Abstract
The key role of the p53 protein in tumor suppression is highlighted by its frequent mutation in human cancers and by the completely penetrant cancer predisposition of p53 null mice. Beyond providing definitive evidence for the critical function of p53 in tumor suppression, genetically engineered mouse models have offered numerous additional insights into p53 function. p53 knock-in mice expressing tumor-derived p53 mutants have revealed that these mutants display gain-of-function activities that actively promote carcinogenesis. The generation of p53 knock-in mutants with alterations in different domains of p53 has helped further elucidate the cellular and biochemical activities of p53 that are most fundamental for tumor suppression. In addition, modulation of p53 post-translational modification (PTM) status by generating p53 knock-in mouse strains with mutations in p53 PTM sites has revealed a subtlety and complexity to p53 regulation. Analyses of mouse models perturbing upstream regulators of p53 have solidified the notion that the p53 pathway can be compromised by means other than direct p53 mutation. Finally, switchable p53 models that allow p53 reactivation in tumors have helped evaluate the potential of p53 restoration therapy for cancer treatment. Collectively, mouse models have greatly enhanced our understanding of physiological p53 function and will continue to provide new biological and clinical insights in future investigations.
Keywords: p53, Tumor suppression, Mouse models
1. Introduction
The p53 protein plays a fundamental role in tumor suppression, a notion underscored by the observation that p53 is mutated in over half of all human cancers of a wide variety of types [1,2]. Further support for the pivotal function of p53 in tumor suppression comes from individuals with Li–Fraumeni Syndrome who inherits a mutant p53 allele and are highly cancer-prone, developing a characteristic spectrum of tumors including sarcomas, brain cancers, breast cancers, and adrenocortical carcinomas [3]. Finally, as described below, definitive evidence for p53’s critical role in tumor suppression came from the generation of p53 knockout mice, as these mice were found to develop cancers at 100% frequency[4–6].
p53 is a sensor for cellular stresses such as DNA damage, hypoxia, or oncogenic signaling [1]. In the presence of such stress signals, p53 is post-translationally modified, resulting in displacement of Mdm2 and Mdm4, negative regulators of p53, and consequent p53 stabilization and activation [7–10]. Upon activation, p53 can trigger specific anti-proliferative responses, including transient cell-cycle arrest, permanent arrest known as cellular senescence, or apoptosis [1]. The temporary cell-cycle arrest response is particularly well studied in response to DNA damage and allows DNA repair before progression through the cell cycle, thus minimizing the propagation of potentially deleterious mutations – a role that led p53 to be named the “guardian of the genome” [11]. In contrast, apoptosis and cellular senescence, the latter of which triggers an innate immune response, are terminal cell fates that cause complete elimination of damaged or premalignant cells [12]. p53 drives these responses primarily by serving as a transcriptional activator that induces programs of gene expression important for each p53 response, although p53 also has some non-transcriptional activities [1,13]. Additionally, typical of a protein that drives different responses upon exposure to diverse stimuli, the p53 protein is regulated by a variety of post-translational modifications in response to stress signals [14,15].
While numerous insights into p53 function and regulation have come from in vitro studies, mouse models have been necessary to delineate the in vivo significance of these observations in physiological contexts. Given that p53 displays cell type-dependent and context-specific mechanisms of action, analyses in the mouse are useful for revealing the full complexity of its action in different settings. Moreover, the mouse affords the possibility to examine the process of tumorigenesis in the proper tissue microenvironment. In this review, we detail a variety of p53 mouse models that have advanced our understanding of various aspects of p53 biology, including how p53 tumor mutants not only promote loss-of-function effects but also gain-of-function phenotypes to fuel carcinogenesis, how p53 acts mechanistically as a tumor suppressor, and how post-translational modifications fine-tune p53 activities. Investigations into p53 function using mouse models have greatly illuminated our understanding of this critical tumor suppressor.
2. Knockout models: when the guardian lets its guard down
While the frequent mutations of p53 in human cancers suggested that p53 inactivation may be causal for tumorigenesis, unequivocal evidence for the importance of p53 in tumor suppression came through the generation of p53 null mice. p53 null mice were generated by three different laboratories, in each case through disruption of the p53 sequence-specific DNA binding domain [4–6]. Surprisingly, these studies demonstrated that these mice, for the most part, do not display embryonic lethality. However, p53−/− mice displayed decreased survival compared to wild-type mice, with the majority of mice succumbing to tumors between 2 and 9 months of age, primarily CD4+CD8+ T-cell lymphomas and some sarcomas. Moreover, p53+/− mice displayed decreased survival and accelerated tumor development compared to wild-type mice – although not to the extent observed in p53−/− mice – with spontaneous tumors developing with a median latency of ~18 months [16]. p53+/− mice displayed a different tumor spectrum from p53−/− mice, with a preponderance of diverse sarcomas (greater than 50% of all tumors), including fibrosarcomas, osteosarcomas, rhabdomyosarcomas, hemangiosarcomas, anaplastic sarcomas, and lyomyosarcomas, some lymphomas (25–35% of cases), and some adenocarcinomas (~9–11% of cases) [4,5]. The reduced incidence of thymic lymphomas in the p53+/− mice is thought to relate to a limited developmental window for these tumors to arise, before thymic involution, and inadequate time for Loss of Heterozygosity (LOH) to occur during this period [16]. While some tumors developing in p53+/− mice underwent LOH, some did not, indicating that p53 can be haploinsufficient for tumor suppression [17]. Importantly, the propensity of p53+/− mice to develop cancer, and particularly to develop sarcomas, is broadly similar to that of Li–Fraumeni Syndrome patients, suggesting that p53+/− mice can serve as a model for this syndrome. Notably, these studies also revealed some variability in precise phenotypes between different models, which is likely related to the different genetic backgrounds examined, with different enrichment for 129/Sv or C57BL/6 backgrounds. Indeed, the importance of genetic background in modifying the phenotype of p53 loss was underscored by studies of p53+/− mice on a BALB/c background, which revealed a dramatic predisposition to breast cancer [18]. In addition, tissue-specific ablation of p53, using p53 conditional knockout mice to circumvent the early lethality from lymphomas and sarcomas typical of constitutive p53 knockout mice, has supported the idea that p53 loss can be sufficient to promote other cancer types, such as epithelial cancers. For example, targeted inactivation of p53 in the mammary gland results in mammary tumors [19,20], and ablation of p53 in the cervical epithelium, in combination with estrogen treatment, promotes cervical cancer [20]. In addition, somatic inactivation of p53 in mature B cells results in IgM+ mature peripheral B-cell lymphomas [21]. Collectively, these studies demonstrated conclusively that p53 deficiency in numerous tissues causes cancer. Moreover, beyond the spontaneous cancer predisposition characterizing p53+/− mice and p53−/− mice, crossing nearly any other tumor-prone strain onto a p53-deficient background enhances tumor predisposition in that strain, underscoring how generally p53 impedes tumorigenesis in many contexts [22].
3. Can a p53 mutation be worse than no p53 at all?
Interestingly, unlike the scenario with p53 knockout mice where no p53 protein is produced, approximately 75% of p53 mutations found in human tumors are missense mutations, suggesting that there may be some selective advantage for retaining the mutant allele rather than losing p53 completely [23,24]. p53 is typically mutated in the DNA binding domain (DBD), most commonly at various hotspot sites (R175, G245, R248, R249, R273, R282) [23,25]. Depending on the consequence of the mutations, p53 DBD mutants can be divided into two categories: contact mutants, which harbor mutations in p53 residues that directly contact the DNA duplex, or structural mutants, which carry mutations that disrupt p53 protein conformation, both of which interfere with p53 binding to its response element [25,26]. From cell culture assays, such p53 point mutants were proposed to promote tumorigenesis by exerting dominant-negative, inhibitory effects on the wild-type p53 protein, as well as by manifesting novel, oncogenic gain-of-function (GOF) activities. To examine the mechanisms by which the point mutants might enhance tumor development, and to re-create models that more accurately recapitulate human cancers, initial experiments focused on generating transgenic mice overexpressing p53 tumor-derived mutants in various mouse tissues [27–29]. These transgenic strains exhibited an enhanced cancer predisposition, and provided support for dominant-negative action of p53 point mutants, as A135V promoted cancer in combination with a wild-type p53 allele but not with a p53 null allele [29]. In contrast, initial evidence for p53 mutant GOF activity came primarily from cell culture and xenograft studies [30].
