampled and snap frozen in liquid nitrogen within 10 minutes of resection and then stored at −80°C until studied. The rest of the specimen was sent for standard pathological evaluation. Further analysis on tumour prognostic index markers was completed, and we studied the expression of Ki-67, p21, bcl2 and p53 for all patients.
Patients were followed until April 2011. All patients were referred to the oncology department for standard treatment and follow-up, and no patient received neoadjuvant therapy. We assessed 1-, 3- and 5-year survival and overall survival.
Telomerase activity
We determined telomerase activity using a real-time polymerase chain reaction (rtPCR) assay (Quantitative Telomerase Detection [QTD] kit, US Biomax) according to the manufacturer’s instructions. Tissue samples (20 ± 2 mg) were washed with phosphate buffered saline and homogenized with a glass homogenizer in 1 × lysis buffer. After 30 minutes of incubation on ice, the lysates were centrifuged at 12 000g for 30 minutes at 4°C, and the supernatant was rapidly frozen and stored at −80°C. We used part of this extract to determine the total protein concentration with the Bradford protein assay (Bio-Rad Laboratories) and bovine serum albumin as a standard. The rest of this extract was diluted with lysis buffer to a concentration of 1 μg/μL. We used the diluted sample as a template for the rtPCR assay. Measuring the increase in fluorescence caused by the binding of SYBR Green I dye to double-stranded DNA monitored direct detection of the PCR product. The rtPCR master mix comprised 12.5 μL of the QTD premix containing telomere primers, 1 μL of tissue extract, template controls or heat-inactivated extracts and 11.5 μL of water, for a final volume of 25 μL. After a 20-minute incubation at 25°C, telomere templates were formed by adding 6 base repeats to the primer with the activity of telomerase. The reaction mixture was heated at 90°C for 10 minutes to activate the HotStart DNA polymerase and then subjected to 40 PCR cycles at 95°C for 30 seconds, 60°C for 30 seconds and 72°C for 30 seconds. In order to verify specificity and identity of the PCR products, the reaction ended using a melt curve analysis in which the temperature was increased from 55°C to 95°C at a linear rate of 0.2°C/second. Data collection was performed both during annealing and extension, with 2 measurements at each step, and at all times during melt curve analysis. We conducted all PCR experiments on the Mx3000P rtPCR thermal cycler using the software version 2.00 (Stratagene). For estimation of telomerase activity, we used a positive control (TSR template control) to generate a standard curve, consisting of 8 serial dilutions ranging from 0.5 μg/μL (300 000 molecules/reaction) to 6.4 × 10−6 μg/μL. The analysis of each sample consisted of 2 assays: one with a sample extract and one with a heat-treated sample extract, which served as a negative control since telomerase is a heat-sensitive enzyme. Control samples were incubated at 85°C for 10 minutes before the telomerase activity assay. To control for mosaicism, all 4 samples were analyzed in 5 patients at the initial phase of the study. Since no significant difference was found among the samples in telomerase activity we continued with the analysis of a single sample (cancer and normal tissue) per patient.
Pathology
For every patient, we selected 1 paraffin block with tumour tissue for the immunohistochemical detection of MLH1, Ki-67, bcl2, p53 and p21 protein expression. We used 4 μm sections from paraffin-embedded tissue blocks of primary tumour specimens. We then performed immunoperoxidase staining in 3 steps using a Dako kit. First, the specimens were dewaxed in xylene and rehydrated with graded alcohols. Endogenous peroxidase was blocked with H2O2 0.3% in Tris buffer (pH 7.60) for 15 minutes. Before application of the primary antibody, sections were immersed in 10 mM citrate buffer, rinsed in Tris-buffered saline (TBS) and heated in a microwave oven (650–800 W) for 30 minutes. In order to reduce nonspecific binding of antisera, sections were washed with TBS before application of the primary antibodies: anti-human MutL protein homologue-1, clone E 505 (ready for use, Dako), anti-Ki-67 (MIB-1Ab, dilution 1:80 Dako), anti-p53 (DO-7, dilution 1:100 Biogenex), anti p21 (dilution 1:40 Dako) and anti-bcl2 (dilution 1:10, Biogenex). We used diaminobenzidine as a chromogen followed by slight hematoxylin counterstaining. Tumours with known Ki-67, p53 and p21 status were used as positive controls, whereas a normal lymph node served the same purpose for bcl2. Preimmune rabbit serum was used as a negative control to test for nonspecific staining.
Two pathologists, blinded to clinical information, independently evaluated immunoreactivity by assessing the percentage of positively immunostained tumour cells. Discrepancies were resolved by consensus. Specifically, MLH1 expression was scored as positive if 10% of cells were found positive. Ki-67 expression was scored as low if less than 20% of cells were found positive, moderate if 20%–50% were positive and high if more than 50% were positive. We classified p53 and p21 expression as negative if less than 5% of cells were positive, low if 5%–30% were positive, moderate if 31%–60% were positive and high if more than 60% were positive. Immunoreactivity for bcl2 was evaluated according to the percentage of tumour cells with positive cytoplasmic staining. We used a cut-off below 5% of positive tumour cells to define negative cases. Strong positive staining was seen in infiltrating lymphocytes within the tumour stroma.
The selection of the cutoff values for each particular marker was based on corresponding categorization from other studies. The chosen thresholds were previously found as best showing prognostic clinicopathological and molecular correlations of the examined markers.12–14
Statistical analysis
Statistical analysis was performed with the IBM SPSS statistics software version 19. Results are expressed as means ± standard errors of the mean. Paired and unpaired t tests or 1-way analysis of variance with Bonferroni post hoc analysis were used where applicable. We performed the χ2 test for the analysis of nonparametric data in 2 × 2 tables. Kaplan–Meyer plots were generated based on overall patient survival. We used Pearson and Spearman correlation as applicable for analysis of correlations.
Results
Patients
We included 49 patients (21 women) with a median age of 74 (range 49–87) years with biopsy-confirmed CRC in our study. The demographic, clinical and pathological characteristics of participants are presented in Table 1. Twelve patients were lost during follow-up. For the remaining 37 patients, 1-, 3- and 5-year survival and overall survival were reviewed.
Table 1.
