Background: Seven antibodies that detect cytosolic and nuclear O-GlcNAc were examined for recognition of O-GlcNAc on cell surface glycoproteins.
Results: Three antibodies recognized cell surface O-GlcNAc and also bound terminal β-GlcNAc on N-glycans.
Conclusion: With careful controls, cell surface O-GlcNAc can be detected using antibodies CTD110.6, 18B10.C7, and 9D1.E4.
Significance: Antibodies to monitor cell surface O-GlcNAcylated proteins were characterized.
Keywords: Antibodies, Flow Cytometry, Mutant, O-GlcNAc, O-GlcNAcylation, CHO, GlcNAc-terminating N-Glycans, Lec1, Lec8
Abstract
The transfer of N-acetylglucosamine (GlcNAc) to Ser or Thr in cytoplasmic and nuclear proteins is a well known post-translational modification that is catalyzed by the O-GlcNAc transferase OGT. A more recently identified O-GlcNAc transferase, EOGT, functions in the secretory pathway and transfers O-GlcNAc to proteins with epidermal growth factor-like (EGF) repeats. A number of antibodies that detect O-GlcNAc in cytosolic and nuclear extracts have been described previously. Here we compare seven of these antibodies (CTD110.6, 10D8, RL2, HGAC85, 18B10.C7(#3), 9D1.E4(#10), and 1F5.D6 (#14) for detection of the O-GlcNAc modification on extracellular domains of membrane or secreted glycoproteins that may also carry various N- and O-glycans. We found that CTD110.6 binds not only to O-GlcNAc on proteins but also to terminal β-GlcNAc on the complex N-glycans of Lec8 Chinese hamster ovary (CHO) cells that lack UDP-Gal transporter activity and express GlcNAc-terminating, complex N-glycans. We show that CTD110.6, #3, and #10 antibodies can be used to detect cell surface glycoproteins bearing O-GlcNAc. Cell surface glycoproteins recognized by CTD110.6 antibody included NOTCH1 that possesses many EGF repeats with a consensus site for EOGT. Knockdown of CHO Eogt reduced binding of CTD110.6 to Lec1 CHO cells, and expression of a human EOGT cDNA increased the O-GlcNAc signal on Lec1 cells and the extracellular domain of NOTCH1. Thus, with careful controls, antibodies CTD110.6 (IgM), #3 (IgG), and #10 (IgG) can be used to detect membrane and secreted proteins modified by O-GlcNAc on EGF repeats.
Introduction
Secreted and cell surface glycoproteins carry a wide spectrum of N- and O-glycans that are synthesized during transit from the endoplasmic reticulum (ER),2 through Golgi compartments, to the cell surface (1). A recent addition to the complement of glycans is the transfer of O-GlcNAc to proteins with epidermal growth factor-like (EGF) repeats by the EGF-repeat specific O-GlcNAc-transferase termed EOGT (2–7). An EGF repeat of Drosophila Notch extracellular domain was first identified as a substrate in Drosophila S2 cells (2), and NOTCH1 is a substrate in human HEK-293T kidney cells (4). The consensus for recognition of an EGF repeat by EOGT is predicted to be C5XXGXS/TGXXC6 based on a large group of actual and potential substrates (8). EOGT is localized to the ER and thus is physically segregated from OGT which transfers O-GlcNAc to Ser or Thr residues in a multitude of proteins in the cytoplasm and nucleus (9).
O-GlcNAcylated proteins are often identified using the monoclonal antibody (mAb) CTD110.6 (10). Six other anti-O-GlcNAc mouse mAbs have been described: HGAC85 (11), RL2 (12), 10D8 (13), and three IgG mAbs, 18B10.C7(#3), 9D1.E4(#10), and 1F5.D6(#14) (14). The peptides and more complex O-GlcNAcylated antigens used to obtain these mAbs are summarized in Table 1. The specificity of the anti-O-GlcNAc mAbs for terminal O-linked GlcNAc versus other forms of exposed GlcNAc, or related sugars such as GalNAc, has not been extensively explored. However, it has been reported that mAb CTD110.6 binds not only to O-GlcNAc but also to the unsubstituted, N-glycan chitobiose core (GlcNAcβ1,4GlcNAcβ1Asn) induced by glucose deprivation (15) and to unsubstituted GlcNAcβ1,4Man on α-dystroglycan (16).
TABLE 1.
Anti-O-GlcNAc antibodies used in this study
All antibodies are mouse monoclonal.
| Name | Antigen | Isotype | Ref. | Source |
|---|---|---|---|---|
| CTD110.6 | YSPTS(O-GlcNAc)PSK | IgM | (10) | Sigma-Aldrich |
| 10D8 | N-Acetylglucosamine | IgM | (13) | Santa Cruz Biotechnology |
| RL2 | Pore complex-lamina fraction purified from rat liver nuclear envelopes | IgG1 | (12) | Abcam |
| HGAC85 | Heat-killed, pepsin-treated group A streptococci | IgG3 | (11) | Thermo Fisher Scientific |
| 18B10.C7 (#3) | GSTPVS (β-O-GlcNAc)SANM | IgG1 | (14) | Geert-Jan Boons |
| Epitope: PVS (β-O-GlcNAc)SA | ||||
| 9D1.E4 (#10) | GSTPVS (β-O-GlcNAc)SANM | IgG1 | (14) | Geert-Jan Boons |
| Epitope: full-length antigen | ||||
| 1F5.D6 (#14) | GSTPVS (β-O-GlcNAc)SANM | IgG2a | (14) | Geert-Jan Boons |
| Epitope: VS (β-O-GlcNAc)S |
Here we compare the specificity of 7 mAbs to detect O-GlcNAc on Ser/Thr in peptides or proteins, with the aim of identifying mAbs that recognize O-GlcNAc on EGF repeats of glycoproteins. At comparatively low concentrations, all mAbs detected proteins with O-GlcNAc in cell lysates, and CTD110.6 detected terminal β-GlcNAc on complex N-glycans. At higher mAb concentrations the mAbs #3, #10, and #14 also detected terminal β-GlcNAc on N-glycans. By comparing binding to Lec1, Lec2, and Lec8 mutant CHO cells (Table 2), we found that IgM mAb CTD110.6 and IgG mAbs #3 and #10 were the only antibodies to specifically detect O-GlcNAc on cell surface glycoproteins. CTD110.6 immunoprecipitated NOTCH1 from cell lysate and detected biotinylated NOTCH1 following affinity purification. CTD110.6 and #10 detected the same major cohort of biotinylated cell surface glycoproteins in Lec1 CHO cells.
TABLE 2.
CHO glycosylation mutants used in this study
The N-glycans expressed on glycoproteins in each mutant are depicted in Fig. 3A.
EXPERIMENTAL PROCEDURES
Antibodies
The anti-O-GlcNAc antibodies used in these experiments are described in Table 1. Anti-O-GlcNAc CTD110.6 mAb (O7764), anti-FLAG (M2) Ab (F1804), anti-mouse IgM-agarose (A4540), and anti-alkaline phosphatase Ab (PLAP; 8B6) conjugated to agarose (A2080) were from Sigma-Aldrich. Anti-O-GlcNAc 10D8 mAb (sc-81483), anti-PLAP (L-19) Ab (sc-15065), anti-PDGFR-α Ab (C20; sc-338), and anti-goat IgG conjugated to horseradish peroxidase (HRP) (sc-2020) were from Santa Cruz Biotechnology. Anti-O-GlcNAc CTD110.6 (ab246879), RL2 mAb (ab2739), anti-β-actin Ab (ab6276), anti-human EOGT Ab (ab69389), mouse IgG1 (ab91353), and mouse IgG2 (ab91361) were from Abcam. Anti-O-GlcNAc HGAC85 mAb (MA1-076), NeutrAvidinTM-HRP (31030), anti-mouse IgG conjugated to HRP (31444), protein G beads, and NeutraAvidin-agarose resin (SA-agarose) were from Thermo Fisher Scientific Inc. Affinity-purified anti-O-GlcNAc mAbs #3 (0.86 mg/ml), #10 (0.59 mg/ml), and #14 (0.97 mg/ml) were kindly provided by Dr. Geert-Jan Boons, University of Georgia, Athens, GA. Mouse anti-protein disulfide isomerase mAb (1D3, SPA-891) was from Stressgen, Kampenhout, Belgium. Anti-human NOTCH3 Ab (5E1) was kindly provided by Dr. Anne Joutel (17). Anti-mouse NOTCH1 Ab (AF5267) and anti-sheep IgG conjugated to HRP (HAF016) were from R&D Systems. Anti-HA (16B12) (MMS-101R) and anti-Myc (9E10) Abs (MMS-150P) were from Covance, Princeton, NJ. Anti-mouse IgM conjugated to HRP (115-035-075) and fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG (115-096-146) were from Jackson ImmunoResearch Laboratories. Cy5-conjugated anti-mouse IgM (072-02-18-03) was from KPL, Gaithersburg, MD.
