Abstract
Several mycoplasma species have been shown to form biofilms that confer resistance to antimicrobials and which may affect the host immune system, thus making treatment and eradication of the pathogens difficult. The present study shows that the biofilms formed by two strains of the human pathogen Mycoplasma pneumoniae differ quantitatively and qualitatively. Compared with strain UAB PO1, strain M129 grows well but forms biofilms that are less robust, with towers that are less smooth at the margins. A polysaccharide containing N-acetylglucosamine is secreted by M129 into the culture medium but found in tight association with the cells of UAB PO1. The polysaccharide may have a role in biofilm formation, contributing to differences in virulence, chronicity and treatment outcome between strains of M. pneumoniae. The UAB PO1 genome was found to be that of a type 2 strain of M. pneumoniae, whereas M129 is type 1. Examination of other M. pneumoniae isolates suggests that the robustness of the biofilm correlates with the strain type.
Introduction
Biofilms are sessile communities of micro-organisms encased within a matrix of polysaccharide, protein and lipid that protect the microbes from harsh interactions with the environment, including the host immune response and antimicrobials (Costerton et al., 1999; Mohamed & Huang, 2007; Vu et al., 2009). It is generally accepted that most infectious diseases are caused by pathogens that have the capacity to form biofilms (Donlan, 2001). Many species of mycoplasma including Mycoplasma pneumoniae form biofilms (García-Castillo et al., 2008; Kornspan et al., 2011; McAuliffe et al., 2006; Simmons et al., 2007). This wall-less pathogen has a significant impact on human health, causing atypical pneumonia and accounting for 100 000 hospitalizations in the United States annually (Waites et al., 2008). Difficulties in eradicating mycoplasmas from their human or animal host have been attributed to the ability of the bacteria to evade host immunity through mechanisms such as formation of a biofilm on the epithelial surface. Identification of the factors that affect biofilm formation may lead to a better understanding of the mechanisms that contribute to the virulence of mycoplasmas.
M. pneumoniae strains are divided into types 1 and type 2 based on the nucleic acid sequence of the P1 adhesin gene (Su et al., 1990; Ursi et al., 1994). We show here that two strains of M. pneumoniae, one a type 1 strain and the other a type 2, form biofilms that differ qualitatively and quantitatively. We also find a difference in the distribution of an N-acetylglucosamine (GlcNAc)-containing polysaccharide within the intercellular matrix. Polysaccharides that contain GlcNAc contribute to virulence and to the structure of the biofilms produced by a diverse number of microbial species (Branda et al., 2005; Conlon et al., 2002; Kropec et al., 2005; Toledo-Arana et al., 2005), possibly including M. pneumoniae.
Methods
Growth of mycoplasma strains for biofilm studies.
M. pneumoniae strain UAB was isolated by throat swab in 1980 at the University of Alabama at Birmingham (UAB) from a patient with pneumonia and has been passaged five times in culture medium. The type 1 strains M129 (ATCC 29342) and PI 1428 (ATCC 29085) and the type 2 strain FH (ATCC 15531) were obtained from the American type Culture Collection (ATCC). The mycoplasmas were grown in SP-4 broth (Tully, 1983) or mycoplasma broth (MB) (Davidson et al., 1988) containing ampicillin at a final concentration of 50 µg ml−1 and phenol red at 0.01 % as a pH indicator. Starter stocks were made by scraping biofilms and disrupting them by vortexing and sonicating. The cells were stored in SP-4 containing 10 % glycerol at −80 °C. Some of the stocks were thawed and assayed for c.f.u. counts.
Biofilm adherence assays.
Tissue culture flasks having a surface area of 25 cm2 were inoculated with 105 or 106 c.f.u. of mycoplasma in 5 ml of medium. The cultures were incubated for up to 5 days without changing the medium. The number of planktonic c.f.u. and the c.f.u. growing as biofilms were determined by assaying cell culture supernatants (planktonic cells) or attached cells (biofilms) that had been scraped and gently sonicated to disrupt cell aggregates, as previously described (Simmons & Dybvig, 2003, 2007). The percentage of the mycoplasmas growing as biofilms was determined by dividing the number of c.f.u. in biofilms by the total number of c.f.u. in the flask. For each time point the flasks were grown in triplicate.
Fluorescence microscopy of biofilms.
Cultures inoculated with 105 c.f.u. were grown as biofilms for up to 10 days on glass coverslips as previously described with the medium changed when the colour turned orange, indicating a change in pH (Simmons et al., 2007). The biofilms were fixed with neutral buffered formalin [4.0 % formaldehyde, 0.4 % sodium dihydrogen orthophosphate, 0.65 % disodium hydrogen orthophosphate (pH 7.0)] for 48 h, followed by three washes for 1 h each in PBS. The lectin Griffonia simplicifolia lectin-I (GS-I) conjugated to DyLight 488 or fluorescein isothiocyanate (EY Laboratories) was used for fluorescence microscopy. Nucleic acids were stained with Hoechst 33342 (20 µg ml−1), propidium iodide (1.0 µg ml−1) or Syto 64 (500 ng ml−1; Invitrogen) and observed by epifluorescence microscopy (Leica HC Microscope) or confocal microscopy (Leica SP1 UV Confocal Laser Scanning Microscope; High Resolution Imaging Facility, UAB). Some biofilms were stained with only propidium iodide or Syto 64 prior to examination.
For staining with Congo red, the biofilms were fixed as described above and incubated in an aqueous solution containing 0.1 % Congo red for 30 min at room temperature (Simmons et al., 2007; Slifkin & Cumbie, 1988). Red fluorescence was observed using 350 nm excitation and 715 nm emission filters.
Some of the biofilms were imaged prior to fixing using a Pentax K-10 SLR camera equipped with an SMC-DA 18–55 mm lens and a 2× close-up lens. After imaging, they were fixed as described above, stained with Syto 64 and examined by epifluorescence microscopy.
Three-dimensional rendering of the cavities within the biofilm towers.
Biofilms stained with Syto 64 were imaged by confocal microscopy to produce stacks of images showing the cross sections of the towers. Black areas within the cross sections of the towers represented areas that did not contain mycoplasma cells and were defined as cavities, and regions external to the towers that did not contain cells were considered background. Using ImageJ (National Institutes of Health, version 1.42g), the brightness and contrast of the images were adjusted such that the value for background equalled zero and the brightness of the cells in the towers was sufficient to make the margins between the cells and the cavities apparent. This resulted in images where the brightness of both the background and the cavities had values of zero. The images were thresholded to delimit the edges of the cavities, converted to binary images and inverted such that the cavities and the background, which had a brightness value of zero before, now had a value of 255. The colour flood tool of ImageJ was used to convert the background area of each image back to a brightness value of zero. These manipulations resulted in stacks of images where the cavities within the towers were represented by pixels with a brightness of 255 while all other areas of the images had values of zero. This distinguished the background regions of the images and the biomass of the biofilm from the cavities in the towers. These stacks of images were used to render three-dimensional (3D) images of the cavities using the software application Daime (version 1.1) (Daims et al., 2006). Series or families of cavities that were connected were identified in the towers by automatic 3D thresholding-based segmentation of the image stacks.