Patient demographic, clinical and pathological characteristics
| Characteristic | No. |
|---|---|
| Patients | |
| Cancer tissues | 49 |
| Adjacent normal tissues | 49 |
| Sex | |
| Male | 28 |
| Female | 21 |
| Smoking | |
| Yes | 12 |
| No | 34 |
| BMI | |
| < 25 | 21 |
| ≥ 25 | 25 |
| Dukes staging system | |
| B | 25 |
| C | 19 |
| D | 5 |
| Histology grade | |
| High | 4 |
| Medium | 41 |
| Low | 4 |
| Tumour location | |
| Right colon | 8 |
| Left colon | 24 |
| Rectum | 17 |
BMI = body mass index.
Telomerase activity in tumours compared with normal adjacent tissue
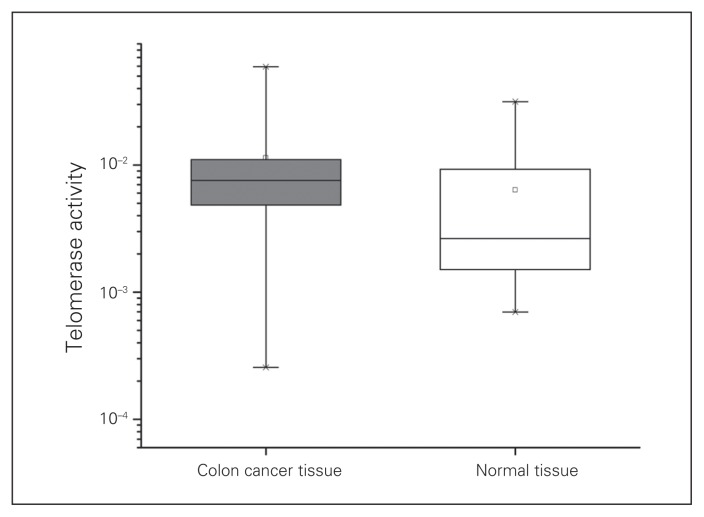
We identified telomerase activity in all tumour samples and in all their adjacent normal mucosa samples. The mean value of telomerase activity copies was significantly higher in CRC samples than the corresponding adjacent normal tissue (1.14 × 10−2/μg of protein v. 0.64 × 10−2, p = 0.006; Fig. 1). A total of 83% of the colorectal tumour samples were found to express higher telomerase activity, while only 17% expressed less telomerase activity than matched adjacent mucosa. Interestingly, only 57% of the patients whose tumour samples expressed less telomerase activity than the adjacent mucosa experienced loss of MLH1 expression, whereas 35% of patients whose tumour samples expressed higher telomerase activity had MLH1 loss; however, the numbers were too small to reach statistical significance.
Fig. 1.
Telomerase activity in colorectal cancer tissues and corresponding adjacent normal tissues in 49 patients with colon cancer (horizontal line: median; box: mean; □: 1%–99% range).
Telomerase activity and clinicopathological features
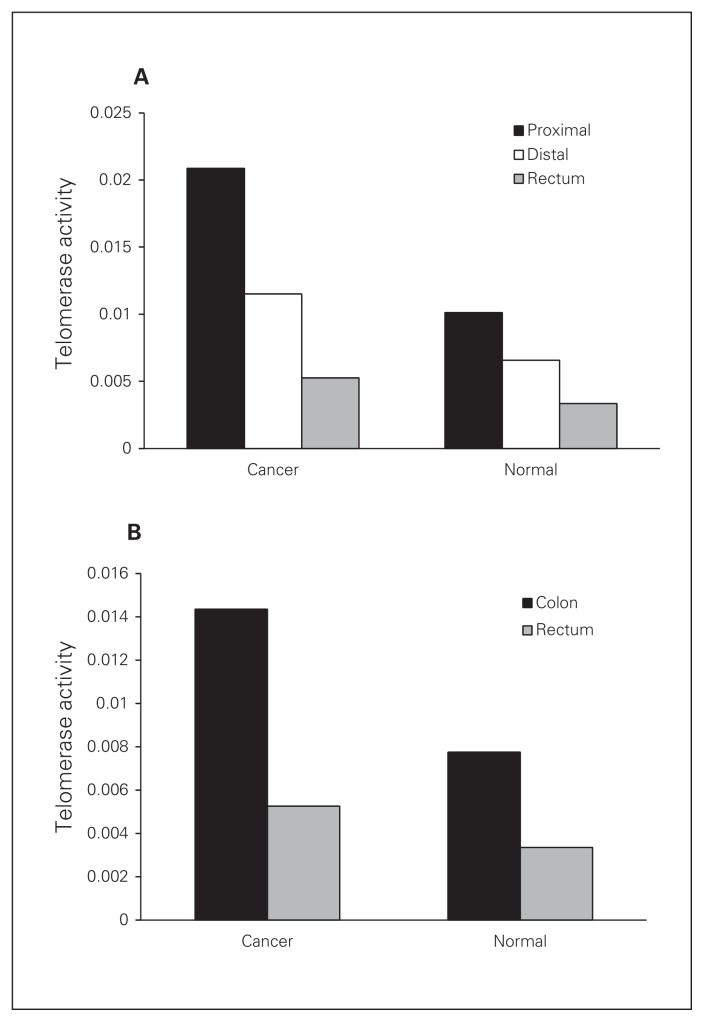
When we analyzed the associations between clinicopatho-logical factors and expression level of telomerase activity, we found no significant difference in relation to sex, age, smoking history, body mass index and histology grade (Table 2). Interestingly, normal adjacent tissue of Dukes stage A/B tumours had significantly higher telomerase activity than normal adjacent tissue of stage C/D tumours (p = 0.005). A similar result was obtained when tumours were analyzed based on the tumour-node-metastasis (TNM) system, with stage I/II tumours having higher telomerase activity than stage III/IV tumours (p = 0.029). Table 2 shows the correlation between telomerase activity expression level and clinicopathological parameters for the patients with CRC. When tumours were classified as right-sided (cecum, ascending colon, transverse colon) and left-sided (splenic flexure, descending colon, sigmoid, rectum), we found that right-sided cancers expressed higher telomerase activity than the left-sided cancers. When tumours were grouped by right colon (cecum, ascending colon, transverse colon), left colon (splenic flexure, descending colon, sigmoid) and rectal cancers, we observed a gradual, significant decrease in human telomerase reverse transcriptase (hTERT) activity (Fig. 2A), with the rectal tumours having the lowest hTERT activity that was significantly different from the proximal tumours. Finally when tumours were grouped as colon or rectal cancers, we observed a significantly higher level of telomerase activity in colon than rectal tumours (p = 0.012). The same increase was evident when normal tissue of patients with colon cancer was compared with normal tissue of those with rectal cancer (p = 0.035; Fig. 2B).
Table 2.