Cell Lines and Cell Culture
Pro−5 parent CHO and CHO glycosylation mutants derived from Pro−5, Lec1.3C (18), Lec2.6A (19), and Lec8.3D (20) were cultured in suspension with α-MEM containing 10% fetal calf serum (FCS) at 37 °C. Lec2 and Lec8 cells stably expressing mouse Manic Fringe (Mfng) tagged with alkaline phosphatase (AP) were generated and characterized previously (21, 22). Lec2/vector.1.1, Lec2/Lfng-AP.A7, Lec2/Mfng-AP.D1, Lec8/vector.3.2, Lec8/Lfng-AP.A1, and Lec8/Mfng-AP.H3 cloned lines were cultured with α-MEM containing 10% FCS and 400 μg/ml active G418 (Gemini Bio Products, West Sacramento, CA) at 37 °C.
Plasmids
The plasmid, cFLAG-pcDNA3, was kindly provided by Stephen Smale (University of California, CA) (23). Mouse Mfng cDNA was fused with the HA epitope YPYDVPDYALKV at the C terminus and inserted into cFLAG-pcDNA3 between HindIII and KpnI sites to produce pcDNA3/Mfng-HA-FLAG (sequence in italics is the actual HA epitope and the last three amino acids are from the construct). Mouse pMirb/Mfng-AP was previously described (21). pCS2+/Notch1-Myc was a kind gift of Raphael Kopan (University of Cincinnati, Cincinnati, OH). Human EOGT was subcloned into pCR3.1 using NotI and XhoI digestion of pCASP/huEOGT (7), a kind gift of Reto Muller (Albert Einstein College of Medicine, New York, NY).
Transient Expression of Mfng-AP and Mfng-HA-FLAG
Cells (5 × 105) were plated on a 6-well plate. Next day, the cells were transfected with vector, pMirb/Mfng-AP or pcDNA3/Mfng-HA-FLAG using polyethylenimine (PEI), a kind gift of Robert Haltiwanger (Stony Brook University, Stony Brook, NY). Plasmid (3 μg) and 10 μl of PEI were diluted separately with 50 μl of 150 mm NaCl, then mixed together and incubated for 10 min at room temperature. DNA-PEI complexes were added to cells in a 6-well plate in fresh medium.
SDS-PAGE and Western Blot Analysis
Cells were washed twice with phosphate-buffered saline (PBS) and lysed using 0.5 ml of RIPA buffer (SDS−); Millipore) with protease inhibitor mixture Complete, mini-EDTA-free (Roche Applied Science). After incubation on ice for 20 min, lysate was centrifuged at 12,000 × g at 4 °C for 15 min. The protein concentration of the supernatant was determined using the DC Protein Assay (Bio-Rad). Proteins (∼20 μg) were separated by SDS-PAGE using a 7.5 or 10% gel and transferred to a polyvinylidine difluoride (PVDF) membrane at 50 mA overnight. To detect O-GlcNAc, the blot was incubated in 5% BSA (Fraction V, Sigma-Aldrich) in Tris-buffered saline, 0.05% Tween 20 (TBS-T) for 1 h at room temperature and then incubated in anti-O-GlcNAc mAbs as follows: CTD110.6 or 10D8 at room temperature for 1 h followed by incubation with anti-mouse IgM conjugated to HRP at room temperature for 1 h; or anti-O-GlcNAc RL2 or HGAC85 mAbs at room temperature for 1 h followed by anti-mouse IgG conjugated to HRP; or anti-O-GlcNAc mAb #3, #10, or #14 in the cold room overnight following by anti-mouse IgG conjugated to HRP. To detect other proteins, blots were blocked with 5% milk in TBS-T for 1 h at room temperature and then incubated with anti-PLAP (L-19) Ab followed by incubation with anti-goat IgG conjugated to HRP or anti-HA Ab followed by incubation with anti-mouse IgG conjugated to HRP. Bands were visualized using enhanced chemiluminescence (Pierce) Western blotting substrate 32209, 34807 (West Pico), or 34094 (West Femto) from Thermo Fisher Scientific.
Affinity Purification of MFNG-AP and MFNG-HA-FLAG
Cells expressing pMirb/Mfng-AP cDNA were cultured in suspension in 10 ml of serum-free medium, CHO-SEMII (12052-098; Invitrogen), for 3 days. The medium was collected after removal of cells by centrifugation at 1200 rpm for 3 min at room temperature. The supernatant was filtered through a 0.2-μm filter (Millipore) and diluted to 13 ml by adding 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, and protease inhibitor mixture Complete mini-EDTA-free. Anti-human PLAP(8B6)-conjugated agarose (10-μl bed volume) was added, and the mixture was rocked at 4 °C overnight. The MFNG-AP beads were washed five times with 1 ml of wash buffer (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100), subjected to glycosidase treatments, and/or heated at 95 °C for 5 min in SDS-PAGE buffer followed by Western blot analysis.
Secreted MFNG-HA-FLAG was affinity-purified from conditioned medium following transient transfection for 2 days. To obtain intracellular MFNG-HA-FLAG, transfected cells were washed twice with 1.5 ml of PBS, pH 7.4 (with 1 mm CaCl2, MgCl2, and MnCl2) and lysed using 0.3 ml of RIPA buffer (SDS−) with protease inhibitor mixture Complete mini-EDTA-free. After incubation on ice for 20 min, lysate was centrifuged at 12,000 × g at 4 °C for 15 min. Lysate and conditioned medium were agitated with 2 μg of anti-FLAG (M2) antibody at 4 °C for 6 h, 10 μl (bed volume) of protein G beads (Thermo Fisher Scientific) were added, and the incubation was continued overnight. The beads were washed with 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 1% Triton X-100. SDS-polyacrylamide gel loading buffer was added, and the samples were heated at 95 °C for 5 min before SDS-PAGE and Western blot analysis.
Glycosidase Treatments
To remove N-glycans, MFNG-AP captured on beads was incubated in 10 μl of glycoprotein denaturing buffer (New England Biolabs) or 50 mm sodium phosphate buffer containing 10% SDS, pH 7.5, at 95 °C for 10 min. N-Glycans were released by adding reaction buffer to 30 μl containing 500 units of peptide N-glycosidase F (PNGase F; New England Biolabs) and incubating at 37 °C for 2 h. To remove terminal GlcNAc, MFNG-AP beads were suspended in 50 mm sodium citrate, pH 4.5, and incubated with 10 units of recombinant β-N-acetylhexosaminidase (New England Biolabs) at 37 °C for 2 h. After glycosidase treatments, beads were added directly to 6 μl of 6× SDS-PAGE loading buffer and heated at 95 °C for 5 min.
Flow Cytometry
Cells (4 × 105) were washed with 0.5 ml of PBS (metal ion-free), pH 7.4, and fixed with 100 μl of 4% paraformaldehyde for 15 min with rocking. The cells were washed twice with 0.5 ml of PBS, then twice with 1 ml of PBS and then 1 ml of binding buffer consisting of Hanks' buffered salt solution, pH 7.4, 1 mm CaCl2, 1% (v/v) BSA (Fraction V), and 0.05% NaN3. The cells were incubated with 50 μl of binding buffer containing 2 μg of mouse IgM or anti-O-GlcNAc mAb CTD110.6 (2 μg) or 10D8 (0.5 μg) on ice for 20 min. Cells were washed twice with 0.5 ml of binding buffer and incubated with 50 μl of binding buffer containing 0.5 μg Cy5-conjugated anti-mouse IgM antibody. The cells were also incubated with 50 μl of binding buffer containing 2 μg of mouse IgG1, mouse IgG2, anti-O-GlcNAc mAb RL2, #3, #10, or #14 on ice for 20 min. After washing, cells were incubated with 0.75 μg of FITC-conjugated anti-mouse IgG antibody on ice for 20 min. Cells were washed twice with 0.5 ml of binding buffer and then analyzed using a FACScan flow cytometer (BD Biosciences).
Swainsonine Treatment and Lectin Binding
Cells were cultured in suspension in 10 ml of α-MEM containing 10% FCS and 5 μg/ml swainsonine (Sigma-Aldrich) for 4 days. For flow cytometry, cells (2 × 105) were washed twice with 0.5 ml of PBS (with cations), pH 7.4, and once with 1 ml of binding buffer. Washed cells were incubated with 50 μl of binding buffer containing 1 μg of fluorescein-labeled L-PHA (Phaseolus vulgaris leukoagglutinin; Vector, Burlingame, CA) or 2 μg of CTD110.6 mAb, followed by incubation with 50 μl of binding buffer containing 0.5 μg of Cy5-conjugated anti-mouse IgM. After washing, flow cytometry was performed using the FACScan flow cytometer.
Treatment with PUGNAc
Cells cultured in 10 ml of α-MEM with 10% FCS and 5 mm GlcNAc were treated with 100 μm O-(2-acetamido-2-deoxy-d-glucopyranosylidene) amino-N-phenylcarbamate (PUGNAc; Sigma-Aldrich) in 0.1% dimethyl sulfoxide or the same amount of dimethyl sulfoxide solution. After culturing at 37 °C for 18 h, cells were biotinylated as described below.
RNAi Knockdown of CHO Eogt
Lec1 cells were transfected with the vector pSUPER (Oligoengine, Seattle, WA), or Eogt siRNA PS1233 or PS1239 using Lipofectamine 2000 (Invitrogen). The siRNAs in pSUPER were a kind gift from Reto Muller (Albert Einstein College Medicine, New York) and targeted the coding region of CHO Eogt: PS1233 5′-GATCCGCAAGCTGACTTTGGATATTTCAAGAGAATATCCAAAGTCAGCTTGCTTTTTTGGAAA and PS1239 5′-GATCCATGTGACCTCATTGTTGAATTCAAGAGATTCAACAATGAGGTCACATTTTTTTGGAAA. The next day, the medium was replaced with α-MEM containing 10% FCS and 5 μg/ml puromycin for selection. After 16 days, flow cytometry was performed on the surviving cell population after fixation with 4% paraformaldehyde and incubation with mAb CTD110.6 as described above.
