Background: SLC25A29 encoded by the human SLC25A29 gene was suggested to be a mitochondrial carnitine/acylcarnitine- or ornithine-like transporter.
Results: The recombinant, purified SLC25A29 transports arginine, lysine, and to a lesser extent ornithine and histidine but not carnitine and acylcarnitines.
Conclusion: SLC25A29 is a mitochondrial transporter for basic amino acids.
Significance: Another member of the mitochondrial carrier superfamily responsible for 12 monogenic diseases has been thoroughly characterized.
Keywords: Amino Acid Transport, Liposomes, Mitochondrial Transport, Recombinant Protein Expression, Reconstitution of Membrane Transporters, SLC25 Family, SLC25A29, Basic Amino Acids, Membrane Transport Protein, Mitochondrial Carrier
Abstract
The human genome encodes 53 members of the solute carrier family 25 (SLC25), also called the mitochondrial carrier family, many of which have been shown to transport carboxylates, amino acids, nucleotides, and cofactors across the inner mitochondrial membrane, thereby connecting cytosolic and matrix functions. In this work, a member of this family, SLC25A29, previously reported to be a mitochondrial carnitine/acylcarnitine- or ornithine-like carrier, has been thoroughly characterized biochemically. The SLC25A29 gene was overexpressed in Escherichia coli, and the gene product was purified and reconstituted in phospholipid vesicles. Its transport properties and kinetic parameters demonstrate that SLC25A29 transports arginine, lysine, homoarginine, methylarginine and, to a much lesser extent, ornithine and histidine. Carnitine and acylcarnitines were not transported by SLC25A29. This carrier catalyzed substantial uniport besides a counter-exchange transport, exhibited a high transport affinity for arginine and lysine, and was saturable and inhibited by mercurial compounds and other inhibitors of mitochondrial carriers to various degrees. The main physiological role of SLC25A29 is to import basic amino acids into mitochondria for mitochondrial protein synthesis and amino acid degradation.
Introduction
SLC25A29, encoded by the human SLC25A29 gene, is a member of the mitochondrial carrier family (1, 2). The previously characterized members of this family transport nucleotides, amino acids, carboxylic acids, coenzymes, and inorganic anions across the mitochondrial inner membrane. The importance of mitochondrial carriers is demonstrated by their wide distribution in all eukaryotes, their role in numerous metabolic pathways and cell functions, and the identification of several diseases caused by alterations of their genes (1–6). SLC25A29 is localized to mitochondria (7, 8), expressed in several tissues, including heart, brain, liver, and kidney (7, 8), and induced after partial hepatectomy or fasting (7). In 2003, Sekoguchi et al. (7) reported that murine Slc25a29 functions as carnitine/acylcarnitine transporter-like (CACL) because a low palmitoylcarnitine transport activity (pmol/30 min) was found in Escherichia coli and in mitochondria from NIH3T3 cells both expressing SLC25A29. Later, Camacho and Rioseco-Camacho (8) concluded that the human and mouse SLC25A29 proteins transport ornithine into mitochondria and designated them ornithine transporter isoform 3 (ORNT3) because their overexpression rescues the defect in radioactive ornithine incorporation into cellular protein in cultured fibroblasts from patients with hyperornithinemia-hyperammonemia-homocitrullinuria (HHH)2 syndrome. As the carnitine/acylcarnitine and ornithine carriers of multiple organisms are distinct subfamilies of mitochondrial carriers, which are characterized by different substrates and different structural hallmarks (5), we decided to perform a more in-depth study on the activity of SLC25A29.
In this study, we provide direct evidence that SLC25A29 is a mitochondrial transporter for basic amino acids with a preference for arginine and lysine. SLC25A29 was overexpressed in E. coli, and the gene product was purified, reconstituted in phospholipid vesicles, and shown to transport arginine, lysine, ornithine, and histidine with high specificity by both counter-exchange and uniport mechanisms. Carnitine and acylcarnitines were not transported by SLC25A29. A main function of SLC25A29 is to catalyze the entry of arginine, lysine, and histidine into mitochondria for mitochondrial protein synthesis.
EXPERIMENTAL PROCEDURES
Materials
Radioactive compounds were purchased from PerkinElmer, Inc., Scopus Research BV, Veenendaal, The Netherlands, and Hartmann Analytic, Braunschweig, Germany. d-Homoarginine was purchased from Molekula Ltd., Dorset, UK. All other compounds were purchased from Sigma-Aldrich.
Sequence Search and Analysis
Protein databases for metazoa, fungi, and plants were screened with the protein sequences of SLC25A29, the human ornithine carriers ORC1 and ORC2, the yeast ornithine carrier Ort1p, the human and yeast carnitine/acylcarnitine carriers CAC (SLC25A20) and Crc1p, respectively, and the Arabidopsis (Arabidopsis thaliana) basic amino acid carriers BAC1 and BAC2, using BLASTP. Multiple sequence alignments were made with ClustalW, and phylogenetic trees were constructed by the neighbor-joining method with MEGA5 (9).
Construction of the Expression Plasmids
The coding sequence for SLC25A29 (NM_001039355) was amplified by PCR from the cDNA of human neuroblastoma SH-SY5Y cells (ATCCID®: CRL-2266TM). The oligonucleotide primers were synthesized corresponding to the extremities of the coding sequence with additional NdeI and EcoRI sites. The coding sequences for SLC25A20 (NM_000387) and SLC25A24 (NM_213651) were amplified from human liver cDNA and human testis cDNA, respectively, as described previously (10–12). The amplified products were cloned into the pRUN (SLC25A29), pMW7 (SLC25A20), and pcDNA3 (SLC25A24) expression vectors and transformed into E. coli TOP10 (SLC25A29 and SLC25A20), and E. coli DH5α (SLC25A24) cells. Transformants were selected on ampicillin (100 μg/ml) and screened by direct colony PCR and by restriction digestion of purified plasmids. The sequences of the inserts were verified.
