Significance
Plants sense foliar shade and neighbors who may become competitors for light. Shade-sensitive species elongate in response to both situations to enhance access to unfiltered sunlight, which is known as the shade avoidance response (SAR). During neighbor detection, plants have access to plenty of light (energy resources), whereas in true shade, light resources are scarce. Our analysis of the molecular mechanisms underlying SAR under these contrasting conditions shows that light intensity balances the production and sensitivity of the growth hormone auxin. In foliar shade, the production of auxin is reduced, whereas the downstream sensitivity to the auxin signal is enhanced. This hints at a resource-aware signaling where the strength of the hormonal signal is tuned to the available resources.
Keywords: regulatory network model, auxin signaling, auxin biosynthesis, phytochrome B
Abstract
Plants such as Arabidopsis thaliana respond to foliar shade and neighbors who may become competitors for light resources by elongation growth to secure access to unfiltered sunlight. Challenges faced during this shade avoidance response (SAR) are different under a light-absorbing canopy and during neighbor detection where light remains abundant. In both situations, elongation growth depends on auxin and transcription factors of the phytochrome interacting factor (PIF) class. Using a computational modeling approach to study the SAR regulatory network, we identify and experimentally validate a previously unidentified role for long hypocotyl in far red 1, a negative regulator of the PIFs. Moreover, we find that during neighbor detection, growth is promoted primarily by the production of auxin. In contrast, in true shade, the system operates with less auxin but with an increased sensitivity to the hormonal signal. Our data suggest that this latter signal is less robust, which may reflect a cost-to-robustness tradeoff, a system trait long recognized by engineers and forming the basis of information theory.
Being photoautotrophic and inescapably exposed to their environment, plants have developed sophisticated ways to adapt to their surroundings and secure access to light (1). For example, when grown in close proximity to neighboring plants, many species develop elongated stems and smaller leaves, a behavior called the shade avoidance response (SAR) (Fig. 1A) (2). This response increases their chance of reaching out to the sunlight above other plants and thus constitutes a competitive advantage (3). Committing additional resources to upward growth is so crucial that it happens at the expense of other functions, such as defense against pest and pathogens (4). An appropriate allocation of resources is vital for the plant, especially during its early and vulnerable developmental stage (5).
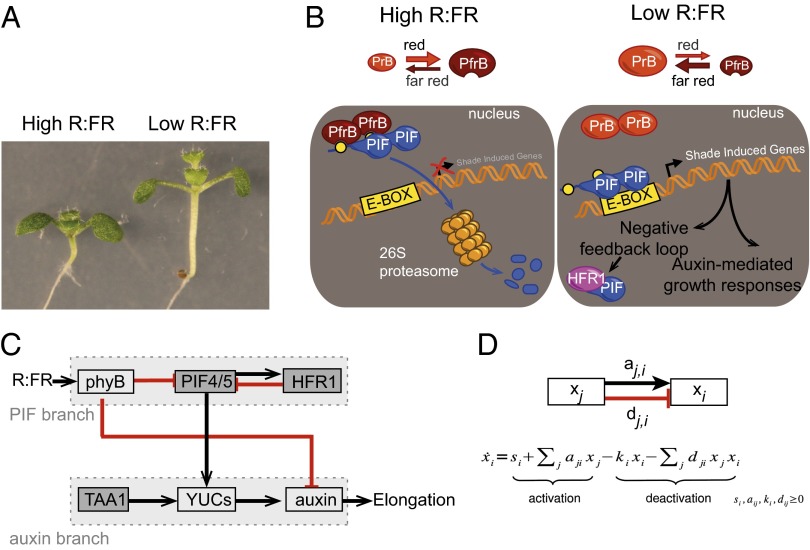
Fig. 1.
Regulation of the SAR. (A) Eight-day-old seedlings grown under high or low R:FR conditions. (B) Main components regulating the SAR. (C and D) Network model and equation used for modeling. Arrows are inhibitory (red) or positive (black).
The SAR is triggered not only by a reduction in the amount of light but also by specific modifications of its spectrum due to plant properties. Photosynthetic pigments absorb red (R) and blue (B) light, whereas plants scatter far red light (FR), leading to a reduction of the R:FR ratio in their vicinity. Under a foliar canopy, access to exploitable light [the photosynthetically active radiation (PAR)] is reduced, and plants sense both a low level of PAR and a low R:FR ratio. Due to FR scattering, a low R:FR ratio can also occur without a decrease in light resources when a plant is surrounded by nonshading neighbors (potential future competitors for light), a feature termed neighbor detection (2). Both neighbor detection and foliar shade lead to similar growth responses characterized in seedlings by the elongation of the embryonic stem (hypocotyl). However, it remains poorly understood how this can be achieved either in light-limiting conditions (true shade) or when plants retain access to the full solar spectrum (neighbor detection). To investigate how the R:FR ratio is transduced in these two contexts, we analyzed the effect of low R:FR in high vs. low PAR using combined computational and biological approaches. As both pathways require the hormone auxin and the transcription factors phytochrome interacting factor (PIF)4 and PIF5, we concentrated our analysis on these regulators of the SAR (6–8), leaving out other regulators such as PIF7, whose role have only been described in one of those conditions (9).
