Significance
Nitrogen nodules formed by the symbiosis of rhizobial microbes and legume roots are essential for fixation of nitrogen in the environment. As part of the symbiosis that leads to nodule formation, a series of changes in gene expression of the Rhizobium must occur. The protein NolR is a global regulator of rhizobial genes for symbiosis and nodulation. Here, we describe the three-dimensional structure of this transcription factor in unliganded and DNA bound forms. These structures show how NolR recognizes asymmetric DNA binding sites and reveal a previously unknown mechanism for conformational switching that alters the energetics of interaction to accommodate variable DNA sequences. Two models for the role of NolR in the regulation of nodulation and symbiosis genes are also proposed.
Keywords: transcription factor, protein structure
Abstract
The symbiosis between rhizobial microbes and host plants involves the coordinated expression of multiple genes, which leads to nodule formation and nitrogen fixation. As part of the transcriptional machinery for nodulation and symbiosis across a range of Rhizobium, NolR serves as a global regulatory protein. Here, we present the X-ray crystal structures of NolR in the unliganded form and complexed with two different 22-base pair (bp) double-stranded operator sequences (oligos AT and AA). Structural and biochemical analysis of NolR reveals protein–DNA interactions with an asymmetric operator site and defines a mechanism for conformational switching of a key residue (Gln56) to accommodate variation in target DNA sequences from diverse rhizobial genes for nodulation and symbiosis. This conformational switching alters the energetic contributions to DNA binding without changes in affinity for the target sequence. Two possible models for the role of NolR in the regulation of different nodulation and symbiosis genes are proposed. To our knowledge, these studies provide the first structural insight on the regulation of genes involved in the agriculturally and ecologically important symbiosis of microbes and plants that leads to nodule formation and nitrogen fixation.
The symbiosis between rhizobial bacteria from the Rhizobium, Sinorhizobium, Mesorhizobium, Azorhizobium, and Bradyrhizobium genera and leguminous plants leads to the formation of root nodules (1, 2). These plant organs are specialized for nitrogen fixation and assimilation and are of major ecological and agricultural importance. For example, nitrogen-fixing nodules account for one quarter of total nitrogen fixed globally each year (3). The development of nitrogen-fixing nodules by rhizobia involves a variety of interactions between the plant and microbe; however, at the center of this process are a set of nod (nodulation) genes required for the synthesis of oligosaccharide-nodulation factors, for determining host-plant specificity, and for optimizing the efficiency of symbiosis (4–6). Successful interaction between the rhizobium and host plant requires expression of both positive and negative transcriptional control of genes related to nodulation and symbiosis (7, 8).
Expression of nod genes in rhizobium is regulated by flavonoids released from the host plant in conjunction with the positive activator NodD, which is a member of the LysR-type transcriptional regulator (LTTR) family of proteins (9, 10). Extensive analyses reveal that, in response to small molecules produced by the plant, the rhizobial NodD protein binds to a cis-acting element—the nod box—located upstream of genes required for nodulation (11–16). In addition to the positive control provided by NodD, negative regulation of the nod regulon in Rhizobium meliloti and other rhizobia by NolR occurs (17, 18).
In rhizobia, NolR modulates expression of the NodD activator protein, the core nod genes, and multiple genes involved in symbiosis (17, 19–24). Based on amino acid sequence homology, NolR was proposed to be a helix-turn-helix family member that binds to a nonpalindromic consensus motif —(A/T)TTAG-N9-A(T/A) (17). NolR is well-conserved across multiple Sinorhizobium and Rhizobium species (23). Originally, NolR was identified as a negative regulator of nodulation that bound to overlapping transcription initiation sites in the nodD1 and nodA promoters and at the nodD2 promoter (17). Differential regulation of nod genes by NolR suggested that only the genes related to the synthesis of the core Nod factor structure were controlled by this transcription factor (22). Interaction of NolR with the target DNA site led to reduced levels of Nod factors (22). Control of Nod factor production by NolR may also aid in optimization of nodulation specificity, as NolR binding sequences were later found in the promoter regions of nodABC, nodD1, ttsI-nodD2, nolR, hesB, and nodZ (23). Transcript levels of nolR are high in free-living rhizobia and in the bacteroid but are down-regulated by luteolin, a nod gene inducer (24). Subsequent studies implicated NolR as a global regulatory factor that responds to environmental factors to fine-tune a range of symbiotic signals, not just the genes required for nodulation, and that the absence or presence of NolR affects symbiotic interactions with host plants (19, 20, 23, 24). For example, NolR represses expression of the type III secretion system ttsI gene, which is required for secretion of nodulation outer proteins (nops) that are beneficial for symbiosis of Sinorhizobium fredii with some soybean cultivars (24). The molecular basis for NolR recognition of nonpalindromic DNA target sites and subsequent control of nodulation and symbiosis genes is unclear.
To understand how NolR functions as a global regulator of nodulation, we used a combination of X-ray crystallography, thermodynamic analysis of protein–DNA interactions, and site-directed mutagenesis. The structures of NolR in the unbound form and in complex with two different 22-base pair (bp) double-stranded DNA fragments (oligos AT and AA) reveal a homodimeric protein adopting a winged helix-turn-helix fold. These structures suggest a previously unknown mechanism for conformational switching of a key glutamine side-chain to accommodate DNA-sequence variation in nonpalindromic operator sites without a loss of interaction affinity but with altered thermodynamic contributions to binding. Models for the role of NolR in the regulation of nodulation and symbiosis genes are proposed.
Results
Overall Structure of NolR.
NolR from S. fredii USDA191 was expressed as an N-terminally His-tagged protein in Escherichia coli and purified by nickel-affinity and size-exclusion chromatography. The His-tag was removed by thrombin digestion for crystallization. NolR migrated as a dimeric 26-kDa species (monomer Mr ∼ 13 kDa) by size-exclusion chromatography. Crystals of uncomplexed native NolR and selenomethionine (SeMet)-substituted NolR in complex with a 22-bp oligonucleotide duplex corresponding to the consensus NolR DNA binding motif (oligo AT) were obtained and optimized for data collection (Table S1). The 3D structure of SeMet-substituted NolR bound to oligo AT was determined using single-wavelength anomalous dispersion (SAD) phasing. The resulting model was used to solve the structure of unliganded NolR by molecular replacement.