Correlation between telomerase activity expression and clinicopathological parameters for patients with colorectal cancer
| Variable | Normal mucosa | Cancer tissue | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| No. | Mean ± SD | p value | No. | Mean ± SD | p value | |
| Sex | 0.52 | 0.66 | ||||
|
| ||||||
| Male | 28 | 0.0126 ± 0.0037 | 28 | 0.0055 ± 0.0014 | ||
|
| ||||||
| Female | 21 | 0.0105 ± 0.0029 | 21 | 0.0070 ± 0.0019 | ||
|
| ||||||
| Smoke | 0.88 | 0.72 | ||||
|
| ||||||
| Yes | 19 | 0.0118 ± 0.0026 | 19 | 0.0066 ± 0.0017 | ||
|
| ||||||
| No | 30 | 0.0126 ± 0.0060 | 30 | 0.0055 ± 0.0019 | ||
|
| ||||||
| BMI | 0.09 | 0.34 | ||||
|
| ||||||
| < 25 | 22 | 0.0073 ± 0.0012 | 22 | 0.0052 ± 0.0011 | ||
|
| ||||||
| ≥ 25 | 27 | 0.0151 ± 0.0040 | 27 | 0.0075 ± 0.0021 | ||
|
| ||||||
| Dukes stage | 0.57 | 0.011 | ||||
|
| ||||||
| A | 0 | — | 0 | — | ||
|
| ||||||
| B | 25 | 0.0140 ± 0.0039 | 25 | 0.0104 ± 0.023 | ||
|
| ||||||
| C | 19 | 0.0095 ± 0.0035 | 19 | 0.0040 ± 0.0012 | ||
|
| ||||||
| D | 5 | 0.0098 ± 0.0010 | 5 | 0.0024 ± 0.0008 | ||
|
| ||||||
| A+B | 25 | 0.0140 ± 0.0039 | 0.34 | 25 | 0.0103 ± 0.0024 | 0.005 |
|
|
|
|||||
| C+D | 24 | 0.00096 ± 0.0028 | 24 | 0.0037 ± 0.0044 | ||
|
| ||||||
| TNM stage | ||||||
|
| ||||||
| I | 6 | 0.0027 ± 0.0009 | 6 | 0.0047 ± 0.0016 | ||
|
| ||||||
| II | 19 | 0.0114 ± 0.0021 | 19 | 0.0155 ± 0.0035 | ||
|
| ||||||
| III | 18 | 0.0040 ± 0.0012 | 18 | 0.0106 ± 0.0035 | ||
|
| ||||||
| IV | 6 | 0.0034 ± 0.0010 | 6 | 0.0081 ± 0.0017 | ||
|
| ||||||
| I–II | 25 | 0.0097 ± 0.0018 | 0.029 | 25 | 0.0133 ± 0.0028 | 0.18 |
|
|
|
|||||
| III–IV | 24 | 0.0038 ± 0.0009 | 24 | 0.0099 ± 0.0026 | ||
|
| ||||||
| Grade | 0.89 | 0.21 | ||||
|
| ||||||
| Low | 4 | 0.0096 ± 0.0039 | 4 | 0.0123 ± 0.0045 | ||
|
| ||||||
| Moderate | 41 | 0.0120 ± 0.0028 | 41 | 0.0057 ± 0.0013 | ||
|
| ||||||
| High | 4 | 0.0088 ± 0.0018 | 4 | 0.0045 ± 0.0024 | ||
|
| ||||||
| Tumour site | 0.09 | 0.20 | ||||
|
| ||||||
| Right | 8 | 0.0209 ± 0.089 | 8 | 0.0101 ± 0.0036 | ||
|
| ||||||
| Left | 7 | 0.0092 ± 0.0015 | 7 | 0.0081 ± 0.0032 | ||
|
| ||||||
| Sigmoid | 16 | 0.0133 ± 0.0040 | 16 | 0.0054 ± 0.0016 | ||
|
| ||||||
| Rectum | 18 | 0.0053 ± 0.0012 | 18 | 0.0034 ± 0.0012 | ||
|
| ||||||
| Total* | 49 | 0.0114 ± 0.0023 | 49 | 0.0064 ± 0.0012 | ||
|
| ||||||
| Right colon | 15 | 0.0151 ± 0.0046 | 0.09 | 15 | 0.0092 ± 0.0024 | 0.043 |
|
|
|
|||||
| Left colon | 34 | 0.0089 ± 0.0092 | 34 | 0.0043 ± 0.0010 | ||
|
| ||||||
| Colon | 31 | 0.0144 ± 0.0032 | 0.012 | 31 | 0.0078 ± 0.0016 | 0.035 |
|
|
|
|||||
| Rectum | 18 | 0.0053 ± 0.0012 | 18 | 0.0034 ± 0.0012 | ||
|
| ||||||
| Right colon | 8 | 0.0209 ± 0.0089 | 0.047 | 8 | 0.0101 ± 0.0036 | 0.13 |
|
|
|
|||||
| Left colon | 23 | 0.0115 ± 0.0023 | 23 | 0.0066 ± 0.0016 | ||
|
|
|
|||||
| Rectum | 18 | 0.0053 ± 0.0012 | 18 | 0.0033 ± 0.0012 | ||
|
| ||||||
| Ki-67 expression | 0.022 | 0.66 | ||||
|
| ||||||
| Negative | 3 | 0.0033 ± 0.0018 | — | 0.0050 ± 0.0025 | ||
|
| ||||||
| Positive | 46 | 0.0119 ± 0.0024 | — | 0.0065 ± 0.0013 | ||
|
| ||||||
| p53 expression | 0.12 | 0.037 | ||||
|
| ||||||
| Negative | 16 | 0.0173 ± 0.0058 | — | 0.0103 ± 0.0033 | ||
|
| ||||||
| Positive | 33 | 0.0093 ± 0.0022 | — | 0.0048 ± 0.0010 | ||
BMI = body mass index; SD = standard deviation; TNM = tumour-node-metastasis.
p = 0.006 for cancer versus normal p value.
Fig. 2.