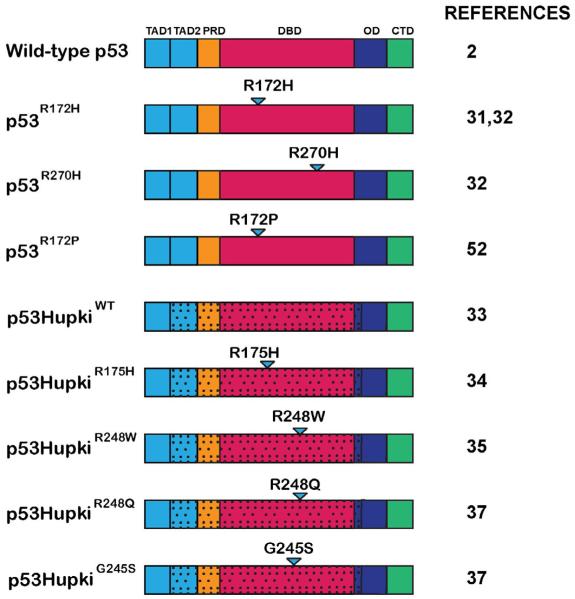
Ultimately, more physiological p53 knock-in mutant models were generated, in which the endogenous p53 mouse allele was replaced with orthologs of specific human p53 hotspot mutants, allowing expression of the mutant alleles at physiological levels and with proper spatial and temporal regulation (Fig. 1). In the first of these studies, mutant knock-in mice expressing p53R172H (corresponding to the human p53R175H conformation mutant) were generated. It was found that p53R172H/−, p53R172H/R172H, and P53−/− cohorts displayed identical survival curves and developed similar tumor spectra [31]. p53+/− and p53R172H/+ mice also exhibited similar survival curves and tumor spectra, but the p53R172H/+ mice developed tumors that metastasized frequently, suggesting potential GOF activity of the p53R172H mutant. In another study analyzing both the p53R172H and p53R270H mutants (corresponding to human p53R175H and the p53R273H contact mutant), p53R270H/+ and p53R172H/+ mice exhibited similar survival curves to p53+/− mice, but developed different tumor types from the p53+/− mice [32]. Specifically, p53R270H/+ mice exhibited an increase in metastatic carcinomas and B-cell lymphomas relative to p53+/− mice, and p53R172H/+ mice developed osteosarcomas that were much more metastatic than in p53+/− mice. In addition, p53R270H/− and p53R172H/− mice displayed similar survival curves to p53−/− mice but also developed some novel tumor types. p53R270H/− mice developed an increased incidence of invasive, metastatic carcinomas relative to p53−/− mice, while p53R172H/− mice displayed an enhanced predisposition to both invasive, metastatic carcinomas and hemangiosarcomas relative to p53−/− mice. Collectively, these studies support a GOF mechanism for p53 mutants, as they induced either a broadened tumor spectrum or conferred increased metastatic potential to tumors relative to p53 nullizygosity. These results were supported by accompanying cell culture assays in which p53R172H/R172H mouse embryonic fibroblasts (MEFs) displayed enhanced proliferation compared to p53−/− MEFs [31]. Additionally, HrasV12;p53R172H/R172H MEFs formed more colonies in soft-agar transformation assays than Hras-V12;p53−/− MEFs, bolstering the notion that p53 mutants display GOF activity. The mechanism for GOF activity in these contexts appears to be through mutant p53 binding and inhibiting the p53-related transcription factors p63 and p73, which can have tumor suppressor activity themselves. Overall, these were landmark studies for substantiating the significance of the GOF idea in vivo.
Fig. 1.
p53 tumor-derived mutants examined in knock-in mouse models. The p53 tumor-derived mutants studied in mouse models and described in this review. Wild-type p53 is shown for reference. p53 functional domains: TAD1 = transactivation domain 1; TAD2 = transactivation domain 2; PRD = proline rich domain; DBD = DNA-binding domain; OD = oligomerization domain; CTD = C-terminal domain. Triangles denote mutated amino acid. Stippling indicates regions of p53 replaced with human sequences in the HUPKI mouse models.
To more accurately study the human p53 hotspot mutations, HUPKI (humanized p53 knock-in) mouse models were generated [33]. In these mice, murine p53 exons 4–9 were replaced with the corresponding human sequences, including specific human tumor mutations. For example, a mouse strain expressing the p53R175H structural mutant, corresponding to murine p53R172H, was generated and assessed for GOF activity [34]. p53hupkiR175H/R175H mice displayed similar survival rates to p53−/− mice, but exhibited a broader tumor spectrum, with the development of peripheral lymphomas and germ cell tumors. These data further support the notion that p53 hotspot mutations, and in this case the human R175H hotspot mutation, confer GOF activity by enhancing the development of new tumor types. Another p53 hotspot HUPKI mouse model, expressing the p53R248W contact mutant, has supported this idea [35]. Specifically, p53hupkiR248W/− mice displayed a similar survival curve to p53−/− mice, but succumbed to a broader spectrum of tumors, with the development of peripheral lymphomas. The GOF activity of these p53 mutants was attributed to their ability to interact with Mre11, blocking the binding of the Mre11-Rad50-Nbs1 complex to double-strand breaks and preventing proper activation of the Atm cascade. This interference with DNA damage response signaling in turn resulted in defective cell-cycle checkpoints and genetic instability.
It has been proposed that the different p53 tumor-derived mutants may have varying degrees of GOF activity [36]. Indeed, additional insight into p53 GOF properties and their unique capacities has come to light with the development of additional HUPKI hotspot mouse models, expressing the contact mutant p53R248Q or the conformation mutant, p53G245S [37]. The p53hupkiG245S/− mice exhibited similar tumor latency and survival rates to p53−/− mice. In contrast, p53hupkiR248Q/− mice displayed diminished survival and decreased tumor latency relative to p53−/− mice, indicating a more dramatic GOF than any previously studied p53 hotspot mouse model. Additionally, both p53hupkiR248Q/− and p53hupkiG245S/− mice developed a slightly broader spectrum of tumors, with the appearance of more sarcoma subtypes, carcinomas, and germ cell tumors. Moreover, lymphoma cells from both p53hupkiR248Q/− and p53hupkiG245S/− mice displayed enhanced Akt signaling relative to p53−/− cells, suggesting a potential mechanism underlying GOF. Interestingly, thymic lymphoma cells from p53hupkiR248Q/− mice proliferated faster than those from p53−/− mice, while those from p53hupkiG245S/− mice did not, demonstrating a potentially contact mutant-specific GOF phenotype. The p53hupkiR248Q/− mice also displayed an expansion of hematopoietic and mesenchymal stem cell populations relative to p53hupkiG245S/− or p53−/− counterparts, which may contribute to the rapid tumor onset observed. Consistent with this notion, a similar expansion of mammary stem cells was coupled with a propensity for tumor development in a p53hupkiR175H mammary mouse model [38]. Notably, the p53R248Q is the most potent GOF mutant reported to date, as it is the first hotspot mutant shown to promote decreased survival relative to p53 deficiency. This side-by-side comparison of p53hupkiR248Q and p53hupkiG245S mice, along with previous studies of p53hupkiR175H and p53hupkiR248W mice, support the assertion that the different p53 hotspot mutations can promote varying degrees of oncogenic GOF activity [34,35,37].
Together, these studies demonstrate that the p53 tumor-derived mutants both lose wild-type p53 tumor suppressor activity and acquire oncogenic GOF properties in vivo. Particularly compelling for establishing a GOF phenotype has been the comparison between p53M/− or p53M/M mice and p53−/− mice (where M denotes p53 mutant), which has indeed shown that the M alleles enhance tumorigenicity in vivo, in the absence of a wild-type p53 allele. Furthermore, it is becoming clear that different p53 hotspot mutants promote different GOF phenotypes. As additional p53 hotspot mutant mouse models are generated, we hope to better elaborate the mutation-specific GOF phenotypes, as well as broader, category-specific (i.e. contact mutant versus conformation mutant) GOF phenotypes and the molecular underpinnings of these phenotypes. Acquiring a better understanding of how each mutant acts to promote cancer could lead to more effective therapeutic targeting of p53 mutants, depending on the specific mutations and the associated conformation of the encoded proteins.
4. Unveiling p53 domain functions
The p53 protein is a transcriptional activator that induces a plethora of diverse target genes involved in different responses [2]. Typical of transcriptional activators, the protein comprises multiple functional domains, including two transcriptional activation domains (TADs), a so-called proline-rich domain (PRD), a sequence-specific DNA-binding domain (DBD), and an oligomerization domain (OD) through which p53 monomers interact to form tetramers. To better understand the contribution of each domain to p53 functions in vivo, a variety of p53 knock-in mouse strains expressing mutants altered in different p53 domains have been engineered (Fig. 2). These studies have helped delineate mechanisms of p53 action in cell-cycle arrest, senescence, apoptosis, and ultimately tumor suppression.
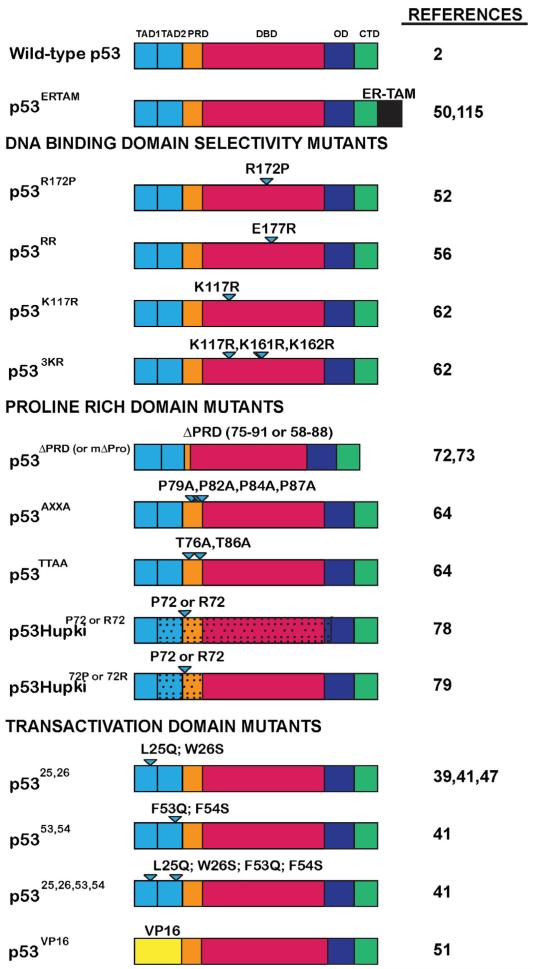
Fig. 2.
p53 domain mutants investigated in knock-in mouse models. The p53 domain mutants described in this review are categorized according to the functional domains targeted. Wild-type p53 is shown for reference. p53 functional domains: TAD1 = transactivation domain 1; TAD2 = transactivation domain 2; PRD = proline rich domain; DBD = DNA-binding domain; OD = oligomerization domain; CTD = C-terminal domain. Triangles denote mutated amino acids. ERTAM = tamoxifen-inducible estrogen receptor ligand-binding domain; VP16-herpes simplex viral protein 16 transactivation domain. RR is so-called because of the E180R mutation adjacent to the unmutated residue R181. Even though it is a tumor-derived mutant, p53R172P is listed here again because it has been used for structure–function analysis of p53. Stippling indicates regions of p53 replaced with human sequences in the HUPKI mouse models.