Current knowledge regarding the interplay between PIF4/5 and auxin during the SAR can be summarized into a simplified model shown in Fig. 1B. The R:FR ratio is perceived by the phytochrome B (phyB) photoreceptor that shifts between an inactive (PrB) and active (PfrB) form. The active form interacts with and inactivates the PIFs, which are positive regulators of the SAR. In high R:FR, phyB is active and targets the PIFs for phosphorylation/degradation, thus repressing the activation of the shade avoidance program (7, 9). In the vicinity of other plants, the low R:FR converts phyB into its inactive form, and the PIFs are free to activate gene expression. In particular, PIFs modulate the auxin pathway, as well as the activation of a negative feedback loop involving the transcription factor hypocotyl in far red 1 (HFR1) (10, 11). In low R:FR auxin is quickly produced by the tryptophan aminotransferase of arabidopsis 1 (TAA1)-YUCCA (YUC) pathway in the cotyledons (embryonic leaves). It is then transported to the hypocotyl to induce its elongation (12, 13).
We modeled this regulation by a network model and rely on it to generate different hypotheses that were experimentally validated to untangle the interaction between the PIFs and the auxin pathways. This combination of computational modeling with experimental validation led us to uncover that HFR1 regulates auxin levels independently of PIF4 and PIF5 and that the intensity of the auxin signal and its downstream sensitivity depend on the light intensity, i.e., on the availability of resources.
Results
Model Assessment.
The network model (Fig. 1C) has the R:FR ratio as single input and the hypocotyl elongation as the single output. Molecular activities are represented by nodes that are connected to each other by arrows representing positive or negative effects. The network is modeled by a dynamical system, where the state of each node is determined by the equation in Fig. 1D at steady state. The corresponding node is set to zero when the activity is null, for example, in a mutant (Materials and Methods).
The network model was first tested in one condition: true shade (low R:FR and low PAR). To do so, we determined hypocotyl length of seedlings grown for 4 d in high R:FR before being transferred to high or low R:FR for an additional 4 d. The elongation during these last 4 d was used as an experimental read-out corresponding to the elongation node of the network model (Fig. S1). This protocol was performed with the WT (Col) and the following genotypes pif4pif5, hfr1, a taa1 allele called sav3-2, hfr1pif4pif5, hfr1taa1, and pif4pif5taa1 (Materials and Methods).
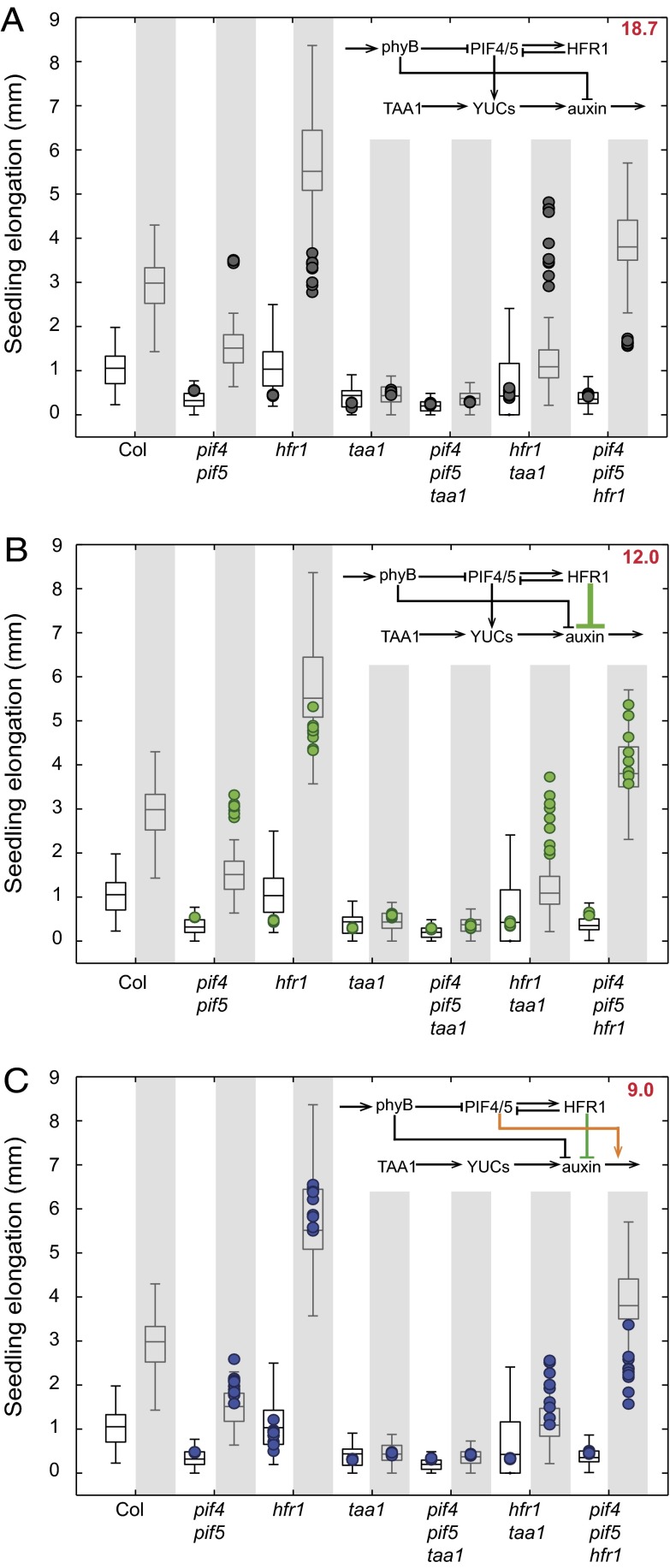
