Abstract
Protein tyrosine phosphatases such as PTPN6 can be downregulated in various neoplasms. PTPN6 expression by immunohistochemistry in 40 diffuse large B-cell lymphoma (DLBCL) tumors was lost or suppressed in 53% (21/40). To elucidate the molecular mechanisms of PTPN6 suppression we performed a comprehensive epigenetic analysis of PTPN6 promoter 2 (P2). None of the DLBCL primary tumors (0/37) had PTPN6 hypermethylation on the CpG1 island using methylation specific PCR, pyrosequencing, and high-resolution melting assays. However, hypermethylation in 57% (21/37) of cases was found in a novel CpG island (CpG2) in P2. PTPN6 gene suppression was reversed by 5-aza-deoxycytidine (5-Aza), a DNA methyltransferase inhibitor, and the histone deacetylase inhibitor (HDACi) LBH589. LBH589 and 5-Aza in combination inhibited DLBCL survival and PTPN6 hypermethylation at CpG2. The role of histone modifications was investigated with a chromatin-immunoprecipitation assay demonstrating that PTPN6 P2 is associated with silencing histone marks H3K27me3 and H3K9me3 in DLBCL cells but not normal B-cells. DZNep, a histone methyltransferase inhibitor, decreased the H3K27me3 mark while HDACi LBH589 increased the H3K9Ac mark within P2 resulting in re-expression of PTPN6. These studies have uncovered novel epigenetic mechanisms of PTPN6 suppression and suggest that PTPN6 may be a potential target of epigenetic therapy in DLBCL.
Keywords: DLBCL, PTPN6, CpG methylation, histone methylation, LBH589, Azacytidine
Introduction
The standard chemo-immunotherapy treatment of diffuse large B-cell lymphoma (DLBCL)(1, 2) cures approximately 60% of patients; new approaches are needed to improve the outcome for the other 40%.(3) In designing these new treatments, it is vital to identify novel tumor targets that will enable clinicians to individualize therapy based on tumor signal pathway activation profiles. DLBCL is characterized by over-expression of several signaling pathways such as NF-kB and JAK2/STAT3(4–7) due to various mechanisms such as genetic mutations or through cytokine deregulation. In addition, other mechanisms such as epigenetic silencing of protein tyrosine phosphatases such as protein tyrosine phosphatase 6 (PTPN6, also known as SHP1), may also contribute to various pathway deregulation.(8) PTPN6 acts as a negative regulator of several signal transduction pathways by dephosphorylating the receptor-associated kinases such as Src and JAK/STAT family kinases.(9, 10) Loss of PTPN6 contributes to JAK3/STAT3 activation in ALK+ anaplastic large cell T-cell lymphoma cells.(11) Deregulation of these pathways (STAT3, Src etc) can promote cell growth leading to tumor development of various kinds of tumors.
The PTPN6 gene in humans is encoded by 17 exons and has 2 Src homology 2 (SH2) domains, which are required for binding to phosphorylated tyrosine residues.(12) The PTPN6 gene also has two promoter regions that are 7 kb apart. The longer region (PTPN6 1A or P1) is expressed primarily in non-hematopoietic cells whereas the shorter region (PTPN6 1B or P2) is expressed only in cells of hematopoietic lineage.(12–14) PTPN6 expression driven by P1 in non-hematopoietic cell such as epithelial or neuronal cells is low compared with the expression regulated by P2 in hematopoietic cells.(12, 15) Decreased expression of PTPN6 due to DNA hypermethylation of CpG island in the PTPN6 P2 promoter has been reported in various hematological malignancies.(16–19) The aims of these studies were focused on whether PTPN6 was lost in DLBCL primary samples and cell lines, understanding the role of various epigenetic mechanisms including CpG hypermethylation and histone modifications in PTPN6 loss, and whether PTPN6 in DLBCL can be re-expressed with epigenetic therapy.
Material and Methods
Patient Samples and Cell Lines
Tissue from formalin fixed paraffin-embedded (FFPE) tumors from 40 untreated DLBCL participating in a clinical trial (N0489; NCT00301821) were utilized for immunohistochemistry (IHC) studies of PTPN6.(20) All patients signed informed consent approved by the institutional review board at each participating site in N0489. Tissue microarrays (TMA) were constructed and included 10 benign tonsil controls from the Predolin Biobank at the Mayo Clinic. For the studies of methylation, DNA from fresh or frozen DLBCL tumor cells was required. Since this type of sample was not available from the N0489 trial we obtained frozen cells from 40 other DLBCL patient samples from the University of Iowa/Mayo Lymphoma SPORE Biospecimen Core. All SPORE patients also provided informed consent for use of their tumor tissue for research. B-cells from normal controls were isolated from peripheral blood mononuclear cells (n=3) using CD19 microbeads (Miltenyi Biotec, Auburn, CA). The DLBCL human cell lines OCILy3 (Ly3), OCILy10 (Ly10), and SUDHL2 (DHL2) used in this study were a kind gift from Dr. Louis Staudt (NCI, Bethesda).(4) They were grown in 20% human serum in Iscove’s Modified Dulbecco’s medium (IMDM).
Quantitative Reverse Transcriptase-Polymerase Chain Reaction for PTPN6 (qRT-PCR)
Total RNA was extracted from DLBCL patient samples (n=9) and CD19 normal B cells (n=3) using RNeasy Mini Kit (Qiagen, Valencia, CA) and cDNAs were synthesized using SuperScript® III First-Strand Synthesis SuperMix (Invitrogen, Carlsbad, CA). QRT-PCR was performed on CFX96TM real-time PCR detection systems. Primers and probe sets are from Integrated DNA Technologies (Coralville, IA). The forward primer for PTPN6 is 5′-CAC CAT CAT CCA CCT CAA GT-3′; its reverse primer is 5′-TCT CAC GCA CAA GAA ACGTC-3′. The probe used for PTPN6 was 5′-/56-FAM/CGC TGA ACT/ZEN/GCT CCG ATC CCA/3IABkFQ/-3′. GAPDH was used as an internal control. Its forward primer is 5′ GAAGGTGAAGGTCGGAGTC 3′; its reverse primer is 5′ GAAGATGGTGATGGGATTTC 3′. The probe used was 5′/HEX/CAAGCTTCCCGTTCTCAGCC/3IABKFQ/3′. The PCR program was: 95°C for 15 minutes, 40 cycles of 95 °C for 10 seconds and 60 °C for 30 seconds. Data analysis was performed using delta-delta-CT method.