4.1. Tinkering with the transactivation domains
Although p53 has well-characterized activity as a transcriptional activator, in vitro studies have suggested that p53 has additional biochemical activities, such as the ability to repress transcription and to induce apoptosis through direct action at the mitochondria [1,2]. Thus, to directly assess the contribution of transcriptional activation to p53 functions in vivo, transcriptional activation-deficient mutant mice were generated. Initially, knock-in mice were generated that expressed p5325,26, with mutations L25Q and W26S in the first TAD [39], as this mutant had been reported to be transcriptionally inactive in reporter assays [40]. Analysis of p5325,26/25,26 cells showed that p5325,26 is indeed severely compromised for inducing most classical p53 target genes such as p21, Noxa, Puma, and Perp, both in response to DNA damage and oncogenic signals, indicating that the first TAD is essential for the transactivation of most known p53 target genes [39,41]. However, p5325,26 retained the capacity to activate a small set of p53 target genes, such as Bax. In terms of biological activity, p5325,26 was unable to induce G1 cell-cycle arrest or apoptosis in response to acute DNA damage signals in both MEFs in vitro and radiosensitive tissues in vivo. These observations indicate that TAD1 and efficient transcriptional activation of p53 target genes are important for mounting responses to acute DNA damage, consistent with the known requirements for p21 and Noxa, Puma, and Perp for DNA damage-induced cell-cycle arrest and apoptosis, respectively [42]. Interestingly, however, p5325,26 can promote apoptosis in oncogene-expressing MEFs in response to serum deprivation or hypoxia, indicating that apoptosis in response to non-genotoxic stresses occurs without full p53 transactivation [39]. Intriguingly, upon examination of the p5325,26 mutant in tumor suppression, it was discovered that it efficiently suppresses the development of tumors derived from multiple different lineages and driven by different oncogenic events, such as Kras-driven non-small-cell lung cancer and Eμ-myc triggered lymphomas [41,43]. Thus despite the inability to efficiently activate most canonical p53 target genes, p5325,26 suppresses cancer effectively, suggesting that either its residual transcriptional activation function or an alternate p53 biochemical activity accounts for its ability to suppress cancer.
To distinguish these possibilities, mice expressing a quadruple mutant, p5325,26,53,54, with alterations in both the first and second p53 TADs (L25Q; W26S; F53Q; F54S) were generated [41], as this mutant was reported to be transcriptionally inactive in vitro [44–46]. To control for the effect of the mutations in TAD2 alone, knock-in mice expressing the p5353,54 mutant, with F53Q and F54S alterations, were also generated. Microarray studies of homozygous mutant cells derived from these mice revealed that p5325,26,53,54 is incapable of transactivating p53 target genes, displaying a gene expression profile indistinguishable from that of p53−/− cells, while the p5353,54 displayed no compromise in transcriptional activity compared to wild-type p53 [41]. The activity of the p5325,26,53,54 mutant in tumor suppression was examined in multiple mouse cancer models, and it was found to be completely unable to suppress carcinogenesis. These findings indicate that transcriptional activation is indeed essential for tumor suppression in a variety of different tumor types such as lung cancer and lymphomas [41,43,47]. Not surprisingly, given its intact transcriptional potential, the p5353,54 mutant retained full tumor suppression activity [41]. Importantly, since the p5325,26 mutant retains tumor suppression activity, yet only robustly transactivates a limited set of p53 targets, analysis of genes efficiently induced by both wild-type p53 and p5325,26 has helped uncover a set of novel direct p53 target genes whose expression is tightly associated with tumor suppression. Indeed, knockdown of these genes in allograft assays have demonstrated that some of these targets display tumor suppressor activity. Thus, these findings together suggest that these tumor suppression-associated target genes may represent new key components of the p53 tumor suppression network. In addition, the finding that TAD1 is essential for p53 responses to acute DNA damage, but not for tumor suppression indicates that the ability of p53 to promote responses to acute DNA damage is dispensable for tumor suppression, a conclusion supported by several independent studies [48–50].
Another study also examined whether transcriptional activation is sufficient for p53 function [51]. Knock-in mice were generated expressing a chimeric p53 protein, in which the first 80 amino acids of the amino-terminus comprising the two p53 TADs were replaced with the TAD of the Herpes Simplex Virus VP16 protein. This p53VP16 chimera was proficient for DNA binding and transcriptional activation on all p53 target genes tested due to the heterologous TAD, but lacked other potential amino-terminal p53 functions. Studies in MEFs revealed that p53VP16 can efficiently induce cell-cycle arrest and cellular senescence, but not apoptosis in response to different DNA damaging agents. These findings suggest that while transcriptional activity may suffice for p53 to trigger arrest responses, eliciting an apoptotic response may require functions other than transcriptional activation, such as the ability of p53 to act at the mitochondria.
4.2. Dissecting DNA binding function: parsing roles of p53-induced cell-cycle arrest and apoptosis in tumor suppression
The majority of mutations found within the p53 DBD abolish sequence-specific DNA binding. However, several different mutations within the p53 DBD have been reported to selectively affect DNA binding to specific p53 target genes, but not others, helping to further illuminate mechanisms of p53 action. In the first of these studies, knock-in mice expressing p53R172P (also known as p53515G→C; [52]), the ortholog of the human tumor-derived mutant p53R175P, were generated. Previous studies had shown that the p53R175P mutant can bind and activate the cell-cycle arrest target gene p21 and drive cell-cycle arrest, but it cannot induce the apoptotic target gene Bax or apoptosis [53]. Similarly, studies of cells derived from the p53R172P knock-in mice revealed that p53R172P retains partial cell-cycle arrest function, associated with some p21 induction, but that it cannot induce apoptosis in response to DNA damaging agent treatment [52]. Interestingly, despite a defective apoptotic response, p53R172P/R172P mice exhibited delayed spontaneous tumorigenesis compared to p53−/− mice, with a reduction of the T-cell lymphomas typically seen in p53−/− mice. Subsequently, however, these mice developed other types of lymphomas and sarcomas. These p53R172P/R172P tumors were genomically stable, unlike the aneuploid tumors that develop in p53−/− mice, suggesting that p53R172P may suppress tumorigenesis through the induction of cell-cycle arrest and the inhibition of genomic instability. Additional studies of the p53R172P knock-in mice on a background deficient for the p21 cyclin dependent kinase inhibitor, to more specifically query the importance of cell-cycle arrest for the observed tumor suppression activity, showed that p21 loss accelerated tumorigenesis [54]. Importantly, this observation suggests that p53R172P indeed suppresses tumor development by inducing cell-cycle arrest and maintaining genome stability. Together, these data strongly implicate cell-cycle arrest as a critical facet of p53-mediated tumor suppression, in at least some tumor types. The inability of p53R172P to suppress the development of late-onset tumors, however, also supports a role for p53 apoptotic function in tumor suppression.
Cooperative DNA binding by p53 can be regulated by the charged residues E180 and R181 within the human p53 DBD [55]. The importance of these residues and of cooperative binding to certain p53 response elements for tumor suppression is suggested by the mutation of these residues in sporadic human tumors and in Li–Fraumeni patients. Moreover, in vitro studies demonstrated that the E180R mutation reduced cooperative p53 binding to DNA, specifically affecting binding to p53 response elements in pro-apoptotic genes. To define the physiological effect of reduced cooperative p53 DNA binding, knock-in mice were generated in which the mouse p53 E177 residue (corresponding to human p53 E180) was changed to arginine (E177R; termed p53RR) [56]. While the p53RR mutant can activate p21 and cell-cycle arrest in response to DNA damage signals, the p53RR mutant is defective in inducing pro-apoptotic target genes and apoptosis in DNA-damage-treated or serum-deprived cells. Aging p53RR/RR mice manifested decreased survival relative to wild-type littermates, and were highly cancer-prone, indicating a deficiency in p53RR tumor suppression function, although not to the extent observed with p53−/− mice. Indeed, early-onset T-cell lymphomas typical of p53−/− mice were significantly inhibited in p53RR/RR mice, indicating that p53RR retains tumor suppression function in some settings, like p53R172P. Interestingly, p53RR can activate p53 metabolic target genes such as Gls2 and Dram upon DNA damage and can limit glycolysis and ROS accumulation. Thus, despite a deficit in activating apoptotic target genes and apoptosis, this cooperativity mutant still retains the capacity to suppress tumorigenesis in some tissues, potentially attributable to its ability to induce cell-cycle arrest and/or regulate metabolism. Again, the fact that these mutant mice ultimately succumb to tumors suggests that apoptosis is an important component of p53-mediated tumor suppression in some tissues.
Recently, p53 knock-in mice carrying acetylation site mutations that selectively affect expression of some p53 target genes were generated. p53 modification by acetylation occurs in response to various stresses, including DNA damage and oxidative stress, and the murine p53 DBD is acetylated at three sites (K117, K161, K162), including two that are conserved in human p53 (K120, K164)[57,58]. Human cell culture studies implicated p53 K120 in the induction of apoptosis target genes, but not cell-cycle arrest target genes [59,60]. Combined mutation of K120 and K164 compromised the upregulation of apoptosis and cell-cycle arrest target genes as well as both p53-mediated apoptosis and cell-cycle arrest [61]. To clarify the in vivo role of acetylation of the p53 DBD for different p53 functions, two p53 acetylation site mutant knock-in mouse strains were generated, p53K117R and p533KR [62]. Analysis of cells derived from p53K117R mice, carrying the acetylation site mutation that renders p53 defective for triggering apoptosis in cultured cells in vitro, confirmed that p53K117R could not induce Puma or apoptosis in response to DNA damage, suggesting that acetylation at K117 is critical for p53-dependent apoptosis. In contrast, p53K117R is still competent to induce p21 and cell-cycle arrest in DNA damage-treated cells, as well as to trigger cellular senescence. Analysis of aging p53K117R/K117R cohorts indicated that these mice are resistant to early-onset spontaneous tumor development, again supporting the role of p53 cell-cycle arrest function in tumor suppression. To completely compromise apoptosis and cell-cycle arrest, p533KR knock-in mice were generated, in which three acetylation sites in the DBD were mutated (K117R, K161R, K162R) [62]. Cells derived from the p533KR/3KR mice indeed failed to undergo cell-cycle arrest and apoptosis in response to DNA damage, or cellular senescence in response to oncogenic HRasV12. Intriguingly, the p533KR/3KR mice were also resistant to early-onset spontaneous tumor formation, indicating that this mutant still retains tumor suppressor function despite lacking apoptosis, cell-cycle arrest, and senescence capabilities. Of note, the p533KR mutant maintains the capacity to activate the metabolic p53 target genes Gls2 and Tigar as well as to restrain glucose uptake, glycolysis, and ROS accumulation. This study therefore suggests that p53’s ability to regulate metabolism may play a role in p53-mediated tumor suppression.