Rather than estimating or optimizing the parameters as it is usually done (14), we sample them from a distribution determined from the biological data. This parameter distribution is used to predict the hypocotyl elongation in a given condition. To evaluate the network model, a leave-one-mutant-out cross-validation procedure was applied, and the mean prediction error was used as the model score. This procedure makes the model evaluation independent from a particular choice of parameters (which are hardly accessible), takes into account the intrinsic variability of the biological data, and avoids overfitting (Materials and Methods). This procedure was applied to test the ability of our initial model to predict the elongation of seedlings in response to high or low R:FR, when trained on all other mutants and the WT elongation data. This analysis showed that the elongation of many mutants was not properly predicted (Fig. 2A), hinting at some weakness in the model.
Fig. 2.
Observed and predicted elongation of seedlings grown in low light intensity in normal (white background) and low R:FR (shaded background). Box plots correspond to observed experimental data, and color dots indicate elongations predicted by the model (10 nondeterministic predictions are made for each condition). A, B, and C correspond to predictions of the same data (identical box plots) by different models, indicated in the upper right corner (colored dots change across panels). The mean prediction error is indicated in the upper right corner.
HFR1 Inhibits Auxin Production, Whereas PIF4 and PIF5 Regulate Auxin Sensitivity in True Shade.
To increase the prediction accuracy of the model, we tried to add edges to our network. Looking at the elongation data, we noticed that the hfr1pif4pif5 mutant differs from the pif4pif5 mutant, a fact that cannot be accounted for from the literature or in our present network, as it assumes that HFR1 acts through PIF4 and PIF5 (10). The best improvements we found was adding a negative edge from HFR1 to auxin or to the YUCs, the simulations being unable to significantly distinguish between both scenarios. This result suggests that HFR1 (directly or indirectly) inhibits the production of auxin in a pathway parallel to PIF4 and PIF5 (Fig. 2B).
This new edge significantly increased the network prediction accuracy; however, some mutants were still poorly predicted, especially the pif4pif5 double mutant (Fig. 2B). In a previous paper, we reported that PIF4 and PIF5 control auxin production but also sensitivity (8). We thus tested whether the model predicted that PIF4 and PIF5 increased auxin sensitivity rather than production or both. As sensitivity cannot be described with the equation in Fig. 1D, we model it as a product between PIF4/5 and auxin activities (Materials and Methods). This link rather than the PIF4/PIF5-YUC link provided a strong improvement in the prediction accuracy (Fig. 2C and Fig. S2).
Taken together, the results of our network simulation suggest that (i) HFR1 inhibits auxin production, (ii) HFR1 also acts independently of PIF4 and PIF5, and (iii) PIF4 and PIF5 regulate auxin sensitivity rather than production in low light intensity.