(A) Telomerase activity between right colon (cecum, ascending colon, transverse colon), left colon (splenic flexure, descending colon, sigmoid) and rectal cancers and their adjacent normal mucosa tissues (p = 0.006). (B) Comparison of telomerase activity between colon and rectal cancers and their adjacent normal mucosa tissues (p = 0.012 for colon v. rectal cancer, and p = 0.035 for normal tissue adjacent to colon v. rectal cancer). Results are means ± standard errors of the mean
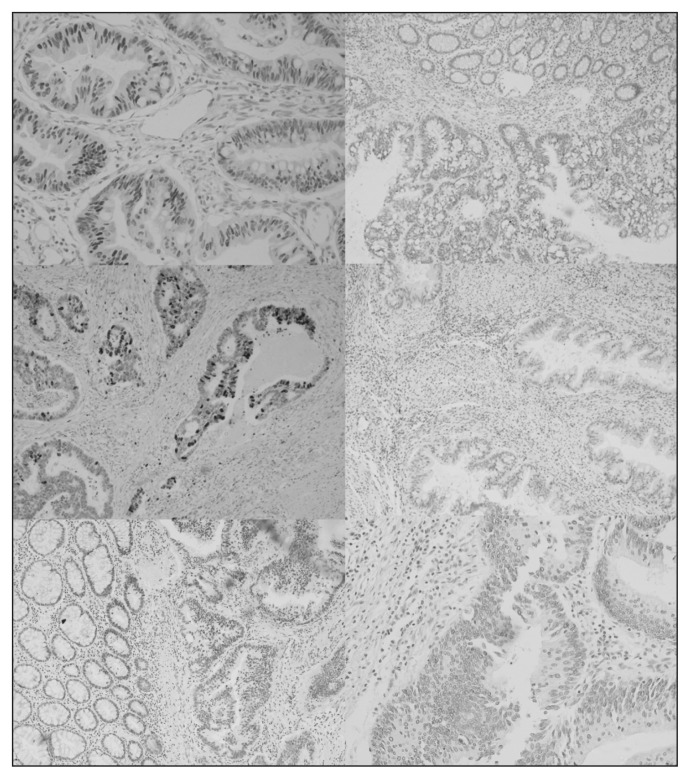
We found that p53-negative tumours had higher telomerase activity than p53-positive tumours, but this result showed a trend toward significance (0.0173 ± 0.0058 v. 0.0093 ± 0.0022 telomerase activity copies, p = 0.06). Interestingly, p53 expression in the cancer tissue negatively correlated with the telomerase activity in the adjacent normal tissue (p = 0.003). Furthermore, colon cancers were p53-positive in 56.3% of patients, while rectal cancers were more commonly positive for p53 (88.2%, χ2 = 5.165, p = 0.022). Typical p53 immunostaining is shown in Fig. 3. There was no correlation between telomerase activity and tumour localization with Ki-67, bcl2 or p21 expression (Table 3).
Fig. 3.
Colorectal cancer specimens with typical immunostainings. (Top left) highly positive nuclear p53 immunoreactivity (×200) compared with (top right) a negative tumour. (Middle left) positive nuclear p21 immunoreactivity (×200) compared with a (middle right) a negative sample. (Bottom left) positive MLH1 Immunoreactivity (×200) compared with (bottom right) a negative sample.
Table 3.
Immunohistochemistry results for p53, bcl2, Ki-67 and p21 in colon and rectal cancer tissues
| Marker | Group; positive tissues, no. (%) | p value | |
|---|---|---|---|
| Colon cancer, n = 32 | Rectal cancer, n = 17 | ||
| p53 | 18 (56.3) | 15 (88.2) | 0.023 |
| bcl2 | 7 (21.9) | 6 (35.3) | 0.25 |
| Ki-67 | 29 (90.6) | 17 (100) | 0.27 |
| p21 | 17 (53.1) | 10 (58.8) | 0.47 |
Microsatellite instability
Eighteen patients experienced loss of MLH1 expression, indicative of MSI. There was no difference in the levels of telomerase activity between MLH1-positive and negative samples (Table 4). Furthermore there were no significant correlations between MLH1 loss and age, sex and smoking history. Colon cancer samples displayed loss of MLH1 only in 8 of 32 patients (25%), whereas rectal cancer samples indicated loss of MLH1 in 10 of 17 patients (59%; p = 0.029). There was no correlation between MLH1 expression and p53, p21 and bcl2 expression by tumour samples, whereas, surprisingly, there was a correlation between MLH1 expression and expression of high Ki-67 levels.
Table 4.
Correlation of MLH1 expression with telomerase activity and clinicopathological features in patients with colon cancer
| Factor | MLH1, negative | MLH1, positive | p value |
|---|---|---|---|
| Telomerase activity | 0.0125 ± 0.0028 | 0.0088 ± 0.0039 | 0.47 |
| Age, yr | 71.71 | 70.28 | 0.62 |
| Sex | 0.45 | ||
| Male | 11 | 17 | |
| Female | 7 | 11 | |
| Smoke | 0.69 | ||
| Yes | 3 | 9 | |
| No | 14 | 22 | |
| Duke stage | 0.048* | ||
| A | — | — | |
| B | 13 | 12 | |
| C | 5 | 14 | |
| D | 1 | 4 | |
| TNM stage | 0.019† | ||
| I | 4 | 2 | |
| II | 9 | 10 | |
| III | 4 | 14 | |
| IV | 1 | 5 | |
| Tumour site | 0.09 | ||
| Right | 1 | 7 | |
| Left | 3 | 5 | |
| Sigmoid | 4 | 12 | |
| Rectum | 10 | 7 | |
| Proximal | 4 | 12 | 0.12 |
| Distal | 14 | 19 | |
| Colon | 8 | 24 | 0.029 |
| Rectum | 10 | 7 | |
| Right | 1 | 7 | 0.046 |
| Left | 7 | 17 | |
| Rectum | 10 | 7 | |
| p53 expression | 0.34 | ||
| Positive | 12 | 22 | |
| Negative | 6 | 8 | |
| bcl2 expression | 0.57 | ||
| Positive | 5 | 8 | |
| Negative | 14 | 22 | |
| Ki-67 expression | 0.033‡ | ||
| Low | 4 | 3 | |
| Moderate | 9 | 8 | |
| High | 5 | 18 | |
| p21 expression | 0.40 | ||
| Positive | 9 | 17 | |
| Negative | 9 | 14 |
TNM = tumour-node-metastasis.
χ2 test for trend p < 0.05; the group had fewer than 5 patients.
χ2 test for trend p < 0.02; the group had fewer than 5 patients.
χ2 test for trend p = 0.033; the group had fewer than 5 patients.