4.3. Pulling apart the proline-rich domain
The p53 proline-rich domain (PRD) is found between residues 61-94 or 55-88 of the human and mouse proteins, respectively [63]. The human PRD contains numerous prolines including 5 PXXP repeats, where P represents proline and XX represents any amino acid. However, the precise sequence of the domain is not highly conserved between species, with the mouse PRD containing only two PXXP repeats and the guinea pig containing none, questioning the precise functional role of the PRD [64]. In vitro cell culture studies indicated that activation of p53-mediated apoptosis is dependent on the PRD, while induction of p21 and cell-cycle arrest is not[65–68]. Furthermore, the PRD has been implicated in p53 protein stabilization, which is thought to be regulated through the prolyl isomerase, Pin1 [69]. In vitro cell culture studies had shown that p53 becomes phosphorylated in response to stress signals, allowing more avid interaction with Pin1, which in turn engenders a conformational change in p53 that decreases Mdm2 accessibility and stabilizes p53 [69–71]. To understand the role of the PRD in apoptosis and p53 protein stabilization in a physiological setting, p53ΔPRD knock-in mice were generated expressing a p53 protein lacking amino acids 75–91 of the PRD, which comprise the two PXXP motifs plus two additional prolines [72]. Surprisingly, in contrast to the previous in vitro studies, p53ΔPRD was completely deficient in inducing cell-cycle arrest in response to DNA damage, possibly due to weak transactivation of p21, but remained competent to induce apoptosis in oncogene-expressing MEFs. Analysis of aging p53ΔPRD/ΔPRD mice revealed that they were relatively resistant to early-onset tumorigenesis, although they eventually succumbed to some tumors. These findings were supported by the generation of another PRD-deficient mouse strain, expressing p53mΔPro, which lacks amino acids 58–88 [73]. These aging p53mΔPro/mΔPro mice were found to develop very few T-cell lymphomas but ultimately did succumb to late-onset B cell lymphomas [73]. Much like the previous study, these experiments suggested that p53mΔPro is compromised for inducing p53-mediated cell-cycle arrest in response to DNA damage signals but showed that p53mΔPro is also defective in inducing apoptotic target genes and triggering apoptosis in response to genotoxic stress [74]. Thus, these observations suggest that the PRD has a context-dependent role in tumor suppression in suppressing early-onset spontaneous cancers.
To refine our understanding of the PRD, additional mouse strains were generated. First, to examine the contribution of Pin1 binding to p53-mediated tumor suppression, p53TTAA knock-in mice were generated, in which the Pin1 binding site was compromised by mutating threonines 76 and 86 to alanines, rendering these residues unphosphorylatable, an essential step for Pin1 binding. Second, to query the importance of the proline motifs, p53AXXA mice were engineered with alterations in the two PXXP motifs (P79A, P82A, P84A and P87A) in the PRD [64]. Mice from both strains were born at Mendelian ratios and had no developmental abnormalities. Moreover, allograft tumor assays performed with oncogene-expressing mutant MEFs derived from these mice demonstrated that both mutants still display intact tumor suppression function, indicating that the PXXP motifs and the Pin1 binding site within the PRD are dispensable for p53 tumor suppressor function. Thus, the PRD plays a role in tumor suppression in some settings, but independently of the PXXP motifs and Pin1 binding.
Within the human p53 PRD, codon 72 is polymorphic, encoding either proline or arginine. Cell culture experiments on these polymorphic p53 variants suggest that they can have different biological activities in terms of their abilities to activate p53 target genes, promote cell-cycle arrest, and localize to mitochondria [75–77]. To understand the contributions of each of these polymorphisms to p53 function, p53hupkiP72 and p53hupkiR72 mice were generated [78,79]. In response to DNA damage, p53P72 and p53R72 protein stabilization [78] and transactivation of the p53 targets Mdm2 and Puma were equivalent. However, p53P72 more efficiently induced p21 expression, cell-cycle arrest, and senescence in response to oncogenic signals and DNA-damage than p53R72. p53hupkiP72/P72 thymocytes also displayed increased apoptosis relative to p53hupkiR72/R72 thymocytes upon DNA-damage as well as increased transactivation of select p53 targets, indicating a functional difference between polymorphic variants. Interestingly, most genes preferentially transactivated by p53P72 include those with roles in inflammation, and these were found to be coordinately regulated by NF-kB. Surprisingly, p53hupkiP72/− and p53hupkiR72/− mice displayed similar survival profiles and spontaneous tumor spectra. In another set of experiments, p53hupki72P/72P and p53hupki72R/72R mice were made by simply humanizing exon 4 mouse p53. Interestingly, in this model, p53 apoptotic target genes, including Perp, Puma, and Noxa (but not the cell-cycle arrest target gene p21), were all induced to a greater extent by DNA damage in p53hupki72R/72R cells than in p53hupki72P/72P cells. Accordingly, DNA-damage-treated p53hupki72R/72R keratinocytes and intestinal cells displayed increased apoptosis relative to p53hupki72P/72P counterparts. Nonetheless, UVB-treated p53hupki72P/72P and p53hupki72R/72R mice developed skin cancer with similar latencies and multiplicities [79]. Collectively, the data from these two studies indicate that the p53P72 and p53R72 mutants display similar tumor suppressor activity despite each mutant exhibiting augmented p53 function in certain cellular assays.
The aforementioned studies interrogating the functional significance of different p53 domains using knock-in mouse model studies have provided key new insights into p53 and illustrate the importance of performing functional studies in an in vivo setting. Through analysis of the p53 TADs, studies have indicated that transcriptional activation by p53 is important for acute DNA-damage-induced cell-cycle arrest and apoptosis, as well as for tumor suppression. However, the TAD requirements are different in these contexts: TAD1 is essential for responses to acute DNA damage, while dispensable for tumor suppression. These findings in turn have suggested quite surprisingly that p53 responses to acute DNA damage, which have been a major focus of studies on p53 over the years, are dispensable for tumor suppression. Dissecting the DNA binding domain through analysis of mice expressing mutants that selectively compromise p53-mediated apoptosis but not cell-cycle arrest has revealed that cell-cycle arrest activity is associated with suppression of early-onset spontaneous tumors while apoptosis suppresses the development of late-onset tumors. Moreover, evidence suggests that p53 can suppress cancer in some contexts without any activity in acute stress-induced cell-cycle arrest and apoptosis, highlighting the potential role for additional, emerging functions of p53, such as regulating metabolism, in tumor suppression. Finally, the proline-rich domain plays a context-specific role in tumor suppression but not through Pin1 binding or through the PXXP motifs. Thus, while in vitro studies can provide an initial hint of the function of a particular p53 domain, defining the precise role of a domain relies on examination in an in vivo context, in the proper tissue microenvironment, and where cell-type-specific roles can be uncovered. Continued examination of mouse models will further decipher the many mysteries regarding the mechanisms of p53 action in tumor suppression.
5. Post-translational modifications – modifying the p53 function du jour
p53 responds to diverse cellular stresses by triggering various responses, and therefore must be tightly regulated by post-translational modifications (PTMs) [15]. p53 is regulated by a number of different PTMs, including phosphorylation, acetylation, ubiquitination, sumoylation, neddylation, and methylation. These modifications regulate different aspects of p53 activity, including protein stabilization and target gene activation. Here we describe knock-in models with alterations in p53 post-translational modification sites to better understand their contribution to p53 function in vivo (Fig. 3).
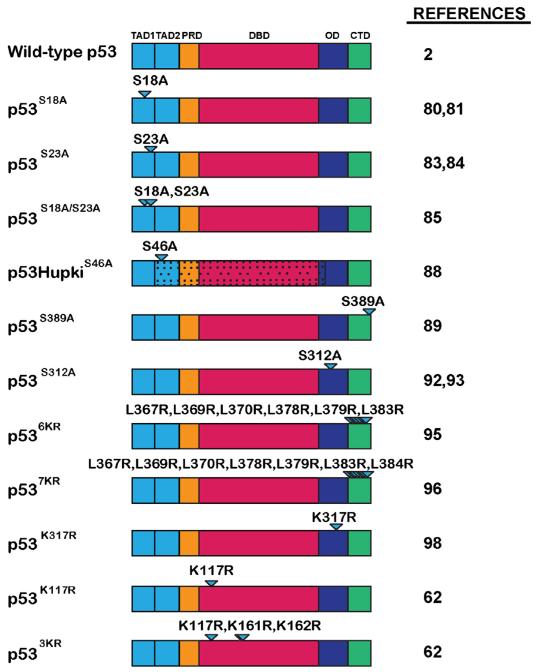
Fig. 3.
p53 post-translational modification site mutants investigated in knock-in mouse models. The p53 post-translational modification site mutants described in this review. Wild-type p53 is shown for reference. p53 functional domains: TAD1 = transactivation domain 1; TAD2 = transactivation domain 2; PRD = proline rich domain; DBD = DNA-binding domain; OD = oligomerization domain; CTD = C-terminal domain. Triangles denote mutated amino acids. Stippling indicates regions of p53 replaced with human sequences in the HUPKI mouse models.
In vitro studies showed that serine 15 in human p53 (serine 18 in mouse p53) becomes phosphorylated by the ATM kinase in response to DNA damage, and that this phosphorylation plays a critical role in stabilizing p53 in response to DNA damage, through the displacement of the negative regulator Mdm2 [8,14]. To interrogate the role of serine 18 phosphorylation in vivo, p53S18A knock-in mice were generated in which serine 18 was mutated to a non-phosphorylatable alanine [80,81]. Surprisingly, analysis of MEFs and thymocytes from p53S18A/S18A mice revealed that p53S18A protein stabilization in response to DNA damage is not significantly compromised, indicating that serine 18 phosphorylation is not essential for disruption of the p53-Mdm2 interaction. Instead, the ability of p53S18A to activate various target genes was impaired. Moreover, while DNA-damage-induced cell-cycle arrest was intact in MEFs, there were partial defects in DNA-damage-induced apoptosis. In addition, one study found that unlike p53−/− mice, p53S18A/S18A mice were not prone to tumorigenesis [80], although another study observed that these mice succumbed to enhanced late-onset tumor development relative wild-type mice [82]. Together, these data suggest that p53 serine 18 phosphorylation does not contribute to p53 protein stability, but rather contributes to p53 target gene activation and, as a consequence, p53-dependent apoptosis and late-onset tumor suppression.