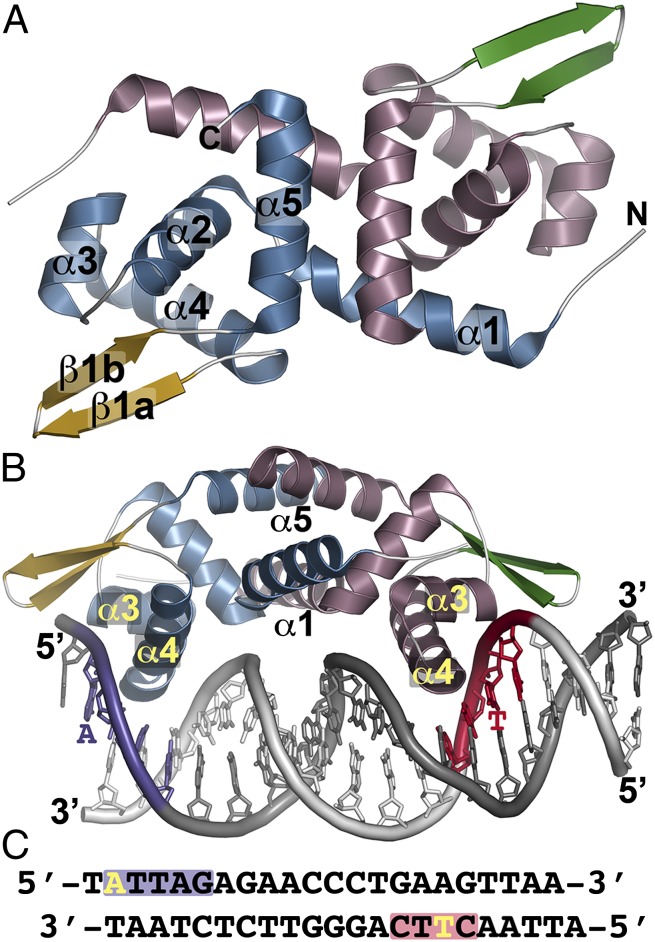
The overall structure of NolR reveals that the protein is a winged helix-turn-helix transcription factor (25) (Fig. 1 A and B). Two α-helices (α1 and α5) of each monomer form the coiled-coil dimerization interface of NolR. A triangular set of α-helices (α2–α4) positions α3 (residues 45–52) and α4 (residues 55–69) as the helix-turn-helix motif for interaction with the DNA major groove, and the “wing,” a two-stranded antiparallel β-sheet (β1a and β1b), extends into the minor groove (Fig. 1B). For cocrystallization, the conserved NolR operator site from R. meliloti was used (Fig. 1C) (17). The operator contains two conserved motifs with variable positions that can be either A or T. Comparison of the free and bound forms of NolR indicates that the structure changes little upon DNA binding with a 0.433-Å root-mean-square deviation (rmsd) for 196 Cα atoms in the homodimer. The crystal structure reveals that the NolR dimer binds to position residues of α4 from one monomer to contact the first sequence block (Fig. 1 B and C, purple) on the 5′ strand of the operator and that the same helix of the second monomer interacts with the sequence of the second consensus block (Fig. 1 B and C, red) on the 3′ strand.
Fig. 1.
Overall structure of NolR. (A) The structure of unliganded NolR is shown as a ribbon diagram. Secondary structure features are labeled in monomer A and are differentially colored in each monomer as follows: blue α-helices and gold β-strands in monomer A and rose α-helices and green β-strands in monomer b. This is a “top” view of the dimeric structure. (B) Structure of NolR in complex with the 22-bp oligo AT duplex. Secondary structure features are colored as in A. The view is rotated ∼90° relative to A to present a “side” view of interaction with DNA. α-Helices forming the dimer interface (α1 and α5) and the helix-turn-helix motif (α3–α4) are labeled. Consensus-motif regions of oligo AT that contact NolR are colored purple and red with key nucleotides indicated. (C) Sequence of oligo AT. The purple and red boxes correspond to the regions of the consensus motif highlighted in B. The yellow A and T indicate nucleotides that are variable in the target DNA sequences of NolR.
Sequence and structural comparisons identify NolR as a member of the ArsR/SmtB family of transcription factors (26–29). NolR shares 22–40% amino acid sequence identity with BigR, HlyU, CadC, CzrA, NmtR, and SmtB (Fig. S1A). Moreover, the secondary structure features forming the helix-turn-helix motifs of these proteins are highly conserved (Fig. S1A). A search of the Protein Data Bank using the DALI server (30) identifies BigR, HylU, and CadC as the closest structural relatives of NolR (Fig. S1 B–D) with Z-scores of 14.4–14.9 and 2.0–2.7 Å rmsd for 95–97 Cα atoms (26, 31–33). The major differences between these proteins occur in the length and positioning of the N-terminal α-helical region at the dimerization interface (Fig. S1 B–D). Residues of the regulatory metal-binding sites found in other ArsR/SmtB family members are missing from NolR (26–29, 31–33).
Asymmetric Operator Site Recognition by NolR.
The structure of NolR complexed with DNA (Fig. 1 B and C) provides detailed information on how this homodimeric protein recognizes an asymmetric operator site to regulate expression of nodulation and symbiosis genes. Electron density for oligo AT DNA bound to NolR was well-defined (Fig. S2A). NolR binds to the operator, with the α4 helix of each monomer positioned 39 Å apart within major grooves of the DNA duplex (Fig. 1B). The molecular surface of NolR along the DNA-binding interface of each monomer is positively charged and provides an interaction surface for phosphate groups of the DNA whereas the opposite side of NolR is largely negative in charge (Fig. S2B). Analysis of the operator DNA geometry using 3D-DART (34) shows that the duplex bends 16.8° from an ideal B-form upon interaction with NolR (Fig. S2C). Each monomer of the NolR dimer interacts with a sequentially distinct half-site on the operator primarily through residues on α4 (Fig. 2).