To determine whether the excessive growth of hfr1 was mediated by an increase in auxin levels, we first grew seedlings in the presence of the polar auxin transport inhibitor NPA, which totally suppressed growth (Fig. 3A). We then determined the sensitivity of hfr1 to the auxin biosynthesis inhibitor l-kynurenine (15). The hfr1 mutant was less affected by l-kynurenine than the WT, suggesting that auxin production is up-regulated in hfr1 (Fig. 3B). This hypothesis was further confirmed by measuring auxin content, which was higher in hfr1 than in the WT (Fig. 3C). To explore how HFR1 regulates auxin content, gene expression quantification using quantitative RT-PCR (qRT-PCR) was performed. YUC2, YUC8, and YUC9, which encode rate-limiting enzymes in auxin synthesis downstream of TAA1, were overexpressed in hfr1 (Fig. 3D and Fig. S3) (16). This is consistent with the finding that auxin levels are also increased in hfr1taa1 compared with taa1 (Fig. 3C). The second prediction from our simulations was that HFR1 represses auxin production independently from PIF4 and PIF5. The normal expression of YUC2, YUC8, and YUC9 in pif4pif5 and the elevated expression of YUC genes in hfr1pif4pif5 support this hypothesis (Fig. 3D and Fig. S3). The important role played by PIF7 during the shade avoidance prompted us to check whether HFR1 may act by inhibiting this member of the PIF family (9). Expression quantification of YUC genes in pif7 and hfr1 and the double mutant showed that elevated YUC expression levels in hfr1 depended partially or totally on PIF7 (Fig. S4). Finally, our third prediction—that PIF4 and PIF5 rather act downstream of auxin production—is consistent with our gene expression data (Fig. 3D and Fig. S3). We thus propose that in low light, PIF4 and PIF5 modulate the low R:FR signal through HFR1 inhibition of auxin production and through their effect on auxin sensitivity.
Fig. 3.
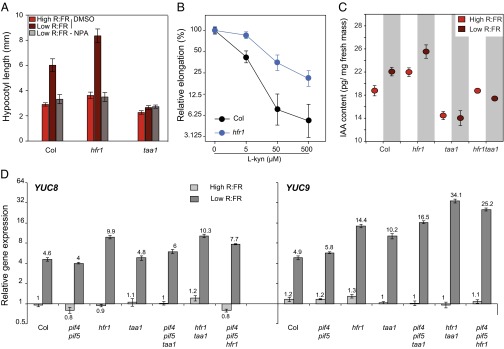
HFR1 inhibits auxin production. (A) Hypocotyl length of seedlings grown with or without the auxin transport inhibitor NPA under low light conditions (see Materials and Methods for seedling growth conditions). Data are mean ± 2 SE, n = 23–29. (B) Relative hypocotyl elongation in low light and in low R:FR in the presence of l-kynurenine, an inhibitor of auxin production. n = 25–34, error bars = 2 SE. (C) Free auxin content in seedlings grown under low light conditions subjected to 1 h of high or low R:FR. Data are mean ± SE (n = 5). (D) Quantitative RT-PCR analysis of two genes participating in the auxin production pathway. Seedlings were collected after 4 d in constant high R:FR light followed by 3 d in low or high R:FR. Data are mean ± SE, n = 3.
Stronger Auxin Production but Weaker Sensitivity in Neighbor Detection than in True Shade.
To determine whether the same regulatory network explains the growth response during neighbor detection, we repeated this experimental protocol but in high PAR. The network providing the best predictions was the one where PIFs induce auxin production and do not influence auxin sensitivity (Fig. 4A and Fig. S5). This result, along with the best-performing network obtained in true shade conditions, suggests a differential role for PIF4/5 in low and high light intensity in response to low R:FR treatment. This hypothesis is consistent with our previously published results that PIF4 and PIF5 have a weaker effect on auxin sensitivity in high than in low light intensity (8). To test whether PIF4 and PIF5 have a differential effect on auxin production depending on PAR, we analyzed the sensitivity of pif4pif5 to yucasin, an inhibitor of YUC enzymes (17), in seedlings grown in high vs. low PAR and subjected to low R/FR. Interestingly, pif4pif5 displayed an increased sensitivity to yucasin only in high PAR, consistent with the hypothesis that PIF4 and PIF5 primarily control YUC-mediated auxin production in this condition (Fig. S6).
Fig. 4.