Immunohistochemistry for PTPN6 expression
The FFPE slides were deparaffinized and endogenous peroxidase activity was quenched by incubation of sections in a solution of 3% hydrogen peroxide and absolute methanol. Antigen retrieval was performed using citrate at pH 6.1. The slides were placed on a Dako Cytomation autostainer with the PTPN6 antibody (Abcam, Cambridge, MA) DAKO advance detection system, and substrate. The slides were reviewed and scored by the study hematopathologist (AD). The expression of PTPN6 was assessed semi-quantitatively as follows; Neg <10%, 10–30% (+, low), 30–80% (++, intermediate), >80% (+++, high).
Methylation-specific polymerase chain reaction (MSP/USMP)
We performed methylation studies by several methods on two different CpG sites within the P2 promoter. A web-based program MethPrimer (http://www.urogene.org/methprimer/)(21) was used to identify CpG islands in P2 of PTPN6. We found 2 CpG islands designated as CpG1 and CpG2 as defined below.
(i) CpG1 methylation by MSP1-PCR1
DNA was extracted from frozen DLBCL tumor cells (n=39) by using Gentra Puregene Cell Kit (Qiagen, Valencia, CA). One μg of DNA was bisulfite-treated with CpGenome Universal DNA Modification Kit from EMD Millipore (Billerica, MA) as per manufacturer instructions. Twenty ng of bisulfite-treated DNA was used as template for the MSP PCR. Primers were synthesized according to Chim et al.(18) Methylated and unmethylated DNA was used as positive and negative controls, respectively, for the methylation studies (EMD Millipore, Billerica, MA). The PCR products were treated with EXOSAP-IT enzyme (Affymetrix, Cleveland, OH) and sequenced.
(ii) CpG2 methylation by MS2P-PCR2
The primers for CpG2 MSP-PCR were: Forward 5′ GTTTTGTTGATGTTTATTTCGAC 3′ Reverse 5′ GAAAATCCTCACACCTTACGAA 3′, the program was: 95°C for 15 minutes, (95 °C for 30 seconds, 61.7 °C for 30 seconds, 72 °C for 30 seconds) 40 cycles, 72 °C for 10 minutes. The primers of U-MSP2 were: Forward 5′ GTTTTGTTGATGTTTATTTTGATGT 3′, Reverse 5′ CAAAAATCCTCACACCTTACAAA 3′, the program was: 95°C for 15 minutes, (95 °C for 30 seconds, 61 °C for 30 seconds, 72 °C for 30 seconds) 40 cycles, 72 °C for 10 minutes.
Methylation analysis by Pyrosequencing
(i) CpG1 pyrosequencing
PCR was run with the bisulfite-treated DNA as template. The primers were as follows: Forward 5′ AGGGTTGTGGTGAGAAATTAATTAG 3′ (with 5′ Biotin-TEG); Reverse 5′ TTACACACTCCAAACCCAAATAATAC 3′. The PCR program was: 95°C for 15 minutes, (95 °C for 30 seconds, 58 °C for 30 seconds, 72 °C for 30 seconds) 35 cycles, 72 °C for 10 minutes. The PCR product was pyrosequenced by the Mayo Clinic Sequencing Core with the reverse primer.(22)
(ii) CpG2 pyrosequencing
The primers were: Forward 5′ TAGTTTTTTGTTAGTTTTGGAGGGA 3′ Reverse 5′ AAAAACAAATACACACTTATCCAAAAAA 3′. The program was: 95°C for 15 minutes, (95 °C for 30 seconds, 58 °C for 30 seconds, 72 °C for 30 seconds) 40 cycles, 72 °C for 10 minutes. Methylation was quantified in the terms of methylation level defined as the mean percent methylated cytosines per CpG.
Methylation analysis by high resolution melting (HRM)
Methylation standards were mixed to produce 100%, 50%, 25%, 10%, 5%, and 0% methylation control samples. Amplification primers, 5′-gcg TGG GTT AGG GAG GGT TG-3′ and 5′-TTA CAC ACT CCA AAC CCA AAT AAT AC-3′, were designed using methprimer (http://www.urogene.org/methprimer/index1.html). PCR reactions were performed using 10 ng bisulfite modified DNA on an LC480 II using the LC480 High Resolution Melting Master Kit reagents (Roche). PCR cycling parameters consisted of an initial denaturation step of 95°C for 10 minutes, 20 cycles of touchdown PCR (95°C for 10 seconds, 65°C–0.5°C/cycle for 15 seconds, 72°C for 10 seconds), and 18 cycles of PCR (95°C for 10 seconds, 55°C for 15 seconds, 72°C for 10 seconds). PCR products were denatured at 95°C for 1 minute and melting profiles generated by cooling samples at 40°C for 1 minute, followed by melting from 65°C to 95°C at 0.02° per second collecting 25 data points every second. Data was analyzed using the Gene Scanning module of the LC480 analysis software. Profiles were normalized using a 5°C window before and after melting. The slope of the melting profile was plotted, and based on control samples, methylated product was shown to melt at 82.5°C and unmethylated at 80.2°C. Methylation was estimated by comparing relative peak size at 80.2°C to that of controls.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed using the ChIP Assay Kit (EMD Millipore Billerica, MA) with antibodies to H3K9me3, H3K27me3, H3K9Ac (Abcam, Cambridge, MA) and IgG as per the manufacturer’s instructions. Briefly, 2 million cells were fixed with 1% formaldehyde and cells were then re-suspended in SDS lysis buffer and immunoprecipitated. The resulting complex was pelleted with protein A agarose, and eluted with elution buffer. Histone-DNA crosslinks were reversed in sodium chloride and DNA was recovered by phenol/chloroform extraction. Immunoprecipitated DNA and input samples were analyzed by PCR. The following primers for the PTPN6 gene promoter were used: 5′-AGTGCCACCCTGCTCTGCTTC-3′ (forward) and the 5′-CAGTTCTGGGGCTGCCACT-3′ (reverse). 5S rRNA gene was used as a control for the ChIP assay.(23)
Treatment with DNA methyltransferase and histone deacetylase inhibitors
DLBCL cells were seeded at a density of 1 million cells/ml in 25 cm2 culture flasks; then treated with 5-azacytidine (Sigma Aldrich) or LBH589 (Novartis Pharmaceuticals) alone or in combination at the indicated concentrations. Fresh media containing 5-azacytidine and/or LBH589 was added every 2 days for 6 days. Cells were harvested at the time points indicated and used for western blot and survival analysis using flow cytometry with Annexin/Propidium Iodide staining.(24)
Results
PTPN6 is lost or silenced in DLBCL tumors
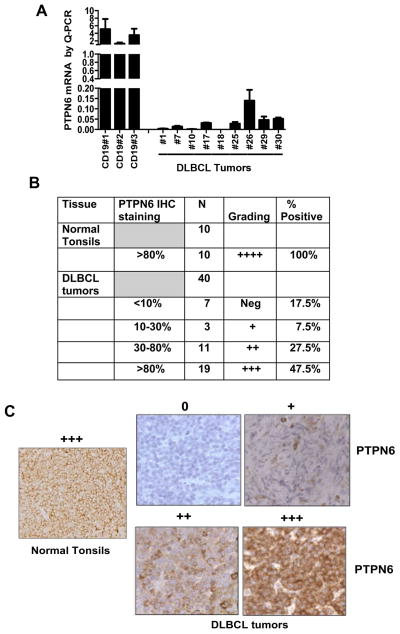
We analyzed PTPN6 mRNA expression in DLBCL (n=9) patient specimens and normal B-cells by QRT-PCR. Decreased expression of PTPN6 mRNA was observed in all the DLBCL patient samples as compared to normal B cells (Figure 1A). To confirm the PTPN6 mRNA expression at the protein level, FFPE DLBCL tumor samples from N0489 clinical trial (n=40) along with normal tonsils (n=10) were stained for the detection of PTPN6 protein by IHC. All normal tonsils (10/10) were strongly positive for PTPN6 (>80%; +++); however, differential expression of PTPN6 staining was found among the DLBCL tumors (Figure 1B–C). PTPN6 expression was completely lost in 17.5% (7/40) of cases (PTPN6 negative); 7.5% (3/40) of cases had very low expression of PTPN6 (10–30%; +); 27.5% (11/40) cases had 30–80% (++) of tumor cells staining positive; and, 47.5% (19/40) cases had >80% (+++) of cells PTPN6 positive. These data, when taken together, confirm that PTPN6 is strongly expressed in normal B-cells and can be lost or suppressed in DLBCL tumors.