Expression of Human EOGT
Lec1 cells were co-transfected with vector or pCR3.1/human EOGT and pCS2+/Notch1-Myc or pCS2+ vector. The next day, cells were washed and biotinylated. Cell lysates were prepared using RIPA buffer (SDS−), and proteins were collected with SA-agarose beads, or anti-Myc antibody and protein G beads, and analyzed by SDS-PAGE and Western blotting. To analyze the effects of overexpression of EOGT on cell surface expression of O-GlcNAc, Lec1 cells were transfected with human pCR3.1/EOGT using Lipofectamine and, the next day were placed in medium containing 600 μg of G418 for 13 days before analysis by flow cytometry using Ab CDT110.6 as above. Human EOGT was detected by Western blot analysis using anti-human EOGT antibody (1:500).
Affinity Purification of Cell Surface Proteins
For biotinylation, cells (1 × 106) were plated on a 10-cm dish. The next day, the monolayer was washed twice with 5 ml of PBS (metal ion-free, pH 8.0) and incubated with 3 ml of 2.5 mm EZ-LinkTM sulfo-NHS-LC-Biotin (Thermo Fisher Scientific) on ice for 20 min with shaking. Cells were washed twice with 3 ml of PBS, pH 8.0, 3 ml of PBS containing 100 mm glycine (PBS/glycine) and then incubated on ice for 5 min with 3 ml of PBS/glycine. After washing twice with 3 ml of PBS, pH 8.0, cells were lysed using 0.5 ml of RIPA buffer (SDS−) containing protease inhibitor mixture Complete mini-EDTA-free. Lysates were collected with a cell scraper and incubated on ice for 15 min followed by centrifugation at 12,000 × g at 4 °C for 15 min. The supernatant (1 ml containing ∼0.7–1 mg of protein in RIPA buffer) was agitated with 50 μl (bed volume) of SA-agarose in the cold room overnight. Alternatively, lysate was incubated with 1–2 μg of anti-Myc mAb or anti-IgG for 3 h at 4 °C and subsequently agitated with protein G-agarose (Pierce) at 4 °C overnight. The SA-agarose or protein G beads were washed five times with 1 ml of 20 mm Tris-HCl, 150 mm NaCl, and 1% Triton X-100 and incubated in SDS-polyacrylamide gel loading buffer at 95 °C for 5 min. Proteins were separated by SDS-PAGE and subjected to Western blot analysis. Blots were blocked with 5% milk in TBS-T for β-actin and PDGFR-α, or with 3% BSA in TBS-T for anti-O-GlcNAc mAbs, and 5% milk for other antibodies in TBS-T for 1 h at room temperature. Blots were probed with NeutrAvidin-HRP (0.2 μg/ml), anti-β-actin Ab (1:5000) followed by incubation with anti-mouse IgG-HRP (0.08 μg/ml), anti-PDGFR-α Ab (0.4 μg/ml), anti-protein disulfide isomerase Ab followed by incubation with anti-rabbit IgG-HRP (0.16 μg/ml).
For immunoprecipitation using CTD110.6 mAb, Lec1 cells were transfected with either pCS2+ vector or pCS2+/Notch1-Myc in medium containing 5 mm GlcNAc. After 48 h, washed cells were lysed in 0.75 ml/plate lysis buffer (20 mm Tris, pH 7.4, 150 mm NaCl, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS with Roche Complete protease inhibitors). Following centrifugation for 15 min at 14,000 rpm at 4 °C, ∼1.5 ml of lysate was mixed with 10 μl of anti-mouse IgM-agarose and allowed to rotate in the cold room for 3 h. The precleared lysate was incubated with 1–5 μg of CTD110.6 mAb overnight at 4 °C before adding 10–20 μl of anti-mouse IgM-agarose for 3 h. Agarose beads were washed four times with lysis buffer, treated with SDS-polyacrylamide gel buffer, heated at 80 °C for 10 min, and separated on a 7.5% SDS-polyacrylamide gel. Following transfer to PVDF membrane, Western analysis was performed with various antibodies.
RESULTS
Antibody CTD110.6 Binds to Terminal β-GlcNAc in Truncated Complex N-Glycans
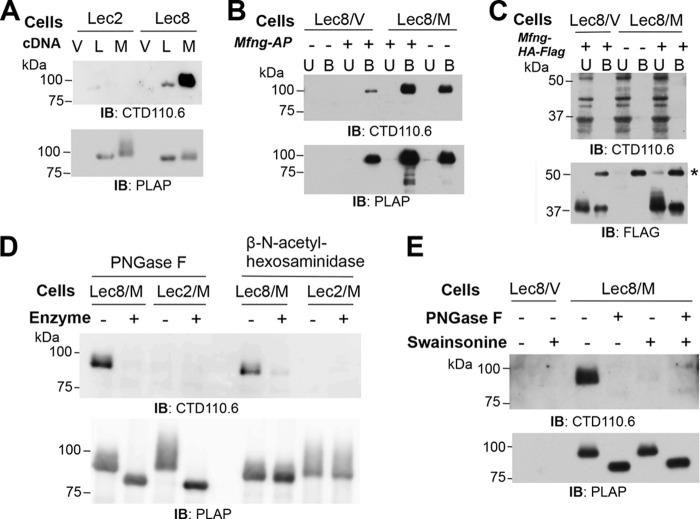
In experiments to investigate post-translational modifications of Lunatic Fringe (LFNG) and Manic Fringe (MFNG) (24, 25), LFNG-AP and MFNG-AP secreted from stable Lec2 or Lec8 CHO transfectants (Table 2) were subjected to Western blot analysis with the anti-O-GlcNAc mAb CTD110.6. Interestingly, both LFNG-AP and MFNG-AP from Lec8 cells bound CTD110.6, whereas the same proteins synthesized in Lec2 cells did not (Fig. 1A). To investigate further, Lec8 cells expressing empty vector (Lec8/V) or stably expressing Mfng-AP (Lec8/M) were transiently transfected with Mfng-AP (Fig. 1B). The transient transfectants generated MFNG-AP recognized by CTD110.6, and signal was increased in MFNG-AP from cells expressing both stable and transient Mfng-AP plasmids (Fig. 1B). To determine whether the antigen recognized by CTD110.6 was on the Mfng coding sequence or the AP tag, Lec8/V and Lec8/M cells were transiently transfected with Mfng-HA-FLAG. Affinity-purified MFNG-HA-FLAG migrated at ∼37 kDa (Fig. 1C). However, there was no signal detected by CTD110.6 in the bound fraction. Thus, the CTD110.6 signal on MFNG-AP was associated with the AP tag.
FIGURE 1.
Antibody CTD110.6 binds to terminal β-GlcNAc in complex N-glycans. A, Western blot (IB) analysis of LFNG-AP and MFNG-AP affinity-purified using anti-PLAP antibody-conjugated agarose from conditioned medium of Lec2/vector (Lec2/V), Lec2/Lfng-AP (Lec2/L), Lec2/Mfng-AP (Lec2/M), Lec8/vector (Lec8/V), Lec8/Lfng-AP (Lec8/L), or Lec8/Mfng-AP (Lec8/M) CHO cells. After separation on a 7.5% SDS-polyacrylamide gel and transfer, PVDF membranes were incubated at room temperature for 1 h with mAbs CTD110.6 (0.4 μg/ml) and anti-mouse IgM antibody-HRP (1.6 μg/ml), or anti-human PLAP (L-19) antibody (1 μg/ml) and anti-goat IgG antibody-HRP (0.08 μg/ml). Representative results are of blots from three independent experiments. B, Lec8/V and Lec8/M cells transiently transfected with pMirb/Mfng-AP. MFNG-AP affinity-purified using anti-FLAG (M2) antibody and protein G beads from conditioned medium were analyzed on a 7.5% SDS-polyacrylamide gel and detected by Western blot analysis as in A. U, Unbound; B, Bound. Representative results are of blots from three independent experiments. C, MFNG-HA-FLAG collected from conditioned medium of Lec8/V and Lec8/M cells transiently expressing pcDNA3/Mfng-HA-FLAG using anti-FLAG (M2) beads, separated in a 7.5% SDS-polyacrylamide gel, and subjected to Western blot analysis with CTD110.6 as in A or anti-FLAG (M2) antibody (1 μg/ml) and anti-mouse IgG antibody-HRP (0.08 μg/ml). Representative results are of blots from three independent experiments. D, MFNG-AP collected from conditioned medium of Lec8/M and Lec2/M cells treated with PNGase F or β-N-acetylhexosaminidase followed by Western blot analysis. E, Lec8/V and Lec8/M cells cultured with or without 5 μg/ml swainsonine for 4 days. MFNG-AP collected from conditioned media was analyzed by SDS-PAGE and Western blot analysis as in A, before and after treatment with PNGase F.