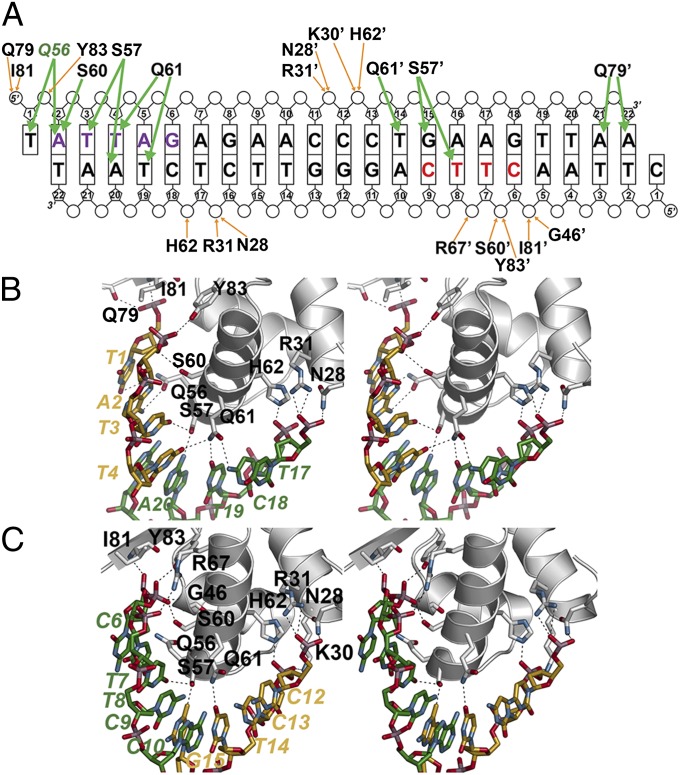
Fig. 2.
NolR and asymmetric operator binding. (A) Schematic showing NolR-oligo AT DNA contacts. The bases are labeled and shown as rectangles, with phosphate and ribose groups drawn as circles and pentagons, respectively. Residues from chain B of the homodimer are noted with an apostrophe after the amino acid number. Orange arrows indicate backbone contacts, and green arrows show base-specific interactions. The two halves of the NolR consensus target sequence that interact with NolR are highlighted with purple and red color, as in Fig. 1 B and C. Gln56 is colored green to emphasize its role in consensus-motif recognition. (B) Stereoview of protein–DNA interactions in the first half-site. Protein side-chains are from chain A. Nucleotides from 5′ and 3′ strands are colored gold and green, respectively, and are labeled. (C) Stereoview of protein–DNA interactions in the second half-site. Protein side-chains are from chain B. Nucleotides from 5′ and 3′ strands are colored gold and green, respectively, and are labeled.
Within the first block of the consensus sequence (Fig. 2A, purple), two clusters of residues from chain A form extensive interactions with the phosphate backbone of each DNA chain (Fig. 2 A and B). Gln79, Ile81, and Tyr83 hydrogen bond with T1 and A2 of the 5′ strand. On the 3′ strand, Asn28, Arg31, and His62 contact the phosphates of T17' and C18'. Gln56, Ser57, Ser60, and Gln61 provide hydrogen-bond interactions with T1, A2, T3, and T4 of the 5′ strand. Ser57 and Gln61 hydrogen bond with T19' and A20' of the complementary strand. At the second half site, (Fig. 2A, red) a similar set of residues from monomer B provides additional protein–DNA contacts (Fig. 2 A and C). Gly46, Arg67, Ile81, and Tyr83 of chain B interact with the phosphate groups of C6', T7', and T8' on the 3′ strand. In the second site, Ser60 from α4 hydrogen bonds to the phosphate of T7' instead of a nucleotide base. Interactions with the 5′-strand phosphates of C12 and C13 are contributed by Asn28, Lys30, Arg31, and His62. The β-sheet wing of monomer B binds in the minor groove to place Gln79, which is on the loop between the two β-strands, within hydrogen-bond distance of A21 and A22 of the 5′ strand (Fig. 2A and Fig. S3). Ser57 of monomer B provides bridging contacts with the nucleotide bases of T8' on the 3′ strand and G15 of the 5′ strand. These interactions are similar to those observed in chain A. The side-chain of Gln61 hydrogen bonds to T14 of the 5′ strand. Interestingly, the side-chain of Gln56 in chain B adopts a conformation that flips the amide group away from T7' of the 3′ strand. In contrast, the side-chain of Gln56 in the first site is oriented toward the A2 adenine ring.
Movement of Gln56 in Recognition of Variable DNA Sequence.
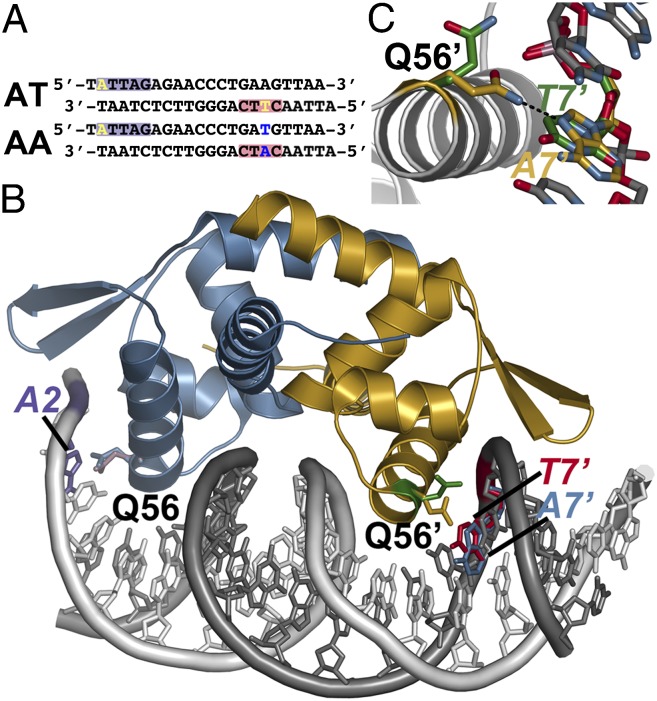
The shift in position of Gln56 in each half-site of the oligo AT DNA, either to interact with A2 in the first site or away from T7' in the second site, suggested that movement of this residue may play a role in the previously observed recognition by NolR of variable operator sites (17–24). To examine the possible role of Gln56 as a conformational switch in DNA binding, NolR was crystallized with oligo AA DNA (Fig. 3A). The oligonucleotides used to form this duplex maintain the sequence of the first half-site but substitute a T for A17 and an A for T7' on the 5′ and 3′ strands, respectively. The 3.0-Å resolution crystal structure of NolR in complex with oligo AA was solved by molecular replacement (Table S1). The overall structures of NolR with either DNA bound are nearly identical with a 0.2-Å rmsd for 196 Cα atoms in the homodimer. Comparison of the orientation of Gln56 in the DNA binding sites of each structure show that the interaction with A2 in the first half-site was identical (Fig. 3B and Fig. S4). In contrast, the side-chain of Gln56 in the second site, which was oriented away from T7' in the NolR•oligo AT crystal structure, flips to position the amide group for interaction with A7' in the NolR•oligo AA complex (Fig. 3 B and C). Movement of Gln56 to accommodate the variable A/T positions in each half-site of the NolR consensus DNA binding sequence provides a mechanism for recognition of diverse operator sites of genes involved in nodulation and symbiosis of rhizobia and plants (17).