Biovolume determinations.
Stacks of images were acquired from biofilms in which the nucleic acids had been stained with propidium iodide or Syto 64. The images were thresholded and analysed using the ImageJ voxel counter plugin (National Institutes of Health, http://rsbweb.nih.gov/ij/plugins/voxel-counter.html). The biovolume was calculated by determining the number of thresholded pixels in each stack and converting them to the volume of the biomass expressed in µm3. The density of the biofilm is expressed as the amount of biovolume per cm2 of glass substrate. Significance was determined by Student’s t test (SigmaStat, version 3.11) with P≤0.001 considered significant.
Analysis of glycomoieties by GC.
The mycoplasmas were grown as described (Bolland et al., 2012) and harvested by centrifugation at 8000 g, and the spent culture medium was collected. The cells were washed three times in PBS and lysed by sonication (full power for 30 s at 90 % duty cycle on a Branson Sonifier 450). The lysates were heated to 80 °C for 15 min to denature proteins and digested overnight at 37 °C with 25 µg DNase I and 25 µg RNase A followed by digestion overnight again at 37 °C with 25 µg proteinase K. The spent culture medium and the lysates were dialysed against deionized H2O using dialysis cassettes with a 2 kDa cut-off as previously described to remove small molecules resulting from digestion and monosaccharides that were not incorporated into polysaccharides (Daubenspeck et al., 2009). GC analysis was performed on the macromolecules remaining after dialysis.
The monosaccharide composition of the samples was determined by GC analysis of the trimethylsilyl derivatives of the sugar methyl glycosides and using known standards as described (Daubenspeck et al., 2009). For comparison of the amount of polysaccharide produced by the strains, the lysates were quantified prior to proteinase K treatment by using the BCATM Protein Assay kit (Pierce) and normalized to 300 µg of total protein.
Genomic analysis.
The genomes of strains UAB PO1 and PI 1428 were sequenced and M129 resequenced using the Illumina HiSeq 2000 platform using 50 bp paired-end reads. A de novo assembly was generated for all strains using Abyss 1.3.4 (Birol et al., 2009) and examined with Abyss-Explorer (Nielsen et al., 2009). FastQ reads were corrected and filtered with Quake 0.3 (Kelley et al., 2010) yielding a total sequenced base count of approximately 203 Mbp (M129), 429 Mbp (PI 1428) and 700 Mbp (UAB PO1). The UAB PO1 and PI 1428 genomes were also assembled using in-house software (unpublished) to generate a reference-based assembly that utilized as input Mauve-based multiple alignments and Burrows–Wheeler-generated short read alignments (Darling et al., 2010; Li & Durbin, 2009). The resulting referenced-based contigs were compared with the de novo-based assembly. Inconsistencies between the assemblies were resolved manually. Long-range PCR was not used to resolve gaps, leaving the final PI 1428 assembly with 93 contigs and the UAB PO1 assembly with 17 contigs over 200 bp in length. The resequencing of M129 used a similar approach but utilized only the original M129 Sanger-based sequence (GenBank U00089.2) for the reference assembly, allowing gaps to be filled to generate a single contig. The whole genome shotgun projects have been deposited at DDBJ/EMBL/GenBank under accession numbers CP003913, ANAA00000000 and ANAB00000000 for strains M129-B7, UAB PO1 and PI 1428, respectively.
Annotations for UAB PO1 were based on our resequenced genome of M. pneumoniae M129 and involved both automated and manual components. ORFs were identified initially using the NCBI ORF Finder tool, and each potential gene product of 50 amino acids or more was subjected to analysis. Putative proteins were annotated by a combination of blastp homology, output from the Conserved Domain Database, automated annotation by RAST (Aziz et al., 2008) and literature on the protein or its close homologues. Start sites were adjusted to match those of homologues where possible. The list of genes was compared with previous annotations and omissions, which were due to either atypical start codons or incorrect calls in the older annotations. ORFs with particularly unusual codon usage and/or small size that lacked homologues in other organisms, or ORFs that exhibited substantial overlap with better-established genes, were not counted as genes. Regions of the chromosome were identified as pseudogenes if they appeared to code for a single protein but had interruptions, including frameshifts, nonsense codons or truncations. Genes were renumbered based on their linear order in the genome with a note field in the GenBank record indicating the name of the gene in the original M129 (U00089.2) annotation, if applicable. BEDTools (Quinlan & Hall, 2010) was utilized to discover novel genes and differences between the UAB PO1 and the M129 genomes, which were then inspected manually.
The genomic sequences were aligned and analysed for regions of divergence. Single nucleotide polymorphisms (SNPs) were identified using Mauve (version 2.3.1 build 173; http://gel.ahabs.wisc.edu/mauve/) (Darling et al., 2010). Analysis of the divergent regions was refined using the local clustal w multiple alignment tool provided with the BioEdit multiple sequence editor (version 7.0.5.3) and blast (bl2seq; National Center for Biotechnology Information). The effects of mutations and rearrangements on the predicted amino acid sequences of the coding regions were analysed using Artemis (version 14.0.0; Wellcome Trust Sanger Institute). Promoter sequences were analysed using bprom (Prediction of Bacterial Promoters; http://linux1.softberry.com). The genome reference sequences for M. pneumoniae strains M129 (GenBank U00089.2), FH (GenBank NC_017504) and 309 (GenBank NC_016807) were used in all of the comparisons. Repetitive elements RepMP4c (GenBank FJ603707.1 and FJ603725.1) and RepMP2/3c (GenBank FJ603698.1 and FJ603716.1) within the P1 adhesin gene and the RepMP5c element (GenBank HQ698773.1 and HQ698781.1) within the MPN_142 gene were used to identify M. pneumoniae strain types (Spuesens et al., 2009).