Correlations with patient survival
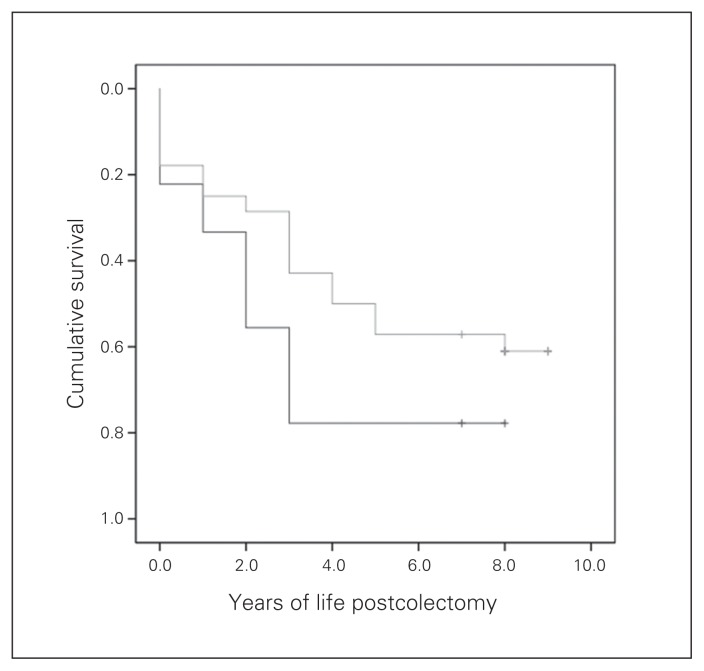
In our study group, 1-, 3- and 5-year survival was 85.7%, 65.3% and 55.1%, respectively. There was no correlation between telomerase activity and 1-, 3- or 5-year survival. There was, however, a trend toward a negative correlation between p53 expression and survival, and p53-positive patients tended to survive longer than p53-negative patients (Fig. 4). The p53-positive patients were estimated to live an average of 5.0 (95% confidence interval [CI] 3.7–6.4) years, and p53-negative patients were estimated to live an average of 3.0 (95% CI 1.1–4.9) years; but we did not consider these results to be significant owing to the small number of patients in each group. There was no correlation between bcl2, p21, Ki-67 and MLH1 expression and patient survival.
Fig. 4.
Kaplan–Meyer plot of survival of patients with p53-positive (grey line) and p53-negative (black line) tumours.
Discussion
The aim of this study was to identify the role of telomerase activity levels in the prognosis and localization of CRC and try to identify clinicopathological factors related to its levels. We have demonstrated that telomerase activity is increased in colon cancer tissue compared with adjacent normal tissue and that there are differences between right and left colon cancers. Moreover, there was an inverse association between telomerase and p53 expression. Although telomerase activity did not correlate with patient survival, p53 expression in colon cancer samples was linked to increased patient survival.
Telomeres are TTAGGG tandem repeated sequences of DNA present at the end of chromosomes that are important for ensuring the complete replication of chromosomal ends and for protecting chromosomal termini from fusion and degradation.15 After a finite number of replications, the telomere reaches a critical length, the senescence checkpoint, and either cell-proliferation arrest or cell apoptosis and death occurs (physiologic aging). However, if the cell escapes or avoids this checkpoint, CIN or end-to-end fusion of the chromosome occurs, which may contribute to cell death or carcinogenesis.16 Telomere length is normally maintained by an enzyme called telomerase. The telomerase complex consists of hTERT, telomerase associate protein (TP1) and an RNA template for telomeric DNA synthesis (hTR).17–19 Human telomerase reverse transcriptase is usually considered to reflect telomerase activity and telomere length. Telomere length and hTERT expression were significantly correlated in normal and cancerous colon tissue.20,21 However, telomerase activity does not always correlate with hTERT in colon cancer22 possibly because of the presence of hTERT in infiltrating lymphocytes in normal mucosa. Therefore, the measurement of hTERT alone may overestimate the actual presence of telomerase within both normal and cancerous bowel epithelial cells.23
Reports on telomerase activity and expression in non-cancerous tissue seem to be controversial. A number of studies have revealed that telomerase activity is almost absent in normal somatic cells16 and that it is highly expressed in almost all human tumours but not in adjacent normal cells.24,25 On the other hand, other investigators report that telomerase activity is invariably present in all normal human cells and can be considered as a marker of cellular proliferation.26–28 Results on the human colon are also conflicting. No hTERT was found in normal mucosa in patients with colon cancer, a result opposite to ours.29,30 In accordance with our study, Nowak and colleagues27 found telomerase activity in normal mucosa taken 5 cm from the tumour, and the levels were higher than those of mucosa taken 10 cm from the tumour. This may be because of the presence of residual cancer cells or tumour-activated normal cells closer to the tumour or to the improvement of the sensitivity of the molecular techniques used for the study of telomerase activity in recent years.
Results of reported studies on telomerase levels in cancerous tissues compared with normal tissues are also controversial. Cancer tissue has been reported to have lower telomerase than the corresponding normal colonic tissue;20,21 however, in a series31 of 53 patients with colorectal carcinomas and 9 patients with adenomas, patients were reported to have higher levels of hTERT in cancerous than normal tissue. The study reported no difference between cancer and adenomas.31 Moreover, in the largest series published so far, hTERT mRNA expression was significantly higher in cancerous tissues than adenomas or normal tissue of 140 patients.32 Further support comes from studies in patients at high risk for CRC because of long-standing inflammatory bowel disease. Overexpression of hTERT in nonaffected colorectal mucosa has been reported in patients with inflammatory bowel disease.33,34 Especially in those with ulcerative colitis, hTERT overexpression has been associated with induction of CIN,34 possibly an effect of oxidative damage secondary to inflammation.35 Our results are in accordance with those of most of the aforementioned studies, as we found telomerase activity to be present in all normal colon tissue samples and in colon cancer tissue samples. Unfortunately, we did not have adenoma samples that could further enlighten the differences in telomerase activity and MLH1 expression in the various stages of CRC carcinogenesis.
Recently, Rampazzo and colleagues10 have studied the association between telomere shortening, genetic instability and site of tumour origin in patients with CRC.10 They found that telomeres were significantly shorter in CRCs than adjacent normal tissue and that telomere length did not differ with tumour progression or p53 status. Interestingly, the telomere length was different depending on tumour location, with rectal cancers having the longest telomeres, but telomerase expression did not differ with tumour location. Rampazzo and colleagues10 did not study telomerase activity. Although their results may appear to conflict with ours and could, at least in part, be attributed to differences in the study population, it is possible that the efficiency of telomerase to elongate telomeres is different depending on CRC location. Further studies are needed to identify the reason for this phenomenon, as it could provide interesting new information regarding cancer biology.