Lec8 CHO cells lack UDP-galactose transporter activity in the Golgi and generate N-glycans that terminate in GlcNAc (Table 2 and Refs. 20, 26). In contrast, Lec2 CHO cells lack CMP-sialic acid (CMP-NeuAc) transporter activity and generate N-glycans terminating in Gal residues (Table 2 and Refs. 19, 27). To determine whether the CTD110.6 mAb was detecting terminal β-GlcNAc on the truncated complex N-glycans synthesized by Lec8 cells, MFNG-AP purified from Lec8/Mfng and Lec2/Mfng conditioned media were treated with PNGase F to remove N-glycans, or with β-N-acetylhexosaminidase to remove terminal GlcNAc residues. Only MFNG-AP from Lec8/Mfng medium was recognized by CTD110.6, and both enzyme treatments eliminated the signal (Fig. 1D). These results suggested that CTD110.6 binds to terminal β-GlcNAc residues on complex N-glycans. Consistent with this interpretation, the presence of terminal Gal on the N-glycans from Lec2/Mfng cells prevented binding of CTD110.6 (Fig. 1, A and D). This result suggests that CTD110.6 does not recognize substituted GlcNAc in the branches or chitobiose core of Lec2 N-glycans.
To provide further evidence for the recognition of terminal β-GlcNAc on N-glycans by CTD110.6, complex N-glycan synthesis was inhibited by treatment of Lec8/Mfng cells with swainsonine. Swainsonine is an inhibitor of Golgi α-mannosidase II (28, 29) and generates hybrid N-glycans with a single terminal GlcNAc in Lec8 cells rather than complex N-glycans with multiple terminal β-GlcNAc residues (26). The detection of MFNG-AP from Lec8/Mfng cells by CTD110.6 was very weak after culture in swainsonine (Fig. 1E), but this band was eliminated by treatment with PNGase F. The data are consistent with detection of a terminal GlcNAc on hybrid N-glycans. Therefore, antibody CTD110.6 recognizes not only O-GlcNAc attached to protein (10), the unmodified chitobiose core of N-glycans (15), and GlcNAc attached to O-mannose on dystroglycan (16), but also terminal β-GlcNAc on truncated complex or hybrid N-glycans. The combined data suggest that the N-glycans predicted to be present on MFNG itself did not terminate in β-GlcNAc on MFNG from Lec8 cells.
Specificity of Other Anti-O-GlcNAc Antibodies
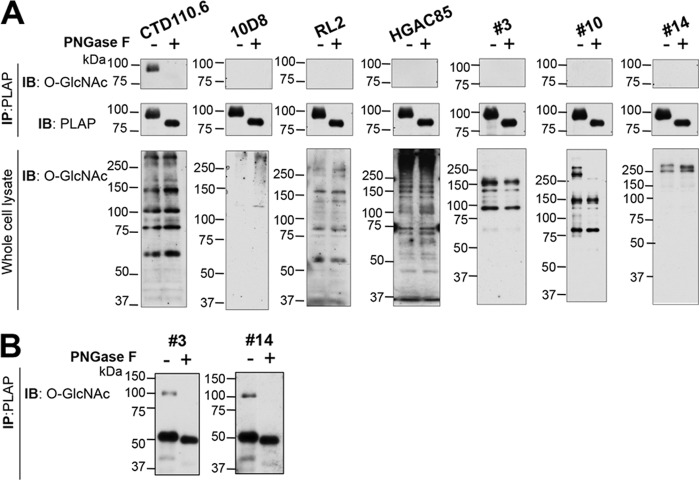
To determine whether other anti-O-GlcNAc mAbs cross-react with terminal β-GlcNAc on complex N-glycans, MFNG-AP was collected from Lec8/Mfng conditioned medium and subjected to Western blot analyses using seven anti-O-GlcNAc mAbs. All mAbs detected bands in whole cell lysates that contain cytosolic and nuclear O-GlcNAcylated proteins (Fig. 2A, lysate), but the profiles with the various mAbs were quite different. This shows that detection or isolation of the full complement of O-GlcNAcylated proteins in a cell or tissue by immunological methods would be difficult. At a concentration of 0.4–4 μg/ml, only CTD110.6 detected β-GlcNAc on MFNG-AP (Fig. 2A). The mAbs #3 and #14 also recognized β-GlcNAc on MFNG-AP but at a concentration of 8.6 or 9.7 μg/ml, respectively (Fig. 2B), and mAb #10 detected MFNG-AP at 1.2 μg/ml (data not shown). Thus, mAbs #3 and #14 required 10-fold, and mAb #10 2-fold, more mAb to detect terminal β-GlcNAc on MFNG-AP compared with O-GlcNAc on proteins in a lysate (Fig. 2B). However, the relative specificity or avidity of each mAb for O-GlcNAc versus β-GlcNAc cannot be inferred from these experiments because of the complex nature of the antigens. Interestingly, whereas all MFNG-AP bands shifted after PNGase F treatment as expected, in cell lysates the loss of faint bands at ∼100 kDa was observed only for mAbs #3 and #10 (Fig. 2A, lysate). Two bands at >250 kDa detected by #10 were also removed by PNGase F treatment. However, no PNGase F-sensitive species were observed in Lec8 whole cell lysate for CTD110.6, RL2, or HGAC85 mAbs (Fig. 2A, lysate), indicating that substrates of OGT represent the major O-GlcNAcylated proteins and obscure the loss of bands sensitive to PNGase F for these mAbs.
FIGURE 2.
Detection of GlcNAc-terminated, complex N-glycans by other anti-O-GlcNAc antibodies. A, MFNG-AP affinity-purified from Lec8/M conditioned medium (upper) or Lec8/M whole cell lysate (lower) was separated in a 10% SDS-polyacrylamide gel, transferred to PVDF membranes, and incubated with anti-O-GlcNAc antibodies (top and bottom), or anti-human PLAP (L-19) antibody (middle), for 1 h at room temperature. O-GlcNAcylated proteins were detected with CTD110.6 (0.4 μg/ml) or 10D8 (0.2 μg/ml) and anti-mouse IgM antibody-HRP (1.6 μg/ml), RL2 (3 μg/ml), HGAC85 (4 μg/ml), #3 (0.86 μg/ml), #10 (0.59 μg/ml), or #14 (0.97 μg/ml) and anti-mouse IgG-HRP (0.08 μg/ml). Representative results are from two preparations. B, mAbs #3 (8.6 μg/ml) and #14 (9.7 μg/ml) were incubated at room temperature for 1 h followed by anti-mouse IgG-HRP (0.08 μg/ml). IP, immunoprecipitate; IB, immunoblot.
CTD110.6 Recognizes Both O-GlcNAc and Terminal β-GlcNAc on Cell Surface Glycoproteins
The ability of CTD110.6 to detect surface O-GlcNAc on EGF repeats of the extracellular domain of membrane proteins was examined by flow cytometry. Lec1 mutant CHO cells that lack MGAT1 (GlcNAcT-I; EC 2.4.1.101) and fail to synthesize complex and hybrid N-glycans were used (Table 2 and Refs. 18, 27). All N-glycans on Lec1 glycoproteins are oligomannosyl, with the major species being the substrate of MGAT1, Man5GlcNAc2Asn (Fig. 3A). O-GalNAc glycans do not contain GlcNAc in Lec1 cells (27, 30), and O-fucose and O-mannose glycans have very minor amounts of terminal GlcNAc (16, 21), not detected by MALDI-TOF mass spectrometry (27). Lec1 cells did not bind the plant lectin L-PHA that recognizes complex N-glycans as expected, but they did bind mAb CTD110.6 (Fig. 3B). The latter binding should reflect O-GlcNAcylated, EGF-containing glycoproteins of the CHO cell surface.
FIGURE 3.
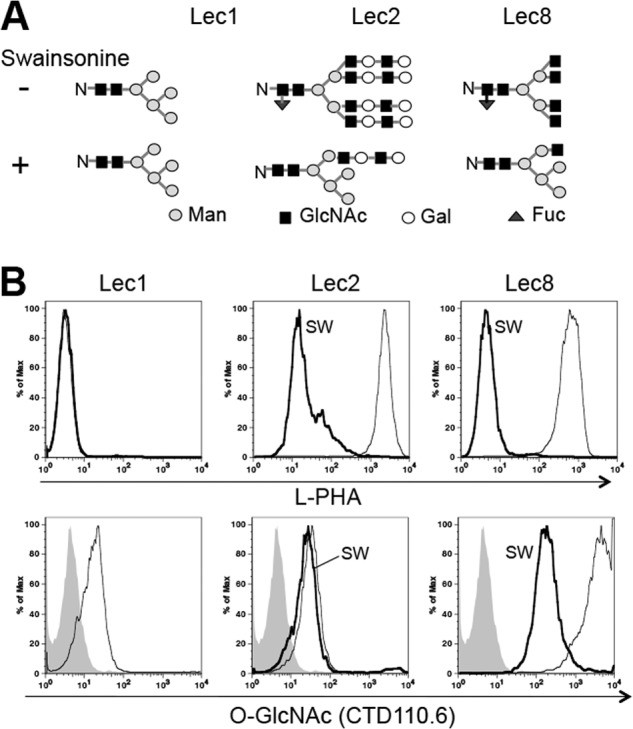
CTD110.6 recognizes both O-GlcNAc and terminal β-GlcNAc on cell surface glycoproteins. A, N-glycans of glycoproteins synthesized in CHO glycosylation mutants cultured in the presence (predicted (28, 29)), or absence (26, 27), of swainsonine. B, binding of L-PHA (upper) and mAb CTD110.6 (lower) to fixed cells. Lec2 and Lec8 cells were cultured with or without 5 μg/ml swainsonine (SW) for 4 days. Cells (4 × 105) were incubated with 20 μg/ml fluorescein-labeled L-PHA or 40 μg/ml CTD110.6 on ice for 20 min followed by 10 μg/ml Cy5-conjugated anti-mouse IgM antibody. Gray profiles, secondary Abs; thin line, control cells; bold line, swainsonine-treated cells. Representative results are from two independent experiments.