Results
Analysis of the mycoplasmas growing planktonically and in biofilms
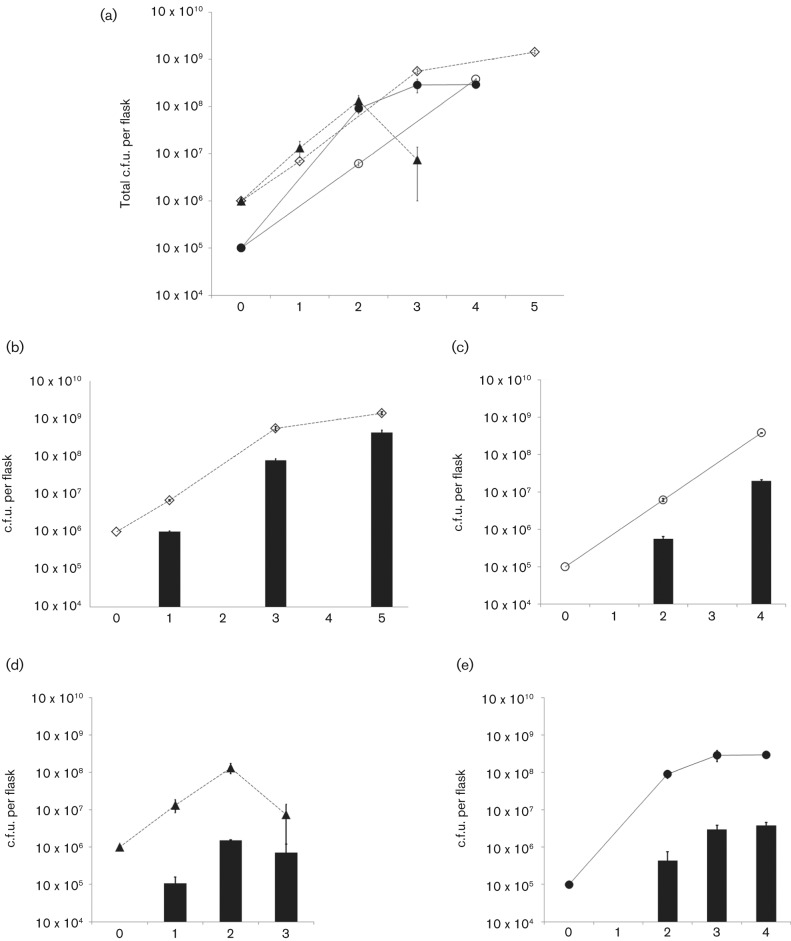
Analysis of the percentage of the mycoplasmas within biofilms as compared with the number of planktonic cells and the overall growth of the cultures revealed differences between strains M129 and UAB PO1. Fig. 1 shows the results of three experiments. Initially, both strains grew at about the same rate with an approximately log increase in c.f.u. per day. M129 grew for 2 or 3 days before entering the stationary phase when the flasks were initially inoculated with 105 or 106 c.f.u., respectively (Fig. 1a). M129 grew robustly as planktonic cells, reaching a titre of 108 c.f.u. per flask with greater than 98 % of the c.f.u. growing in suspension. The one exception to this was on day 3 of the cultures that had been initiated with 106 c.f.u. per flask. These flasks experienced a 94 % drop in c.f.u. of the planktonic cells and a 53 % drop in biofilm c.f.u., indicating that the cells had entered a death phase. UAB PO1 was able to grow for 3 or 4 days before reaching stationary phase, with a titre of 109 c.f.u. per flask (Fig. 1a). A greater proportion of UAB PO1 c.f.u., ranging from about 5 to 30 %, was in a biofilm (Fig. 1b, c) as compared with strain M129 (Fig. 1c, d), which had less than 2 % of c.f.u. attached to the flasks. In another experiment performed in triplicate using growth conditions identical to the experiments shown in Fig. 1, UAB PO1 reached 1.3×109 c.f.u. per flask (sem 1.1×108 c.f.u.) with 5.3×108 c.f.u. (39.7 %, sem 8.3×107 c.f.u.) locked in the biofilms.
Fig. 1.
Analysis of planktonic and biofilm-locked mycoplasmas in culture. (a) Total number of c.f.u. recovered from the cultures of strains M129 and UAB PO1 when inoculated at either 105 or 106 c.f.u. per flask; the data represent the results of three experiments. UAB PO1 106 c.f.u. (◊) and M129 106 c.f.u. (▴) represent cultures that were inoculated with 106 c.f.u. of the indicated strain. UAB PO1 105 c.f.u. (○) and M129 105 c.f.u. (•) represent cultures that were inoculated with 105 c.f.u.; n = 3 for each time point of each experiment except for day 1 of the M129 cultures that were inoculated with 106 c.f.u., for which n = 6. (b–e) The number of c.f.u. of the mycoplasmas attached in biofilms (bar graphs) relative to the total number of c.f.u. recovered from the flasks (line graphs). (b, d) The c.f.u. recovered from cultures inoculated with 106 c.f.u. of strains UAB PO1 and M129, respectively. (c, e) The c.f.u. recovered from cultures inoculated with 105 c.f.u. of strains UAB PO1 and M129, respectively. Error bars, sem.
Structural differences in the biofilms of UAB PO1 and M129
The biofilms formed by M129 and UAB PO1 differed qualitatively and quantitatively. Biofilms were grown for up to 10 days, changing the medium when indicated by pH to prevent cells from entering the death phase. Gross differences in the structures of the biofilms were apparent to the naked eye. The UAB PO1 biofilms were more robust. A macroscopic view of the unfixed biofilms on glass coverslips showed tower structures in UAB PO1 that were not obvious in M129 unless the biofilms were observed microscopically (Fig. 2a, b). Epifluorescence microscopy revealed that the towers of UAB PO1 were clearly distinct from the honeycombed regions (Fig. 2c, e). The M129 biofilms had towers, but the honeycombed region contained few mycoplasmas between the towers (Fig. 2b, d, f). At no time during the 10 days of growth did the biofilms of M129 appear similar to those of UAB PO1.
Fig. 2.
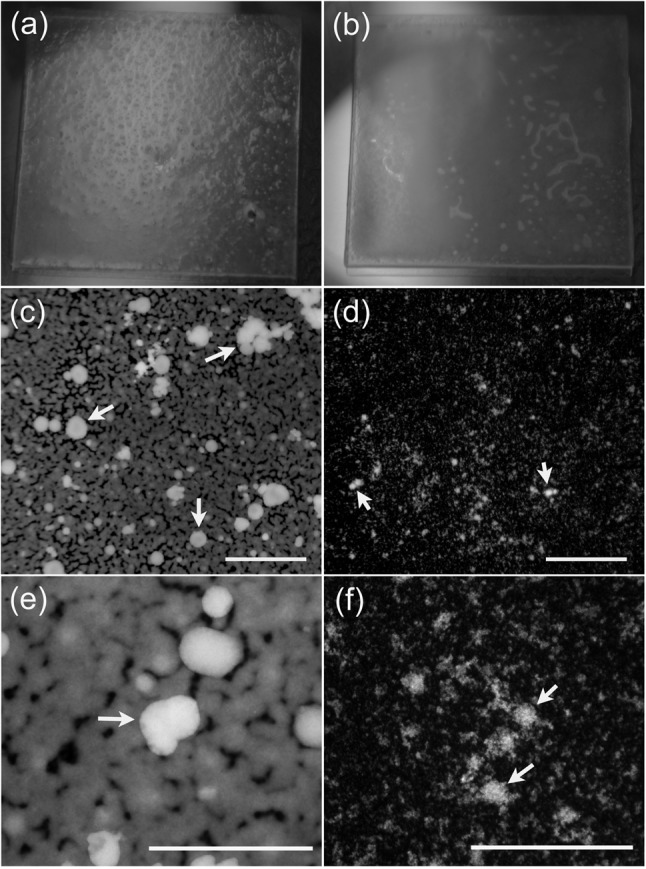
Structure of M. pneumoniae biofilms. (a, b) Macroscopic structure of a biofilm grown for 10 days on 18 mm ×18 mm glass coverslips prior to fixing. The biofilm formed by UAB PO1 (a) has a granular surface with numerous tower structures penetrating through to the surface of the biofilm, whereas that formed by M129 (b) has a smooth surface. (c–f) Fluorescence micrographs of the biofilms of UAB PO1 (c, e) and M129 (d, f) stained with propidium iodide. These low-power (×40) fluorescence images reveal the smooth margins of the biofilms in the honeycombed regions. The mycoplasmas between the towers are difficult to discern with the lower-power images of M129. (e, f) Higher-power (×400) images of biofilms formed by UAB PO1 and M129, respectively, revealing the towers and the honeycombed region of strain M129. Arrows indicate the towers. Bars, 400 µm (c, d) and 200 µm (e, f).