An additional controversy arises when telomerase is examined in conjunction with colon cancer stage, pathology and patient survival. We found significantly lower telomerase activity in the normal tissue of patients with Dukes stage C or D than in those with stage A or B disease. This finding was replicated when patients were classified according to disease stage based on the American Joint Committee on Cancer (AJCC) TNM staging system. Although stage C and D colon cancer specimens had lower telomerase activity than stage A and B specimens and AJCC TNM stages III and IV specimens had lower telomerase activity than stage I and II specimens, the result in both cases was not statistically significant. We also found no correlation of telomerase activity with either 3- or 5-year survival. However, others have reported that telomerase activity tended to be higher in the larger, less differentiated late-stage Dukes C and D samples,36 while Union for International Cancer Control stage I tumours showed shorter telomeres and lower telomerase activity than stage IV samples. In another series of studies, hTERT mRNA expression correlated significantly with the histological grade, while patients with higher values had poorer survival than patients with lower values.20,21,37 Poorer prognosis in patients with high telomerase activity was also reported by others, but there was no correlation with other clinicopathological factors.38 An explanation for these differences may be that in the human bowel, telomere length is inversely related with age until 60–70 years. Beyond this age telomere length is positively related with age,39 and differences in age between our series and others may account for the discrepancies, although analysis based on age groups did not lead to any useful conclusions (data not shown). Alternatively, our results might imply that in early stages normal epithelium might be more susceptible to new telomerase-driven tumour formation while in late stages the extent of the systemic illness may decrease this potency or that there might be a less powerful regeneration process of the normal colonic epithelium in late stages of colon cancer.
Although several clinical and molecular data support the distinct nature of colon and rectal cancers, CRC is considered a relatively homogeneous disease. However, rectal cancer treatment is differentiated, since radiation therapy is indicated in locally advanced rectal tumours only. From a molecular point of view, there have been studies indicating that tumours located in the proximal colon represent a distinct entity with specific clinical and pathological characteristics.8 Although a recent study from the Cancer Genome Atlas Network reported that colon and rectal cancer have considerably similar patterns of genomic alteration, this conclusion was reached after excluding hypermutated cancers (accounting for 16% of their specimens) that included cancers with high MSI, usually with hypermethylation and MLH1 silencing.40 Our results, although in some disagreement with the Cancer Genome Atlas Network study, are valuable because they reflect a real life sample from a real world hospital. Furthermore, it has been reported that it is possible that both high telomerase, by conferring cellular immortality, and low telomerase, by promoting CIN, increase the risk of cancer.41 Our results indicate that high telomerase activity in the colon is associated with the development of cancer, whereas lower telomerase activity in the rectum is associated with increased loss of MLH1 expression signifying increased MSI.
Further molecular differences related to colon cancer localization that have been identified include overexpression of p53 protein mostly in left-sided tumours (43%–60% v. 16%–23% in right-sided tumours).10,42 In agreement with these findings, in our series just over half of the colon cancer samples expressed p53, while 88.2% of rectal cancer samples were p53-positive. DNA damage, oncogene activation and hypoxia triggers p53 protein accumulation and transcriptional transactivation of target genes including Bax.43 It also transrepresses the antiapoptotic bcl2 gene.44
Several studies have addressed the issue of telomerase/p53 interactions, and it seems that telomerase and p53 counteract each other. Several reports have shown that telomerase activity is inhibited when p53 is overexpressed in human cancer cells.45,46 Moreover, the hTERT gene has been shown to be downregulated on activation of wild-type p53.47 Inactivation of p53 in human mammary cells led to reactivation of telomerase,48 while short dysfunctional or near-dysfunctional telomeres sensitized these cells to p53-dependent signals for growth arrest.49 There is also evidence that the p53 family member p73 regulates hTERT, since p73 overexpression downregulates hTERT.50 Conversely, hTERT seems to suppress p53-mediated antiapoptotic response by inducing basic fibroblast growth factor.51 It has also been shown that constitutive hTERT expression inhibits wild-type p53-dependent apoptosis in colon carcinoma cells, and this effect is independent of telomerase activity since a telomerase inactive hTERT mutant was equally effective in antagonizing p53-induced apoptosis.52 This finding is consistent with the report that hTERT can promote cell survival independently of its enzymatic activity.53 In a study of 43 patients with CRC, alteration of p53 was found in 44.19% of patients, and mutations of p53 were positively associated with hTERT expression.54 Moreover, telomerase suppression (hence short telomeres) and p53 absence have been reported to be associated with abnormalities in chromosomes of colon cancer cells.55 In a recent report, patients with ulcerative colitis demonstrated low p53 expression and short telomeres in low-grade dysplasia, while p53 and telomere length were progressively increased in high-grade dysplasia, indicating an inverse association between the 2 in these preneoplastic conditions.56 These findings are in agreement with our finding of an inverse association between p53 and hTERT in patients with colon cancer.
Finally, we found that p53 expression was associated with increased patient survival. It is unclear whether there is an association between this finding and telomerase activity, since we found no correlation of telomerase activity levels with patient survival. Further studies are needed to clarify the importance of such an interaction.
Conclusion
Our findings further support the concept of multiple molecular entities within the spectrum of CRC. The differences that we and others10 have identified in telomerase activity in colon and rectal cancers may be related to the diversity of carcinogenesis and disease progression mechanisms in these distinct locations that can be attributed to their discrete embryology, anatomy, physiology and molecular biology. Furthermore, it would be interesting to examine whether p53 expression is in any way related to the response of patients to specific chemotherapeutic agents.
Footnotes
Competing interests: None declared.
Contributors: G.D. Ayiomamitis, G. Notas and E. Kouroumallis designed the study. G.D. Ayiomamitis, A. Zaravinos, A. Zizi-Sermetzoglou, M. Georgiadou and O. Sfakianaki acquired the data, which G. Notas analyzed. G.D. Ayiomamitis and G. Notas wrote the article, which all authors reviewed and approved for publication.