To investigate further, binding of CTD110.6 was assayed before and after treatment with swainsonine for 4 days to convert complex to hybrid N-glycans (Fig. 3A). Swainsonine treatment reduced L-PHA binding to both Lec2 and Lec8 cells, as expected (Fig. 3B). Binding of CTD110.6 mAb to swainsonine-treated Lec8 cells was also markedly reduced by swainsonine, but remained greater than binding to Lec1 CHO cells, presumably because of a terminal β-GlcNAc on Lec8 hybrid N-glycans (Fig. 3A). By contrast, Lec2 cells bound CTD110.6 similarly, both before and after treatment with swainsonine (Fig. 3B). In Lec2 cells, complex and hybrid N-glycans terminate in Gal residues that block access to GlcNAc (Fig. 1D). Importantly, this shows that CHO cells generate few, if any, truncated complex or hybrid N-glycans with terminal β-GlcNAc residues, consistent with N-glycan structural studies (27). Therefore, CTD110.6 binding to Lec2 and wild type CHO cells reflects cell surface glycoproteins with O-GlcNAcylated EGF repeats. Evidence that the substituted chitobiose core in high mannose N-glycans (Man5–9GlcNAc2Asn) is not recognized by CTD110.6 is presented in Fig. 6A.
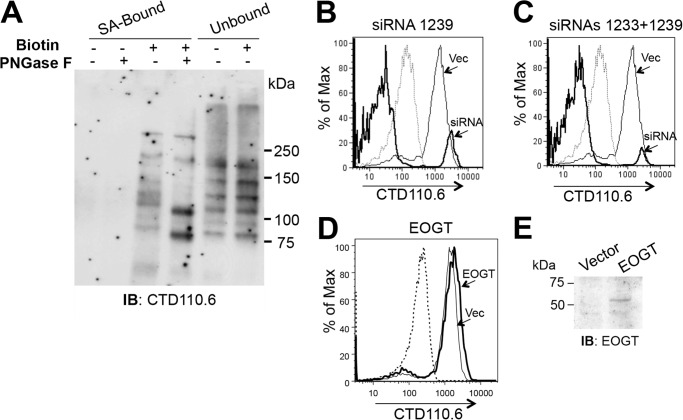
FIGURE 6.
Specificity of CTD110.6 for O-GlcNAc on cell surface glycoproteins. A, Lec1 cells were biotinylated, separated into SA-Bound and Unbound fractions on SA-agarose, and analyzed before and after treatment with PNGase F by SDS-PAGE and Western blotting (IB) using the CTD110.6 mAb. B, flow cytometry profile shows CTD110.6 mAb of fixed Lec1 cells expressing vector (thin line) or siRNA 1239 (bold line). Dashed line is isotype control. Representative results are from two independent experiments. C, results are same as B except the bold line is for Lec1 cells treated with a mixture of siRNA 1239 and 1233 (1:1). Representative results are from two independent experiments. D, Lec1 cells expressing vector control (thin line) or a human EOGT cDNA (bold line) were fixed and analyzed by flow cytometry with CTD110.6 mAb. Dashed line is isotype control. Representative results are from three independent experiments. E, Lec1 cells expressing vector control or human EOGT cDNA were analyzed by Western blotting using anti-EOGT antibody (1:500 at 4 °C overnight). Representative results are from two independent experiments.
Three Antibodies Detect O-GlcNAc on the Cell Surface
To compare the abilities of the different antibodies to detect cell surface O-GlcNAc, Lec1 cells were fixed with 4% paraformaldehyde, and half the cells were permeabilized with 0.1% Triton X-100. Each population was assayed for binding of anti-O-GlcNAc mAbs by flow cytometry. The IgM isotype control for mAbs CTD110.6 and 10D8 gave higher nonspecific binding than the IgG1 and IgG2a isotype controls of the other four mAbs (Fig. 4, top panel). All anti-O-GlcNAc mAbs bound to the surface of Lec1 cells (Fix), albeit to different levels. With the exception of mAb #14, binding was increased after cell permeabilization (Perm) (Fig. 4).
FIGURE 4.
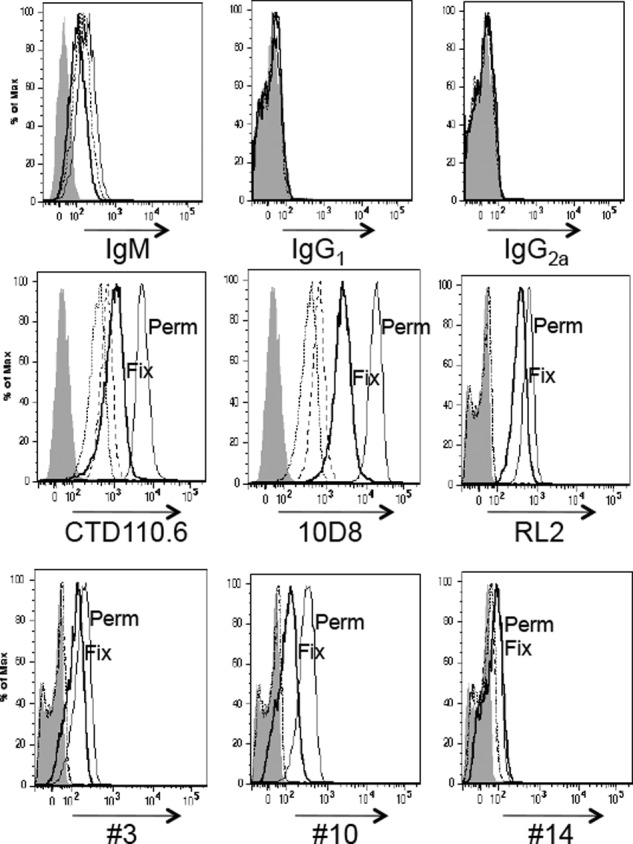
Binding of anti-O-GlcNAc antibodies to Lec1 cells. Lec1 cells (4 × 105) fixed with 4% paraformaldehyde (Fix) or fixed and permeabilized with 0.1% Triton X-100 (Perm) were incubated with 10 μg/ml 10D8 or 40 μg/ml other anti-O-GlcNAc antibodies including isotype control Abs followed by incubation with 10 μg/ml secondary Ab. Gray profiles, secondary Ab; short dashed line, isotype control for Fix; long dashed line, isotype control for Perm; bold solid line, anti-O-GlcNAc mAb for Fix; thin solid line, anti-O-GlcNAc mAb for Perm. Representative results are from two independent experiments.
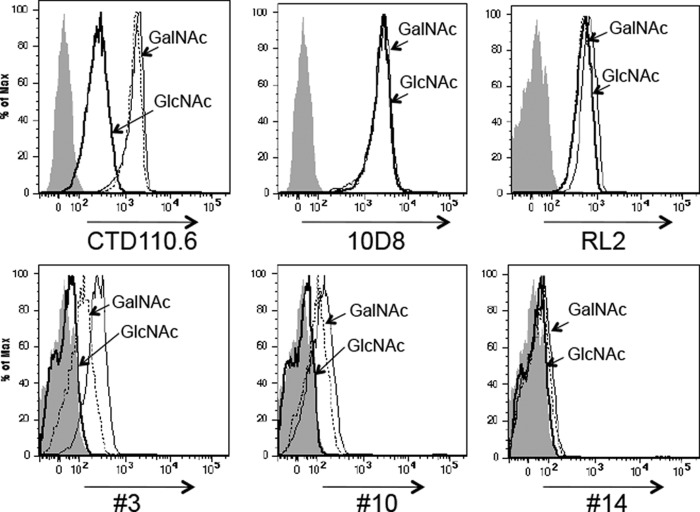
To determine that cell surface binding was due to O-GlcNAc, inhibition of mAb binding by 5 mm GlcNAc versus 5 mm GalNAc was examined (Fig. 5). Binding by antibodies CTD110.6, #3, and #10 was inhibited by 5 mm GlcNAc, but not 5 mm GalNAc, and was thus specific for recognition of O-GlcNAc. However, binding of antibodies 10D8, RL2, and #14 was not inhibited by the presence of 5 mm GlcNAc. Therefore, only antibodies CTD110.6, #3, and #10 should be used for cell surface detection of O-GlcNAc.
FIGURE 5.
GlcNAc competes for cell surface binding of only some anti-O-GlcNAc mAbs. Lec1 cells (4 × 105) fixed with 4% paraformaldehyde were incubated with 10 μg/ml 10D8 mAb or 40 μg/ml each of the other anti-O-GlcNAc mAbs (CTD110.6, RL2, #3, #10, and #14) in the presence of 5 mm GlcNAc (bold line) or 5 mm GalNAc (dotted line), or no sugar (thin line) on ice for 20 min followed by 10 μg/ml secondary Ab. Gray profiles, secondary Ab alone. Representative results are from four independent experiments.