Confocal microscopic analysis of M129 and UAB PO1 biofilms revealed substantial differences in robustness and texture (Figs 3a, b and 4). The honeycombed regions of the biofilm of UAB PO1 were robust and juxtaposed to the towers (Fig. 4a, g). The towers of UAB PO1 were larger and more mucoid with smoother margins (Fig. 4a, c, e, g). The biofilms of M129 contained distinct towers but lacked a well-defined honeycombed region (Fig. 4b, d, f). The towers and their margins appeared to be granular (Fig. 4h), and individual cells attached to the glass were apparent in the region adjacent to the towers (Fig. 4b, h).
Fig. 3.
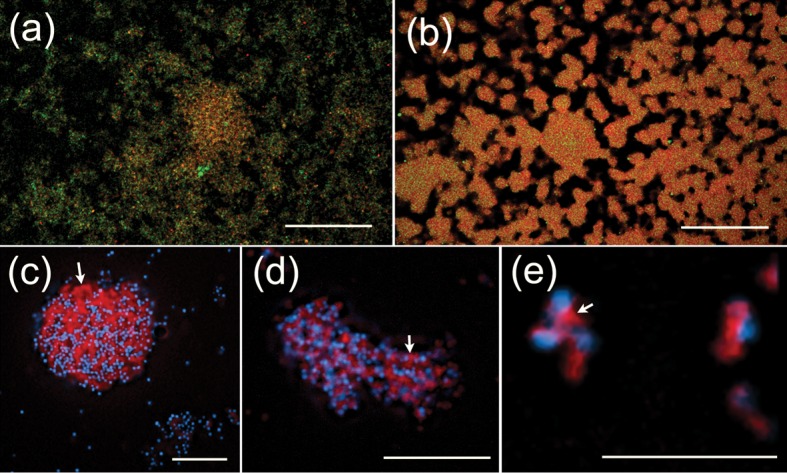
Production of an extracellular matrix. (a, b) Binding of the lectin GS-I (green) to biofilms of M129 (a) and UAB PO1 (b). The DNA in the biofilms is shown in red and areas of colocalization of GS-I and DNA are shown in orange-red. (c–e) Fluorescence microscopy at ×1600 magnification of M129 stained with Congo red. The DNA of the mycoplasmas is shown in cyan and the Congo red, which binds matrix material between the cells (white arrows), is shown in red. Cell clusters or biofilms shown in all panels were grown for 7 days in SP-4 (a, b) or MB (c–e). Bars, 50 µm (a, b), 20 µm (c, d) and 5 µm (e).
Fig. 4.
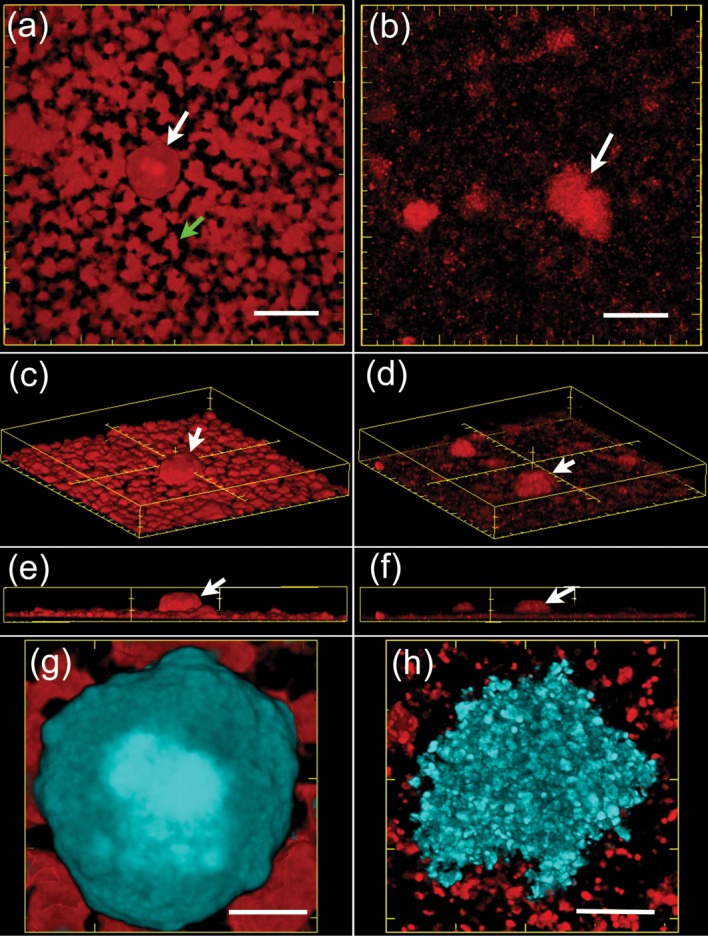
Biofilms formed by strains UAB PO1 and M129. 3D reconstructions made from confocal microscopic images of biofilms grown for 7 days and stained with Syto 64. (a, c, e, f) Biofilms of UAB PO1, and (b, d, f, h) biofilms of M129. (a, b) Overhead views, (c, d) views looking down on the biofilm from an angle, and (e, f) side views. Examples of towers are indicated by white arrows. The honeycombed region of the biofilm formed by UAB PO1 is shown by the green arrow. (g, h) A close up, overhead view of the towers of UAB PO1 and M129, respectively. The towers are pseudocoloured in cyan while the honeycomb-associated mycoplasmas are shown in red. Bars, 40 µm (a, b), with major and minor ticks of 50 and 10 µm, respectively; 10 µm (g, h).