Figure 1. Evaluation of PTPN6 expression in DLBCL tumors.
(A) PTPN6 expression by QRT-PCR in cryopreserved DLBCL tumor cells from 9 patients and CD19+ B cells from 3 normal controls. (B) Table summarizing the expression of PTPN6 protein by immunohistochemistry in 40 DLBCL tumors and 10 normal tonsils. (C) Representative PTPN6 staining in paraffin-embedded tissues from DLBCL tumors (magnification X400) and normal tonsils (magnification X200).
CpG1 island are not hypermethylated in PTPN6 promoter 2
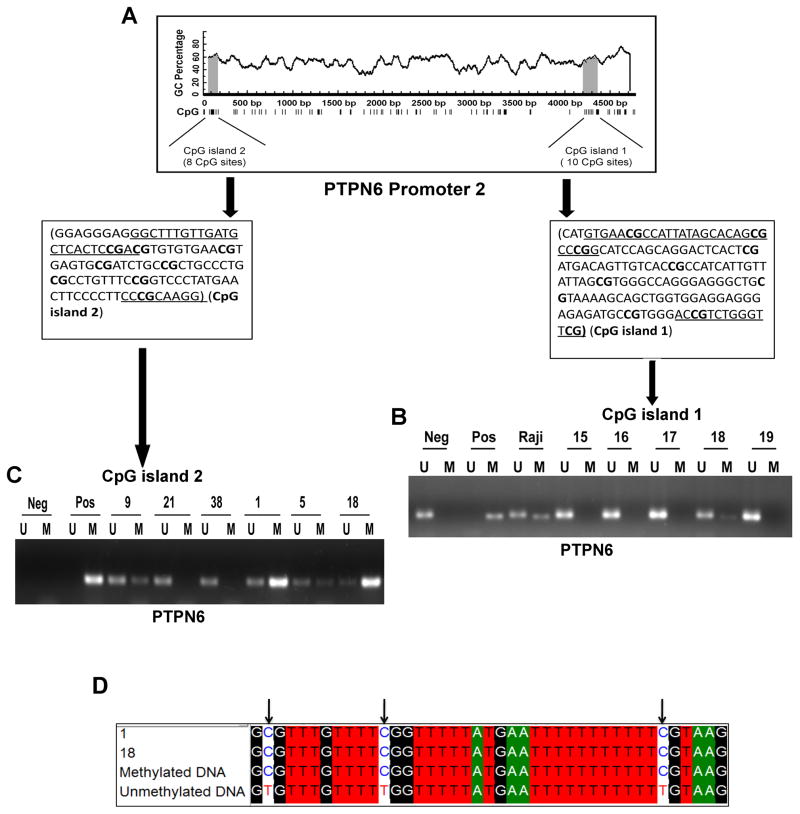
Promoter methylation has been found to be an important mechanism regulating PTPN6 expression in peripheral T-cell lymphomas and multiple myeloma.(17, 18, 25) DLBCL patient samples were analyzed for PTPN6 methylation by MSP1/USMP1 PCR by use of previously published PCR primers(17) that encompass the CpG1 region of PTPN6 P2 (Figure 2A). CpG1 hypermethylation by MSP PCR was detected in the tumor cells from only one patient (#18) (1/38; 2.6%) (Figure 2B) which after further review had a neuroendocrine carcinoma (vide infra). None of the DLBCL cell lines (Ly3, DHL2, Ly10) along with CD19+ B cells tested showed hypermethylation of PTPN6 at CpG1 (data not shown). Since the MSP PCR technique yields qualitative rather than quantitative data it is unable to provide information about the degree of methylation at specific CpG1 sites. In order to quantify methylation, pyrosequencing was performed on the same DLBCL samples and methylation level was generated for CpG1 sites in the PTPN6 promoter 2.(26, 27) Cases with <10% methylation were categorized as unmethylated; cases >10% methylation were low (10–25%), intermediate (25–40%) and high methylation (>40%). Table 1 shows the average percent methylation of CpG1 sites in the DLBCL patients and cell lines. The pyrosequencing analysis was consistent with MSP PCR analysis and demonstrated that again only patient sample #18 was highly hypermethylated (76%) at CpG1 (Table 1). CD19+ normal B cells were unmethylated (9.4%) whereas the Raji Burkitt lymphoma cell line (positive control for PTPN6 methylation) was highly methylated (86%) at CpG1 (data not shown).(18)
Figure 2. PTPN6 promoter 2 methylation at CpG1and CpG2 sites by MSP/UMSP PCR in primary DLBCL samples.
(A) Schematic diagram showing location of CpG1 and CpG2 islands within PTPN6 promoter 2. (B) Methylation analysis of the CpG1 island using MSP1/UMSP1 primers in tumors (n=38). Representative results are shown. (C) Methylation analysis of the CpG2 island using MSP2/UMSP2 primers confirms the CpG2 pyrosequencing data. (D) Sequencing results of CpG2 methylated PCR products.
Table 1. Pyrosequencing for the detection of CpG1 and CpG2 hypermethylation in the PTPN6 promoter 2.