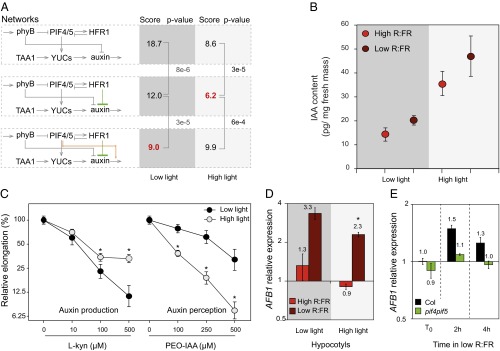
Auxin sensitivity and production depends on the light intensity. (A) Modeling suggests different networks for low and high light intensity (best score in red, P values obtained by a t test) (B) Free auxin content in seedlings grown under high or low light intensity subjected to 1 h of low R:FR or kept in high R:FR as a control. Data are mean ± SE (n = 5). Low light intensity data already presented in ref. 8. (C) Relative hypocotyl elongation of WT seedlings in low R:FR in the presence of inhibitor of auxin synthesis (l-kynurenine) or perception (PEO-IAA). n= 22–32, error bars = 2 SE. *Significant difference (t test, P < 0.01). (D) AFB1 expression in hypocotyls of 7-d-old WT seedlings grown in high and low light intensity, high R:FR, or subjected for 2 h to low R:FR. In low R:FR, AFB1 is more expressed in low than in high light (*t test, P < 0.05). Data are mean ± 2 SE (n = 3 × 40 seedlings). (E) AFB1 expression in entire seedlings of 7-d-old seedlings grown in low light intensity, high R:FR, and subjected to 2 or 4 h of low R:FR. Data are mean ± 2 SE (n = 3 × 40 seedlings).
More generally, the difference between the best-performing network in both conditions hints at a modulation of auxin production vs. sensitivity dependent on light intensity. We thus propose that for low R:FR signaling, in low light intensity, auxin sensitivity is enhanced, whereas auxin production is stronger in high light intensity. This modulation would suggest an adaptive signaling depending on the availability of resources. As photosynthesis is less productive in low light, we hypothetized that less auxin would be produced (18, 19). Consequently the SAR would involve lower levels of auxin that would be compensated at least partially by a higher sensitivity.
Supporting our hypothesis, we measured more auxin in the aerial part of the plant in high than in low light intensity (Fig. 4B). Another observation points to elevated auxin levels in high light that involves both TAA1-dependent and -independent pathways. In our conditions, the taa1 mutant reacted to the low R:FR treatment in high but not in low light intensity, a response that was inhibited by the auxin perception inhibitor α-(phenyl ethyl-2-one)-IAA (PEO-IAA) (Fig. S7).
The effect of light intensity on auxin production and sensitivity was further validated by the differential effect of competitive inhibitors (Fig. 4C). On one hand, the auxin biosynthesis inhibitor l-kynurenine was more efficient to inhibit hypocotyl elongation under low than under high light conditions, whereas the auxin perception inhibitor PEO-IAA was more efficient in high than in low light intensity. This observation is consistent with more auxin production under high PAR (Fig. 4B), whereas in low PAR, auxin sensitivity is enhanced. The mechanisms underlying auxin sensitivity are presumably multifactorial; however, the effect of PEO-IAA suggested a possible role for auxin receptors (Fig. 4C). Our previous ChIP-seq analysis identified AFB1, a gene coding for an auxin receptor, as a potential PIF5 target gene (8). We reasoned that to control auxin sensitivity of hypocotyl growth, this gene should be expressed in hypocotyls. We thus analyzed expression of AFB1 in dissected seedlings grown in high or low PAR and transferred into low R:FR. Interestingly, low R:FR led to up-regulation of AFB1 expression in hypocotyls, whereas in cotyledons, this response was marginal (Fig. 4D and Fig. S8). Moreover, low R/FR-mediated AFB1 expression was significantly stronger when seedlings were grown in low than in high PAR (Fig. 4D). Finally, we showed that in seedlings grown in low PAR, low R/FR-mediated AFB1 expression largely depended on PIF4 and PIF5 (Fig. 4E).
Collectively, our results indicate that light quantity and thus resource availability determines the amount of auxin produced: in other words, the hormonal signal intensity. A strong signal (high auxin level) is more costly and requires more resources, but is likely to be more robust than a cheaper and weaker signal. To verify whether this tradeoff prediction is supported by our data, we fitted the data to a noise model that distinguishes the measurement noise from the auxin signal read-out noise (SI Materials and Methods). The latter was indeed significantly reduced in high vs. low PAR for Col, (F test, P < 1 × 10−7). This effect was reproduced in an independent dataset (F test, P < 1 × 10−4) but was not observed in pif4pif5 and hfr1pif4pif5 (Fig. S9), in line with a putative role of the PIFs in the modulation of the auxin signal intensity.
Discussion
This work, which integrates modeling and experimental approaches, provides insights both in terms of biology and methodology. Regarding the methodological aspects, it shows that although the model is very coarse, it can provide new insights into a biological system, something that has long been argued by the Boolean network community (20). However, in contrast to standard Boolean networks and their continuous extension (21), our model can make accurate quantitative predictions, which is particularly appropriate for a system with a continuous output such as hypocotyl elongation. The accuracy of the predictions is attributable to the parameter sampling approach that we used. This approach, which is reminiscent of approximate Bayesian computation (Materials and Methods), marginalizes over the parameters and seems to extract the global constraints imposed by the network topology, irrespective of particular parameter values. As a consequence, the coarseness of the model, assuming only linear and bilinear activation and inactivation, does not hamper the precision of the predictions.