Differences were observed in the internal structure of the towers. The towers formed by M129 contained cavities (Fig. 5a–i). Some of the cavities appeared to run throughout the towers and connect the interior with the external medium (arrows). 3D renderings revealed complex networks of interconnected cavities (Fig. 5g–i). The towers of UAB PO1 contained few if any of these cavities (Fig. 5j–l).
Fig. 5.

Cavities in the towers of M. pneumoniae biofilms. Confocal microscopic sections (a–f) of a biofilm of M129 showing cavities within a tower. (a) The biofilm attached to the glass, and (b–f) cross sections of a tower at 6, 9, 14, 18 and 22 µm above the glass surface, respectively. The arrows in (a) point to an area that forms a channel in the bottom section of the tower. The arrows in (b–f) point to voids in the images of selected sections of the towers that represent cavities. (f) A confocal section of the top of the tower; the two arrows indicate where the cavities contact the medium immersing the biofilm. (g–i) A shaded surface rendering of some of the cavities inside the tower formed by M129. (g) Side view of the cavities, and (i) an overhead view. (h) View looking down at the cavities at an angle of 45° above the glass coverslip. (j–l) Cross sections of a biofilm of UAB PO1 at the level of attachment to the glass, and at 14 and 18 µm above the glass, respectively. The white arrows point to void areas in the cross sections that form a continuous cavity that extends from the bottom of the tower to the top of the tower. Bars, 20 µm (a–f, j, k); 10 µm (g–i).
An examination of other strains of M. pneumoniae revealed that M129 and UAB PO1 were not unique. Epifluorescent microscopy revealed that the biofilm of the type 2 strain FH was similar to that of UAB PO1, whereas that of the type 1 strain PI 1428 was similar to M129 (Fig. 6).
Fig. 6.
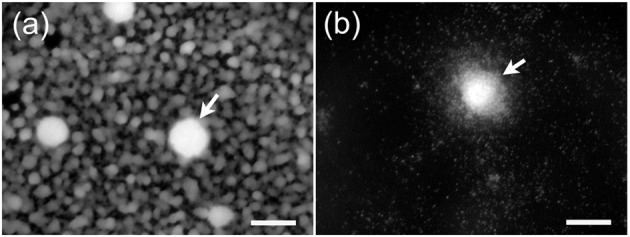
Biofilms formed by M. pneumoniae strains FH and PI 1428. (a) The honeycombed region surrounding the towers (arrows) of the biofilm formed by strain FH. Little biofilm growth is observed around the towers (arrow) formed by PI 1428 (b). Bars, 25 µm.
Biovolume of UAB PO1 biofilm is greater than M129
When the biovolume of the biofilms was determined after 7 days of growth, UAB PO1 produced a heavier biofilm than M129 (Fig. 3a, b). UAB PO1 formed a biofilm with a density of 3.4×108 µm3 per cm2 of glass substrate (n = 10, sem 3.3×107), while the biofilm of M129 had a density of 6.5×107 µm3 cm–2 (n = 10, sem 1.1×107). This difference was significant (P<0.001). As the biovolume measured the thickness of the biofilms based on the staining of the nucleic acids, the total thickness of the matrix may have been underestimated.
Distribution of glycomoieties in biofilms of M129 and UAB PO1
Confocal lectin-fluorescence microscopy indicated differences in the binding of the lectin GS-I to M129 and UAB PO1. In UAB PO1, almost all of the GS-I bound to the cellular mass (Fig. 3b). In contrast, in addition to some co-localization with the cellular mass, much of the GS-I bound to areas away from the cellular mass of M129 (Fig. 3a). Consistent with this observation, in other experiments where M129 was stained with Congo red (Fig. 3c–e), large amounts of matrix were observed between the cells within clusters of mycoplasmas.
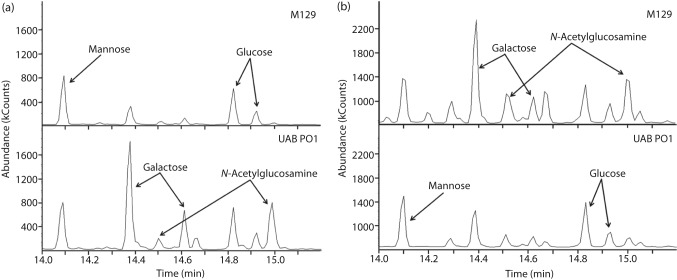
GC analysis of the glycomoieties that were cell-associated or in the spent culture medium revealed differences between the two strains (Fig. 7). Glycomoieties containing the monosaccharides galactose and GlcNAc were major constituents of the UAB PO1 cells with little galactose or GlcNAc found in the spent culture medium. In contrast, small amounts of galactose and GlcNAc were found associated with the cells of M129 while large amounts of these monosaccharides were found in the spent medium. For both strains, the ratio of galactose to GlcNAc was approximately 1 : 1 in both the cellular lysates and the spent medium.
Fig. 7.
GC of polysaccharides from M. pneumoniae. GC analysis shows the peaks for the methyl glycosides of the monosaccharides mannose (shown as a single peak), and glucose, galactose and GlcNAc (shown as double peaks). The chromatograms represent the sugars derived from the polysaccharides associated with the cells (a) or the spent medium (b). The y axis represents the absorbance of the methyl glycosides eluted from the column, and the x axis represents the time for elution.
The monosaccharides mannose and glucose were also detected by GC analysis and found to be associated with the cells of M129 and UAB PO1. Integration of the area under the peaks of the chromatograms indicated the cells of both strains contained about the same amount of mannose and glucose in approximately a 1 : 1 ratio (Fig. 7a). The spent medium from both strains also had comparable amounts of mannose and glucose in a 1 : 1 ratio.
Genomics
As many factors have the potential to influence biofilm formation, we compared the genomic sequences of M129 and UAB PO1. Coverage of the UAB PO1 genome sequence and the resequenced M129 genome was estimated to be greater than 98 %. The reference-based genomic sequences of UAB PO1 (supplementary data file MPN_UAB_PO1.fa) and the resequenced M129 (supplementary data file MPN_M129_resequenced.fas) were about 817 100 and 816 400 bp, respectively, as compared with the M. pneumoniae reference strain M129 (816 394 bp). Numerous differences in the genomes were identified. SNPs calculated from the reference-based sequences are summarized in Table S1 (available with the online version of this paper). All nucleotide positions (ntp) are relative to the reference M129 strain unless otherwise noted.