Bacterial Expression and Purification of SLC25A29, SLC25A20, and SLC25A24
The overproduction of SLC25A29, SLC25A20, and SLC25A24 as inclusion bodies in the cytosol of E. coli was accomplished as described previously (13), except that the host cells were E. coli Rosetta-gami B(DE3) (Novagen), E. coli C0214(DE3) (14), and E. coli M15(pREP4) (Qiagen) (15), respectively. Control cultures with the empty vector were processed in parallel. Inclusion bodies were purified on a sucrose density gradient (13) and washed at 4 °C first with Tris-EDTA buffer (10 mm Tris-HCl, 1 mm EDTA, pH 7.0), then with a buffer containing 3% Triton X-114 (w/v), 1 mm EDTA, 20 mm Na2SO4, and 10 mm HEPES-NaOH, pH 7.2, and lastly with the Tris-EDTA buffer, pH 7.0. Finally, SLC25A29, SLC25A20, or SLC25A24 was solubilized in 1.7% sarkosyl (w/v), and eventual small residues were removed by centrifugation (20,800 × g for 10 min at 4 °C).
Reconstitution of SLC25A29, SLC25A20, or SLC25A24 into Liposomes
The recombinant proteins in sarkosyl were diluted with a buffer containing 3% Triton X-114 (w/v), 1 mm EDTA, 20 mm Na2SO4, and 10 mm HEPES-NaOH, pH 7.2, to a final concentration of 0.4 mg/ml. Then, they were reconstituted into liposomes by cyclic removal of the detergent with a hydrophobic column of Amberlite beads (Fluka), as described in Ref. 16 with some modifications. The composition of the initial mixture used for reconstitution consisted of 7.5 μl of purified SLC25A29, SLC25A20, or SLC25A24 (3 μg of protein), 80 μl of 10% Triton X-114, 100 μl of 10% phospholipids in the form of sonicated liposomes (17), 10 mm substrate (except where otherwise indicated), 0.7 mg of cardiolipin, 20 mm HEPES-NaOH (pH 7.2), and water to a final volume of 700 μl. After vortexing, this mixture was recycled 13 times through the Amberlite column (3.5 × 0.5 cm) pre-equilibrated with a buffer containing 10 mm HEPES-NaOH, pH 7.2, and the substrate at the same concentration as in the starting mixture.
Transport Measurements
External substrate was removed from proteoliposomes on Sephadex G-75 columns pre-equilibrated with 10 mm HEPES-NaOH, pH 7.2 (buffer A). The eluted proteoliposomes were distributed in reaction vessels and used for transport measurements by the inhibitor-stop method (18). Transport at 25 °C was started by adding [3H]arginine or other indicated labeled compounds to substrate-loaded proteoliposomes (exchange) or to empty proteoliposomes (uniport). In both cases, transport was terminated by the addition of 20 mm pyridoxal 5′-phosphate and 0.1 mm mercuric chloride, which in combination inhibit the activity of SLC25A29 rapidly and completely. In controls, the inhibitors were added at the beginning together with the radioactive substrate. Finally, the external radioactivity was removed by a Sephadex G-75 column, and the radioactivity in the proteoliposomes was measured. The experimental values were corrected by subtracting control values. The initial transport rate was calculated from the radioactivity taken up by proteoliposomes after 4 min (in the initial linear range of substrate uptake). Alternatively, the initial transport rate was calculated from the time course of isotope equilibration (16). For efflux measurements, proteoliposomes containing 1 mm arginine or lysine were labeled with 5 μm [3H]arginine or [3H]lysine, respectively, by carrier-mediated exchange equilibration (16). After 50 min, the external radioactivity was removed by passing the proteoliposomes through Sephadex G-75 columns pre-equilibrated with buffer A. Efflux was started by adding unlabeled external substrate or buffer A alone to aliquots of proteoliposomes and terminated by adding the inhibitors indicated above. In some efflux experiments, proteoliposomes were labeled by adding 0.4 mm [3H]carnitine or [32P]phosphate in the initial reconstitution mixture; external radioactivity was removed immediately after their preparation, and efflux was performed as described above.
Other Methods
Proteins were analyzed by SDS-PAGE and stained with Coomassie Blue dye. The amount of pure SLC25A29 was estimated by laser densitometry of stained samples, using carbonic anhydrase as protein standard (19). The identity of purified SLC25A29 was assessed by matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) mass spectrometry of trypsin digests of the corresponding band excised from a Coomassie Blue-stained gel (20). To assay the protein incorporated into liposomes, the vesicles were passed through a Sephadex G-75 column, centrifuged at 300,000 × g for 30 min, and delipidated with organic solvents as described in Capobianco et al. (21). Then, the SDS-solubilized protein was determined by comparison with carbonic anhydrase in SDS gels. The share of incorporated protein was about 20% of the protein added to the reconstitution mixture.
RESULTS
Bacterial Expression of SLC25A29
SLC25A29 was expressed at high levels in E. coli Rosetta-gami B(DE3) cells (Fig. 1, lane 4). The gene product accumulated as inclusion bodies and was purified by centrifugation and washing (Fig. 1, lane 5). The apparent molecular mass of the recombinant protein was 32 kDa (calculated value with initiator methionine, 32.01 kDa). The protein was not detected in bacteria harvested immediately before induction of expression (Fig. 1, lanes 1 and 2) nor in cells harvested after induction but lacking the coding sequence in the expression vector (Fig. 1, lane 3). The identity of the purified protein was confirmed by MALDI-TOF mass spectrometry, and the yield of the purified protein was ∼40 mg/liter of culture.
FIGURE 1.