CTD110.6 Binds Predominantly to O-GlcNAc on the Cell Surface Glycoproteins of Lec1
Lec1 CHO cells do not synthesize complex N-glycans (Table 2 and Fig. 3A), but do contain substituted GlcNAc residues in the core of oligomannosyl N-glycans that might conceivably be recognized by CTD110.6. To investigate this question, binding of CTD110.6 to biotinylated cell surface glycoproteins from Lec1 cells was examined before and after treatment with PNGase F (Fig. 6A). The complement of bands identified by CTD110.6 in the biotinylated fraction (SA-Bound) differed markedly from those in the nonbiotinylated, cytosolic and nuclear fraction (Unbound). After treatment of the SA-Bound fraction with PNGase F, several bands shifted to a lower molecular mass, as expected for cell surface glycoproteins that have lost N-glycans. However, the overall intensity of the signal did not change appreciably, indicating that the N-glycans removed by PNGase F were not responsible for a significant proportion of the CTD110.6 signal.
O-GlcNAc on cell surface and secreted proteins is added by the ER-localized O-GlcNAc transferase EOGT (2–7). To determine whether reduced EOGT led to a reduction in O-GlcNAc on cell surface proteins, RNAi knockdown of CHO Eogt was examined. Following transfection of Lec1 cells and selection for resistance to puromycin for 16 days, vector control and RNAi knockdown transfectant populations were examined for CTD110.6 binding. Lec1 cells transfected with control vector bound CTD110.6 similarly (Fig. 6, B and C). However, the RNAi construct, pSUPER1239, caused a marked reduction in CTD110.6 binding (Fig. 6B). An even smaller fraction of Lec1 cells bound CTD110.6 when a mixture of pSUPER1233 and pSUPER1239 (1:1) was tested (Fig. 6C). Whereas endogenous CHO EOGT could not be detected with available antibodies, quantitative RT-PCR using SYBR Green and β-actin as control showed that Eogt transcripts were reduced 76–78% by the combined siRNA treatment compared with vector control.3 Treatment with siRNA1239 alone reduced Eogt transcripts by 64–70%. Treatment with siRNA1233 alone was least effective, reducing Eogt transcripts by 48–64% but not significantly reducing CTD110.6 binding (data not shown). Reduced binding of CTD110.6 was also observed for parent CHO cells expressing the siRNAs targeting Eogt (data not shown). A small increase in CTD110.6 binding to Lec1 cells was induced by overexpression of a human EOGT cDNA (Fig. 6D). Overexpressed human EOGT was detected by Western blot analysis (Fig. 6E).
O-GlcNAcylated Cell Surface Proteins of CHO Cells
To establish conditions for ultimately identifying the range of O-GlcNAcylated cell surface glycoproteins in a cell, NOTCH1 was transiently expressed in Lec1 CHO cells and examined following treatment with PUGNAc or hexosaminidase, or following overexpression of human EOGT cDNA. PUGNAc is an inhibitor of O-GlcNAcase localized in the cytosol (31). If, as expected, O-GlcNAc is added to NOTCH1-Myc in the secretory pathway, there should be no effect of PUGNAc on CTD110.6 binding to NOTCH1 ECD. Cells were biotinylated and lysed. NOTCH1-Myc was collected with anti-Myc or anti-IgG control beads (first immunoprecipitate), and the unbound fraction was subsequently affinity-purified with SA-agarose (SA-Bound) prior to Western blot analysis (Fig. 7A). One blot was probed with anti-NOTCH1 ECD mAb, stripped, and re-probed with anti-Myc mAb; a second blot was probed with CTD110.6 to detect O-GlcNAc. Fig. 7A shows that Myc-Bound NOTCH1-Myc gave essentially the same signal for CTD110.6 in the presence and absence of PUGNAc, whereas Unbound lysate exhibited numerous bands and the predicted increase in O-GlcNAc signal induced by PUGNAc inhibition of cytosolic O-GlcNAcase. Two unbound proteins >250 kDa (Fig. 7A) had less O-GlcNAc after PUGNAc treatment but did not co-migrate with NOTCH1-Myc and do not reflect endogenous CHO NOTCH1, which is not detected in CHO whole cell lysate (Fig. 7, B–D). Moreover, these two bands were also observed with mAb #10 and were removed by PNGase F treatment (Fig. 2A). Interestingly, NOTCH1 collected on SA beads from lysate that did not bind to Myc beads, had lost the Myc C-terminal tag (migrated faster and was not detected by anti-Myc mAb). This truncated species predominated over full-length NOTCH1-Myc, which was poorly detected in SA-Bound fractions (Fig. 7, A and C). The CTD110.6 signal was equivalent in the presence and absence of PUGNAc for the truncated form of NOTCH1 which contains the NOTCH1 ECD. Thus, PUGNAc did not inhibit the O-GlcNAcylation of NOTCH1.
FIGURE 7.
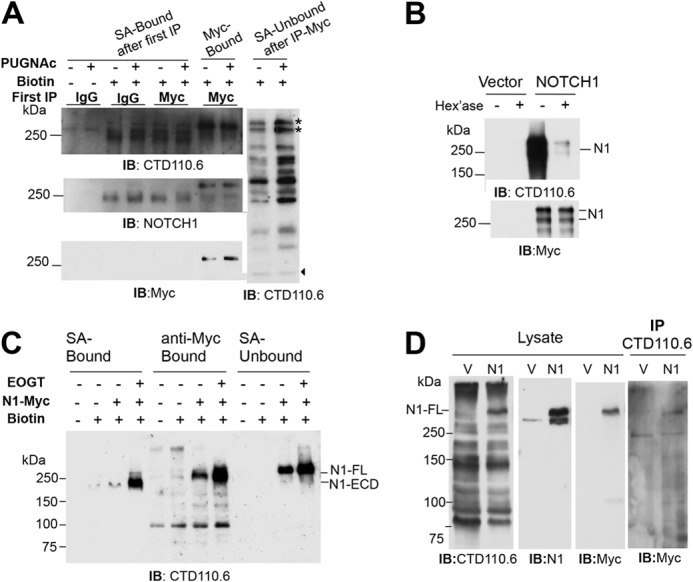
O-GlcNAc on NOTCH1 is detected at the cell surface. A, Lec1 cells transiently expressing NOTCH1-Myc were treated for 18 h with 100 μm PUGNAc (+) or vehicle (dimethyl sulfoxide) (−), biotinylated, lysed, and the lysate was incubated with anti-Myc beads (Myc-Bound) or anti-IgG beads. The fraction that did not bind to anti-Myc or anti-IgG beads was incubated with SA-agarose and gave the (SA-Bound after first immunoprecipitation (IP)) and (SA-Unbound after immunoprecipitate-Myc) fractions. Samples were separated on a 7.5% SDS-polyacrylamide gel, and one set was probed with anti-NOTCH1 ECD mAb (2 μg/ml), stripped, and probed with anti-Myc mAb (1:500). The second set was probed with CTD110.6 (0.4 μg/ml). The stars identify proteins that migrated similarly to but distinct from NOTCH1-Myc. Arrowhead identifies band indicating equal loading. Representative results are from three independent experiments. IB, immunoblot. B, NOTCH1-Myc prepared from transient CHO transfectants as in A was incubated with 10 units of β-N-acetylhexosaminidase at 37 °C for 2 h, separated in a 7.5% SDS-polyacrylamide gel, transferred to PVDF membranes, and incubated with CTD110.6 (0.4 μg/ml) and anti-mouse IgM-HRP (1.6 μg/ml) or anti-Myc 9E10 (1:500) and anti-mouse IgG-HRP (0.08 μg/ml) Abs at room temperature for 1 h. Representative results are from two independent experiments. C, Lec1 cells were co-transfected with vector or pCR3.1/EOGT, and pCS2+/Notch1-Myc. After 24 h, biotinylation was performed; biotinylated proteins were collected on SA-agarose beads or by incubation with anti-Myc antibody and protein G beads, and analyzed by SDS-PAGE and Western blotting with CTD110.6 mAb as in A. D, lysate from Lec1 cells transfected with plasmid pCS2+ (V) or pCS2+/Notch1-Myc (N1) was precleared with anti-IgM-agarose, incubated with mAb CTD110.6 (1 μg) overnight at 4 °C, and collected on mouse anti-IgM-agarose. Lysate (50 μl) and proteins solubilized from beads were separated on a 7.5% SDS-polyacrylamide gel, transferred to membrane, and subjected to Western blot analysis using the indicated antibodies sequentially, in the order CTD110.6, anti-NOTCH1, and anti-Myc antibodies. Representative results are from three experiments.
The O-GlcNAc on NOTCH1-Myc ECD was removed by β-N-acetylhexosaminidase treatment (Fig. 7B). When a human EOGT cDNA was introduced, the O-GlcNAc signal on NOTCH1-Myc was enhanced (Fig. 7C). The combined data show that overexpressed NOTCH1-Myc was a major substrate of CHO EOGT under these conditions. The anti-O-GlcNAc mAb CTD110.6 was able to immunoprecipitate NOTCH1-Myc, although the efficiency was very low (Fig. 7D). The combined results show that CTD110.6 detects biotinylated, O-GlcNAcylated proteins at the cell surface.