Comparison of the major attachment proteins
The DNA sequences were analysed for potential differences that would affect the predicted coding regions of major proteins (adhesins) of the attachment organelle. The P1 operon codes for two major proteins having a role in adherence, the P1 protein and ORF6 gene product, which are encoded by the MPN_141 and MPN_142 genes, respectively. Comparisons of the RepMP elements within the MPN_141 and MPN_142 genes indicated that UAB PO1 is a type 2 strain. The predicted amino acid sequences of the high molecular mass proteins HMW1, HMW2 and HMW3 shared identity in 99.7 % of their positions. The sequence of the P65 gene revealed an 18 aa insertion at positions 162–179 within the acidic proline-rich domain of the UAB PO1 protein, as described by Proft et al. (1995). However, comparison of the P65 protein in strains FH, 309 and PI 1428 revealed polymorphisms in this domain. The P30 protein shared 99.6 % identity, the P200 protein 99.5 %, P24 protein 99.5 %, TopJ protein 99.7 % and P41 protein 99.7 % between their orthologues in the two strains. The promoter region for the operon coding for P65 and HMW1 contained an SNP at position 364 188 of the M129 reference sequence. This SNP was not predicted to affect the −10 or −35 boxes or any transcription factor binding sites. The promoter regions of the other genes discussed above were identical between M129 and UAB PO1. The genes coding for PrkC, a protein kinase involved in stabilizing the attachment proteins (Schmidl et al., 2010), and PrpC, a protein phosphatase within the same operon (Halbedel et al., 2006), were identical between the two strains.
Major insertions/deletions
In addition to the chromosomal differences described above, we detected major rearrangements in five regions of the M. pneumoniae chromosome (Table 1). Only regions 1, 2 and 4 correlated with strain type and biofilm type, of which regions 1 and 2 have been previously described (Musatovova et al., 2008). Region 1, which is missing in UAB PO1, is a segment of the M129 genome from ntp 168 936 to 169 592 that codes for MPN_130 and is flanked by 24 bp direct repeats. Musatovova et al. (2008) identified an orthologue to MPN_130 in the type 2 genomes. This gene, which is referred to as MPN_465 in M129, has very little amino acid sequence similarity to MPN_130. However, a second copy of the gene coding for MPN_130 is present in M129 (MPN_094) and UAB PO1 has this orthologue.
Table 1. Indels between the UAB PO1 and M129 genomes.
| M129 reference ntp for indel region | Indel length (bp) | Presence(+)/absence (−) | Proteins affected and comments | |
| Type 1 strains | Type 2 strains | |||
| Region 1 168 936 to 169 592 | 657 | M129+ PI 1428+ | UAB PO1− FH− 309− | MPN_130; second copy MPN_094 in M129, UAB PO1 has orthologue |
| Region 2 177 772 to 178 646 | 875 | M129+ PI 1428+ | UAB PO1− FH− 309− | MPN_137, MPN_138; fused in type 2 strains; UAB PO1 191 aa long; M129 228 aa long |
| Region 3 249 583 | 72 | M129− PI 1428+ | UAB PO1+ FH+ 309+ | MPN_205; 24 aa insertion |
| Region 4 558 099 to 562 042 | 3944 | M129+ PI 1428+ | UAB PO1− FH− 309− | MPN_457, MPN_458, MPN_459, MPN_t26 |
| Region 5 708 744 | 5883 | M129− PI 1428− | UAB PO1+ FH− 309+ | MPN_586 |
Region 2 is a segment of M129 that is absent in UAB PO1 and affects MPN_137 and MPN_138. In UAB PO1, this absent sequence results in a gene fusion (Musatovova et al., 2008). The fusion gene has the orthologues MPNE_0161 and MPNA1370 in strains FH and 309, respectively, and PI 1428 has orthologues to MPN_137 and MPN_138. A tandem repetitive region was identified at the C-terminal region of this protein. The reference M129, the resequenced M129 and PI 1428 contained seven tandem repeat units of 21 bp in this region while FH, 309 and UAB PO1 had six repeat units. An internal set of seven tandem repeats of 21 bp each, starting at ntp 225 in the gene coding for MPN_138, was absent in the fusion protein.
Region 4 is present in the genome of M129 and includes a 2892 bp segment flanked by two imperfect direct repeats of 525 and 527 bp. This region codes for MPN_457, MPN_458 and MPN_459. It is also present in PI 1428 but absent in UAB PO1, FH and 309.
Discussion
Biofilms provide a structure in which micro-organisms can be protected from host immunity and treatment with antimicrobials (Abdi-Ali et al., 2006; Campos et al., 2004; Costerton et al., 1999; Hoyle et al., 1990; Kropec et al., 2005). The structure of the biofilm has been shown to contribute to the ability of the mycoplasmas encased within it to survive killing by complement and antimicrobial peptides and inhibit the penetration of antibody (Simmons et al., 2007; Simmons & Dybvig, 2007). Contributing to the biofilm’s structure is the ability of micro-organisms to produce polysaccharides that can act as glue holding the biofilm together (Itoh et al., 2005; Sutherland, 2001). It has been shown that polysaccharide produced by Mycoplasma pulmonis affects biofilm formation (Daubenspeck et al., 2009). M. pulmonis forms biofilm structures on mouse tracheal epithelium (Simmons & Dybvig, 2009). Hence, the different characteristics of the biofilms formed by M129 and UAB PO1 may reflect factors that are important for virulence. Nilsson et al. (2010) found no correlation between the genotype of M. pneumoniae and the severity of the disease as measured by whether a patient was hospitalized. While hospitalization may be one marker to differentiate disease severity, it does not necessarily consider other important aspects of infection that might be affected by biofilm formation or genotype, such as extrapulmonary complications, chronicity and efficacy of antibiotic therapy.
The propensity of the mycoplasmas to grow in a biofilm was assessed from the ratio of c.f.u. adhered to the surface versus those growing in suspension. UAB PO1 formed a much heavier biofilm than M129. A far greater percentage of UAB PO1 c.f.u. grew as a biofilm at all time points, suggesting that UAB PO1 is more cohesive. Microscopic analysis yielded information on the structure and the biovolume of the biofilms for growth periods of up to 10 days. Even after heavy growth, the biofilm formed by M129 did not appear similar to the biofilm grown by UAB PO1. The regions between the towers of M129 never developed robustly, and the margins of the towers and honeycombed regions never became smooth or mucoid. We conclude that the structures of the biofilms formed by the two strains are of two distinct types. This is further supported as the biofilms produced by strain PI 1428 are similar to those of M129 and the biofilms produced by strain FH are similar to those of UAB PO1. The structural differences could affect the ability of the mycoplasmas to resist complement and other antimicrobials. The cavities extending throughout the towers and a matrix that holds a biofilm loosely might facilitate the exchange of nutrients between the medium and the internal regions of the biofilms but also might allow more access of antimicrobials and render the mycoplasmas more susceptible to killing.