In vitro studies showed that phosphorylation of human p53 at serine 20 (mouse p53 serine 23) also plays a key part in p53 stabilization in response to DNA damage signals, again through displacement of Mdm2. To define the role of S23 modification in a physiological context, p53S23A knock-in mice were generated [83,84]. While one study did not observe any compromise in DNA-damage-induced p53 stability or activity, another reported various phenotypes. Whereas p53S23A protein levels and apoptosis-inducing activity were diminished in thymocytes upon irradiation, p53S23A displayed normal stabilization and cell-cycle arrest activity in MEFs upon irradiation. In addition, p53S23A/S23A mice displayed decreased survival relative to wild-type mice, but not to the extent observed in p53−/− mice. Moreover, these mice developed B-cell lymphomas rather than the T-cell lymphomas typifying p53−/− mice. These data show that serine 23 phosphorylation in response to DNA-damage contributes to p53 protein stabilization and p53-dependent apoptosis in a cell-type dependent manner as well as to suppression of B-cell lymphomagenesis. Finally, given that phosphorylation of serines 18 and 23 is thought to coordinately lead to p53 stabilization, a p53S18A-S23A mouse knock-in strain harboring mutations in both serines was generated [85]. The responses were very cell-type-dependent. In MEFs, p53S18A-S23A showed normal protein stabilization as well as somewhat impaired transcriptional activity and mildly compromised cell-cycle arrest function upon DNA damage, similar to the p53S18A single mutant, indicating no cooperativity between phosphorylation sites in this context. However, in thymocytes, p53S18A-S23A was inefficiently stabilized, did not transactivate target genes effectively, and was severely impaired in driving apoptosis in response to DNA damage, indicating that these phosphorylation sites are required to promote apoptosis in thymocytes. Additionally, p53S18A-S23A/S18A-S23A mice displayed decreased survival compared to wild-type mice due to succumbing to late-onset spontaneous tumors, a more dramatic phenotype than that observed by the same group upon analysis of p53S18A/S18A mice. Together, these results suggest that serines 18 and 23 act coordinately to promote apoptosis and tumor suppression in some tissues. Moreover, these findings further highlight the idea that retaining cell-cycle arrest is sufficient to inhibit early-onset spontaneous tumors but lack of apoptosis activity can drive later-onset tumors.
Another p53 protein phosphorylation site linked specifically to p53 apoptotic function in vitro is serine 46 of human p53, which is phosphorylated in response to DNA damage, helping to elicit an apoptotic response through the induction of the p53 target gene p53AIP1 [86]. As there is no conserved residue in murine p53 [87], a HUPKI mouse strain with serine 46 mutation (p53hupkiS46A) was generated to characterize the in vivo role of serine 46 phosphorylation in p53-dependent DNA-damage responses [88]. p53hupkiS46A/S46A cells treated with DNA damaging agents displayed decreased induction of apoptotic targets, including Noxa, Puma, and Perp, as well as partially impaired apoptosis compared to counterparts expressing wild-type p53. p53S46A was also slightly compromised in inducing senescence. Thus, this residue is primarily important for p53-mediated apoptosis, but its role in tumor suppression in vivo remains to be determined.
While the aforementioned studies suggest a role for p53 phosphorylation in regulating p53 function upon exposure to DNA double-strand break inducers like gamma-irradiation, in vitro studies indicate that phosphorylation of serine 389 of murine p53 occurs specifically in response to UV-irradiation, but not gamma-irradiation. To examine the significance of this modification in vivo, p53S389A knock-in mice were generated [89]. Relative to wild-type p53, p53S389A displayed decreased DNA-binding and compromised transactivation of such targets as p21, Mdm2, Noxa, and Bax as well as impaired apoptosis in response to UV-irradiation but not in response to double-strand break inducers. In keeping with this specific role in responding to UV-irradiation, chronic UV-irradiation of p53S389A/S389A mice increased skin tumor development relative to wild-type mice. These studies thus demonstrate that serine 389 phosphorylation selectively promotes apoptotic and tumor suppressive programs in response to UV-irradiation.
Human p53 can be phosphorylated at serine 315 (serine 312 in mouse) upon irradiation as well as upon endoplasmic-reticulum stress [90,91]. In vitro data suggested that phosphorylation at this site is involved in target gene activation or cytoplasmic localization. To clarify the role of phosphorylation at this site in a physiological setting, p53S312A knock-in mice were generated[92,93]. However, analysis of cells from these mice revealed that p53 protein stability, transactivation capacity, cell-cycle arrest function, and apoptotic activity were all intact. Furthermore, survival of p53S312A/S312A mice was comparable to p53+/+ mice. However, p53 S312A mutation partially rescues the early embryonic lethality of Mdm4−/− embryos, which is caused by active p53, suggesting that this mutation inactivates p53 function during development [93]. Moreover, p53S312A/S312A mice are more susceptible to radiation-induced tumorigenesis, indicating that phosphorylation at S312 is important for tumor suppression. In addition, p53 phosphorylation at this site has been implicated as critical in ES cells for Nanog repression and ES cell differentiation [94], suggesting a role for serine 315 phosphorylation in stem cells as well.
Beyond the key role of phosphorylation in regulating p53, in vitro studies have demonstrated that p53 is acetylated in response to stress signals such as DNA damage and oxidative stress [57,58]. The murine p53 C-terminus contains multiple lysine residues (K367, K369, K370, K378, K379, K383, and K384) that can be post-translationally modified by ubiquitination, acetylation, neddylation, sumoylation, or methylation. Two knock-in mouse strains sought to address the importance of these residues by mutating all C-terminal lysines to arginines to block any type of modification at those residues. The first knock-in mouse strain, expressing “p536KR” carried six C-terminal lysine mutations (K367R, K369R, K370R, K378R, K379R, K383R) [95], while the second mouse strain, termed “p537KR” had seven mutations, including the aforementioned ones and one at lysine 384 (K384R), which is not conserved in humans [96]. Surprisingly, homozygous p536KR and p537KR mutant cells exhibited rather subtle phenotypes. p53 protein accumulation under basal conditions was similar to that of wild-type p53, an unexpected finding given the envisaged need for these lysines for ubiquitination and destabilization of p53 in unstressed cells. Neither p536KR nor p537KR showed dramatic phenotypes in various p53-dependent assays: p536KR showed a slight compromise in inducing certain p53 target genes and apoptosis in some contexts, while p537KR engaged DNA-damage-induced cell-cycle arrest and apoptotic programs normally. The only phenotypes evident for p537KR were increased stabilization and activation of target genes in response to DNA damage, and a slightly enhanced ability to induce senescence relative to wild-type controls, suggesting the mutant is mildly hyperactive [96]. Consistent with this notion, p537KR impedes hematopoietic stem/progenitor cell proliferation in response to irradiation, rendering p537KR mice highly radiosensitive [97]. However, both p537KR and p536KR were fully competent as tumor suppressors. Thus, together, these data indicate that the modifications at p53’s C-terminus may play a role in fine-tuning the p53 response to DNA damage.
The concurrent mutation of many lysines simultaneously in these strains may have masked the specific roles of PTMs on individual residues. To address the role of single lysines, some studies have examined the consequences of altering individual lysines, such as in the knock-in mouse strain expressing p53K117R, as described above. In addition, p53K317R knock-in mice were made to pursue the in vitro finding that acetylation of human p53 at K320 (corresponding to mouse K317) correlated with enhanced sequence-specific DNA binding and transactivation of p53 targets [98]. However, the p53K317R knock-in mice, defective for acetylation at this residue, surprisingly indicated the opposite. Despite normal p53 protein stabilization in DNA-damage-treated p53K317R/K317R cells, target genes such as Noxa and Pidd were induced to higher levels than in wild-type cells in response to DNA damage and irradiated p53K317R/K317R mice showed increased apoptosis in the small intestine relative to p53+/+ mice. Taken together, the augmentation of both pro-apoptotic gene expression and apoptosis indicates that acetylation at lysine 317 negatively regulates p53 transcriptional activity. This illustration that acetylation on one specific residue can negatively regulate p53 activity, while other acetylation events can positively regulate p53 function, highlights the possibility that the analysis of compound mutants with numerous alterations may obscure the role of a PTM on an individual residue.
Analysis of p53 PTM sites using mutant mouse models has revealed that many of these modifications are required for maximal p53 responses to cellular stresses. In addition, there are context-dependent roles for individual p53 PTMs, in specific cell types or in response to particular stress signals. While some studies have confirmed an expected function for a given PTM based on in vitro studies, there have been some surprises, such as the relatively normal stability of p537KR and p536KR under basal conditions and the efficient stabilization of p53S18A/S23A in some cell types after DNA damage. In addition, we have learned that stress signals can promote both “activating” and “repressing” PTMs that can fine-tune p53 activity in a particular setting. Importantly, the phenotypes of compound PTM site mutant mice may thus represent the net effect of altering both positively- and negatively-regulated p53 PTMs. Future investigations will provide further insight into the specific role of each PTM and how modifications can be manipulated as therapeutic targets for cancer.