Pyrosequencing for CpG1 and CpG2 islands was performed on DNA from 37 primary DLBCL tumors and 3 DLBCL cell lines. Percent CpG1 and CpG2 hypermethylation for cases and controls is reported; positive methylation was defined as >25% methylation.
| DLBCL samples | CpG1 Methylation (%) | CpG2 Methylation (%) | DLBCL samples | CpG1 Methylation (%) | CpG2 Methylation (%) |
|---|---|---|---|---|---|
| 1 | 7.3 | 47.2 | 26 | 7.2 | 22.9 |
| 2 | 4.1 | 41.3 | 27 | 5.0 | 7.7 |
| 3 | 4.7 | 25.4 | 28 | 4.5 | 11.0 |
| 4 | 7.3 | 28.4 | 29 | 8.8 | 19.5 |
| 5 | 3.0 | 83.6 | 30 | 4.6 | 24.4 |
| 6 | 11.3 | 56.7 | 31 | 5.0 | 11.5 |
| 7 | 6.2 | 10.0 | 32 | NA | NA |
| 8 | 6.1 | 62.3 | 33 | 5.6 | 41.3 |
| 9 | 6.6 | 22.9 | 34 | 4.3 | 53.6 |
| 10 | 6.2 | 33.5 | 35 | NA | NA |
| 11 | 12.2 | 43.0 | 36 | 10.6 | 33.8 |
| 12 | 4.3 | 54.3 | 37 | 6.7 | 51.7 |
| 13 | 5.2 | 23.1 | 38 | 4.9 | 20.4 |
| 14 | 4.1 | 15.8 | 39 | 8.6 | 43.1 |
| 15 | 4.9 | 52.4 | 40 | 4.2 | 28.4 |
| 16 | 4.6 | 30.4 | |||
| 17 | 6.6 | 11.8 | Cell lines | ||
| *18 | 76.4 | 83.1 | LY3 | 5.5 | 48.0 |
| 19 | 6.1 | 11.7 | LY10 | 11.2 | 44.1 |
| 20 | 4.8 | 24.9 | DHL2 | 5.8 | 25.8 |
| 21 | 3.4 | 23.1 | |||
| 22 | 10.2 | 47.6 | |||
| 23 | 6.3 | 15.4 | |||
| 24 | 4.1 | 34.2 | |||
| 25 | 4.3 | 27.6 |
Unmethylated (0–10%)
Low methylation (>10–25%)
Intermediate-methylation (>25–40%)
Hypermethylation (>40%)
Sample 18 after repeat pathology review was a neuroendocrine carcinoma-see text
Abbreviations: DLBCL, diffuse large B-cell lymphoma; NA, no DNA available at time of extraction
To confirm the MSP1 PCR and pyrosequencing results, we performed HRM on the same DLBCL DNA samples. HRM is a very sensitive method for the detection of methylation at gene-specific loci.(28) With HRM we again found only 1 sample (patient #18) with high levels of methylation with the peak of the melt profile falling between that of the 50% and 100% methylation controls (Supplemental Figure 1A). Seven additional samples showed low levels of methylation: two samples fell between the 5 and 10% methylation controls; the remaining 5 had less than 5% methylation. Examples of low methylation (#31 and #29) and unmethylated (#7) results are shown in Supplemental Figure 1B relative to standards. Our data conclusively demonstrate that PTPN6 methylation at CpG1 is absent in DLBCL tumors. In fact, this finding led us to re-examine the clinical and pathologic data from case #18 that we found methylated. The tumor was originally classified as DLBCL in the clinical record and our biobank; however, the final pathology diagnosis was a neuroendocrine carcinoma.
Novel PTPN6 Promoter CpG island sites (CpG2) are hypermethylated in DLBCL tumors
The finding that PTPN6 was lost or silenced in many of the clinical samples without finding any CpG1 P2 hypermethylation led us to extend our methylation studies to another CpG island (named CpG2). This CpG2 island is located at the proximal end of the PTPN6 P2 promoter and contains 8 CpG sites (Figure 2A). Hypermethylation of the CpG2 island has not been previously studied for the PTPN6 gene in any hematological malignancy. We designed primers specific for these CpG2 sites and ran the pyrosequencing on the same 37 DLBCL tumors as used above for CpG1 methylation. Using a cut off of >25% (intermediate to high methylation) 57% (21/37) of patient samples were methylated (Table 1). The Ly3, Ly10, and DHL2 DLBCL cell lines were also hypermethylated at CpG2 (Table 1). CpG2 pyrosequencing data was confirmed in 7 DLBCL samples by methylation specific PCR using MSP2/UMSP2 primers (Figure 2C). Sequencing of the methylated MSP products from methylated DNA control and patient #18 and #1 sample showed no change in C to T residues in the CpG1 sites confirming that PTPN6 was indeed hypermethylated in that sample (Figure 2D). Overall, these data demonstrate that PTPN6 hypermethylation occurs at this novel CpG2 site in the PTPN6 P2 promoter in many DLBCL tumors and may explain the loss or silencing of PTPN6 found in our patient samples.