Regarding the biological aspects, our experimental validations made extensive use of drug treatments. This pharmacological approach allows us to deal with the genetic redundancy at the level of auxin biosynthesis and auxin receptor genes. Moreover, it allowed us to challenge auxin signaling or biosynthesis at specific times, which is otherwise only doable with conditional mutants that are, to our knowledge, unfortunately inexistent. Pharmacological experiments indicated that HFR1 inhibits auxin production (Fig. 3 A and B), which was further demonstrated by direct auxin measurements (Fig. 3C). Moreover, we show that in the conditions tested here, HFR1 acts independently from PIF4 and PIF5 (Fig. 3D). In contrast, HFR1 acts partially but not exclusively through PIF7, as the epistatic relationship between hfr1 and pif7 is distinct for the expression of different YUC genes (Fig. S4). We propose that the elevated levels of auxin in hfr1 are the result of the increased expression of the YUC genes in the mutant as it has been reported that overexpression of YUC1 can rescue the short hypocotyl phenotype of taa1 in shade (16). Consistent with this idea, hfr1 partially suppresses the shade phenotype of taa1 (Fig. 2 and Fig. S5).
More globally, our study indicates that the way the low R:FR signal is transduced into auxin signaling pathway depends on the availability of light resources (Fig. 4). Under high light conditions where resources are abundant, plants produce more carbohydrates that may be associated with more auxin production (18, 19). Thus, in response to low R:FR, a strong auxin signal can be produced. However, in a low light environment, the overall auxin production is weaker (Fig. 4B), and thus signal intensity may be reduced. We propose that to compensate for reduced auxin levels due to a lack of resources in low light conditions, the sensitivity to auxin is enhanced, as if the hypocotyl expecting a lower signal, was “listening” more carefully (Fig. S10). How auxin sensitivity is translated in terms of molecular activity is complex and poorly understood. Auxin is perceived by a coreceptor formed by a member of the TIR1/AFB family and an Aux/IAA protein (22, 23). Here we show that in response to shade AFB1 is selectively up-regulated in the hypocotyl, which may contribute to enhanced sensitivity of this organ to auxin (Fig. 4 D and E and Fig. S8). Interestingly, robust AFB1 up-regulation depends on PIF4 and PIF5 and is greater at low than high PAR (Fig. 4 D and E). Moreover, PIF4 and PIF5 directly control the expression of members of the Aux/IAA family (8), which may also contribute to the control of auxin sensitivity. The increased sensitivity to auxin could also be achieved through the brassinosteroids, previously shown to be required for low blue induced shade avoidance (13), and to increase auxin sensitivity (24).
This signal modulation is likely related to the energetic cost of signal transduction, the reduction of which would be advantageous in conditions of low resources even at the cost of its robustness. The difference between signaling cascades in the context of neighbor detection and canopy shade avoidance may thus depend not only on light signals as such, but also on the internal energy status of the plant. The coregulation of hormonal signal production and downstream sensitivity to the same hormone has also been described in the case of insulin (25). The present study begs the question of the optimization by the plant of a tradeoff between cost and robustness of the signal. This tradeoff has long been recognized as fundamental by engineers, and its study laid the foundation of information theory to quantify the amount, cost, and reliability of information transmission (26). Biological systems also face this tradeoff, which was investigated in the case of neural signal transduction (27). Our work on shade avoidance suggests that, in plants, hormonal signaling can also be subjected to this tradeoff. Interestingly, a recent study involving information theoretic measures on mammalian cells showed that transmitting an information through the amplitude of a signal (as is usually assumed) is not the most robust way (28), thus also hinting that more elaborate modes of signaling should also be envisaged in the study of biological systems.
Materials and Methods
Growth Conditions.
For determination of hypocotyl length, ∼40 seeds were plated on 1.6% (wt/vol) agar 1/2 Murashige and Skoog plate on a 180-μm nylon net filter (Millipore). Plates were kept 3 d in the dark and cold before being transferred at 20–21 °C in a Percival AR22L incubator (www.percival-scientific.com/products/arabidopsis-chamber) in constant white light (PAR = 110 µmol/m2/s, R:FR = 12.2 or PAR = 30 µmol/m2/s, R:FR = 13.8). The spectral light composition in the Percival incubator was measured as described in ref. 29. Plates were kept vertically so that seedlings grew along the mesh. After 4 d in the high R:FR ratio, plates were transferred into low R:FR conditions (PAR = 110 µmol/m2/s supplemented with FR = 60 µmol/m2/s, R:FR = 0.7 or PAR = 30 µmol/m2/s, R:FR = 40 µmol/m2/s, R:FR = 0.3) or kept in the same conditions as a control for an additional 4 d. Pictures of the plates were taken at days 4 and 8. Hypocotyl length was measured using the ImageJ software (http://rsbweb.nih.gov/ij/).