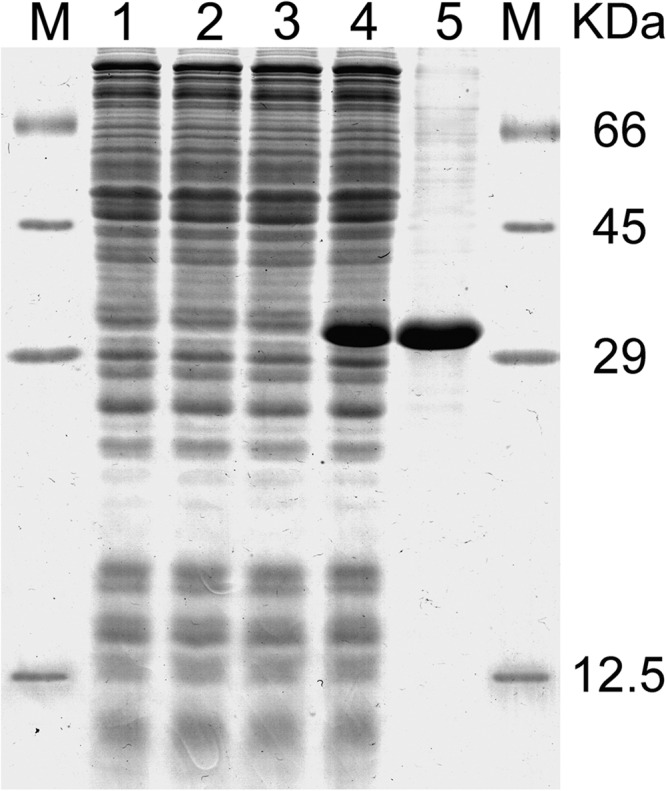
Expression of SLC25A29 in Escherichia coli and its purification. Proteins were separated by SDS-PAGE and stained with Coomassie Blue dye. Molecular mass markers (M) (bovine serum albumin, ovalbumin, carbonic anhydrase, and cytochrome c) are shown on the left and on the right. Lanes 1–4, E. coli Rosetta-gami B(DE3) cells containing the expression vector with (lanes 2 and 4) and without (lanes 1 and 3) the coding sequence of SLC25A29. Samples were taken immediately before (lanes 1 and 2) and 5 h after induction (lanes 3 and 4). The same number of bacteria was analyzed in each sample. Lane 5, purified SLC25A29 (about 20 μg) originated from the bacteria shown in lane 4.
Functional Characterization of Recombinant SLC25A29
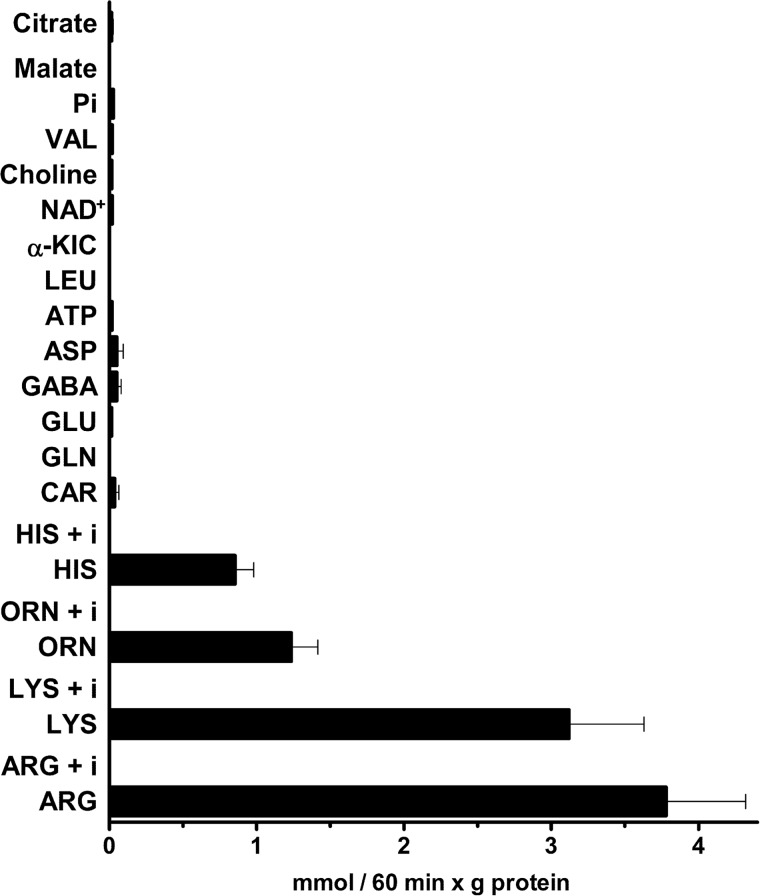
SLC25A29 was reconstituted into liposomes, and its transport activities for a variety of potential substrates were tested in homoexchange experiments (i.e. with the same substrate inside and outside). Using external and internal substrate concentrations of 0.4 and 10 mm, respectively, the reconstituted protein catalyzed the homoexchanges [3H]arginine/arginine and [3H]lysine/lysine efficiently and [3H]ornithine/ornithine and [14C]histidine/histidine to a lesser extent (Fig. 2). All of these homoexchanges were completely inhibited by a mixture of pyridoxal 5′-phosphate and HgCl2 (Arg + i, Lys + i, Orn + i, and His + i) (Fig. 2). However, despite the long incubation period (i.e. 60 min), negligible homoexchange activity was measured for carnitine (Car), Gln, Glu, γ-aminobutyric acid (GABA), Asp, ATP, Leu, α-ketoisocaproic acid, NAD+, choline, Val, Pi, malate, citrate (Fig. 2), oxoglutarate, and sulfate (not shown). Furthermore, no arginine/arginine exchange activity was detected when SLC25A29 had been boiled before incorporation into liposomes or when liposomes were reconstituted with sarkosyl-solubilized material from bacterial cells containing the expression vector either lacking the coding sequence for SLC25A29 or harvested immediately before the induction of expression. Likewise, no such activity was detected in liposomes reconstituted with two unrelated mitochondrial carriers, AGC1 and Sam5p (22, 23). Finally, the [3H]arginine/arginine, [3H]lysine/lysine, [3H]ornithine/ornithine, and [14C]histidine/histidine homoexchanges were nil using pure liposomes, i.e. without incorporated protein (results not shown).