Re-expression of PTPN6 with 5-Aza, a DNA methyltransferase inhibitor
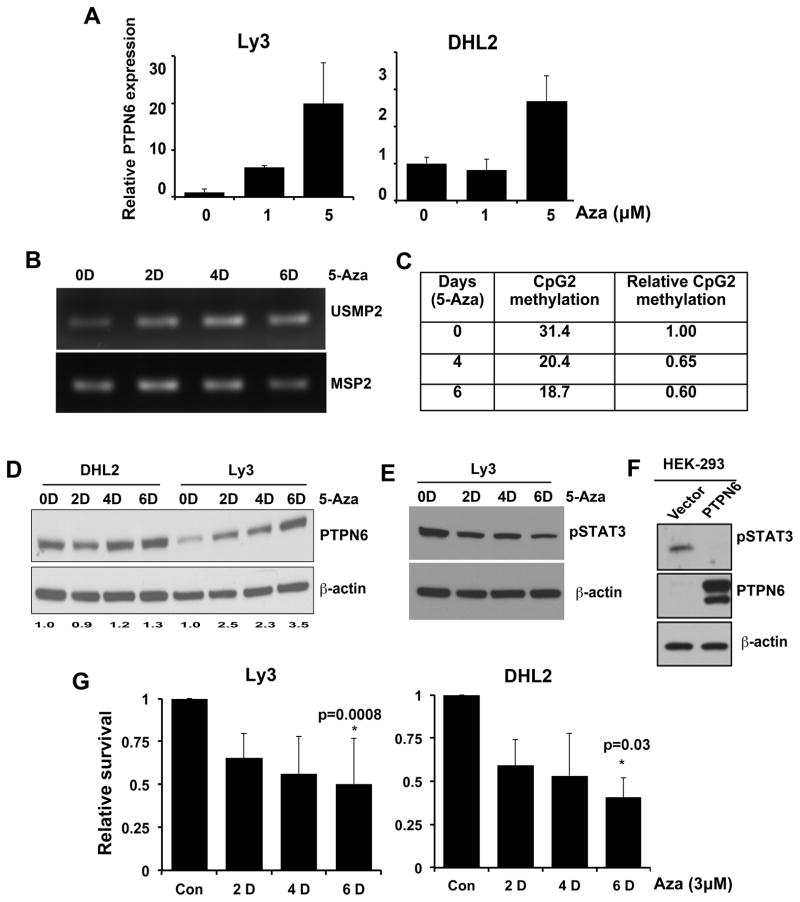
To further confirm the role of DNA methylation in PTPN6 regulation, we investigated PTPN6 re-expression after treatment of Ly3 (high methylation) and DHL2 (intermediate methylation) cells with the DNA methyltransferase inhibitor 5-Azacytidine (5-Aza). 5-Aza treatment increased PTPN6 mRNA expression compared to untreated control in both cell lines (Figure 3A). Ly3 was more amenable to PTPN6 re-expression compared to DHL2 cells most likely because of the higher basal hypermethylation of CpG2 in Ly3 cells (Table 1). In Ly3, treatment with 5-Aza led to a progressive demethylation of PTPN6 that started from day 2 onward, as shown by positive U-MSP with increasing amplification intensity (Figure 3B). 5-Aza-mediated demethylation of CpG2 in Ly3 was confirmed by pyrosequencing demonstrating inhibition of CpG2 methylation by 59% by day 4 (Figure 3C). Interestingly, this progressive CpG2 demethylation of PTPN6 was associated with a parallel re-expression of PTPN6 protein beginning from day 2 onwards specifically in Ly3 cells with modest effect in DHL2 cells at days 4 and 6 (Figure 3D). This resulted in a corresponding down-regulation of activated STAT3 in a similar time dependent manner (Figure 3E). To investigate the influence of ectopic PTPN6 WT and mutants expression on STAT3 activation, we transiently over-expressed vector alone and WT PTPN6 plasmids in the HEK-293 cells. Figure 3F clearly demonstrates overexpression of the PTPN6 in WT PTPN6 transfected cells as compared with vector alone. Interestingly, overexpression of WT PTPN6 was able to completely dephosphorylate STAT3 in these cells (Figure 3F). Treatment of Ly3 and DHL2 cells with 5-Aza (3μM) for 0–6 days significantly (p=0.0008 for Ly3 and p=0.03 for DHL2) reduced the cell survival at day 6 (Figure 3G). Overall, these data suggest that the DNA methyltransferase inhibitor 5-Aza was able to successfully demethylate CpG2 hypermethylation leading to re-expression of PTPN6 mRNA and protein resulting in induction of apoptosis by inhibition of STAT3 signaling.
Figure 3. Effect of 5-Aza on PTPN6 re-expression and DLBCL survival.
(A) Ly3 and DHL2 cells were treated with different concentrations of 5-Aza for 48 hours and PTPN6 expression relative to GAPDH was measured by qRT-PCR. (B) Effect of 5-Aza (3μM) on CpG2 demethylation detected by MSP2/USMP2 PCR in Ly3 cells. (C) Effect of 5Aza (3μM) on CpG2 demethylation by pyrosequencing in Ly3 cells. (D) Effect of 5-Aza at indicated concentrations on PTPN6 protein expression by western blotting. (E) Effect of 5-Aza (3μM) on STAT3 phosphorylation in Ly3 cells by western blotting. (F) WT PTPN6 plasmid along with vector alone was transiently transfected into the HEK-293 cells and STAT3 phosphorylation and PTPN6 overexpression determined by western blotting. (G) Effect of 5-Aza (3μM) on Ly3 and DHL2 cells survival by flow cytometry using Annexin/PI staining. Bars represent mean ± SD from 3 independent experiments. p-value was analyzed by the two-tailed unpaired student’s t-test.
Effect of combined DNA methyltransferase and HDAC inhibitor treatment on PTPN6 expression, CpG2 methylation, and STAT3 signaling
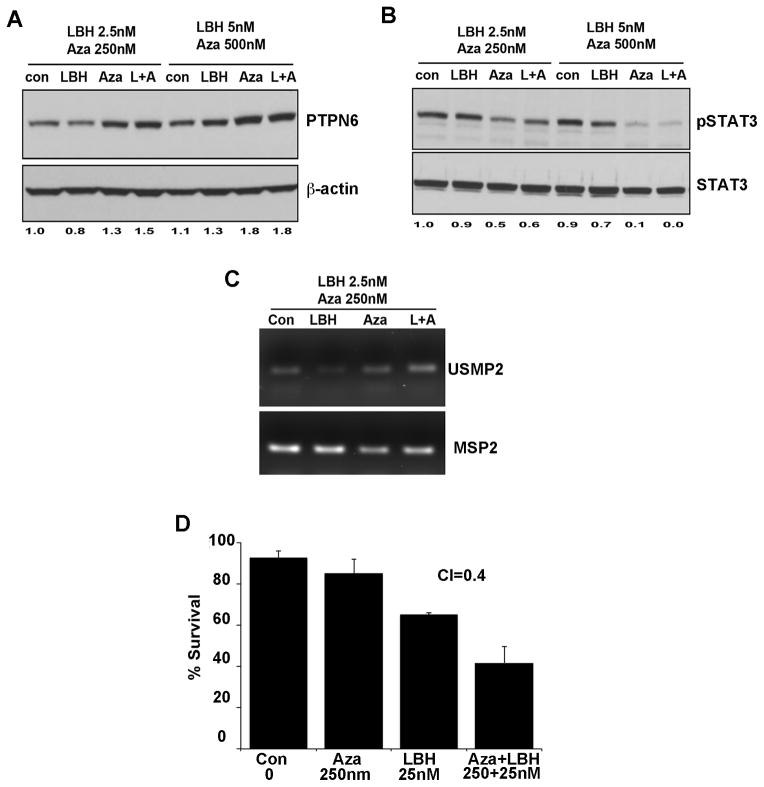
To determine whether the histone deacetylase inhibitor (HDACi) LBH589 can further enhance 5-Aza-induced PTNP6 expression and STAT3 de-phosphorylation, Ly3 cells were treated simultaneously with 2.5 nM LBH and 250 nM 5-Aza or 5 nM LBH and 500 nM 5-Aza for 72 hours (Figure 4A). Both LBH 5 nM and 5-Aza 250 and 500 nM as single agents increased PTPN6 expression; the combination of very low dose LBH (2.5 nM) with 5-Aza 250 nM modestly increased PTPN6 expression over single agent alone. With regard to STAT3 dephosphorylation, 5-Aza was the more active of the two agents (Figure 4B). We next determined the combinatorial effect of 5-Aza and LBH on PTPN6 demethylation at CpG2 island by USMP2/MSP2 PCR and found that the combination modestly increased the CpG2 demethylation over either single agent (Figure 4C). Combined treatment of DHL2 cells with 2.5, 5, 10 and 25 nM of LBH589 with 25, 50, 100 and 250nM of 5-Aza for 72 hours reduced the survival of DHL2 cells more compared to single agent alone. To determine which concentrations were synergistic, we calculated the combination index (CI) by Chou-Talalay equation (29). A strong drug synergy (CI=of 0.4) with the 25 nM LBH and 250 nM of 5-Aza combination was observed (Figure 4D).