Fig. 3.
Conformational switching of Gln56 for recognition of variable DNA target sites by NolR. (A) Sequences of oligos AT and AA. The purple and red boxes correspond to the regions of the consensus motif highlighted in Fig. 1 B and C. The variable position bases are highlighted yellow in oligo AT. Changes at these positions in oligo AA are highlighted in blue. (B) Structure of NolR in complex with the 22-bp oligo AA duplex. Chains A and B of NolR are colored blue and gold, respectively. The 5′ and 3′ strands of oligo AA are colored white and gray, respectively. The orientation of the Gln56 side-chain in each monomer of NolR complexed with either oligo AT (purple and green sticks) or oligo AA (blue and gold sticks) is shown. The positions of A2 (purple) from oligo AA, T7' (red) from oligo AT, and A7' (blue) from oligo AA are shown. (C) Close-up view of Gln56 movement in the second consensus half-site of NolR. The position of Gln56 of chain B and the variable base is shown. The structures observed with NolR complexed with either oligo AT (green) and oligo AA (gold) are shown.
Analysis of DNA Binding by NolR Using Isothermal Titration Calorimetry.
The X-ray crystal structures of NolR bound to oligo AT (Fig. 2) and oligo AA (Fig. 3) provide molecular insight on how changes in the position of Gln56 allows for interaction with operator sites with varied sequence at key positions. To examine NolR binding to varied target sites, isothermal titration calorimetry (ITC) was used to characterize interaction with oligos AT and AA. Binding of each DNA duplex to NolR was observed by ITC (Fig. S5); however, the energetic contributions to protein–DNA interaction varied for each operator sequence (Table 1). Analysis of DNA binding to NolR indicated binding of one dimer per operator site, as observed crystallographically, with comparable affinity (Kd ∼ 0.4 μM). Interestingly, the thermodynamics of interaction were distinct. NolR binding to oligo AT displayed a greater entropic contribution to protein–DNA interaction whereas association with oligo AA was dominated by enthalpic energy. These data indicate that differences in operator site sequence do not alter binding affinity but do change the energetics driving NolR–DNA interaction.
Table 1.
Thermodynamic parameters of DNA binding to wild-type NolR
| DNA | n | Kd, μM | ΔG, kcal⋅mol−1 | ΔH, kcal⋅mol−1 | −TΔS, kcal⋅mol−1 |
| AT | 1.02 ± 0.01 | 0.43 ± 0.06 | −8.69 ± 1.11 | −3.32 ± 0.04 | −5.37 |
| AA | 0.98 ± 0.01 | 0.36 ± 0.03 | −8.78 ± 0.69 | −6.67 ± 0.05 | −2.11 |
Titrations were performed using 22-bp DNA duplexes.
The structures of NolR also implicate key residues as important for DNA binding. To examine the contribution of Arg31, Gln56, Ser57, Ser60, and Gln61, a series of site-directed mutants were generated, expressed, and purified. The Q56A mutation was designed to remove the mobile side-chain from NolR for examination of the energetics of protein–DNA interaction by ITC. Arg31, which provides a charge–charge interaction with the DNA backbone, is invariant across members of the ArsR/SmtB family (Fig. S1A). Ser57, which in NolR is positioned for bridging interactions between each DNA strand, is also highly conserved (Fig. S1A). Ser60 provides either a base contact (site 1) or interacts with the phosphate backbone (site 2) and is invariant in the ArsR/SmtB family (Fig. S1A). Gln61 of NolR is either a glutamine (NolR, BigR, HlyU) or a histidine (CadC, CzrA, NmtR, SmtB) in the ArsR/SmtB family (Fig. S1A).
The NolR R31A, S57A, S60A, and Q61A mutant proteins were soluble and migrated as dimeric species, as observed for wild-type protein; however, each protein displayed a loss of DNA binding based on the lack of heat signal observed by ITC. These results indicate that these residues in NolR provide critical interactions for formation of protein–DNA complexes. In contrast, binding of oligos AT and AA to the NolR Q56A mutant was observed (Fig. S6). Substitution of Gln56 with an alanine results in less than twofold changes in the Kd values for each DNA duplex compared with wild-type NolR (Table 2). Removal of the amide side-chain in the G56A mutant results in comparable enthalpic and entropic contributions to DNA binding of each duplex. This result is consistent with the movement of Gln56 accounting for the energetic differences for variable DNA operator sequences observed with wild-type NolR.
Table 2.
Thermodynamic parameters of DNA binding to the NolR Q56A mutant
| DNA | n | Kd, μM | ΔG, kcal⋅mol−1 | ΔH, kcal⋅mol−1 | −TΔS, kcal⋅mol−1 |
| AT | 1.00 ± 0.01 | 0.26 ± 0.01 | −8.99 ± 0.46 | −3.33 ± 0.01 | −5.66 |
| AA | 1.03 ± 0.01 | 0.82 ± 0.11 | −8.30 ± 1.09 | −3.77 ± 0.07 | −4.53 |
Titrations were performed using 22-bp DNA duplexes.
Discussion
Although the symbiosis between rhizobial microbes and host plants that leads to nitrogen-fixing nodules is an ecologically and agriculturally important process (1–3), the molecular basis underlying the transcriptional regulation of nodulation and symbiosis remains incompletely understood. Nodulation requires induction of nod gene expression; however, efficient symbiosis with host plants occurs only when these genes are expressed in an appropriate quantitative, spatial, and temporal pattern and involves both positive and negative regulation (7, 8, 35). Mutations that alter either positive or negative regulation of nod genes result in aberrant and delayed nodulation phenotypes.
As part of the transcriptional regulation machinery of nodulation and symbiosis in various Rhizobium, NolR was originally identified as a putative helix-turn-helix family member that bound a nonpalindromic consensus motif in the core nodABC gene cluster (17). Later studies broadly implicate NolR as a global regulator of multiple symbiosis-related genes. The crystal structure of NolR is consistent with DNA footprinting studies that mapped the operator site as containing the (A/T)TTAG-N9-A(T/A) consensus motif (17); however, the NolR structures obtained in complex with two different DNA duplexes (Figs. 1–3) more accurately define the interaction sequences in each asymmetric half site as ATTAG on the 5′ strand and CTTC on the 3′ strand. A remarkable feature of NolR is that the homodimer binds to an asymmetric operator site with variable sequences at two positions, one in each half site, which allows versatility in recognition of NolR operator sites of multiple target genes (19, 20, 22–24).