To determine the range of glycoproteins modified with O-GlcNAc on the CHO cell surface, Lec1 CHO cells were biotinylated and subjected to affinity purification and Western blot analysis. The cell surface marker, PDGFR-α, was detected in the biotinylated, but not the nonbiotinylated, SA-Bound fraction (Fig. 8A). Importantly, β-actin and protein disulfide isomerase were not detected in the SA-Bound fraction, showing that biotin did not penetrate the membrane to modify cytosolic or ER proteins, respectively. Several biotinylated proteins were detected by anti-O-GlcNAc antibody CTD110.6 (Fig. 8B). Western blot analysis with anti-O-GlcNAc mAb #10 detected a similar set of O-GlcNAcylated cell surface proteins (Fig. 8C). Several cell surface CHO proteins of <150 kDa appear to be novel substrates of EOGT. The bands of >250 kDa are likely to include Notch receptors. Thus, NOTCH3, with many EGF repeats containing the EOGT consensus site (8), was detected in the SA-Bound fraction at the expected molecular mass (Fig. 8B). Unfortunately, endogenous NOTCH3 could not be affinity-purified with anti-NOTCH3 antibodies to show directly that NOTCH3 carried O-GlcNAc.
FIGURE 8.
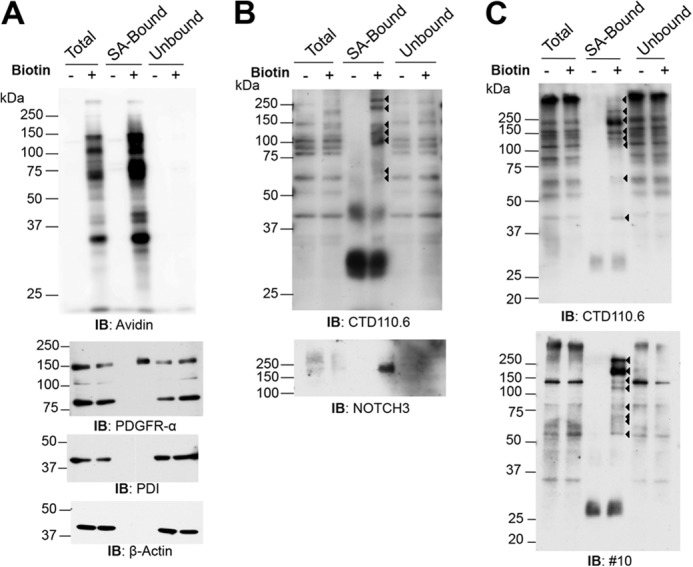
O-GlcNAc is present on a range of CHO cell surface proteins. A, Lec1 cells were biotinylated, lysed, and biotinylated proteins were collected on SA-agarose. SA-Bound and Unbound proteins were separated in a 10% SDS-polyacrylamide gel. The PVDF membranes were incubated with anti-PDGFR-α (2 μg/ml) and anti-rabbit IgG-HRP (1:5000), anti-protein disulfide isomerase (2 μg/ml) or anti-β-actin (1:5000) Abs and anti-mouse IgG-HRP (0.16 μg/ml), or NeutrAvidin-HRP (1:5000) at room temperature for 1 h. Representative results are from four independent experiments. IB, immunoblot. B, samples were the same as in A, but run on a 7.5% SDS-polyacrylamide gel. Immunoblotting was with CTD110.6 (0.4 μg/ml) and anti-mouse IgM antibody-HRP (1.6 μg/ml), or anti-NOTCH3 (5E1) (1:500) and anti-mouse IgG-HRP (0.08 μg/ml) antibodies at room temperature for 1 h. Arrowheads identify O-GlcNAcylated cell surface glycoproteins. C, samples were the same as in A and run on a 10% SDS-polyacrylamide gel. Immunoblotting was with CTD110.6 (0.4 μg/ml) or #10 (0.59 μg/ml) antibodies at room temperature for 1 h. Arrowheads indicate bands present in the biotinylated, SA-Bound fraction. Representative results are from two independent experiments.
DISCUSSION
O-GlcNAcylation was discovered on cytosolic proteins 30 years ago (32). The first identification of O-GlcNAc on the extracellular domain of Drosophila Notch was quite recent (2). The ER-localized EOGT responsible for transferring O-GlcNAc to proteins of the secretory pathway has been identified, and functional roles in Drosophila have been revealed (5, 7). Our finding that secreted Fringe-AP proteins were recognized by mAb CTD110.6 suggested that they may be unexpected substrates of EOGT, although this was unlikely because EOGT was proposed to be specific for EGF repeats of membrane proteins (4, 5, 7, 8). We subsequently determined that CTD110.6 mAb was, in fact, detecting terminal β-GlcNAc on the antennae of complex N-glycans of the AP tag of LFNG-AP and MFNG-AP proteins (Fig. 1). CTD110.6 has also been shown to recognize terminal β-GlcNAc on unsubstituted chitobiose of an N-glycan (15) and on O-mannose linked to α-dystroglycan (16). To identify mAbs that could differentiate between O-GlcNAc linked directly to protein versus terminal β-GlcNAc on an N- or O-glycan, we used CHO mutant cells with and without N-glycans terminating in β-GlcNAc (Table 2) and compared recognition of cell surface glycoproteins by several mAbs raised against O-GlcNAc (Table 1). The mAb CTD110.6 most readily detected terminal β-GlcNAc on MFNG-AP (Fig. 2A). The IgG mAbs #3, #10, and #14 required a higher concentration to detect terminal β-GlcNAc on complex N-glycans compared with O-GlcNAc (Fig. 2). However, comparisons of relative binding to O-GlcNAc versus terminal β-GlcNAc in our experiments were necessarily qualitative because of the complex nature of the antigen mixtures.
For proteins at the surface of intact Lec1 cells (that have no N-glycans terminating in β-GlcNAc), only three mAbs detected O-GlcNAc specifically: CTD110.6, #3, and #10. Binding of each of these mAbs was inhibited by 5 mm GlcNAc but not by 5 mm GalNAc. Glycoproteins with N-glycans from Lec8 CHO cells that terminate in β-GlcNAc resulted in the greatest binding of mAb CTD110.6. Even the presence of a single terminal β-GlcNAc on N-glycans of Lec8 cells treated with swainsonine was detected by CTD110.6 (Figs. 1E and 3B). However, internal, substituted GlcNAc in the core of oligomannosyl N-glycans (Lec1), or complex N-glycans (parent CHO and Lec2 cells) were not detected by CTD110.6 (Figs. 3B and 6A).
The anti-O-GlcNAc mAbs used in this paper were generated from a variety of different antigens (Table 1), and they recognized different subsets of O-GlcNAcylated proteins in cell lysates (Fig. 2A). mAb 10D8 showed the least reactivity in our experiments, and mAb CTD110.6 was the most reactive. The mAbs CTD110.6, #3, #10, and #14 recognized terminal β-GlcNAc on N-glycans (Fig. 2). However, CHO cells, probably like most cells, have very few, if any, truncated N-glycans that terminate in GlcNAc (27). In addition, we show here that GlcNAc substituted with another sugar in an N-glycan is not recognized by the anti-O-GlcNAc mAbs tested. Therefore, O-GlcNAc on cell surface proteins will be the primary target of anti-O-GlcNAc mAbs. Importantly, O-GlcNAc on mouse NOTCH1 ECD can be substituted with a β(1,4)Gal residue (4) which would prevent anti-O-GlcNAc mAb recognition. However, we found that treatment of Lec1 cells with β-galactosidase, or neuraminidase and β-galactosidase, did not increase binding of CTD110.6 (data not shown). Three of the mAbs we examined (CTD110.6, #3, and #10) proved useful for flow cytometry and cell sorting based on cell surface expression of O-GlcNAc. However, careful controls should be employed to determine that GlcNAc-terminating N- or O-glycans are not also being detected in significant amounts.
Biotinylation of cell surface proteins of Lec1 CHO cells allowed pulldown of NOTCH1 as expected (Figs. 6 and 7) and several other proteins recognized by the anti-O-GlcNAc antibody CTD110.6 (Fig. 8). Thrombospondin-1 acquires an O-GlcNAc on its EGF repeat (6), but this protein is not expressed in CHO cells. Because the number of O-GlcNAcylated cell surface proteins are few compared with cytosolic and nuclear proteins with an O-GlcNAc, identifying them efficiently will require capture of tagged O-GlcNAcylated proteins followed by glycoproteomics (8). The subset of EOGT substrates in different cell types will be important to determine for functional studies of this new modification.
Acknowledgments
We thank Subha Sundaram, Huimin Shang, and Margreet Wolfert for excellent technical assistance and all those who provided plasmids, antibodies, and other reagents (see “Experimental Procedures”).
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA095022 from the NCI and R01 GM106417 from the NIGMS (to P. S.).
S. Varshney and P. Stanley, unpublished observations.
- ER
- endoplasmic reticulum
- α-MEM
- α-minimum Eagle's medium
- AP
- alkaline phosphatase
- CHO
- Chinese hamster ovary
- ECD
- extracellular domain
- EGF
- epidermal growth factor-like
- EOGT
- EGF-specific O-GlcNAc transferase
- LFNG
- Lunatic Fringe
- L-PHA
- Phaseolus vulgaris leukoagglutinin
- mAb
- monoclonal antibody
- MFNG
- Manic Fringe
- OGT
- O-GlcNAc transferase
- O-GlcNAc
- O-linked β-d-N-acetylglucosamine
- PDGFR
- platelet-derived growth factor receptor
- PLAP
- placental alkaline phosphatase
- PNGase F
- peptide N-glycanase F
- PUGNAc
- O-(2-acetamido-2-deoxy-d-glucopyranosylidene) amino-N-phenylcarbamate
- RIPA
- radioimmuneprecipitation assay
- SA-agarose
- NeutrAvidin-agarose.