References
- 1.Bufill JA. Colorectal cancer: evidence for distinct genetic categories based on proximal or distal tumor location. Ann Intern Med. 1990;113:779–88. doi: 10.7326/0003-4819-113-10-779. [DOI] [PubMed] [Google Scholar]
- 2.Oh SW, Kim YH, Choi YS, et al. The comparison of the risk factors and clinical manifestations of proximal and distal colorectal cancer. Dis Colon Rectum. 2008;51:56–61. doi: 10.1007/s10350-007-9083-5. [DOI] [PubMed] [Google Scholar]
- 3.Gervaz P, Bucher P, Neyroud-Caspar I, et al. Proximal location of colon cancer is a risk factor for development of metachronous colorectal cancer: a population-based study. Dis Colon Rectum. 2005;48:227–32. doi: 10.1007/s10350-004-0805-7. [DOI] [PubMed] [Google Scholar]
- 4.Papagiorgis P, Oikonomakis I, Karapanagiotou I, et al. The impact of tumor location on the histopathologic expression of colorectal cancer. J BUON. 2006;11:317–21. [PubMed] [Google Scholar]
- 5.Azzoni C, Bottarelli L, Campanini N, et al. Distinct molecular patterns based on proximal and distal sporadic colorectal cancer: arguments for different mechanisms in the tumorigenesis. Int J Colorectal Dis. 2007;22:115–26. doi: 10.1007/s00384-006-0093-x. [DOI] [PubMed] [Google Scholar]
- 6.Lindblom A. Different mechanisms in the tumorigenesis of proximal and distal colon cancers. Curr Opin Oncol. 2001;13:63–9. doi: 10.1097/00001622-200101000-00013. [DOI] [PubMed] [Google Scholar]
- 7.Catalano T, Curia MC, Aceto G, et al. Mutations in the p53 and Ki-ras genes, microsatellite instability and site of tumor origin in colorectal cancer. Oncol Rep. 2005;14:625–31. [PubMed] [Google Scholar]
- 8.Li FY, Lai MD. Colorectal cancer, one entity or three. J Zhejiang Univ Sci B. 2009;10:219–29. doi: 10.1631/jzus.B0820273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–99. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rampazzo E, Bertorelle R, Serra L, et al. Relationship between telomere shortening, genetic instability, and site of tumour origin in colorectal cancers. Br J Cancer. 2010;102:1300–5. doi: 10.1038/sj.bjc.6605644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cukusic A, Skrobot Vidacek N, Sopta M, et al. Telomerase regulation at the crossroads of cell fate. Cytogenet Genome Res. 2008;122:263–72. doi: 10.1159/000167812. [DOI] [PubMed] [Google Scholar]
- 12.Allegra CJ, Parr AL, Wold LE, et al. Investigation of the prognostic and predictive value of thymidylate synthase, p53, and Ki-67 in patients with locally advanced colon cancer. J Clin Oncol. 2002;20:1735–43. doi: 10.1200/JCO.2002.07.080. [DOI] [PubMed] [Google Scholar]
- 13.Baretton GB, Diebold J, Christoforis G, et al. Apoptosis and immunohistochemical bcl-2 expression in colorectal adenomas and carcinomas. Aspects of carcinogenesis and prognostic significance. Cancer. 1996;77:255–64. doi: 10.1002/(SICI)1097-0142(19960115)77:2<255::AID-CNCR6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 14.Hilska M, Collan YU, O Laine VJ, et al. The significance of tumor markers for proliferation and apoptosis in predicting survival in colo-rectal cancer. Dis Colon Rectum. 2005;48:2197–208. doi: 10.1007/s10350-005-0202-x. [DOI] [PubMed] [Google Scholar]
- 15.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–73. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 16.Satyanarayana A, Manns MP, Rudolph KL. Telomeres, telomerase and cancer: an endless search to target the ends. Cell Cycle. 2004;3:1138–50. [PubMed] [Google Scholar]
- 17.Cong YS, Wen J, Bacchetti S. The human telomerase catalytic subunit hTERT: organization of the gene and characterization of the promoter. Hum Mol Genet. 1999;8:137–42. doi: 10.1093/hmg/8.1.137. [DOI] [PubMed] [Google Scholar]
- 18.Wick M, Zubov D, Hagen G. Genomic organization and promoter characterization of the gene encoding the human telomerase reverse transcriptase (hTERT) Gene. 1999;232:97–106. doi: 10.1016/s0378-1119(99)00108-0. [DOI] [PubMed] [Google Scholar]
- 19.Nugent CI, Lundblad V. The telomerase reverse transcriptase: components and regulation. Genes Dev. 1998;12:1073–85. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- 20.Rosenberg R, Gertler R, Stricker D, et al. Telomere length and hTERT expression in patients with colorectal carcinoma. Recent Results Cancer Res. 2003;162:177–81. doi: 10.1007/978-3-642-59349-9_16. [DOI] [PubMed] [Google Scholar]
- 21.Gertler R, Rosenberg R, Stricker D, et al. Telomere length and human telomerase reverse transcriptase expression as markers for progression and prognosis of colorectal carcinoma. J Clin Oncol. 2004;22:1807–14. doi: 10.1200/JCO.2004.09.160. [DOI] [PubMed] [Google Scholar]
- 22.Tahara H, Yasui W, Tahara E, et al. Immuno-histochemical detection of human telomerase catalytic component, hTERT, in human colorectal tumor and non-tumor tissue sections. Oncogene. 1999;18:1561–7. doi: 10.1038/sj.onc.1202458. [DOI] [PubMed] [Google Scholar]
- 23.Kammori M, Kanauchi H, Nakamura K, et al. Demonstration of human telomerase reverse transcriptase in human colorectal carcinomas by in situ hybridization. Int J Oncol. 2002;20:15–21. [PubMed] [Google Scholar]
- 24.Kim NW, Piatyszek MA, Prowse KR, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–5. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 25.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–91. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 26.de Kok JB, Ruers TJ, van Muijen GN, et al. Real-time quantification of human telomerase reverse transcriptase mRNA in tumors and healthy tissues. Clin Chem. 2000;46:313–8. [PubMed] [Google Scholar]
- 27.Nowak J, Januszkiewicz D, Lewandowski K, et al. Activity and expression of human telomerase in normal and malignant cells in gastric and colon cancer patients. Eur J Gastroenterol Hepatol. 2003;15:75–80. doi: 10.1097/00042737-200301000-00013. [DOI] [PubMed] [Google Scholar]