Figure 4. Combinatorial effect of LBH589 and 5-Aza on PTPN6 re-expression/CpG2 demethylation and DLBCL cell survival.
(A–B) Ly3 cells were treated with various concentrations of LBH or 5-Aza as indicated for 72 hours and PTPN6 (A) and pSTAT3 (B) were quantified by western blotting. (C) CpG2 methylation analysis in Ly3 cells treated with 5-Aza and LBH at indicated concentrations for 72 hrs. (D) DHL2 cells were treated with combinations of LBH (range, 2.5 to 25nM) and Aza (range, 25 to 250nM) for 72 hours and survival assessed by Annexin/PI flow cytometry. Results are depicted for the most synergistic (CI=0.4) combination (LBH 25 nM+Aza 250 nM). Bars represent mean ± SD from 3 independent experiments.
PTPN6 promoter 2 is associated with inactivating histone marks
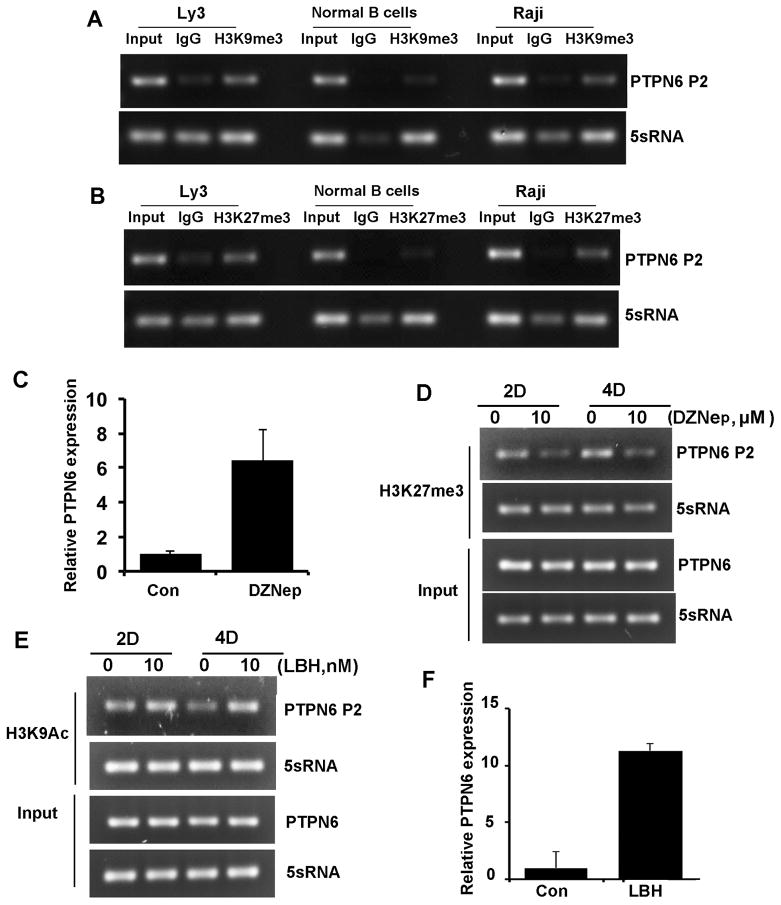
To further investigate epigenetic control of PTPN6 in DLBCL, we proceeded to study whether histone modifications (methylation and acetylation) of the PTPN6 P2 were involved in the regulation of PTPN6 expression. For the studies of histone methylation, ChIP assays were performed in the Ly3 cell line, normal CD19+ cells, and in Raji cells by using antibodies to repressing marks on histones. In Ly3 cells we found the PTPN6 P2 core region to be enriched with trimethylation of lysine 9 on histone H3 (H3K9me3) and lysine 27 on histone H3 (H3K27me3) as compared to CD19+ cells (Figure 5A, B). To confirm that these suppressive histone marks regulate PTPN6 expression, we treated Ly3 cells with the histone methyltransferase inhibitor 3-deazaneplanocin A (DZNep, Sigma-Aldrich). DZNep is an EZH2 histone methyltransferase inhibitor that selectively inhibits the H3K27 trimethylation mark.(30) Treatment with DZNep increased the PTPN6 expression in Ly3 cells in a time dependent manner (Figure 5C). Results of the ChIP assay showed reduced expression of the H3K27me3 histone mark in the DZNep treated cells within the PTPN6 P2. (Figure 5D). To evaluate the role of histone acetylation, we performed the ChIP assay with LBH treatment and demonstrated increased acetylation at lysine 9 on histone H3 (H3K9Ac) (Fig 5E) in PTPN6 P2 without any effect on the H3K27me3 mark (data not shown). Moreover, LBH treatment resulted in increased expression of PTPN6 mRNA (Figure 5F).
Figure 5. PTPN6 promoter 2 histone methylation analysis.
(A–B) ChIP assay demonstrating enrichment of the H3K9me3 and H3K27me3 on PTPN6 promoter 2 in Ly3 DLBCL cells, CD19+ normal B cells and Raji cells. A representative experiment out of three independent experiments is shown. (C) Effect of DZNEP (1μM) on PTPN6 mRNA expression (relative to GAPDH) in Ly3 cells. (D) ChIP assay in Ly3 cells demonstrates the effect of DZNep on HeK27me3 mark associated with PTPN6 promoter 2. (E) Effect of LBH589 on H3K9Ac mark associated with PTPN6 promoter 2 in Ly3 cells. (F) Effect of LBH589 (10 nM) on PTPN6 mRNA (relative to GAPDH) expression by qRT-PCR in Ly3 cells.