6. Looking upstream of p53
In addition to mutations in p53 itself, alterations in other components of the p53 pathway may contribute to tumorigenesis[99,100]. The significance of perturbations in specific upstream regulators of p53 for carcinogenesis has been unequivocally demonstrated using mouse models. As described above, p53 is normally restrained by negative regulators, the best known of which are Mdm2 and Mdm4 [9,10]. Mdm2 binds the p53 transcriptional activation domains and serves as a key negative regulator both by masking the p53 transactivation domains and by serving as an E3 ubiquitin ligase to destabilize p53. The importance of the Mdm2-p53 interaction in restricting p53 activity was initially shown through the generation of Mdm2−/− mice, which displayed early embryonic lethality that was rescued in the backdrop of p53 nullizygosity [9]. Moreover, Mdm2 transgenic mice demonstrated that Mdm2 promotes the development of multiple tumor types[101]. In further support of a role for p53 pathway alterations in human carcinogenesis, Mdm2 is frequently amplified in 30–40% of human sarcomas of various types [99,101,102] as well as in other tumor types [101]. Akin to Mdm2, the Mdm4 protein also interacts with p53 transactivation domains to block their function. Moreover, although not an E3 ubiqutin ligase itself, Mdm4 can hetero-oligomerize with Mdm2 to enhance Mdm2 stability and ligase activity. As with Mdm2 deficiency, Mdm4−/− mice exhibit early embryonic lethality due to unrestrained p53 activity, and viability is restored upon p53 loss [103–105]. Mdm4 transgenic mice have bolstered the idea that modulating p53 pathway components can promote cancer, as these mice more readily developed sarcomas than non-transgenic counterparts [106]. Mdm4 is also amplified in human cancers such as melanomas and retinoblastomas, illustrating the significance of this event in human tumorigenesis [107,108].
Oncogenic signals activate p53 through induction of the p19ARF tumor suppressor, which positively regulates p53 by binding and inhibiting Mdm2 [109–111]. The importance of p19ARF in activating p53 tumor suppressor function is underscored by the observation that p19ARF null mice are predisposed to early-onset spontaneous tumors, including fibrosarcomas and lymphomas [112]. Moreover, frequent mutations or deletions of the Ink4a locus, which comprises both p16 and p19ARF, are found in human cancers. Together, these results are consistent with the important role of p19ARF in activating p53 in response to oncogenic signals as a safeguard against tumorigenesis. Collectively, these studies highlight the fact that the p53 pathway can be compromised not only through p53 mutation but also through disruption of other pathway components.
7. Therapeutic promise of p53 activation
As described above, complete loss of p53 function results in a dramatic predisposition to tumor development. An important follow-up question was whether sustained p53 loss is necessary for tumor maintenance, as has been noted with oncogenes, where tumors display “oncogene addiction” [113]. Given the genomic instability that can ensue after p53 loss, it was plausible that reactivation of p53 would no longer be able to dampen tumor growth – a question of paramount importance for evaluating the therapeutic potential of p53 restoration. Thus, several studies in mouse models have investigated this question by directly evaluating the consequence of p53 reactivation in tumors. In initial studies, three different approaches to p53 restoration were taken. In one case, mice homozygous for a p53LSL-wt allele, in which a floxed transcriptional stop cassette silences the p53 locus, creating a p53 null allele, were aged to allow spontaneous tumor development [114]. Upon activation of a tamoxifen-inducible, ubiquitously expressed Cre recombinase to induce widespread p53 expression, the majority of spontaneous lymphomas and sarcomas were found to regress. p53-driven regression of lymphomas was associated with apoptosis, while regression of sarcomas was associated with cellular senescence. Another study leveraged knock-in mice expressing a fusion protein of p53 to the hormone-binding domain of the estrogen receptor (p53ERTAM, Fig. 2), which remains inactive until the administration of tamoxifen [50,115]. After allowing Eμ-myc;p53ERTAM/− B-cell lymphomas to develop, tamoxifen was used to activate p53, and it was found that tumors underwent efficient regression by p53-mediated apoptosis [116]. Additionally, in a model of hepatocellular carcinoma driven by HRasV12 and expression of an shRNA directed against p53, reactivating p53 by extinguishing the p53 shRNA also caused tumor regression, associated with cellular senescence and tumor clearance through activation of the innate immune system [12]. All of these studies highlighted the therapeutic potential of p53 reactivation in tumors. However, in contrast to these findings, p53 reactivation in a KrasG12D-driven model of non-small-cell lung cancer showed more limited activity in tumor regression [117,118]. Specifically, p53 restoration led to tumor cell elimination only in malignant adenocarcinomas but not adenomas, and this difference was attributable to lack of a sufficiently potent oncogenic signal to activate p53 in the less malignant lesions. These studies therefore provide a cautionary note in terms of the efficacy of tumor eradication by p53 restoration, and indicate that the potential of reactivating wild-type p53 as a therapeutic avenue for treating p53-deficient tumors may be limited to select contexts.
8. Summary
The development of genetically engineered mice expressing p53 mutants has greatly contributed to our understanding of p53 function during tumorigenesis. These studies have established the importance not only of p53 loss-of-function but also p53 gain-of-function properties for promoting carcinogenesis, setting the stage for more detailed unraveling of the pathways underlying these activities. Structure–function analyses have provided new insight into the molecular and cellular mechanisms underlying p53 action in vivo, prompting some revision of models for how p53 acts. Investigation of the roles of p53 post-translational modification in regulating p53 has yielded an enhanced understanding of how stress signals impinge upon p53. Finally, reactivation of p53 in mouse models has shown the potential of p53 restoration in limiting tumorigenesis in some but not all contexts. Although studies of each mouse model have revealed new insight into p53, they also have raised new questions about p53 function that will require additional investigation both in vitro and in vivo. Importantly, having a complete and accurate understanding of p53 biology in vivo will ultimately pave the way for translating our knowledge into improved cancer therapy.
Acknowledgements
We would like to thank Kathryn Bieging and Stephano Mello for critical reading of the manuscript. We apologize to those whose work was not cited due to space constraints. LDA’s laboratory is supported by the DOD, the Leukemia and Lymphoma Society, and the NIH (grants R01 CA140875 and R21 CA169673). PBG is supported by the NIH (grant T32 CA09151).
References
- [1].Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009 May;137:413. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- [2].Brady CA, Attardi LD. p53 at a glance. Journal of Cell Science. 2010 Aug;123:2527. doi: 10.1242/jcs.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Malkin D. Li–Fraumeni syndrome. Genes and Cancer. 2011 Apr;2:475. doi: 10.1177/1947601911413466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Purdie CA, Harrison DJ, Peter A, Dobbie L, White S, Howie SE, et al. Tumour incidence, spectrum and ploidy in mice with a large deletion in the p53 gene. Oncogene. 1994 Feb;9:603. [PubMed] [Google Scholar]
- [5].Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, et al. Tumor spectrum analysis in p53-mutant mice. Current Biology: CB. 1994 Jan;4:1. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- [6].Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992 Mar;356:215. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- [7].Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nature Reviews Cancer. 2004 Oct;4:793. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- [8].Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997 Oct;91:325. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- [9].Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995 Nov;378:203. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- [10].Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends in Cell Biology. 2010 May;20:299. doi: 10.1016/j.tcb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lane DP. Cancer. p53, guardian of the genome. Nature. 1992 Jul;358:15. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- [12].Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007 Feb;445:656. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nature Reviews Molecular Cell Biology. 2008 May;9:402. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- [14].Meek DW, Anderson CW. Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harbor Perspectives in Biology. 2009 Dec;1:a000950. doi: 10.1101/cshperspect.a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nature Reviews Molecular Cell Biology. 2008 Sep;9:702. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- [16].Attardi LD, Donehower LA. Probing p53 biological functions through the use of genetically engineered mouse models. Mutation Research. 2005 Aug;576:4. doi: 10.1016/j.mrfmmm.2004.08.022. [DOI] [PubMed] [Google Scholar]
- [17].Venkatachalam S, Shi YP, Jones SN, Vogel H, Bradley A, Pinkel D, et al. Retention of wild-type p53 in tumors from p53 heterozygous mice: reduction of p53 dosage can promote cancer formation. EMBO Journal. 1998 Aug;17:4657. doi: 10.1093/emboj/17.16.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kuperwasser C, Hurlbut GD, Kittrell FS, Dickinson ES, Laucirica R, Medina D, et al. Development of spontaneous mammary tumors in BALB/c p53 heterozygous mice. A model for Li–Fraumeni syndrome. American Journal of Pathology. 2000 Dec;157:2151. doi: 10.1016/S0002-9440(10)64853-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lin SC, Lee KF, Nikitin AY, Hilsenbeck SG, Cardiff RD, Li A, et al. Somatic mutation of p53 leads to estrogen receptor alpha-positive and negative mouse mammary tumors with high frequency of metastasis. Cancer Research. 2004 May;64:3525. doi: 10.1158/0008-5472.CAN-03-3524. [DOI] [PubMed] [Google Scholar]