Using Congo red and analysis with lectins, a matrix was observed between the cells in the towers, in smaller clusters of cells, and on individual cells attached to the glass. An intercellular matrix containing polysaccharides rich in GlcNAc has been shown to contribute to the adhesion of many biofilms (Lasa, 2006). GC analysis indicated that both M129 and UAB PO1 produce a polymer containing GlcNAc. The results of the GC and fluorescence analyses suggest that polymers containing GlcNAc are not tightly associated with the M129 cell surface and are secreted into the medium. As such, the GlcNAc-based polymer would not tether the cells of the biofilm together efficiently. In this model, the GlcNAc-containing polymer of UAB PO1 is held tightly to the cells and is able to contribute to the intercellular adhesion. This could account for the cavities observed in the towers of the biofilms formed by M129 but not in UAB PO1 and also may account for the smoothness of the towers formed by UAB PO1. The reason(s) that UAB PO1 holds the polysaccharide more tightly to the cell are unknown but might involve a lectin at the surface of the type 2 strains that binds the polysaccharide. Alternatively, the polysaccharide could be held to the cell by a lipid anchor that is absent in M129.
Genomic analysis indicated that M129 and PI 1428 are type 1 strains and FH and UAB PO1 are type 2, suggesting that the strain type of M. pneumoniae determines the biofilm type. Type-specific genetic differences could affect mycoplasma–polysaccharide interactions and hence biofilm structure. The genome rearrangements in regions 3 and 5, which do not correlate with the strain type, could logically be excluded from contributing to the biofilm type. The rearrangements at regions 1 and 4 potentially provide for the production of additional proteins by the type 1 strains. Perhaps one of these proteins interferes with the anchoring of polysaccharide to the cells. In region 2, fusion between the MPN_137 and MPN_138 genes of the type 2 strains may code for a protein involved with anchoring of the polysaccharide. The differences in the tandem repeat regions, which may encode leucine zippers, found in MPN_137, MPN_138 and the fusion protein (Musatovova et al., 2008, 2012) may affect biofilm formation, perhaps through regulatory mechanisms. As the major proteins involved in attachment, such as the P1 and accessory proteins (Atkinson et al., 2008; Balish et al., 2003; Krause, 1998; Waites et al., 2008), were intact and correlated with strain type, it is also possible that the attachment apparatus has a role in anchoring polysaccharide.
Acknowledgements
This work was supported by the National Institutes of Health (Public Health Service grants AI63909, AI64848 and AI93750). Genome sequencing was performed at the Hudson Alpha Institute of Biotechnology and was supported by the UAB Center for Clinical and Translational Science via the NIH National Center for Research Resources through grant number 5UL1 RR025777-03. We thank Elliot Lefkowitz, Portia Caldwell and Donna Crabb for expert technical assistance.
Abbreviations:
- GS-I
Griffonia simplicifolia lectin-I
- ntp
nucleotide position
- SNPs
single nucleotide polymorphisms
Footnotes
A supplementary table and two supplementary data files are available with the online version of this paper.
References
- Abdi-Ali A., Mohammadi-Mehr M., Agha Alaei Y. (2006). Bactericidal activity of various antibiotics against biofilm-producing Pseudomonas aeruginosa. Int J Antimicrob Agents 27, 196–200. 10.1016/j.ijantimicag.2005.10.007 [DOI] [PubMed] [Google Scholar]
- Atkinson T. P., Balish M. F., Waites K. B. (2008). Epidemiology, clinical manifestations, pathogenesis and laboratory detection of Mycoplasma pneumoniae infections. FEMS Microbiol Rev 32, 956–973. 10.1111/j.1574-6976.2008.00129.x [DOI] [PubMed] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., Formsma K., Gerdes S., Glass E. M. & other authors (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75–89. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balish M. F., Santurri R. T., Ricci A. M., Lee K. K., Krause D. C. (2003). Localization of Mycoplasma pneumoniae cytadherence-associated protein HMW2 by fusion with green fluorescent protein: implications for attachment organelle structure. Mol Microbiol 47, 49–60. 10.1046/j.1365-2958.2003.03282.x [DOI] [PubMed] [Google Scholar]
- Birol I., Jackman S. D., Nielsen C. B., Qian J. Q., Varhol R., Stazyk G., Morin R. D., Zhao Y., Hirst M. & other authors (2009). De novo transcriptome assembly with ABySS. Bioinformatics 25, 2872–2877. 10.1093/bioinformatics/btp367 [DOI] [PubMed] [Google Scholar]
- Bolland J. R., Simmons W. L., Daubenspeck J. M., Dybvig K. (2012). Mycoplasma polysaccharide protects against complement. Microbiology 158, 1867–1873. 10.1099/mic.0.058222-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda S. S., Vik S., Friedman L., Kolter R. (2005). Biofilms: the matrix revisited. Trends Microbiol 13, 20–26. 10.1016/j.tim.2004.11.006 [DOI] [PubMed] [Google Scholar]
- Campos M. A., Vargas M. A., Regueiro V., Llompart C. M., Albertí S., Bengoechea J. A. (2004). Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect Immun 72, 7107–7114. 10.1128/IAI.72.12.7107-7114.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon K. M., Humphreys H., O’Gara J. P. (2002). icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J Bacteriol 184, 4400–4408. 10.1128/JB.184.16.4400-4408.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton J. W., Stewart P. S., Greenberg E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- Daims H., Lücker S., Wagner M. (2006). daime, a novel image analysis program for microbial ecology and biofilm research. Environ Microbiol 8, 200–213. 10.1111/j.1462-2920.2005.00880.x [DOI] [PubMed] [Google Scholar]
- Darling A. E., Mau B., Perna N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5, e11147. 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubenspeck J. M., Bolland J. R., Luo W., Simmons W. L., Dybvig K. (2009). Identification of exopolysaccharide-deficient mutants of Mycoplasma pulmonis. Mol Microbiol 72, 1235–1245. 10.1111/j.1365-2958.2009.06720.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson M. K., Lindsey J. R., Parker R. F., Tully J. G., Cassell G. H. (1988). Differences in virulence for mice among strains of Mycoplasma pulmonis. Infect Immun 56, 2156–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan R. M. (2001). Biofilm formation: a clinically relevant microbiological process. Clin Infect Dis 33, 1387–1392. 10.1086/322972 [DOI] [PubMed] [Google Scholar]
- García-Castillo M., Morosini M.-I., Gálvez M., Baquero F., del Campo R., Meseguer M.-A. (2008). Differences in biofilm development and antibiotic susceptibility among clinical Ureaplasma urealyticum and Ureaplasma parvum isolates. J Antimicrob Chemother 62, 1027–1030. 10.1093/jac/dkn337 [DOI] [PubMed] [Google Scholar]