FIGURE 2.
Homoexchange activities of various substrates in proteoliposomes reconstituted with SLC25A29. Transport was initiated by adding radioactive substrate (final concentration, 0.4 mm) to proteoliposomes preloaded internally with the same substrate (concentration, 10 mm). Where indicated (Arg + i, Lys + i, Orn + i, and His + i), the radioactive substrate was added together with 20 mm pyridoxal 5′-phosphate and 0.1 mm HgCl2. The reaction was terminated after 60 min. The data are means ± S.D. of at least three independent experiments. Differences between the arginine/arginine, lysine/lysine, ornithine/ornithine, and histidine/histidine homoexchanges and all the other homoexchanges were significant (p < 0.01, one-way ANOVA). Abbreviations: i, inhibitors (pyridoxal 5′-phosphate and HgCl2); α-KIC, α-ketoisocaproic acid; Pi, phosphate.
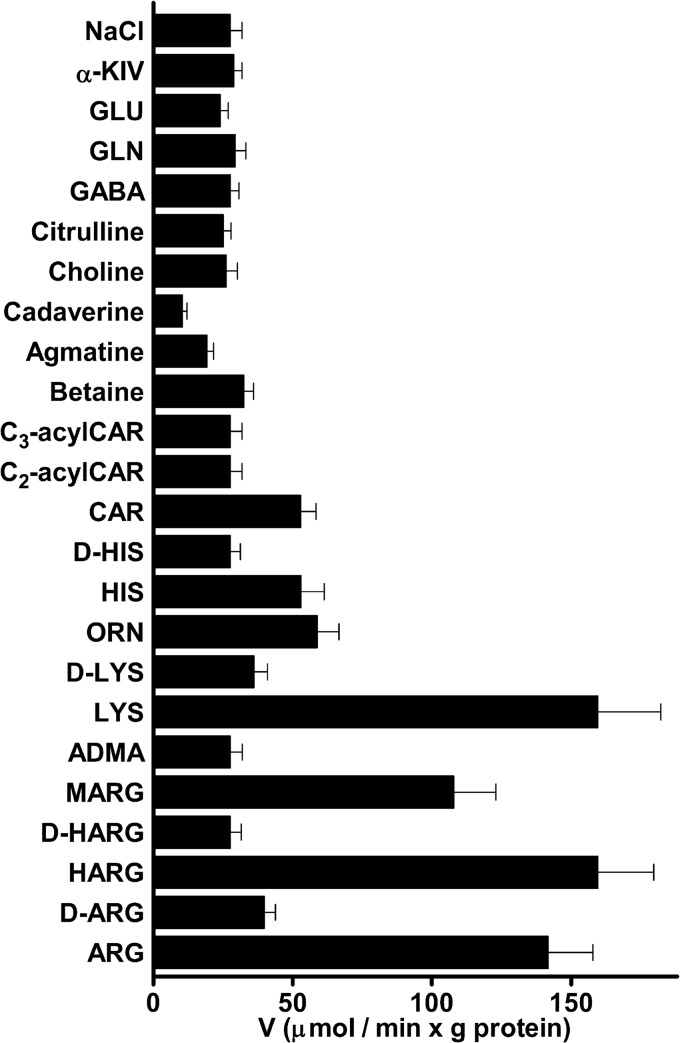
The substrate specificity of SLC25A29 was further investigated by measuring the initial rate of [3H]arginine uptake into proteoliposomes that had been preloaded with a high concentration (10 mm) of various potential substrates (Fig. 3). The highest activities of arginine exchange into proteoliposomes were observed with internal Arg, homoarginine, methylarginine, and Lys. Lower activities were found in the presence of internal Orn, His, and Car, and still lower with internal d-arginine and d-lysine. Virtually no exchange was detected with internal d-homoarginine, asymmetric dimethylarginine, d-histidine, acetylcarnitine, propionylcarnitine, betaine, choline, citrulline, GABA, Gln, Glu, α-ketoisovaleric acid, and NaCl (Fig. 3) and pyruvate, phosphate, sulfate, malate, citrate, α-oxoglutarate, aspartate, methionine, cysteine, valine, leucine, glutathione, S-adenosylmethionine, ADP, ATP, GDP, UDP, UTP, CDP, CTP, NAD+, FAD, coenzyme A, and thiamine diphosphate (not shown). In the presence of these latter substrates, the activity was virtually the same as the unidirectional activity observed without internal substrate (uniport, with NaCl present). Similar results were obtained using [3H]lysine instead of [3H]arginine and unlabeled arginine, homoarginine, lysine, ornithine, and histidine as internal counter-substrates (data not shown). Therefore, the substrate specificity of SLC25A29 is confined to basic amino acids if excluding carnitine (see below). In addition, the results of Fig. 3 show that the decarboxylated analogs of arginine and lysine, i.e. agmatine and cadaverine, inhibited the transport activity of reconstituted SLC25A29.
FIGURE 3.
Dependence of SLC25A29 activity on internal substrate. Liposomes reconstituted with SLC25A29 were preloaded internally with various substrates (concentration, 10 mm). Transport was started by adding 0.4 mm [3H]arginine and terminated after 4 min. The values are means ± S.D. of at least three independent experiments. Differences between the activities of arginine uptake with internal Arg, homoarginine (HARG), methylarginine (MARG), Lys, Orn, His, and Car and the activity with internal NaCl and no substrate were significant (p < 0.01, one-way ANOVA). Differences between the activity of arginine uptake with internal NaCl and the activities with internal cadaverine and agmatine were significant (p < 0.01 and p < 0.05, respectively, one-way ANOVA). Differences between the activities of arginine uptake with internal substrates other than those mentioned above and the activity with internal NaCl are not significant (p > 0.05, one-way ANOVA). Other abbreviations: d-HARG, d-homoarginine; ADMA, asymmetric dimethylarginine; C2-acylCar, acetylcarnitine; C3-acylCar, propionylcarnitine; α-KIV, α-ketoisovaleric acid.