- [20].Shai A, Pitot HC, Lambert PF. p53 loss synergizes with estrogen and papillomaviral oncogenes to induce cervical and breast cancers. Cancer Research. 2008 Apr;68:2622. doi: 10.1158/0008-5472.CAN-07-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gostissa M, Bianco JM, Malkin DJ, Kutok JL, Rodig SJ, Morse HC, 3rd, et al. Conditional inactivation of p53 in mature B cells promotes generation of nongerminal center-derived B-cell lymphomas. Proceedings of the National Academy of Sciences of the United States of America. 2013 Feb;110:2934. doi: 10.1073/pnas.1222570110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Attardi LD, Jacks T. The role of p53 in tumour suppression: lessons from mouse models. Cellular and Molecular Life Sciences: CMLS. 1999 Jan;55:48. doi: 10.1007/s000180050269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nature Reviews Cancer. 2009 Oct;9:701. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- [24].Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Human Mutation. 2007 Jun;28:622. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- [25].Muller PA, Vousden KH. p53 mutations in cancer. Nature Cell Biology. 2013 Jan;15:2. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- [26].Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994 Jul;265:346. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- [27].Lavigueur A, Maltby V, Mock D, Rossant J, Pawson T, Bernstein A. High incidence of lung, bone, and lymphoid tumors in transgenic mice overexpressing mutant alleles of the p53 oncogene. Molecular and Cellular Biology. 1989 Sep;9:3982. doi: 10.1128/mcb.9.9.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lee JM, Abrahamson JL, Kandel R, Donehower LA, Bernstein A. Susceptibility to radiation-carcinogenesis and accumulation of chromosomal breakage in p53 deficient mice. Oncogene. 1994 Dec;9:3731. [PubMed] [Google Scholar]
- [29].Harvey M, Vogel H, Morris D, Bradley A, Bernstein A, Donehower LA. A mutant p53 transgene accelerates tumour development in heterozygous but not nullizygous p53-deficient mice. Nature Genetics. 1995 Mar;9:305. doi: 10.1038/ng0395-305. [DOI] [PubMed] [Google Scholar]
- [30].Dittmer D, Pati S, Zambetti G, Chu S, Teresky AK, Moore M, et al. Gain of function mutations in p53. Nature Genetics. 1993 May;4:42. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- [31].Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li–Fraumeni syndrome. Cell. 2004 Dec;119:861. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- [32].Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li–Fraumeni syndrome. Cell. 2004 Dec;119:847. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- [33].Luo JL, Yang Q, Tong WM, Hergenhahn M, Wang ZQ, Hollstein M. Knock-in mice with a chimeric human/murine p53 gene develop normally and show wild-type p53 responses to DNA damaging agents: a new biomedical research tool. Oncogene. 2001 Jan;20:320. doi: 10.1038/sj.onc.1204080. [DOI] [PubMed] [Google Scholar]
- [34].Liu DP, Song H, Xu Y. A common gain of function of p53 cancer mutants in inducing genetic instability. Oncogene. 2010 Feb;29:949. doi: 10.1038/onc.2009.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Song H, Hollstein M, Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nature Cell Biology. 2007 May;9:573. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- [36].Mello SS, Attardi LD. Not all p53 gain-of-function mutants are created equal. Cell Death and Differentiation. 2013 Jul;20:855. doi: 10.1038/cdd.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hanel W, Marchenko N, Xu S, Yu SX, Weng W, Moll U. Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death and Differentiation. 2013 Jul;20:898. doi: 10.1038/cdd.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lu X, Liu DP, Xu Y. The gain of function of p53 cancer mutant in promoting mammary tumorigenesis. Oncogene. 2013 Jun;32:2900. doi: 10.1038/onc.2012.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Johnson TM, Hammond EM, Giaccia A, Attardi LD. The p53QS transactivation-deficient mutant shows stress-specific apoptotic activity and induces embryonic lethality. Nature Genetics. 2005 Feb;37:145. doi: 10.1038/ng1498. [DOI] [PubMed] [Google Scholar]
- [40].Lin J, Chen J, Elenbaas B, Levine AJ. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes and Development. 1994 May;8:1235. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- [41].Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011 May;145:571. doi: 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lozano G, Zambetti GP. What have animal models taught us about the p53 pathway? Journal of Pathology. 2005 Jan;205:206. doi: 10.1002/path.1704. [DOI] [PubMed] [Google Scholar]
- [43].Jiang D, Brady CA, Johnson TM, Lee EY, Park EJ, Scott MP. Full p53 transcriptional activation potential is dispensable for tumor suppression in diverse lineages. Proceedings of the National Academy of Sciences of the United States of America. 2011 Oct;108:17123. doi: 10.1073/pnas.1111245108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Candau R, Scolnick DM, Darpino P, Ying CY, Halazonetis TD, Berger SL. Two tandem and independent sub-activation domains in the amino terminus of p53 require the adaptor complex for activity. Oncogene. 1997 Aug;15:807. doi: 10.1038/sj.onc.1201244. [DOI] [PubMed] [Google Scholar]
- [45].Venot C, Maratrat M, Sierra V, Conseiller E, Debussche L. Definition of a p53 transactivation function-deficient mutant and characterization of two independent p53 transactivation subdomains. Oncogene. 1999 Apr;18:2405. doi: 10.1038/sj.onc.1202539. [DOI] [PubMed] [Google Scholar]
- [46].Zhu J, Zhou W, Jiang J, Chen X. Identification of a novel p53 functional domain that is necessary for mediating apoptosis. Journal of Biological Chemistry. 1998 May;273:13030. doi: 10.1074/jbc.273.21.13030. [DOI] [PubMed] [Google Scholar]
- [47].Gaidarenko O, Xu Y. Transcription activity is required for p53-dependent tumor suppression. Oncogene. 2009 Dec;28:4397. doi: 10.1038/onc.2009.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hinkal G, Parikh N, Donehower LA. Timed somatic deletion of p53 in mice reveals age-associated differences in tumor progression. PLoS ONE. 2009;4:e6654. doi: 10.1371/journal.pone.0006654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Efeyan A, Garcia-Cao I, Herranz D, Velasco-Miguel S, Serrano M. Tumour biology: policing of oncogene activity by p53. Nature. 2006 Sep;443:159. doi: 10.1038/443159a. [DOI] [PubMed] [Google Scholar]
- [50].Christophorou MA, Ringshausen I, Finch AJ, Swigart LB, Evan GI. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature. 2006 Sep;443:214. doi: 10.1038/nature05077. [DOI] [PubMed] [Google Scholar]
- [51].Johnson TM, Meade K, Pathak N, Marques MR, Attardi LD. Knockin mice expressing a chimeric p53 protein reveal mechanistic differences in how p53 triggers apoptosis and senescence. Proceedings of the National Academy of Sciences of the United States of America. 2008 Jan;105:1215. doi: 10.1073/pnas.0706764105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Liu G, Parant JM, Lang G, Chau P, Chavez-Reyes A, El-Naggar AK. Chromosome stability, in the absence of apoptosis, is critical for suppression of tumorigenesis in Trp53 mutant mice. Nature Genetics. 2004 Jan;36:63. doi: 10.1038/ng1282. [DOI] [PubMed] [Google Scholar]
- [53].Ludwig RL, Bates S, Vousden KH. Differential activation of target cellular promoters by p53 mutants with impaired apoptotic function. Molecular and Cellular Biology. 1996 Sep;16:4952. doi: 10.1128/mcb.16.9.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Barboza JA, Liu G, Ju Z, El-Naggar AK, Lozano G. p21 Delays tumor onset by preservation of chromosomal stability. Proceedings of the National Academy of Sciences of the United States of America. 2006 Dec;103:19842. doi: 10.1073/pnas.0606343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Schlereth K, Heyl C, Krampitz AM, Mernberger M, Finkernagel F, Scharfe M, et al. Characterization of the p53 cistrome – DNA binding cooperativity dissects p53’s tumor suppressor functions. PLoS Genetics. 2013 Aug;9:e1003726. doi: 10.1371/journal.pgen.1003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Timofeev O, Schlereth K, Wanzel M, Braun A, Nieswandt B, Pagenstecher A, et al. p53 DNA binding cooperativity is essential for apoptosis and tumor suppression in vivo. Cell Reports. 2013 May;3:1512. doi: 10.1016/j.celrep.2013.04.008. [DOI] [PubMed] [Google Scholar]
- [57].Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001 Oct;107:137. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- [58].Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009 May;137:609. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Molecular Cell. 2006 Dec;24:841. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Molecular Cell. 2006 Dec;24:827. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- [61].Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008 May;133:612. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012 Jun;149:1269. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Walker KK, Levine AJ. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proceedings of the National Academy of Sciences of the United States of America. 1996 Dec;93:15335. doi: 10.1073/pnas.93.26.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Toledo T, Lee CJ, Krummel KA, Rodewald LW, Liu CW, Wahl GM. Mouse mutants reveal that putative protein interaction sites in the p53 proline-rich domain are dispensable for tumor suppression. Molecular and Cellular Biology. 2007 Feb;27:1425. doi: 10.1128/MCB.00999-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Venot C, Maratrat M, Dureuil C, Conseiller E, Bracco L, Debussche L. The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. EMBO Journal. 1998;17:4668. doi: 10.1093/emboj/17.16.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Baptiste N, Friedlander P, Chen X, Prives C. The proline-rich domain of p53 is required for cooperation with anti-neoplastic agents to promote apoptosis of tumor cells. Oncogene. 2002 Jan;21:9. doi: 10.1038/sj.onc.1205015. [DOI] [PubMed] [Google Scholar]
- [67].Sakamuro D, Sabbatini P, White E, Prendergast GC. The polyproline region of p53 is required to activate apoptosis but not growth arrest. Oncogene. 1997 Aug;15:887. doi: 10.1038/sj.onc.1201263. [DOI] [PubMed] [Google Scholar]
- [68].Berger M, Vogt Sionov R, Levine AJ, Haupt Y. A role for the polyproline domain of p53 in its regulation by Mdm2. Journal of Biological Chemistry. 2001 Feb;276:3785. doi: 10.1074/jbc.M008879200. [DOI] [PubMed] [Google Scholar]
- [69].Wulf GM, Liou YC, Ryo A, Lee SW, Lu KP. Role of Pin1 in the regulation of p53 stability and p21 transactivation, and cell cycle checkpoints in response to DNA damage. Journal of Biological Chemistry. 2002 Dec;277:47976. doi: 10.1074/jbc.C200538200. [DOI] [PubMed] [Google Scholar]
- [70].Zacchi P, Gostissa M, Uchida T, Salvagno C, Avolio F, Volinia S, et al. The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature. 2002 Oct;419:853. doi: 10.1038/nature01120. [DOI] [PubMed] [Google Scholar]