The 3D structures of NolR in complex with oligo AT (Fig. 2) and oligo AA (Fig. 3) provide new insight on protein–DNA interactions that regulate nodulation and symbiosis gene expression. Binding contacts with the phosphate backbone are largely contributed by residues on α2, α3, and the β-wing. These contacts likely drive nonspecific association with the operator. Base-specific interactions come from Gln56, Ser57, Ser60, and Gln61 on α4 and Gln79 on the β-wing (Fig. 2 and Fig. S3). The hydrogen-bond network between residues of α4 in each half-site and the DNA duplex shows conserved interactions for operator binding. Ser57 anchors protein interaction by contacting T3 in the first half-site and T8' in the second half-site (Fig. 2). Ser57 also provides hydrogen bonds with A20' in site one and G15 in site two. Gln61 supports the bridging interactions of Ser57 at both sides of the operator. The amide of Gln61 hydrogen bonds with T19' and T14 in the first and second half-sites, respectively. ITC analysis of the NolR S57A and Q61A mutants, which showed a lack of interaction, confirms these residues role in DNA binding. Comparison of NolR operators in nodD1, nodZ, nolR, nodABC, and ttsI with oligos AT and AA reveals that this central set of interactions would likely be conserved across recognition sites (Table S2). In particular, the high conservation of the first half-site suggests that this site is critical for driving NolR–DNA interaction and that nucleotide variation in the second half-site leads to differences in repression by NolR; however, further studies are required to examine how sequence variations potentially modify NolR binding interactions and/or alter target-gene expression.
In addition to these critical contacts with the core consensus motifs, movement of Gln56 allows for accommodation of variable sequences at A2 and T7' (Fig. 3). The structure of NolR with oligo AT showed that the Gln56 side-chain hydrogen bonds with A2 in the first half-site (Fig. S4) but is flipped away from T7' in the second site (Fig. 3). Substitution of T7' with A7' in oligo AA provides an additional hydrogen bond interaction that orients Gln56 toward the base (Fig. 3). Although NolR binds both operator sequences with comparable affinity, the energetics of protein–DNA interaction differs between the two oligonucleotides (Fig. S5 and Table 1). Interaction with oligo AT displays a larger contribution from entropy compared with oligo AA, but introduction of an additional hydrogen-bond interaction with A7' in oligo AA enhances the enthalpic component of binding. Mutation of Gln56 to an alanine did not significantly alter the affinity of NolR for either oligo AT or oligo AA (Fig. S6 and Table 2) but led to a decreased enthalpic contribution to binding of oligo AA. Movement of Gln56 leads to compensatory energetic effects that maintain the binding affinity of NolR for varied operator sites.
The X-ray crystal structures of NolR in complex with DNA also provide a first view of protein–DNA interaction in the ArsR/SmtB transcription factor family (Fig. S1). Unlike other ArsR/SmtB transcription factors (26–29, 31–33), no global structural changes in the unliganded and DNA bound forms of NolR were observed, which likely results from a lack of metal-binding sites in the dimer interface of NolR. In the ArsR/SmtB proteins, metal binding triggers conformational changes that result in derepression of gene expression by driving repositioning of the helix-turn-helix DNA interaction motif (26–29, 31–33). This movement results in a switch from a “closed” DNA-binding structure to an “open” low-affinity conformation of the homodimer. To date, there is no evidence for other ligand interactions with NolR that trigger conformational changes that alter binding of the operator site.
Previous structural studies of ArsR/SmtB proteins largely focused on the role of metal-dependent conformational changes and not protein–DNA interaction, with one exception. NMR and mutagenesis probed residues in CzrA from Staphylococcus aureus for roles in binding to the palindromic 28-bp czr operator (28). This work suggests that CzrA interaction with target DNA increases protein motion in the allosteric sites and showed essential roles for residues analogous to Gln56, Ser57, Ser60, and Gln61 in DNA binding by CzrA, but did not reveal the details of protein–DNA contacts required for phosphate backbone interactions and/or base specificity.
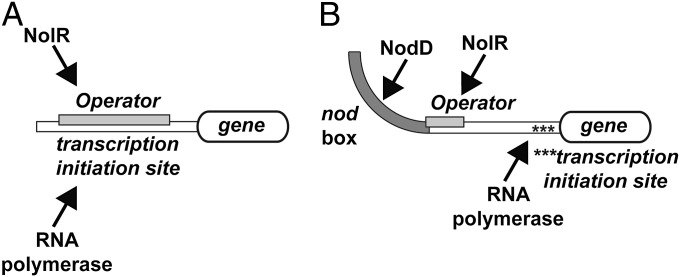
At the molecular level, interaction of NolR with operator sites of genes related to nodulation and symbiosis suggests at least two models for how the repressor functions. The first model of NolR regulation of gene expression is that of binding to an operator within the transcription initiation site of a target gene (Fig. 4A). The NolR binding sites for nodD1, nodZ, and nolR (Table S2) are all examples of this arrangement. NodD1 is the major transcription factor for nodulation and NodZ is an α1,6-fucosyltransferase for Nod-factor assembly (7–9, 14, 35–40). In each gene, a NolR operator is positioned 15–60 bp upstream of the coding region (23). Likewise, the presence of a NolR binding site 35 bp in front of the nolR gene suggests that levels of NolR modulate its own expression. Increased expression of NolR in free-living Rhizobia down-regulates expression of nodD1 and other nodulation/symbiosis genes, such as nodZ (22). In this arrangement, NolR binding to the operator would compete with RNA polymerase in the promoter site (Fig. 4A).
Fig. 4.
Models of NolR regulation of nodulation and symbiosis gene expression. (A) In promoters with overlapping transcription initiation and operator sites, NolR binding prevents RNA polymerase interaction and gene expression. (B) In promoter regions containing upstream nod box sequences for binding of the transcriptional activator NodD, binding of NolR to the operator site may either alter association of NodD to the nod box or alter DNA bending that results from NodD binding to prevent activation of gene expression.