Discussion
We have shown in this study that expression of an important protein tyrosine phosphatase, PTPN6, is suppressed or lost in the 53% of DLBCL tumors as compared to normal B cells. The PTPN6 gene has been shown to be silenced in T cell lymphomas of various types(31–33) but PTPN6 protein expression in the most common lymphoma, DLBCL, has not been reported. Others have shown PTPN6 to be silenced in other unusual types of lymphoma such as adult T-cell leukemia/lymphoma and NK/T-cell lymphomas as well as in acute and chronic leukemias.(17, 25) Several studies in hematological malignancies have shown that promoter hypermethylation of the PTPN6 gene occurs at the known CpG1 site on promoter 2.(18, 25) In DLBCL however we demonstrated that PTPN6 is not methylated at CpG1 sites. These results appear to be conclusive as we performed the experiments on the same samples by 3 different methods (MSP1/USMP1 PCR, HRM, and pyrosequencing) in three different independent laboratories at our institution and found the same results. These results in DLBCL are in contrast with previous reports describing PTPN6 P2 CpG1 methylation to be present in DLBCL.(34) The possible explanation for these discordant results could be the differences in the methylation techniques used, differences in DNA sample source used (frozen vs FFPE), or even geographical differences between DLBCL patient samples (United States vs Africa). In order to gain further mechanistic insight to PTPN6 suppression, we further analyzed the PTPN6 promoter 2 and observed a novel CpG2 island at the proximal end. Pyrosequencing for CpG2 sites on the same 37 DLBCL samples used for CpG1 studies found 57% of cases with CpG2 hypermethylation. To our knowledge this is the first description of methylation at this CpG island on PTPN6 P2. Histone modification also plays an important role in gene silencing by methylating inactivating histone marks. We performed the ChIP assay in DLBCL cell lines and demonstrated that PTPN6 P2 was enriched for the silencing histone marks H3K9me3 and H3K27me3 in DLBCL cell lines but not in normal B cells. These findings support the notion that control of PTPN6 P2 in DLBCL tumors is associated with both DNA promoter methylation (CpG2) and histone methylation/acetylation.
In conclusion, these data provide a comprehensive characterization of the epigenetic mechanisms leading to PTPN6 deregulation in the DLBCL tumors. These findings support the notion that control of PTPN6 P2 in DLBCL tumors is associated with DNA promoter methylation, histone methylation, and histone acetylation. These findings explain why treatment with 5-Aza, DZNep and LBH589 can reactivate PTPN6 expression and function. PTPN6 expression and patterns of methylation and acetylation may prove useful as diagnostic and prognostic markers and provide a rationale to test epigenetic therapy in DLBCL patients.
Supplementary Material
Acknowledgments
This work was supported through Iowa/Mayo Lymphoma Specialized Program of Research Excellence (P50 CA097274) to MG; R01CA127433 to TEW and the Predolin Foundation.
Footnotes
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Authorship: Contributions: TEW interpreted data, wrote the manuscript and provided clinical samples. GH performed the PTPN6 methylation (MSP-PCR and pyrosequencing for CpG1 and CpG2) and ChIP assay. SMO performed high-resolution methylation array. LEW performed PTPN6 IHC staining. MJS performed apoptosis. JJH performed PTPN6 western blotting and PTPN6 Q-PCR. AD scored and reviewed PTPN6 IHC slides. RBD edited the paper. MG designed and supervised the research, interpreted and analyzed the data, finalized the figures and wrote the manuscript.
References
- 1.Habermann TM, Weller EA, Morrison VA, Gascoyne RD, Cassileth PA, Cohn JB, et al. Rituximab-CHOP versus CHOP alone or with maintenance rituximab in older patients with diffuse large B-cell lymphoma. J Clin Oncol. 2006 Jul 1;24(19):3121–3127. doi: 10.1200/JCO.2005.05.1003. [DOI] [PubMed] [Google Scholar]
- 2.Coiffier B, Lepage E, Briere J, Herbrecht R, Tilly H, Bouabdallah R, et al. CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(4):235–242. doi: 10.1056/NEJMoa011795. [DOI] [PubMed] [Google Scholar]
- 3.Witzig TE, Reeder CB, LaPlant BR, Gupta M, Johnston PB, Micallef IN, et al. A phase II trial of the oral mTOR inhibitor everolimus in relapsed aggressive lymphoma. Leukemia. 2011 Feb;25(2):341–347. doi: 10.1038/leu.2010.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam LT, Wright G, Davis RE, Lenz G, Farinha P, Dang L, et al. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-{kappa}B pathways in subtypes of diffuse large B-cell lymphoma. Blood. 2008 Apr 1;111(7):3701–3713. doi: 10.1182/blood-2007-09-111948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding BB, Yu JJ, Yu RY, Mendez LM, Shaknovich R, Zhang Y, et al. Constitutively activated STAT3 promotes cell proliferation and survival in the activated B-cell subtype of diffuse large B-cell lymphomas. Blood. 2008 Feb 1;111(3):1515–1523. doi: 10.1182/blood-2007-04-087734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta M, Maurer MJ, Wellik LE, Law ME, Han JJ, Ozsan N, et al. Expression of Myc, but not pSTAT3, is an adverse prognostic factor for diffuse large B-cell lymphoma treated with epratuzumab/R-CHOP. Blood. 2012 Nov 22;120(22):4400–4406. doi: 10.1182/blood-2012-05-428466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta M, Han JJ, Stenson M, Wellik L, Witzig TE. Regulation of STAT3 by histone deacetylase-3 in diffuse large B-cell lymphoma: implications for therapy. Leukemia. 2012 Jun;26(6):1356–1364. doi: 10.1038/leu.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Shea JJ, Gadina M, Schreiber RD. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell. 2002 Apr;109(Suppl):S121–131. doi: 10.1016/s0092-8674(02)00701-8. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida K, Kharbanda S, Kufe D. Functional interaction between SHPTP1 and the Lyn tyrosine kinase in the apoptotic response to DNA damage. J Biol Chem. 1999 Dec 3;274(49):34663–34668. doi: 10.1074/jbc.274.49.34663. [DOI] [PubMed] [Google Scholar]