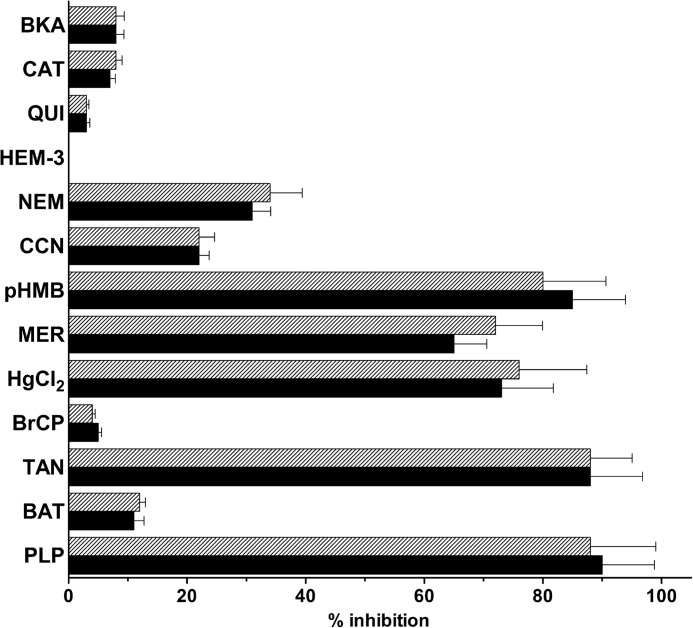
The effects of other mitochondrial carrier inhibitors on the arginine uniport uptake and the arginine/arginine exchange reactions catalyzed by reconstituted SLC25A29 were also examined. Both arginine uniport (hatched bars) and exchange (black bars) were markedly inhibited by pyridoxal 5′-phosphate, tannic acid, HgCl2, mersalyl, and p-hydroxymercuribenzoate and partially by N-ethylmaleimide and α-cyano-4-hydroxycinnamate (Fig. 4). In contrast, little or no inhibition was observed with bathophenanthroline, bromcresol purple, hemicholinium-3, quinine, carboxyatractyloside, and bongkrekic acid.
FIGURE 4.
Effect of inhibitors on the arginine uniport uptake and the arginine/arginine exchange by SLC25A29. Liposomes were reconstituted with SLC25A29 and preloaded internally with 10 mm arginine (black bars) or NaCl and no substrate (hatched bars). Transport was initiated by adding 0.4 mm [3H]arginine and terminated after 4 min. Thiol reagents and α-cyano-4-hydroxycinnamate were added 2 min before the labeled substrate; the other inhibitors were added together with the labeled substrate. The final concentrations of the inhibitors were 10 μm (CAT, carboxyatractyloside; BKA, bongkrekic acid), 20 mm (PLP, pyridoxal 5′-phosphate; BAT, bathophenanthroline), 0.2 mm (pHMB, p-hydroxymercuribenzoate; MER, mersalyl), 1 mm (NEM, N-ethylmaleimide; CCN, α-cyano-4-hydroxycinnamate), 0.1% (TAN, tannic acid), 0.4 mm (HEM-3, hemicholinium-3; QUI, quinine), 0.2 mm (BrCP, bromcresol purple), and 0.1 mm HgCl2. The data expressed as the percentage of inhibition are means ± S.D. of at least three independent experiments. Differences between the percentage of inhibition by pyridoxal 5′-phosphate, tannic acid, HgCl2, mersalyl and p-hydroxymercuribenzoate and the percentage of inhibition by N-ethylmaleimide and α-cyano-4-hydroxycinnamate were significant (p < 0.01, one-way ANOVA). Differences between the percentage of inhibition by N-ethylmaleimide and α-cyano-4-hydroxycinnamate and the percentage of inhibition by bathophenanthroline, bromcresol purple, hemicholinium-3, quinine, carboxyatractyloside, and bongkrekic acid were significant (p < 0.01, one-way ANOVA).
Kinetic Characteristics of Recombinant SLC25A29
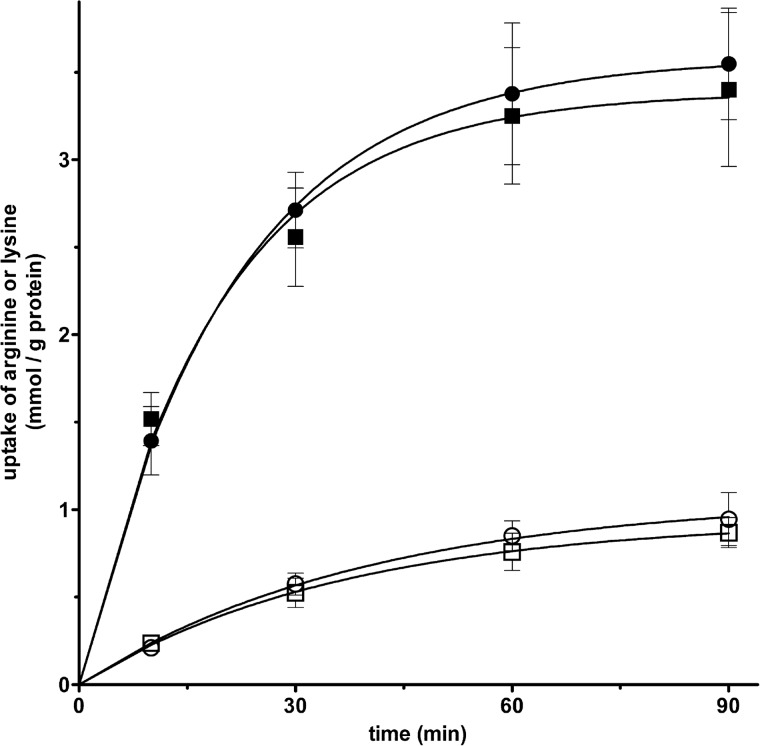
The time courses of uptake by proteoliposomes of 0.4 mm [3H]arginine (● and ○) and 0.7 mm [3H]lysine (■ and □) measured either as exchange (with 10 mm arginine or lysine, respectively, inside the proteoliposomes) or as uniport (without internal substrate) are shown in Fig. 5. All curves fitted a first-order rate equation with rate constants (k) for the exchange (● and ■) and uniport (○ and □) reactions of ∼0.05 and 0.03 min−1, respectively. The initial rates of arginine and lysine uptake (the product of k and intraliposomal quantity of arginine or lysine taken up at equilibrium (16)) were about 145 and 25 μmol/min/g of protein for the exchange and uniport reactions, respectively.