A second model of action involves the binding of NolR to its operator site to alter how NodD either recognizes the nod box or how it interacts with RNA polymerase at the transcription initiation site (Fig. 4B). NolR operator sites are present in the regulatory regions of the nodABC operon and the ttsI gene (Table S2) (23). The role of NodD as a key regulator of nodABC gene expression is well explored at the genetic level (39). Binding of NodD to the nod box sequence 218 bp upstream of nodABC activates gene expression. Similarly, NodD binding to the nod box 295 bp upstream of the ttsI gene controls expression of a protein that in turn activates expression of genes associated with symbiosis (i.e., nopX, nopA, rhcJ and rhcQ) (41–43). Bending of DNA following NodD binding to the nod box has been suggested as important for activating expression of genes downstream (13). Multiple studies suggest that binding of LTTR transcription factors at activator sites alters DNA structure to allow for either direct interaction with RNA polymerase or to enhance RNA polymerase promoter escape by DNA bending (10). In the case of the nodABC gene cluster, the NolR operator overlaps with the nod box (17, 23). Binding of NolR at the operator may either physically compete with NodD at the nod box or alter bending at the promoter to down-regulate expression of downstream nodulation genes (Fig. 4B). The structures of NolR complexed with DNA suggest that the β-wing and α3 would likely sterically interfere with NodD binding at the nod box. It is possible that bending of the operator by NolR (Fig. S2C) either rigidifies the DNA to alter interaction with the promoter or changes the structure of the nod box to modulate NodD interaction. Further studies are needed to fully understand how these opposing transcription factors modulate gene expression.
In conclusion, the structural and thermodynamic studies presented here provide, to our knowledge, the first insights on the molecular foundation for the regulation of gene expression in nodulation and symbiosis and suggest how the global regulatory protein NolR recognizes variable asymmetric operator sites in the promoter regions of diverse rhizobial genes.
Methods
Protein Expression, Purification, and Mutagenesis.
The coding region of nolR was PCR-amplified from S. fredii USDA191 genomic DNA using oligonucleotide primers that included NdeI and XhoI restriction sites, respectively, to facilitate cloning. The PCR product was digested with NdeI and XhoI and ligated into pET-28a. The resulting vector was then transformed into E. coli BL21 (DE3) cells. Transformed E. coli cells were grown at 37 °C in Terrific broth containing 50 μg⋅mL−1 kanamycin until A600nm ∼ 0.6–0.9. After induction with 0.1 mM isopropyl 1-thio-β-d-galactopyranoside, the cultures were grown at 20 °C overnight. Following centrifugation, the cell pellet was suspended in 50 mM Tris (pH 8.0), 500 mM NaCl, 20 mM imidazole, 1 mM β-mercaptoethanol (βME), 10% (vol/vol) glycerol and 1% Tween 20. Sonication was used for cell lysis. After centrifugation, the supernatant was passed over an Ni2+-nitriloacetic acid column. The column was washed with buffer minus Tween 20, and the bound His-tagged protein was eluted using 250 mM imidazole in wash buffer. The N-terminal His-tag was removed by thrombin digestion in dialysis against wash buffer. A mixed Ni2+-nitriloacetic acid/benzamidine Sepharose column was used to remove undigested protein and thrombin. Size-exclusion chromatography was performed using a Superdex-75 26/60 FPLC column equilibrated in 150 mM Tris (pH 8.5), 100 mM NaCl, and 5 mM βME. Fractions corresponding to the protein peak were pooled and concentrated to 10 mg⋅mL−1 by centrifugal filtration. Protein concentration was determined using the Bradford method, with BSA as a standard. SeMet-substituted NolR protein was produced by inhibition of the E. coli methionine biosynthesis pathway (44) and was purified as described above. Site-directed mutants of NolR (R31A, Q56A, S57A, S60A, and Q61A) were generated using the QuikChange PCR method and were expressed and purified using the same methods used with wild-type NolR.
Protein Crystallography.
All NolR protein crystals were grown by the hanging drop vapor diffusion method at 4 °C. For crystallization of NolR in the presence of oligo AT, two 22-bp-long oligonucleotides (5′-dTATTAGAGAACCCTGAAGTTAA-3′ and 5′-dATTAACTTCAGGGTTCTCTAAT-3′) were suspended in purification buffer and annealed. Crystals of SeMet-substituted NolR (10.0 mg⋅mL−1) in complex with oligo AT DNA (1 mM) grew in drops containing a 1:1 mixture of protein and crystallization buffer [20% PEG-3350, 0.1 M sodium citrate/citric acid, and 0.2 M sodium citrate (pH 4.0)]. Crystals of native NolR (10 mg⋅mL−1) were formed in drops of a 1:1 mixture of protein and crystallization buffer [1.6 M sodium phosphate monobasic/0.4 M potassium phosphate dibasic, 0.1 M sodium phosphate dibasic/citric acid (pH 4.2)]. Crystals of native NolR (10 mg⋅mL−1) in complex with 1 mM oligo AA DNA (1 mM) (5′-dTATTAGAGAACCCTGATGTTAA-3′ and 5′-dATTAACATCAGGGTTCTCTAAT-3′) were obtained in conditions similar to that of the NolR oligo AT complex. All crystals were stabilized in crystallization solution with 30% glycerol before flash freezing in liquid nitrogen for data collection at 100 K. All X-ray diffraction data (wavelengh = 0.979 Å) were collected at beamline 19-ID of the Argonne National Laboratory Advanced Photon Source. HKL3000 (45) was used to index, integrate, and scale diffraction data. The structure of SeMet-substituted NolR in complex with oligo AT DNA was determined by SAD phasing. SHELX (46) was used to determine SeMet positions and to estimate initial phases from the peak wavelength dataset. Refinement of SeMet positions and parameters was performed with MLPHARE (47). Solvent flattening using density modification implemented with ARP/wARP (48) was used to build an initial model. Subsequent iterative rounds of manual model building and refinement, which included translation-libration-screen parameter refinement, used COOT (49) and PHENIX (50), respectively. The structures of unliganded NolR and NolR complexed with oligo AA DNA were solved by molecular replacement in PHASER (51) using the SeMet-substituted NolR structure as a search model, with model building and refinement performed with COOT and PHENIX. Waters were added to the unliganded NolR model using default parameters in PHENIX. Crystallographic statistics are summarized in Table S1.
Isothermal Titration Calorimetry.