Further information on the biological material and methods is available in SI Materials and Methods.
Computational Method.
A detailed account of the computational method is provided in SI Materials and Methods. In summary, the network is modeled using the following ordinary differential equation (ODE) system:
where xi is the positive molecular activity of node i, si is a constant activation term, ki is a constant inactivation rate, and aji and dji are the activation and inactivation effects of node j on node i. To model sensitivity, a bilinear activation term aijkxjxk is added to this equation. The network output (elongation) is gated by a sigmoidal function of the form 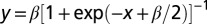 of amplitude β.
of amplitude β.
The network parameter vector θ thus contains one (effective) parameter per node (si/ki), one parameter per edge, and β. To train the network, a parameter sampling approach is taken rather than optimizing the parameters. The training is done by sampling the parameter space such that the distribution of network outputs generated by the distribution of network parameters approximates the distribution of observed elongation data. More formally, if the vector λ describes the network inputs reflecting the experimental conditions (light conditions and inactivated genes), g(θ, λ) is the vector of network outputs for inputs λ and parameter θ, and Ω is the (multidimensional) distribution of observed elongations in the conditions corresponding to λ, then the parameter space is sampled according to
This way, not only the average of observed elongations is taken into account, but also their variability, which also provides additional useful information. Sampling is done using a Markov chain Monte Carlo method (GaA-MCMC) (30), resulting in a distribution for the parameter vector θ. This distribution can then be used to make predictions for a new experimental condition by looking at the corresponding distribution of the network output for this new input. The mean of this distribution is estimated and used as prediction value.
To evaluate a network, a leave-one-mutant-out procedure is followed, whereby the data for all genotypes but one mutant are used to train the parameters (i.e., estimate the distribution of θ) and predict the remaining mutant. This prediction is then compared with the actual observations for that mutant. This whole procedure is repeated 10 times for each mutant to evaluate the robustness of the prediction to random sampling effects. The network score is defined by the average Mahalanobis distance between the observed elongations and the predicted ones.
The model is implemented in C++, and the code is freely available under a GNU general public licence on www.unil.ch/cbg. It uses the CVODE library (31) as the numerical equation solver. The sampling GaA-MCMC algorithm is implemented in MatLab and provided by ref. 30. Generating the simulation data presented in Fig. 2 takes about 2 d on 30 CPUs (2.27 GHz, 256 GB RAM).
Supplementary Material
Acknowledgments
We thank Laure Allenbach and Roger Granbom for technical support; Patricia Hornitschek and Markus Kohnen for starting the crosses with the sav3 mutant; Anupama Goyal and Markus Kohnen for setting up the protocol to extract RNA from dissected seedlings; Ioannis Xenarios for fruitful discussions; Ken-ichiro Hayashi and Tomokazu Koshiba for generously providing Peo-IAA and yucasin; Joanne Chory for the sav3-2 mutant; Ted Farmer, Niko Geldner, Sam Zeeman, and Tim Hohm for critically reading the paper; and Miguel Giraldo for the metaphorical illustration (Fig. S10). This work was supported by the University of Lausanne, a grant from SystemsX.ch “Plant Growth in a Changing Environment” (to C.F. and S.B.), the Swedish Governmental Agency for Innovation Systems, and the Swedish Research Council (K.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320355111/-/DCSupplemental.