REFERENCES
- 1. Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etlzler M. E. (2009) Essentials of Glycobiology, 2nd Edition, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 2. Matsuura A., Ito M., Sakaidani Y., Kondo T., Murakami K., Furukawa K., Nadano D., Matsuda T., Okajima T. (2008) O-Linked N-acetylglucosamine is present on the extracellular domain of Notch receptors. J. Biol. Chem. 283, 35486–35495 [DOI] [PubMed] [Google Scholar]
- 3. Sakaidani Y., Furukawa K., Okajima T. (2010) O-GlcNAc modification of the extracellular domain of Notch receptors. Methods Enzymol. 480, 355–373 [DOI] [PubMed] [Google Scholar]
- 4. Sakaidani Y., Ichiyanagi N., Saito C., Nomura T., Ito M., Nishio Y., Nadano D., Matsuda T., Furukawa K., Okajima T. (2012) O-Linked N-acetylglucosamine modification of mammalian Notch receptors by an atypical O-GlcNAc transferase Eogt1. Biochem. Biophys. Res. Commun. 419, 14–19 [DOI] [PubMed] [Google Scholar]
- 5. Sakaidani Y., Nomura T., Matsuura A., Ito M., Suzuki E., Murakami K., Nadano D., Matsuda T., Furukawa K., Okajima T. (2011) O-Linked-N-acetylglucosamine on extracellular protein domains mediates epithelial cell-matrix interactions. Nat. Commun. 2, 583. [DOI] [PubMed] [Google Scholar]
- 6. Hoffmann B. R., Liu Y., Mosher D. F. (2012) Modification of EGF-like module 1 of thrombospondin-1, an animal extracellular protein, by O-linked N-acetylglucosamine. PLoS One 7, e32762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Müller R., Jenny A., Stanley P. (2013) The EGF repeat-specific O-GlcNAc-transferase Eogt interacts with Notch signaling and pyrimidine metabolism pathways in Drosophila. PLoS One 8, e62835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alfaro J. F., Gong C. X., Monroe M. E., Aldrich J. T., Clauss T. R., Purvine S. O., Wang Z., Camp D. G., 2nd, Shabanowitz J., Stanley P., Hart G. W., Hunt D. F., Yang F., Smith R. D. (2012) Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc. Natl. Acad. Sci. U.S.A. 109, 7280–7285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Butkinaree C., Park K., Hart G. W. (2010) O-Linked β-N-acetylglucosamine (O-GlcNAc): extensive cross-talk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim. Biophys. Acta 1800, 96–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Comer F. I., Vosseller K., Wells L., Accavitti M. A., Hart G. W. (2001) Characterization of a mouse monoclonal antibody specific for O-linked N-acetylglucosamine. Anal. Biochem. 293, 169–177 [DOI] [PubMed] [Google Scholar]
- 11. Turner J. R., Tartakoff A. M., Greenspan N. S. (1990) Cytologic assessment of nuclear and cytoplasmic O-linked N-acetylglucosamine distribution by using anti-streptococcal monoclonal antibodies. Proc. Natl. Acad. Sci. U.S.A. 87, 5608–5612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Snow C. M., Senior A., Gerace L. (1987) Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J. Cell Biol. 104, 1143–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yoshida N., Mortara R. A., Araguth M. F., Gonzalez J. C., Russo M. (1989) Metacyclic neutralizing effect of monoclonal antibody 10D8 directed to the 35- and 50-kilodalton surface glycoconjugates of Trypanosoma cruzi. Infect. Immun. 57, 1663–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Teo C. F., Ingale S., Wolfert M. A., Elsayed G. A., Nöt L. G., Chatham J. C., Wells L., Boons G. J. (2010) Glycopeptide-specific monoclonal antibodies suggest new roles for O-GlcNAc. Nat. Chem. Biol. 6, 338–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Isono T. (2011) O-GlcNAc-specific antibody CTD110.6 cross-reacts with N-GlcNAc2-modified proteins induced under glucose deprivation. PLoS One 6, e18959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ogawa M., Nakamura N., Nakayama Y., Kurosaka A., Manya H., Kanagawa M., Endo T., Furukawa K., Okajima T. (2013) GTDC2 modifies O-mannosylated α-dystroglycan in the endoplasmic reticulum to generate N-acetylglucosamine epitopes reactive with CTD110.6 antibody. Biochem. Biophys. Res. Commun. 440, 88–93 [DOI] [PubMed] [Google Scholar]
- 17. Joutel A., Andreux F., Gaulis S., Domenga V., Cecillon M., Battail N., Piga N., Chapon F., Godfrain C., Tournier-Lasserve E. (2000) The ectodomain of the Notch3 receptor accumulates within the cerebrovasculature of CADASIL patients. J. Clin. Invest. 105, 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen W., Stanley P. (2003) Five Lec1 CHO cell mutants have distinct Mgat1 gene mutations that encode truncated N-acetylglucosaminyltransferase I. Glycobiology 13, 43–50 [DOI] [PubMed] [Google Scholar]
- 19. Eckhardt M., Gotza B., Gerardy-Schahn R. (1998) Mutants of the CMP-sialic acid transporter causing the Lec2 phenotype. J. Biol. Chem. 273, 20189–20195 [DOI] [PubMed] [Google Scholar]
- 20. Oelmann S., Stanley P., Gerardy-Schahn R. (2001) Point mutations identified in lec8 Chinese hamster ovary glycosylation mutants that inactivate both the UDP-galactose and CMP-sialic acid transporters. J. Biol. Chem. 276, 26291–26300 [DOI] [PubMed] [Google Scholar]
- 21. Chen J., Moloney D. J., Stanley P. (2001) Fringe modulation of Jagged1-induced Notch signaling requires the action of β4-galactosyltransferase-1. Proc. Natl. Acad. Sci. U.S.A. 98, 13716–13721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hou X., Tashima Y., Stanley P. (2012) Galactose differentially modulates lunatic and Manic Fringe effects on Delta1-induced NOTCH signaling. J. Biol. Chem. 287, 474–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sanjabi S., Williams K. J., Saccani S., Zhou L., Hoffmann A., Ghosh G., Gerondakis S., Natoli G., Smale S. T. (2005) A c-Rel subdomain responsible for enhanced DNA-binding affinity and selective gene activation. Genes Dev. 19, 2138–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moloney D. J., Panin V. M., Johnston S. H., Chen J., Shao L., Wilson R., Wang Y., Stanley P., Irvine K. D., Haltiwanger R. S., Vogt T. F. (2000) Fringe is a glycosyltransferase that modifies Notch. Nature 406, 369–375 [DOI] [PubMed] [Google Scholar]
- 25. Brückner K., Perez L., Clausen H., Cohen S. (2000) Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature 406, 411–415 [DOI] [PubMed] [Google Scholar]
- 26. Kawar Z. S., Haslam S. M., Morris H. R., Dell A., Cummings R. D. (2005) Novel poly-GalNAcβ1–4GlcNAc (LacdiNAc) and fucosylated poly-LacdiNAc N-glycans from mammalian cells expressing β1,4-N-acetylgalactosaminyltransferase and α1,3-fucosyltransferase. J. Biol. Chem. 280, 12810–12819 [DOI] [PubMed] [Google Scholar]
- 27. North S. J., Huang H. H., Sundaram S., Jang-Lee J., Etienne A. T., Trollope A., Chalabi S., Dell A., Stanley P., Haslam S. M. (2010) Glycomics profiling of Chinese hamster ovary cell glycosylation mutants reveals N-glycans of a novel size and complexity. J. Biol. Chem. 285, 5759–5775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Foddy L., Feeney J., Hughes R. C. (1986) Properties of baby hamster kidney (BHK) cells treated with swainsonine, an inhibitor of glycoprotein processing: comparison with ricin-resistant BHK cell mutants. Biochem. J. 233, 697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Foddy L., Hughes R. C. (1986) Interactions of lectins with normal, swainsonine-treated and ricin-resistant baby hamster kidney BHK cells. Carbohydr. Res. 151, 293–304 [DOI] [PubMed] [Google Scholar]
- 30. Aguilan J. T., Sundaram S., Nieves E., Stanley P. (2009) Mutational and functional analysis of Large in a novel CHO glycosylation mutant. Glycobiology 19, 971–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Haltiwanger R. S., Grove K., Philipsberg G. A. (1998) Modulation of O-linked N-acetylglucosamine levels on nuclear and cytoplasmic proteins in vivo using the peptide O-GlcNAc-β-N-acetylglucosaminidase inhibitor O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino-N-phenylcarbamate. J. Biol. Chem. 273, 3611–3617 [DOI] [PubMed] [Google Scholar]
- 32. Torres C. R., Hart G. W. (1984) Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes: evidence for O-linked GlcNAc. J. Biol. Chem. 259, 3308–3317 [PubMed] [Google Scholar]