NolR protein was dialyzed overnight in 150 mM Tris (pH 8.5), 100 mM NaCl, 5 mM βME, 5 mM MgCl2, and 5% glycerol at 4 °C. Synthetic double-stranded DNA (oligo AT, AA, and TT) were prepared in the same buffer. ITC experiments were performed using a VP-ITC calorimeter (Microcal) at 4 °C. Data obtained from the titrations were analyzed using a single-site binding model: 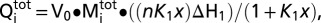 in which
in which 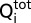 is the total heat after the ith injection, V0 is the calorimetric cell volume,
is the total heat after the ith injection, V0 is the calorimetric cell volume, 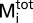 is the concentration of protein in the cell after the ith injection, ΔH is the corresponding enthalpy change to NolR•DNA binding, n is the number of nucleotide binding sites, and K is the equilibrium binding constant. Estimates of Kobs and ΔH were obtained by fitting the experimental data using Origin software (Microcal). Values for the change in free energy (ΔG) were calculated using ΔG = −RTln(Kobs), where R is the gas constant (1.9872 cal⋅K−1⋅mol−1) and T is absolute temperature. Changes in entropy (ΔS) were calculated using ΔG = ΔH − TΔS. Kd was calculated as 1/Kobs.
is the concentration of protein in the cell after the ith injection, ΔH is the corresponding enthalpy change to NolR•DNA binding, n is the number of nucleotide binding sites, and K is the equilibrium binding constant. Estimates of Kobs and ΔH were obtained by fitting the experimental data using Origin software (Microcal). Values for the change in free energy (ΔG) were calculated using ΔG = −RTln(Kobs), where R is the gas constant (1.9872 cal⋅K−1⋅mol−1) and T is absolute temperature. Changes in entropy (ΔS) were calculated using ΔG = ΔH − TΔS. Kd was calculated as 1/Kobs.
Supplementary Material
Acknowledgments
Portions of this research were carried out at the Argonne National Laboratory Structural Biology Center of the Advanced Photon Source, a national user facility operated by the University of Chicago for the Department of Energy Office of Biological and Environmental Research (DE-AC02-06CH11357).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 4OMY (SeMet NolR•oligo AT DNA), 4OMZ (unliganded NolR), and 4ON0 (NolR•oligo AA DNA)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402243111/-/DCSupplemental.
References
- 1.Desbrosses GJ, Stougaard J. Root nodulation: A paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe. 2011;10(4):348–358. doi: 10.1016/j.chom.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Kondorosi E, Mergaert P, Kereszt A. A paradigm for endosymbiotic life: Cell differentiation of Rhizobium bacteria provoked by host plant factors. Annu Rev Microbiol. 2013;67:611–628. doi: 10.1146/annurev-micro-092412-155630. [DOI] [PubMed] [Google Scholar]
- 3.Masson-Boivin C, Giraud E, Perret X, Batut J. Establishing nitrogen-fixing symbiosis with legumes: How many rhizobium recipes? Trends Microbiol. 2009;17(10):458–466. doi: 10.1016/j.tim.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Horvath B, et al. Organization, structure and symbiotic function of Rhizobium meliloti nodulation genes determining host specificity for alfalfa. Cell. 1986;46(3):335–343. doi: 10.1016/0092-8674(86)90654-9. [DOI] [PubMed] [Google Scholar]
- 5.Long SR. Rhizobium symbiosis: Nod factors in perspective. Plant Cell. 1996;8(10):1885–1898. doi: 10.1105/tpc.8.10.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. How rhizobial symbionts invade plants: The Sinorhizobium-Medicago model. Nat Rev Microbiol. 2007;5(8):619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kondorosi E, et al. Positive and negative control of nod gene expression in Rhizobium meliloti is required for optimal nodulation. EMBO J. 1989;8(5):1331–1340. doi: 10.1002/j.1460-2075.1989.tb03513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loh J, Stacey G. Nodulation gene regulation in Bradyrhizobium japonicum: A unique integration of global regulatory circuits. Appl Environ Microbiol. 2003;69(1):10–17. doi: 10.1128/AEM.69.1.10-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Göttfert M, et al. At least two nodD genes are necessary for efficient nodulation of alfalfa by Rhizobium meliloti. J Mol Biol. 1986;191(3):411–420. doi: 10.1016/0022-2836(86)90136-1. [DOI] [PubMed] [Google Scholar]
- 10.Maddocks SE, Oyston PC. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology. 2008;154(Pt 12):3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 11.Fisher RF, Brierley HL, Mulligan JT, Long SR. Transcription of Rhizobium meliloti nodulation genes: Identification of a nodD transcription initiation site in vitro and in vivo. J Biol Chem. 1987;262(14):6849–6855. [PubMed] [Google Scholar]
- 12.Fisher RF, Egelhoff TT, Mulligan JT, Long SR. Specific binding of proteins from Rhizobium meliloti cell-free extracts containing NodD to DNA sequences upstream of inducible nodulation genes. Genes Dev. 1988;2(3):282–293. doi: 10.1101/gad.2.3.282. [DOI] [PubMed] [Google Scholar]
- 13.Fisher RF, Long SR. Interactions of NodD at the nod Box: NodD binds to two distinct sites on the same face of the helix and induces a bend in the DNA. J Mol Biol. 1993;233(3):336–348. doi: 10.1006/jmbi.1993.1515. [DOI] [PubMed] [Google Scholar]
- 14.Winsor BAT. A nod at differentiation: The nodD gene product and initiation of Rhizobium nodulation. Trends Genet. 1989;5(7):199–201. doi: 10.1016/0168-9525(89)90079-6. [DOI] [PubMed] [Google Scholar]
- 15.Peck MC, Fisher RF, Long SR. Diverse flavonoids stimulate NodD1 binding to nod gene promoters in Sinorhizobium meliloti. J Bacteriol. 2006;188(15):5417–5427. doi: 10.1128/JB.00376-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peck MC, Fisher RF, Bliss R, Long SR. Isolation and characterization of mutant Sinorhizobium meliloti NodD1 proteins with altered responses to luteolin. J Bacteriol. 2013;195(16):3714–3723. doi: 10.1128/JB.00309-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondorosi E, et al. Identification of NolR, a negative transacting factor controlling the nod regulon in Rhizobium meliloti. J Mol Biol. 1991;222(4):885–896. doi: 10.1016/0022-2836(91)90583-r. [DOI] [PubMed] [Google Scholar]
- 18.Li F, Hou B, Hong G. Symbiotic plasmid is required for NolR to fully repress nodulation genes in Rhizobium leguminosarum A34. Acta Biochim Biophys Sin (Shanghai) 2008;40(10):901–907. [PubMed] [Google Scholar]