References
- 1.Kami C, Lorrain S, Hornitschek P, Fankhauser C. Current Topics in Developmental Biology. Amsterdam: Elsevier; 2010. [DOI] [PubMed] [Google Scholar]
- 2.Casal JJ. Photoreceptor signaling networks in plant responses to shade. Annu Rev Plant Biol. 2013;64:403–427. doi: 10.1146/annurev-arplant-050312-120221. [DOI] [PubMed] [Google Scholar]
- 3.Hautier Y, Niklaus PA, Hector A. Competition for light causes plant biodiversity loss after eutrophication. Science. 2009;324(5927):636–638. doi: 10.1126/science.1169640. [DOI] [PubMed] [Google Scholar]
- 4.Ballaré CL, Mazza CA, Austin AT, Pierik R. Canopy light and plant health. Plant Physiol. 2012;160(1):145–155. doi: 10.1104/pp.112.200733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yanovsky MJ, Casal JJ, Whitelam GC. Phytochrome A, phytochrome B and HY4 are involved in hypocotyl growth responses to natural radiation in Arabidopsis: Weak de-etiolation of the phyA mutant under dense canopy. Plant Cell Environ. 1995;18(7):788–794. [Google Scholar]
- 6.Keller MM, et al. Cryptochrome 1 and phytochrome B control shade-avoidance responses in Arabidopsis via partially independent hormonal cascades. Plant J. 2011;67(2):195–207. doi: 10.1111/j.1365-313X.2011.04598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53(2):312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- 8.Hornitschek P, et al. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 2012;71(5):699–711. doi: 10.1111/j.1365-313X.2012.05033.x. [DOI] [PubMed] [Google Scholar]
- 9.Li L, et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012;26(8):785–790. doi: 10.1101/gad.187849.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 2009;28(24):3893–3902. doi: 10.1038/emboj.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sessa G, et al. A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 2005;19(23):2811–2815. doi: 10.1101/gad.364005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tao Y, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133(1):164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keuskamp DH, Pollmann S, Voesenek LACJ, Peeters AJM, Pierik R. Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc Natl Acad Sci USA. 2010;107(52):22740–22744. doi: 10.1073/pnas.1013457108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Middleton AM, Farcot E, Owen MR, Vernoux T. Modeling regulatory networks to understand plant development: Small is beautiful. Plant Cell. 2012;24(10):3876–3891. doi: 10.1105/tpc.112.101840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He W, et al. A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell. 2011;23(11):3944–3960. doi: 10.1105/tpc.111.089029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Won C, et al. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci USA. 2011;108(45):18518–18523. doi: 10.1073/pnas.1108436108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishimura T, et al. Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis. Plant J. 2014;77(3):352–366. doi: 10.1111/tpj.12399. [DOI] [PubMed] [Google Scholar]
- 18.Lilley JLS, Gee CW, Sairanen I, Ljung K, Nemhauser JL. An endogenous carbon-sensing pathway triggers increased auxin flux and hypocotyl elongation. Plant Physiol. 2012;160(4):2261–2270. doi: 10.1104/pp.112.205575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sairanen I, et al. Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis. Plant Cell. 2012;24(12):4907–4916. doi: 10.1105/tpc.112.104794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang R-S, Saadatpour A, Albert R. Boolean modeling in systems biology: An overview of methodology and applications. Phys Biol. 2012;9(5):055001. doi: 10.1088/1478-3975/9/5/055001. [DOI] [PubMed] [Google Scholar]
- 21.Mendoza L, Xenarios I. A method for the generation of standardized qualitative dynamical systems of regulatory networks. Theor Biol Med Model. 2006;3:13. doi: 10.1186/1742-4682-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calderon-Villalobos LI, Tan X, Zheng N, Estelle M. Auxin perception—Structural insights. Cold Spring Harb Perspect Biol. 2010;2(7):a005546. doi: 10.1101/cshperspect.a005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calderón Villalobos LIA, et al. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol. 2012;8(5):477–485. doi: 10.1038/nchembio.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vert G, Walcher CL, Chory J, Nemhauser JL. Integration of auxin and brassinosteroid pathways by auxin response factor 2. Proc Natl Acad Sci USA. 2008;105(28):9829–9834. doi: 10.1073/pnas.0803996105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bergman RN. Lilly lecture 1989. Toward physiological understanding of glucose tolerance. Minimal-model approach. Diabetes. 1989;38(12):1512–1527. doi: 10.2337/diab.38.12.1512. [DOI] [PubMed] [Google Scholar]
- 26.Shannon CE. A mathematical theory of communication. Bell Syst Tech J. 1948;27:379–423. [Google Scholar]
- 27.Laughlin SB, de Ruyter van Steveninck RR, Anderson JC. The metabolic cost of neural information. Nat Neurosci. 1998;1(1):36–41. doi: 10.1038/236. [DOI] [PubMed] [Google Scholar]
- 28.Uda S, et al. Robustness and compensation of information transmission of signaling pathways. Science. 2013;341(6145):558–561. doi: 10.1126/science.1234511. [DOI] [PubMed] [Google Scholar]
- 29.Dornbusch T, et al. Measuring the diurnal pattern of leaf hyponasty and growth in Arabidopsis - a novel phenotyping approach using laser scanning. Funct Plant Biol. 2012;39(11):860–869. doi: 10.1071/FP12018. [DOI] [PubMed] [Google Scholar]
- 30.Müller CL, Sbalzarini IF. 2010. Gaussian adaptation as a unifying framework for continuous black-box optimization and adaptive Monte Carlo sampling. IEEE World Congress on Evolutionary Computation, July 18–23, 2010, Barcelona (IEEE, Piscataway, NJ), pp 2594–2601.
- 31.Cohen SD, Hindmarsh AC. CVODE, a stiff/nonstiff ODE solver in C. Comput Phys. 1996;10(2):138–143. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.