FIGURE 5.
Kinetics of [3H]arginine or [3H]lysine transport in liposomes reconstituted with SLC25A29. [3H]arginine (0.4 mm) or [3H]lysine (0.7 mm) was added to proteoliposomes containing 10 mm arginine (●) or lysine (■) (exchange) or 10 mm NaCl and no substrate (uniport, (○) for [3H]arginine or (□) for [3H]lysine). The data are means ± S.D. of three independent experiments. Differences between arginine and lysine exchanges and uniport uptakes were significant (p < 0.01, one-way ANOVA).
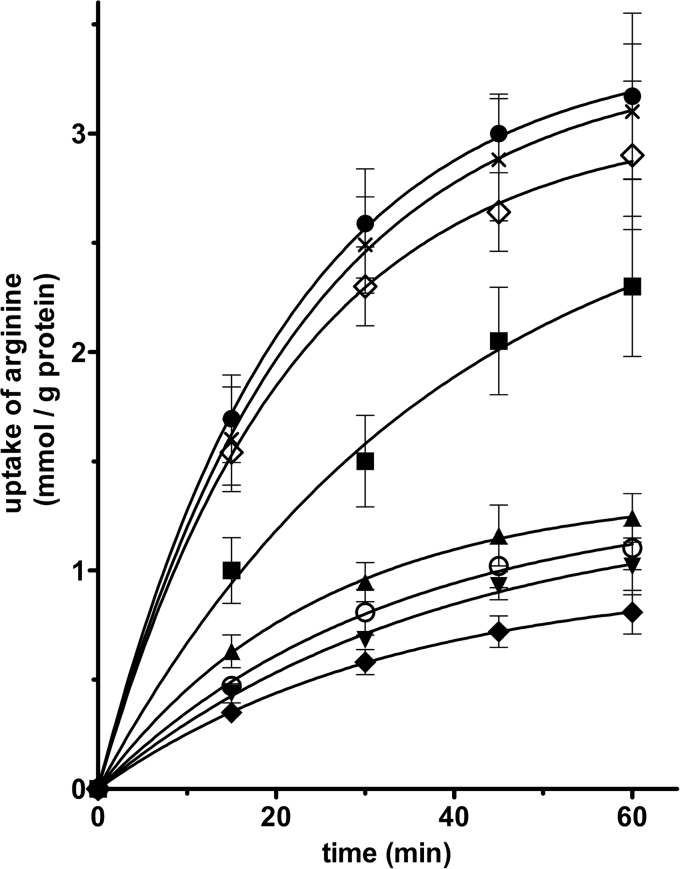
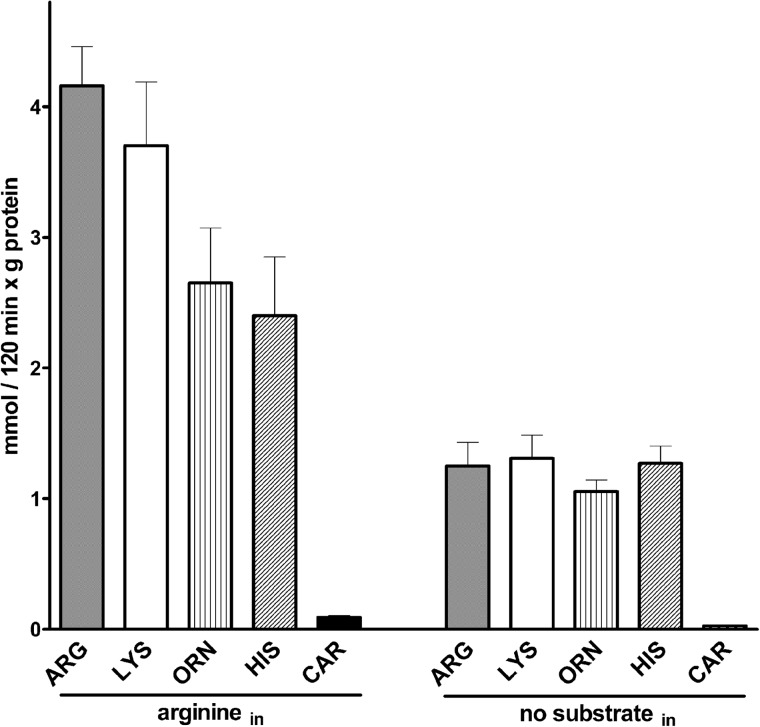
The results of Figs. 3 and 5 indicate that SLC25A29 catalyzes a substantial unidirectional transport (uniport) of arginine and lysine besides the arginine/arginine and lysine/lysine exchanges. Fig. 6 compares the time courses of [3H]arginine uptake by SLC25A29-reconstituted liposomes containing no substrate (uniport) or potential substrates of SLC25A29. The amount of labeled arginine taken up by SLC25A29-reconstituted liposomes in the presence of internal arginine (●), lysine (Χ), homoarginine (♢), or methylarginine (■) is greater than that entering in the presence of internal ornithine (▴), histidine (○), or carnitine (▾) and in the absence of any internal substrate (♦). These results indicate that arginine, lysine, homoarginine, and methylarginine are exchanged by SLC25A29 efficiently, whereas ornithine, histidine, and carnitine are exchanged rather poorly. To verify whether the latter substrates are transported by SLC25A29, we set out to measure the uptake of [3H]ornithine, [14C]histidine, [3H]carnitine, [3H]lysine, and [3H]arginine into proteoliposomes containing arginine (exchange) or no substrate (uniport) (Fig. 7). It was found that after a 120-min incubation period, labeled ornithine (line bars), histidine (hatched bars), lysine (white bars), and arginine (gray bars) entered the SLC25A29-reconstituted liposomes either in exchange with arginine or as unidirectional transport, whereas [3H]carnitine (black bars) did not enter with or without internal substrate. Furthermore, under both conditions, the uptake of labeled ornithine, histidine, lysine, and arginine into pure liposomes was virtually nil even after 120-min incubation (results not shown). Therefore, ornithine and histidine are transported by SLC25A29, although much less efficiently than labeled lysine and arginine, whereas [3H]carnitine is not.