- 19.Chen H, et al. Identification of nolR-regulated proteins in Sinorhizobium meliloti using proteome analysis. Electrophoresis. 2000;21(17):3823–3832. doi: 10.1002/1522-2683(200011)21:17<3823::AID-ELPS3823>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 20.Chen H, Gao K, Kondorosi E, Kondorosi A, Rolfe BG. Functional genomic analysis of global regulator NolR in Sinorhizobium meliloti. Mol Plant Microbe Interact. 2005;18(12):1340–1352. doi: 10.1094/MPMI-18-1340. [DOI] [PubMed] [Google Scholar]
- 21.Cren M, Kondorosi A, Kondorosi E. An insertional point mutation inactivates NolR repressor in Rhizobium meliloti 1021. J Bacteriol. 1994;176(2):518–519. doi: 10.1128/jb.176.2.518-519.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cren M, Kondorosi A, Kondorosi E. NolR controls expression of the Rhizobium meliloti nodulation genes involved in the core Nod factor synthesis. Mol Microbiol. 1995;15(4):733–747. doi: 10.1111/j.1365-2958.1995.tb02381.x. [DOI] [PubMed] [Google Scholar]
- 23.Vinardell JM, et al. NolR regulates diverse symbiotic signals of Sinorhizobium fredii HH103. Mol Plant Microbe Interact. 2004;17(6):676–685. doi: 10.1094/MPMI.2004.17.6.676. [DOI] [PubMed] [Google Scholar]
- 24.López-Baena FJ, et al. Regulation and symbiotic significance of nodulation outer proteins secretion in Sinorhizobium fredii HH103. Microbiology. 2008;154(Pt 6):1825–1836. doi: 10.1099/mic.0.2007/016337-0. [DOI] [PubMed] [Google Scholar]
- 25.Brennan RG. The winged-helix DNA-binding motif: Another helix-turn-helix takeoff. Cell. 1993;74(5):773–776. doi: 10.1016/0092-8674(93)90456-z. [DOI] [PubMed] [Google Scholar]
- 26.Guimarães BG, et al. Plant pathogenic bacteria utilize biofilm growth-associated repressor (BigR), a novel winged-helix redox switch, to control hydrogen sulfide detoxification under hypoxia. J Biol Chem. 2011;286(29):26148–26157. doi: 10.1074/jbc.M111.234039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eicken C, et al. A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins. J Mol Biol. 2003;333(4):683–695. doi: 10.1016/j.jmb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 28.Arunkumar AI, Campanello GC, Giedroc DP. Solution structure of a paradigm ArsR family zinc sensor in the DNA-bound state. Proc Natl Acad Sci USA. 2009;106(43):18177–18182. doi: 10.1073/pnas.0905558106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osman D, Cavet JS. Bacterial metal-sensing proteins exemplified by ArsR-SmtB family repressors. Nat Prod Rep. 2010;27(5):668–680. doi: 10.1039/b906682a. [DOI] [PubMed] [Google Scholar]
- 30.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye J, Kandegedara A, Martin P, Rosen BP. Crystal structure of the Staphylococcus aureus pI258 CadC Cd(II)/Pb(II)/Zn(II)-responsive repressor. J Bacteriol. 2005;187(12):4214–4221. doi: 10.1128/JB.187.12.4214-4221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kandegedara A, Thiyagarajan S, Kondapalli KC, Stemmler TL, Rosen BP. Role of bound Zn(II) in the CadC Cd(II)/Pb(II)/Zn(II)-responsive repressor. J Biol Chem. 2009;284(22):14958–14965. doi: 10.1074/jbc.M809179200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishi K, et al. Crystal structure of the transcriptional activator HlyU from Vibrio vulnificus CMCP6. FEBS Lett. 2010;584(6):1097–1102. doi: 10.1016/j.febslet.2010.02.052. [DOI] [PubMed] [Google Scholar]
- 34.van Dijk M, Bonvin AMJJ. 3D-DART: A DNA structure modelling server. Nucleic Acids Res. 2009;37(Web Server issue):W235-9. doi: 10.1093/nar/gkp287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knight CD, Rossen L, Robertson JG, Wells B, Downie JA. Nodulation inhibition by Rhizobium leguminosarum multicopy nodABC genes and analysis of early stages of plant infection. J Bacteriol. 1986;166(2):552–558. doi: 10.1128/jb.166.2.552-558.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horvath B, Bachem CW, Schell J, Kondorosi A. Host-specific regulation of nodulation genes in Rhizobium is mediated by a plant-signal, interacting with the nodD gene product. EMBO J. 1987;6(4):841–848. doi: 10.1002/j.1460-2075.1987.tb04829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Honma MA, Ausubel FM. Rhizobium meliloti has three functional copies of the nodD symbiotic regulatory gene. Proc Natl Acad Sci USA. 1987;84(23):8558–8562. doi: 10.1073/pnas.84.23.8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mulligan JT, Long SR. A family of activator genes regulates expression of Rhizobium meliloti nodulation genes. Genetics. 1989;122(1):7–18. doi: 10.1093/genetics/122.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roche P, et al. The common nodABC genes of Rhizobium meliloti are host-range determinants. Proc Natl Acad Sci USA. 1996;93(26):15305–15310. doi: 10.1073/pnas.93.26.15305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stacey G, et al. nodZ, a unique host-specific nodulation gene, is involved in the fucosylation of the lipooligosaccharide nodulation signal of Bradyrhizobium japonicum. J Bacteriol. 1994;176(3):620–633. doi: 10.1128/jb.176.3.620-633.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viprey V, Del Greco A, Golinowski W, Broughton WJ, Perret X. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol Microbiol. 1998;28(6):1381–1389. doi: 10.1046/j.1365-2958.1998.00920.x. [DOI] [PubMed] [Google Scholar]
- 42.Krishnan HB, et al. Extracellular proteins involved in soybean cultivar-specific nodulation are associated with pilus-like surface appendages and exported by a type III protein secretion system in Sinorhizobium fredii USDA257. Mol Plant Microbe Interact. 2003;16(7):617–625. doi: 10.1094/MPMI.2003.16.7.617. [DOI] [PubMed] [Google Scholar]
- 43.Deakin WJ, Marie C, Saad MM, Krishnan HB, Broughton WJ. NopA is associated with cell surface appendages produced by the type III secretion system of Rhizobium sp. strain NGR234. Mol Plant Microbe Interact. 2005;18(5):499–507. doi: 10.1094/MPMI-18-0499. [DOI] [PubMed] [Google Scholar]
- 44.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229(1):105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 45.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 46.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64(Pt 1):112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 47.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 8):965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 2003;374:229–244. doi: 10.1016/S0076-6879(03)74011-7. [DOI] [PubMed] [Google Scholar]
- 49.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.