- Halbedel S., Busse J., Schmidl S. R., Stülke J. (2006). Regulatory protein phosphorylation in Mycoplasma pneumoniae. A PP2C-type phosphatase serves to dephosphorylate HPr(Ser-P). J Biol Chem 281, 26253–26259. 10.1074/jbc.M605010200 [DOI] [PubMed] [Google Scholar]
- Hoyle B. D., Jass J., Costerton J. W. (1990). The biofilm glycocalyx as a resistance factor. J Antimicrob Chemother 26, 1–5. 10.1093/jac/26.1.1 [DOI] [PubMed] [Google Scholar]
- Itoh Y., Wang X., Hinnebusch B. J., Preston J. F., III, Romeo T. (2005). Depolymerization of β-1,6-N-acetyl-d-glucosamine disrupts the integrity of diverse bacterial biofilms. J Bacteriol 187, 382–387. 10.1128/JB.187.1.382-387.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D. R., Schatz M. C., Salzberg S. L. (2010). Quake: quality-aware detection and correction of sequencing errors. Genome Biol 11, R116. 10.1186/gb-2010-11-11-r116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornspan J. D., Tarshis M., Rottem S. (2011). Adhesion and biofilm formation of Mycoplasma pneumoniae on an abiotic surface. Arch Microbiol 193, 833–836. 10.1007/s00203-011-0749-y [DOI] [PubMed] [Google Scholar]
- Krause D. C. (1998). Mycoplasma pneumoniae cytadherence: organization and assembly of the attachment organelle. Trends Microbiol 6, 15–18. 10.1016/S0966-842X(97)01168-2 [DOI] [PubMed] [Google Scholar]
- Kropec A., Maira-Litran T., Jefferson K. K., Grout M., Cramton S. E., Götz F., Goldmann D. A., Pier G. B. (2005). Poly-N-acetylglucosamine production in Staphylococcus aureus is essential for virulence in murine models of systemic infection. Infect Immun 73, 6868–6876. 10.1128/IAI.73.10.6868-6876.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasa I. (2006). Towards the identification of the common features of bacterial biofilm development. Int Microbiol 9, 21–28. [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuliffe L., Ellis R. J., Miles K., Ayling R. D., Nicholas R. A. J. (2006). Biofilm formation by mycoplasma species and its role in environmental persistence and survival. Microbiology 152, 913–922. 10.1099/mic.0.28604-0 [DOI] [PubMed] [Google Scholar]
- Mohamed J. A., Huang D. B. (2007). Biofilm formation by enterococci. J Med Microbiol 56, 1581–1588. 10.1099/jmm.0.47331-0 [DOI] [PubMed] [Google Scholar]
- Musatovova O., Kannan T. R., Baseman J. B. (2008). Genomic analysis reveals Mycoplasma pneumoniae repetitive element 1-mediated recombination in a clinical isolate. Infect Immun 76, 1639–1648. 10.1128/IAI.01621-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musatovova O., Kannan T. R., Baseman J. B. (2012). Mycoplasma pneumoniae large DNA repetitive elements RepMP1 show type specific organization among strains. PLoS ONE 7, e47625. 10.1371/journal.pone.0047625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen C. B., Jackman S. D., Birol I., Jones S. J. (2009). ABySS-Explorer: visualizing genome sequence assemblies. IEEE Trans Vis Comput Graph 15, 881–888. 10.1109/TVCG.2009.116 [DOI] [PubMed] [Google Scholar]
- Nilsson A. C., Björkman P., Welinder-Olsson C., Widell A., Persson K. (2010). Clinical severity of Mycoplasma pneumoniae (MP) infection is associated with bacterial load in oropharyngeal secretions but not with MP genotype. BMC Infect Dis 10, 39. 10.1186/1471-2334-10-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft T., Hilbert H., Layh-Schmitt G., Herrmann R. (1995). The proline-rich P65 protein of Mycoplasma pneumoniae is a component of the Triton X-100-insoluble fraction and exhibits size polymorphism in the strains M129 and FH. J Bacteriol 177, 3370–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidl S. R., Gronau K., Hames C., Busse J., Becher D., Hecker M., Stülke J. (2010). The stability of cytadherence proteins in Mycoplasma pneumoniae requires activity of the protein kinase PrkC. Infect Immun 78, 184–192. 10.1128/IAI.00958-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons W. L., Dybvig K. (2003). The Vsa proteins modulate susceptibility of Mycoplasma pulmonis to complement killing, hemadsorption, and adherence to polystyrene. Infect Immun 71, 5733–5738. 10.1128/IAI.71.10.5733-5738.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons W. L., Dybvig K. (2007). Biofilms protect Mycoplasma pulmonis cells from lytic effects of complement and gramicidin. Infect Immun 75, 3696–3699. 10.1128/IAI.00440-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons W. L., Dybvig K. (2009). Mycoplasma biofilms ex vivo and in vivo. FEMS Microbiol Lett 295, 77–81. 10.1111/j.1574-6968.2009.01592.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons W. L., Bolland J. R., Daubenspeck J. M., Dybvig K. (2007). A stochastic mechanism for biofilm formation by Mycoplasma pulmonis. J Bacteriol 189, 1905–1913. 10.1128/JB.01512-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slifkin M., Cumbie R. (1988). Congo red as a fluorochrome for the rapid detection of fungi. J Clin Microbiol 26, 827–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spuesens E. B. M., Oduber M., Hoogenboezem T., Sluijter M., Hartwig N. G., van Rossum A. M., Vink C. (2009). Sequence variations in RepMP2/3 and RepMP4 elements reveal intragenomic homologous DNA recombination events in Mycoplasma pneumoniae. Microbiology 155, 2182–2196. 10.1099/mic.0.028506-0 [DOI] [PubMed] [Google Scholar]
- Su C. J., Chavoya A., Dallo S. F., Baseman J. B. (1990). Sequence divergency of the cytadhesin gene of Mycoplasma pneumoniae. Infect Immun 58, 2669–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland I. W. (2001). Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147, 3–9. [DOI] [PubMed] [Google Scholar]
- Toledo-Arana A., Merino N., Vergara-Irigaray M., Débarbouillé M., Penadés J. R., Lasa I. (2005). Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J Bacteriol 187, 5318–5329. 10.1128/JB.187.15.5318-5329.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully J. G. (1983). New laboratory techniques for isolation of Mycoplasma pneumoniae. Yale J Biol Med 56, 511–515. [PMC free article] [PubMed] [Google Scholar]
- Ursi D., Ieven M., van Bever H., Quint W., Niesters H. G., Goossens H. (1994). Typing of Mycoplasma pneumoniae by PCR-mediated DNA fingerprinting. J Clin Microbiol 32, 2873–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu B., Chen M., Crawford R. J., Ivanova E. P. (2009). Bacterial extracellular polysaccharides involved in biofilm formation. Molecules 14, 2535–2554. 10.3390/molecules14072535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites K. B., Balish M. F., Atkinson T. P. (2008). New insights into the pathogenesis and detection of Mycoplasma pneumoniae infections. Future Microbiol 3, 635–648. 10.2217/17460913.3.6.635 [DOI] [PMC free article] [PubMed] [Google Scholar]