FIGURE 6.
Time courses of [3H]arginine uptake by SLC25A29-reconstituted liposomes containing no substrate or potential substrates of SLC25A29. Proteoliposomes were preloaded internally with 10 mm arginine (●), lysine (Χ), homoarginine (♢), methylarginine (■), ornithine (▴), histidine (○), carnitine (▾), and 10 mm NaCl and no substrate (♦). Transport was initiated by adding 0.4 mm [3H]arginine and terminated at the indicated times. The data are means ± S.D. of at least three independent experiments. Differences between arginine uptake (initial rate and after 60 min) with internal arginine, lysine, homoarginine, and methylarginine and arginine uptake with internal ornithine, histidine, and carnitine were significant (p < 0.01, one-way ANOVA). Differences in arginine uptake with internal ornithine, histidine, and carnitine were not significant (p > 0.05, one-way ANOVA).
FIGURE 7.
Uptake by exchange or uniport of labeled basic amino acids and carnitine into SLC25A29-reconstituted liposomes. Proteoliposomes were preloaded internally with 10 mm arginine (left) or 10 mm NaCl and no substrate (right). Transport was initiated by adding 1 mm [3H]arginine (gray bars), [3H]lysine (white bars), [3H]ornithine (lined bars), [14C]histidine (hatched bars), or [3H]carnitine (black bars) and terminated after 120 min. The data are means ± S.D. of at least three independent experiments.
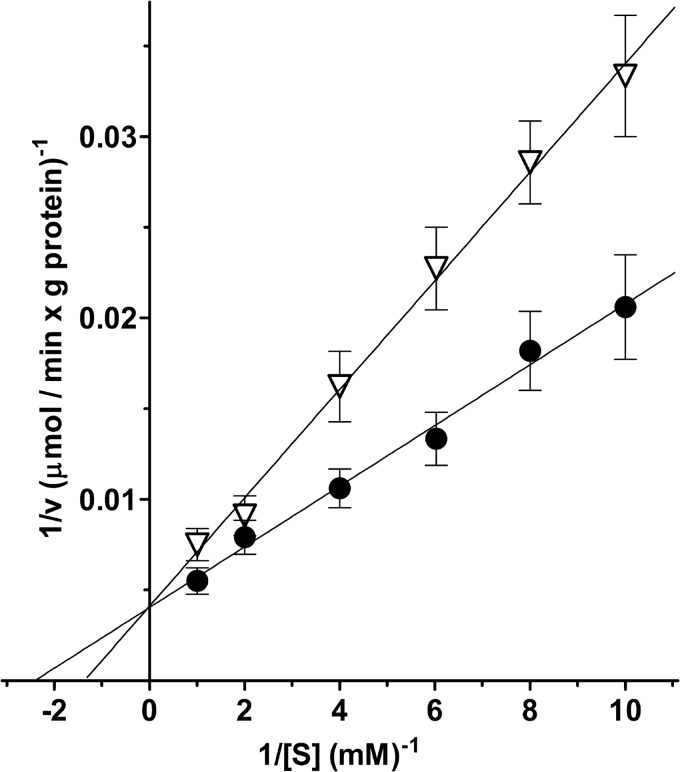
The kinetic constants of SLC25A29 were determined from the initial transport rate of the homoexchanges arginine/arginine and lysine/lysine at various external labeled substrate concentrations and a constant internal substrate concentration of 10 mm. Linear functions were obtained in double-reciprocal plots for both homoexchanges. The lines intersected the ordinate close to a common point (Fig. 8). For arginine (●) and lysine (▿), the transport affinities (Km) were 0.42 ± 0.04 and 0.71 ± 0.10 mm, respectively. The average Vmax value was 237 ± 48 μmol/min/g of protein. Ornithine, histidine, homoarginine, methylarginine, cadaverine, and agmatine were competitive inhibitors of the [3H]arginine/arginine exchange as they increased the apparent Km without changing the Vmax (not shown). The Ki values of these compounds for SLC25A29 are the following: 1.6 ± 0.2 mm (ornithine), 2.7 ± 0.3 mm (histidine), 0.45 ± 0.07 mm (homoarginine), 1.0 ± 0.2 mm (methylarginine), 4.8 ± 0.7 mm (cadaverine), and 6.2 ± 1.0 mm (agmatine). By contrast, the [3H]arginine/arginine exchange, measured as described in the legend for Fig. 3, was virtually unaffected by carnitine, acetylcarnitine, and propionylcarnitine up to 10 mm. Importantly, the Km values of arginine and lysine uptake in SLC25A29-reconstituted liposomes containing no substrate, i.e. determined in uniport experiments as Lineweaver-Burk plots, were 0.47 ± 0.04 and 0.69 ± 0.06 mm, respectively. These values are similar to those evaluated from arginine/arginine and lysine/lysine homoexchanges (see above). By contrast, the average Vmax of arginine and lysine uptake measured as uniport is 3.1-fold lower than that derived from homoexchanges, i.e. 77 ± 11 μmol/min/g of protein.