- 10.Jiao H, Berrada K, Yang W, Tabrizi M, Platanias LC, Yi T. Direct association with and dephosphorylation of Jak2 kinase by the SH2-domain-containing protein tyrosine phosphatase SHP-1. Mol Cell Biol. 1996 Dec;16(12):6985–6992. doi: 10.1128/mcb.16.12.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han Y, Amin HM, Frantz C, Franko B, Lee J, Lin Q, et al. Restoration of shp1 expression by 5-AZA-2′-deoxycytidine is associated with downregulation of JAK3/STAT3 signaling in ALK-positive anaplastic large cell lymphoma. Leukemia. 2006 Sep;20(9):1602–1609. doi: 10.1038/sj.leu.2404323. [DOI] [PubMed] [Google Scholar]
- 12.Banville D, Stocco R, Shen SH. Human protein tyrosine phosphatase 1C (PTPN6) gene structure: alternate promoter usage and exon skipping generate multiple transcripts. Genomics. 1995 May 1;27(1):165–173. doi: 10.1006/geno.1995.1020. [DOI] [PubMed] [Google Scholar]
- 13.Paling NR, Welham MJ. Tyrosine phosphatase SHP-1 acts at different stages of development to regulate hematopoiesis. Blood. 2005 Jun 1;105(11):4290–4297. doi: 10.1182/blood-2004-08-3271. [DOI] [PubMed] [Google Scholar]
- 14.Wu C, Sun M, Liu L, Zhou GW. The function of the protein tyrosine phosphatase SHP-1 in cancer. Gene. 2003 Mar 13;306:1–12. doi: 10.1016/s0378-1119(03)00400-1. [DOI] [PubMed] [Google Scholar]
- 15.Tsui FW, Martin A, Wang J, Tsui HW. Investigations into the regulation and function of the SH2 domain-containing protein-tyrosine phosphatase, SHP-1. Immunologic research. 2006;35(1–2):127–136. doi: 10.1385/IR:35:1:127. [DOI] [PubMed] [Google Scholar]
- 16.Oka T, Yoshino T, Hayashi K, Ohara N, Nakanishi T, Yamaai Y, et al. Reduction of hematopoietic cell-specific tyrosine phosphatase SHP-1 gene expression in natural killer cell lymphoma and various types of lymphomas/leukemias : combination analysis with cDNA expression array and tissue microarray. Am J Pathol. 2001 Oct;159(4):1495–1505. doi: 10.1016/S0002-9440(10)62535-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oka T, Ouchida M, Koyama M, Ogama Y, Takada S, Nakatani Y, et al. Gene silencing of the tyrosine phosphatase SHP1 gene by aberrant methylation in leukemias/lymphomas. Cancer Res. 2002 Nov 15;62(22):6390–6394. [PubMed] [Google Scholar]
- 18.Chim CS, Fung TK, Cheung WC, Liang R, Kwong YL. SOCS1 and SHP1 hypermethylation in multiple myeloma: implications for epigenetic activation of the Jak/STAT pathway. Blood. 2004 Jun 15;103(12):4630–4635. doi: 10.1182/blood-2003-06-2007. [DOI] [PubMed] [Google Scholar]
- 19.Chim CS, Wong AS, Kwong YL. Epigenetic dysregulation of the Jak/STAT pathway by frequent aberrant methylation of SHP1 but not SOCS1 in acute leukaemias. Ann Hematol. 2004 Aug;83(8):527–532. doi: 10.1007/s00277-004-0843-1. [DOI] [PubMed] [Google Scholar]
- 20.Micallef IN, Maurer MJ, Wiseman GA, Nikcevich DA, Kurtin PJ, Cannon MW, et al. Epratuzumab with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone chemotherapy in patients with previously untreated diffuse large B-cell lymphoma. Blood. 2011 Oct 13;118(15):4053–4061. doi: 10.1182/blood-2011-02-336990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002 Nov;18(11):1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 22.Ronaghi M. Pyrosequencing sheds light on DNA sequencing. Genome Res. 2001 Jan;11( 1):3–11. doi: 10.1101/gr.11.1.3. [DOI] [PubMed] [Google Scholar]
- 23.Lynch M, Chen L, Ravitz MJ, Mehtani S, Korenblat K, Pazin MJ, et al. hnRNP K binds a core polypyrimidine element in the eukaryotic translation initiation factor 4E (eIF4E) promoter, and its regulation of eIF4E contributes to neoplastic transformation. Mol Cell Biol. 2005 Aug;25(15):6436–6453. doi: 10.1128/MCB.25.15.6436-6453.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta M, Dillon SR, Ziesmer SC, Feldman AL, Witzig TE, Ansell SM, et al. A proliferation-inducing ligand mediates follicular lymphoma B-cell proliferation and cyclin D1 expression through phosphatidylinositol 3-kinase-regulated mammalian target of rapamycin activation. Blood. 2009 May 21;113(21):5206–5216. doi: 10.1182/blood-2008-09-179762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koyama M, Oka T, Ouchida M, Nakatani Y, Nishiuchi R, Yoshino T, et al. Activated proliferation of B-cell lymphomas/leukemias with the SHP1 gene silencing by aberrant CpG methylation. Lab Invest. 2003 Dec;83(12):1849–1858. doi: 10.1097/01.lab.0000106503.65258.2b. [DOI] [PubMed] [Google Scholar]
- 26.Dupont JM, Tost J, Jammes H, Gut IG. De novo quantitative bisulfite sequencing using the pyrosequencing technology. Anal Biochem. 2004 Oct 1;333(1):119–127. doi: 10.1016/j.ab.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003 Jul;35(1):146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 28.Candiloro IL, Mikeska T, Dobrovic A. Assessing combined methylation-sensitive high resolution melting and pyrosequencing for the analysis of heterogeneous DNA methylation. Epigenetics. 2011 Apr;6(4):500–507. doi: 10.4161/epi.6.4.14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Advances in enzyme regulation. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 30.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007 May 1;21(9):1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Q, Raghunath PN, Vonderheid E, Odum N, Wasik MA. Lack of phosphotyrosine phosphatase SHP-1 expression in malignant T-cell lymphoma cells results from methylation of the SHP-1 promoter. Am J Pathol. 2000 Oct;157(4):1137–1146. doi: 10.1016/S0002-9440(10)64629-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Migone TS, Cacalano NA, Taylor N, Yi T, Waldmann TA, Johnston JA. Recruitment of SH2-containing protein tyrosine phosphatase SHP-1 to the interleukin 2 receptor; loss of SHP-1 expression in human T-lymphotropic virus type I-transformed T cells. Proc Natl Acad Sci U S A. 1998 Mar 31;95(7):3845–3850. doi: 10.1073/pnas.95.7.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khoury JD, Rassidakis GZ, Medeiros LJ, Amin HM, Lai R. Methylation of SHP1 gene and loss of SHP1 protein expression are frequent in systemic anaplastic large cell lymphoma. Blood. 2004 Sep 1;104(5):1580–1581. doi: 10.1182/blood-2004-03-1151. [DOI] [PubMed] [Google Scholar]
- 34.Amara K, Trimeche M, Ziadi S, Laatiri A, Hachana M, Korbi S. Prognostic significance of aberrant promoter hypermethylation of CpG islands in patients with diffuse large B-cell lymphomas. Ann Oncol. 2008 Oct;19(10):1774–1786. doi: 10.1093/annonc/mdn374. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.