- 28.Yasui W, Tahara E, Tahara H, et al. Immunohistochemical detection of human telomerase reverse transcriptase in normal mucosa and pre-cancerous lesions of the stomach. Jpn J Cancer Res. 1999;90:589–95. doi: 10.1111/j.1349-7006.1999.tb00787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan P, Saraga EP, Bouzourene H, et al. Expression of telomerase genes correlates with telomerase activity in human colorectal carcinogenesis. J Pathol. 2001;193:21–6. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH728>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 30.Boldrini L, Faviana P, Gisfredi S, et al. Evaluation of telomerase in the development and progression of colon cancer. Int J Mol Med. 2002;10:589–92. [PubMed] [Google Scholar]
- 31.Saleh S, Lam AK, Ho YH. Real-time PCR quantification of human telomerase reverse transcriptase (hTERT) in colorectal cancer. Pathology. 2008;40:25–30. doi: 10.1080/00313020701716425. [DOI] [PubMed] [Google Scholar]
- 32.Niiyama H, Mizumoto K, Sato N, et al. Quantitative analysis of hTERT mRNA expression in colorectal cancer. Am J Gastroenterol. 2001;96:1895–900. doi: 10.1111/j.1572-0241.2001.03890.x. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalo V, Petit A, Castellvi-Bel S, et al. Telomerase mRNA expression and immunohistochemical detection as a biomarker of malignant transformation in patients with inflammatory bowel disease. Gastroenterol Hepatol. 2010;33:288–96. doi: 10.1016/j.gastrohep.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 34.O’Sullivan JN, Bronner MP, Brentnall TA, et al. Chromosomal instability in ulcerative colitis is related to telomere shortening. Nat Genet. 2002;32:280–4. doi: 10.1038/ng989. [DOI] [PubMed] [Google Scholar]
- 35.Risques RA, Lai LA, Brentnall TA, et al. Ulcerative colitis is a disease of accelerated colon aging: evidence from telomere attrition and DNA damage. Gastroenterology. 2008;135:410–8. doi: 10.1053/j.gastro.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshida R, Kiyozuka Y, Ichiyoshi H, et al. Change in telomerase activity during human colorectal carcinogenesis. Anticancer Res. 1999;19:2167–72. [PubMed] [Google Scholar]
- 37.Gertler R, Rosenberg R, Stricker D, et al. Prognostic potential of the telomerase subunit human telomerase reverse transcriptase in tumor tissue and nontumorous mucosa from patients with colorectal carcinoma. Cancer. 2002;95:2103–11. doi: 10.1002/cncr.10939. [DOI] [PubMed] [Google Scholar]
- 38.Tatsumoto N, Hiyama E, Murakami Y, et al. High telomerase activity is an independent prognostic indicator of poor outcome in colorectal cancer. Clin Cancer Res. 2000;6:2696–701. [PubMed] [Google Scholar]
- 39.O’Sullivan J, Risques RA, Mandelson MT, et al. Telomere length in the colon declines with age: A relation to colorectal cancer? Cancer Epidemiol Biomarkers Prev. 2006;15:573–7. doi: 10.1158/1055-9965.EPI-05-0542. [DOI] [PubMed] [Google Scholar]
- 40.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudolph KL, Millard M, Bosenberg MW, et al. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat Genet. 2001;28:155–9. doi: 10.1038/88871. [DOI] [PubMed] [Google Scholar]
- 42.Gervaz P, Bucher P, Morel P. Two colons — two cancers: paradigm shift and clinical implications. J Surg Oncol. 2004;88:261–6. doi: 10.1002/jso.20156. [DOI] [PubMed] [Google Scholar]
- 43.Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 44.Miyashita T, Harigai M, Hanada M, et al. Identification of a p53-dependent negative response element in the bcl-2 gene. Cancer Res. 1994;54:3131–5. [PubMed] [Google Scholar]
- 45.Kusumoto M, Ogawa T, Mizumoto K, et al. Adenovirus-mediated p53 gene transduction inhibits telomerase activity independent of its effects on cell cycle arrest and apoptosis in human pancreatic cancer cells. Clin Cancer Res. 1999;5:2140–7. [PubMed] [Google Scholar]
- 46.Kanaya T, Kyo S, Hamada K, et al. Adenoviral expression of p53 represses telomerase activity through down-regulation of human telomerase reverse transcriptase transcription. Clin Cancer Res. 2000;6:1239–47. [PubMed] [Google Scholar]
- 47.Xu D, Wang Q, Gruber A, et al. Downregulation of telomerase reverse transcriptase mRNA expression by wild type p53 in human tumor cells. Oncogene. 2000;19:5123–33. doi: 10.1038/sj.onc.1203890. [DOI] [PubMed] [Google Scholar]
- 48.Stampfer MR, Garbe J, Nijjar T, et al. Loss of p53 function accelerates acquisition of telomerase activity in indefinite lifespan human mammary epithelial cell lines. Oncogene. 2003;22:5238–51. doi: 10.1038/sj.onc.1206667. [DOI] [PubMed] [Google Scholar]
- 49.Beliveau A, Yaswen P. Soothing the watchman: telomerase reduces the p53-dependent cellular stress response. Cell Cycle. 2007;6:1284–7. doi: 10.4161/cc.6.11.4298. [DOI] [PubMed] [Google Scholar]
- 50.Beitzinger M, Oswald C, Beinoraviciute-Kellner R, et al. Regulation of telomerase activity by the p53 family member p73. Oncogene. 2006;25:813–26. doi: 10.1038/sj.onc.1209125. [DOI] [PubMed] [Google Scholar]
- 51.Jin X, Beck S, Sohn YW, et al. Human telomerase catalytic subunit (hTERT) suppresses p53-mediated anti-apoptotic response via induction of basic fibroblast growth factor. Exp Mol Med. 2010;42:574–82. doi: 10.3858/emm.2010.42.8.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rahman R, Latonen L, Wiman KG. hTERT antagonizes p53-induced apoptosis independently of telomerase activity. Oncogene. 2005;24:1320–7. doi: 10.1038/sj.onc.1208232. [DOI] [PubMed] [Google Scholar]
- 53.Cao Y, Li H, Deb S, et al. TERT regulates cell survival independent of telomerase enzymatic activity. Oncogene. 2002;21:3130–8. doi: 10.1038/sj.onc.1205419. [DOI] [PubMed] [Google Scholar]
- 54.Boldrini L, Faviana P, Gisfredi S, et al. Regulation of telomerase and its hTERT messenger in colorectal cancer. Oncol Rep. 2004;11:395–400. [PubMed] [Google Scholar]
- 55.Pantic M, Zimmermann S, El Daly H, et al. Telomere dysfunction and loss of p53 cooperate in defective mitotic segregation of chromosomes in cancer cells. Oncogene. 2006;25:4413–20. doi: 10.1038/sj.onc.1209486. [DOI] [PubMed] [Google Scholar]
- 56.Risques RA, Lai LA, Himmetoglu C, et al. Ulcerative colitis-associated colorectal cancer arises in a field of short telomeres, senescence, and inflammation. Cancer Res. 2011;71:1669–79. doi: 10.1158/0008-5472.CAN-10-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]