- [71].Zheng H, You H, Zhou XZ, Murray SA, Uchida T, Wulf G, et al. The prolyl isomerase Pin1 is a regulator of p53 in genotoxic response. Nature. 2002 Oct;419:849. doi: 10.1038/nature01116. [DOI] [PubMed] [Google Scholar]
- [72].Toledo F, Krummel KA, Lee CJ, Liu CW, Rodewald LW, Tang M, et al. A mouse p53 mutant lacking the proline-rich domain rescues Mdm4 deficiency and provides insight into the Mdm2-Mdm4-p53 regulatory network. Cancer Cell. 2006 Apr;9:273. doi: 10.1016/j.ccr.2006.03.014. [DOI] [PubMed] [Google Scholar]
- [73].Slatter TL, Ganesan P, Holzhauer C, Mehta R, Rubio C, Williams G, et al. p53-mediated apoptosis prevents the accumulation of progenitor B cells and B-cell tumors. Cell Death and Differentiation. 2010 Mar;17:540. doi: 10.1038/cdd.2009.136. [DOI] [PubMed] [Google Scholar]
- [74].Campbell HG, Mehta R, Neumann AA, Rubio C, Baird M, Slatter TL, et al. Activation of p53 following ionizing radiation, but not other stressors, is dependent on the proline-rich domain (PRD) Oncogene. 2013 Feb;32:827. doi: 10.1038/onc.2012.102. [DOI] [PubMed] [Google Scholar]
- [75].Salvioli S, Bonafé M, Barbi C, Storci G, Trapassi C, Tocco F, et al. p53 codon 72 alleles influence the response to anticancer drugs in cells from aged people by regulating the cell cycle inhibitor p21WAF1. Cell Cycle. 2005 Sep;4:1264. doi: 10.4161/cc.4.9.1978. [DOI] [PubMed] [Google Scholar]
- [76].Dumont P, Leu JI, Della Pietra AC, 3rd, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nature Genetics. 2003 Mar;33:357. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- [77].Sullivan A, Syed N, Gasco M, Bergamaschi D, Trigiante G, Attard M, et al. Polymorphism in wild-type p53 modulates response to chemotherapy in vitro and in vivo. Oncogene. 2004 Apr;23:3328. doi: 10.1038/sj.onc.1207428. [DOI] [PubMed] [Google Scholar]
- [78].Frank AK, Leu JI, Zhou Y, Devarajan K, Nedelko T, Klein-Szanto A, et al. The codon 72 polymorphism of p53 regulates interaction with NF-{kappa}B and transactivation of genes involved in immunity and inflammation. Molecular and Cellular Biology. 2011 Mar;31:1201. doi: 10.1128/MCB.01136-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Zhu F, Dollé ME, Berton TR, Kuiper RV, Capps C, Espejo A, et al. Mouse models for the p53 R72P polymorphism mimic human phenotypes. Cancer Research. 2010 Jul;70:5851. doi: 10.1158/0008-5472.CAN-09-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Chao C, Hergenhahn M, Kaeser MD, Wu Z, Saito S, Iggo R, et al. Cell type- and promoter-specific roles of Ser18 phosphorylation in regulating p53 responses. Journal of Biological Chemistry. 2003 Oct;278:41028. doi: 10.1074/jbc.M306938200. [DOI] [PubMed] [Google Scholar]
- [81].Sluss HK, Armata H, Gallant J, Jones SN. Phosphorylation of serine 18 regulates distinct p53 functions in mice. Molecular and Cellular Biology. 2004 Feb;24:976. doi: 10.1128/MCB.24.3.976-984.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Armata HL, Garlick DS, Sluss HK. The ataxia telangiectasia-mutated target site Ser18 is required for p53-mediated tumor suppression. Cancer Research. 2007 Dec;67:11696. doi: 10.1158/0008-5472.CAN-07-1610. [DOI] [PubMed] [Google Scholar]
- [83].MacPherson D, Kim J, Kim T, Rhee BK, Van Oostrom CT, DiTullio RA, et al. Defective apoptosis and B-cell lymphomas in mice with p53 point mutation at Ser 23. EMBO Journal. 2004 Sep;23:3689. doi: 10.1038/sj.emboj.7600363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Wu Z, Earle J, Saito S, Anderson CW, Appella E, Xu Y. Mutation of mouse p53 Ser23 and the response to DNA damage. Molecular and Cellular Biology. 2002 Apr;22:2441. doi: 10.1128/MCB.22.8.2441-2449.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Chao C, Herr D, Chun J, Xu Y. Ser18 and 23 phosphorylation is required for p53-dependent apoptosis and tumor suppression. EMBO Journal. 2006 Jun;25:2615. doi: 10.1038/sj.emboj.7601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 2000 Sep;102:849. doi: 10.1016/s0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- [87].Saito S, Yamaguchi H, Higashimoto Y, Chao C, Xu Y, Fornace AJ, Jr, et al. Phosphorylation site interdependence of human p53 post-translational modifications in response to stress. Journal of Biological Chemistry. 2003 Sep;278:37536. doi: 10.1074/jbc.M305135200. [DOI] [PubMed] [Google Scholar]
- [88].Feng L, Hollstein M, Xu Y. Ser46 phosphorylation regulates p53-dependent apoptosis and replicative senescence. Cell Cycle. 2006 Dec;5:2812. doi: 10.4161/cc.5.23.3526. [DOI] [PubMed] [Google Scholar]
- [89].Bruins W, Zwart E, Attardi LD, Iwakuma T, Hoogervorst EM, Beems RB, et al. Increased sensitivity to UV radiation in mice with a p53 point mutation at Ser389. Molecular and Cellular Biology. 2004 Oct;24:8884. doi: 10.1128/MCB.24.20.8884-8894.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Qu L, Huang S, Baltzis D, Rivas-Estilla AM, Pluquet O, Hatzoglou M, et al. Endoplasmic reticulum stress induces p53 cytoplasmic localization and prevents p53-dependent apoptosis by a pathway involving glycogen synthase kinase-3beta. Genes and Development. 2004 Feb;18:261. doi: 10.1101/gad.1165804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Blaydes JP, Luciani MG, Pospisilova S, Ball HM, Vojtesek B, Hupp TR. Stoichiometric phosphorylation of human p53 at Ser315 stimulates p53-dependent transcription. Journal of Biological Chemistry. 2001 Feb;276:4699. doi: 10.1074/jbc.M003485200. [DOI] [PubMed] [Google Scholar]
- [92].Lee MK, Tong WM, Wang ZQ, Sabapathy K. Serine 312 phosphorylation is dispensable for wild-type p53 functions in vivo. Cell Death and Differentiation. 2011 Feb;18:214. doi: 10.1038/cdd.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Slee EA, Benassi B, Goldin R, Zhong S, Ratnayaka I, Blandino G, et al. Phosphorylation of Ser312 contributes to tumor suppression by p53 in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2010 Nov;107:19479. doi: 10.1073/pnas.1005165107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nature Cell Biology. 2005 Feb;7:165. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- [95].Feng L, Lin T, Uranishi H, Gu W, Xu Y. Functional analysis of the roles of posttranslational modifications at the p53C terminus in regulating p53 stability and activity. Molecular and Cellular Biology. 2005 Jul;25:5389. doi: 10.1128/MCB.25.13.5389-5395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Krummel KA, Lee CJ, Toledo F, Wahl GM. The C-terminal lysines fine-tune P53 stress responses in a mouse model but are not required for stability control or transactivation. Proceedings of the National Academy of Sciences of the United States of America. 2005 Jul;102:10188. doi: 10.1073/pnas.0503068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Wang YV, Leblanc M, Fox N, Mao JH, Tinkum KL, Krummel K, et al. Fine-tuning p53 activity through C-terminal modification significantly contributes to HSC homeostasis and mouse radiosensitivity. Genes and Development. 2011 Jul;25:1426. doi: 10.1101/gad.2024411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Chao C, Wu Z, Mazur SJ, Borges H, Rossi M, Lin T. Acetylation of mouse p53 at lysine 317 negatively regulates p53 apoptotic activities after DNA damage. Molecular and Cellular Biology. 2006 Sep;26:6859. doi: 10.1128/MCB.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Marine JC, Jochemsen AG. Mdmx as an essential regulator of p53 activity. Biochemical and Biophysical Research Communications. 2005 Jun;331:750. doi: 10.1016/j.bbrc.2005.03.151. [DOI] [PubMed] [Google Scholar]
- [100].Sherr CJ. Divorcing ARF and p53: an unsettled case. Nature Reviews Cancer. 2006 Sep;6:663. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- [101].Jones SN, Hancock AR, Vogel H, Donehower LA, Bradley A. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 1998 Dec;95:15608. doi: 10.1073/pnas.95.26.15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992 Jul;358:80. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- [103].Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nature Genetics. 2001 Sep;29:92. doi: 10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- [104].Finch RA, et al. mdmx is a negative regulator of p53 activity in vivo. Cancer Research. 2002 Jun;62:3221. [PubMed] [Google Scholar]
- [105].Migliorini D, Lazzerini Denchi E, Danovi D, Jochemsen A, Capillo M, Gobbi A, et al. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Molecular and Cellular Biology. 2002 Aug;22:5527. doi: 10.1128/MCB.22.15.5527-5538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Xiong S, Pant V, Suh Y, Van Pelt CS, Wang Y, Valentin-Vega YA, et al. Spontaneous tumorigenesis in mice overexpressing the p53-negative regulator Mdm4. Cancer Research. 2010 Sep;70:7148. doi: 10.1158/0008-5472.CAN-10-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Gembarska A, Luciani F, Fedele C, Russell EA, Dewaele M, Villar S, et al. MDM4 is a key therapeutic target in cutaneous melanoma. Nature Medicine. 2012 Aug;18:1239. doi: 10.1038/nm.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Laurie NA, Donovan SL, Shih CS, Zhang J, Mills N, Fuller C, et al. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006 Nov;444:61. doi: 10.1038/nature05194. [DOI] [PubMed] [Google Scholar]
- [109].Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proceedings of the National Academy of Sciences of the United States of America. 1998 Jul;95:8292. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Pomerantz J, Schreiber-Agus N, Liégeois NJ, Silverman A, Alland L, Chin L, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998 Mar;92:713. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- [111].Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S, et al. The alternative product from the human CDKN2A locus, p14 (ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO Journal. 1998 Sep;17:5001. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997 Nov;91:649. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- [113].Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009 Mar;136:823. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007 Feb;445:661. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- [115].Christophorou MA, Martin-Zanca D, Soucek L, Lawlor ER, Brown-Swigart L, Verschuren EW, et al. Temporal dissection of p53 function in vitro and in vivo. Nature Genetics. 2005 Jul;37:718. doi: 10.1038/ng1572. [DOI] [PubMed] [Google Scholar]
- [116].Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006 Dec;127:1323. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- [117].Junttila MR, Karnezis AN, Garcia D, Madriles F, Kortlever RM, Rostker F, et al. Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature. 2010 Nov;468:567. doi: 10.1038/nature09526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Feldser DM, Kostova KK, Winslow MM, Taylor SE, Cashman C, Whittaker CA, et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature. 2010 Nov;468:572. doi: 10.1038/nature09535. [DOI] [PMC free article] [PubMed] [Google Scholar]