FIGURE 8.
Lineweaver-Burk plots of the [3H]arginine/arginine and [3H]lysine/lysine exchanges in liposomes reconstituted with SLC25A29. [3H]arginine (●) or [3H]lysine (▿) was added at the concentrations indicated to proteoliposomes containing 10 mm arginine (●) or 10 mm lysine (▿). The reaction time was 4 min. The data are means ± S.D. of four independent experiments.
Efflux of Arginine and Lysine from SLC25A29-reconstituted Liposomes
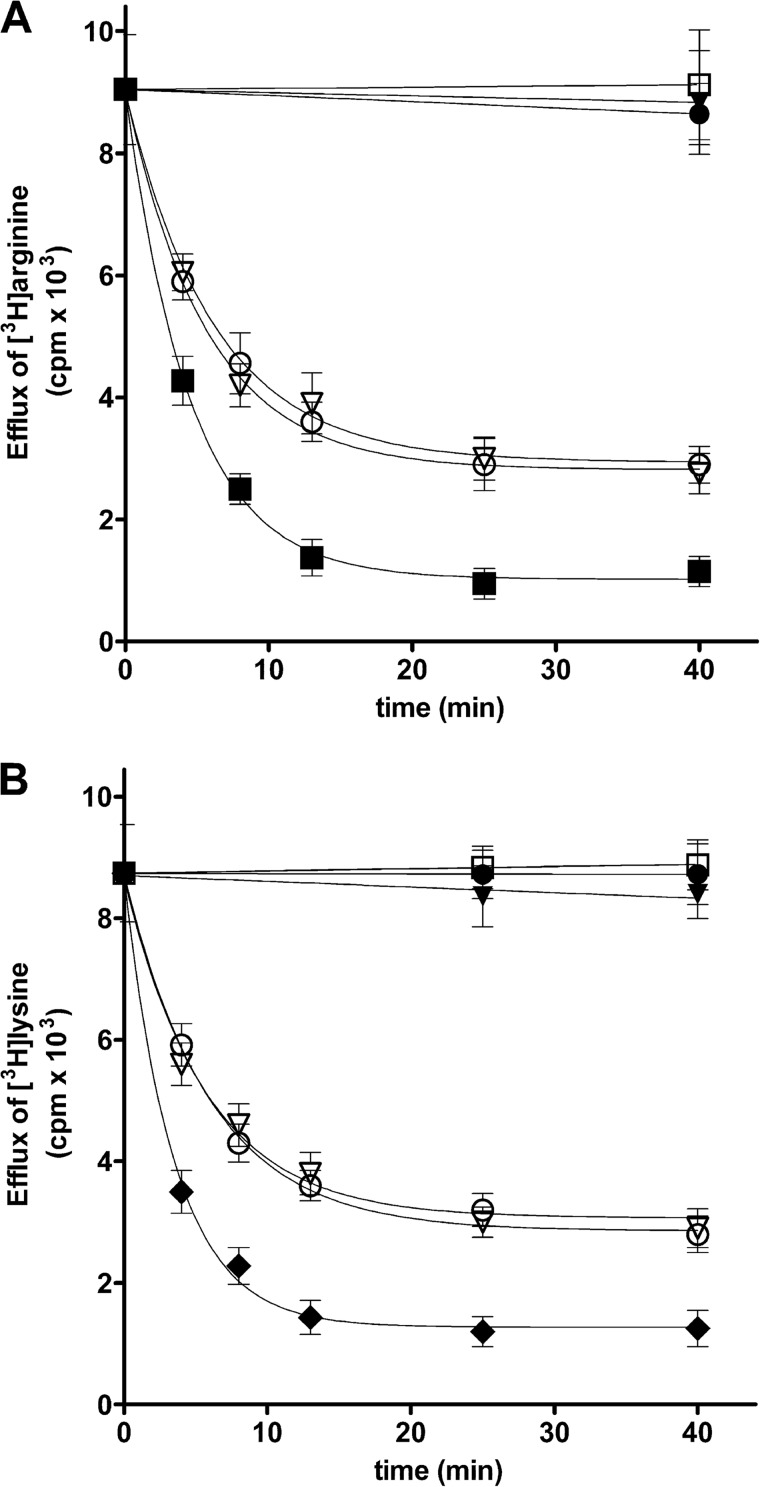
The uniport mode of transport by SLC25A29 was further investigated by measuring the efflux of [3H]arginine and [3H]lysine from preloaded active proteoliposomes because it provides a more convenient assay for unidirectional transport (16). As shown in Fig. 9, A and B, a substantial efflux of [3H]arginine and [3H]lysine from SLC25A29-reconstituted liposomes was observed with the addition of buffer A alone (○), i.e. in the absence of external substrate. Notably, in the presence of palmitoylcarnitine (▿), the rate and extent of arginine and lysine efflux were the same as those with buffer alone, indicating that palmitoylcarnitine does not exchange with arginine or lysine in liposomes reconstituted with SLC25A29. By contrast, a greater efflux of arginine and lysine occurred upon the addition of unlabeled arginine (■) and lysine (♦), respectively. Both effluxes, i.e. with and without external substrate, were prevented completely if the inhibitors pyridoxal 5′-phosphate and HgCl2 (□, ▾, and ●) were present from the beginning of the proteoliposome incubation (time 0). The finding that the amount of internal [3H]arginine and [3H]lysine did not decrease by the combined addition of palmitoylcarnitine and the inhibitors at time 0 as compared with the inhibitors alone shows that palmitoylcarnitine, up to 4 mm, does not behave as a detergent under these experimental conditions. Similar results were obtained using miristoylcarnitine, octanoylcarnitine, and propionylcarnitine instead of palmitoylcarnitine (data not shown).
FIGURE 9.
Efflux of [3H]arginine or [3H]lysine from liposomes reconstituted with SLC25A29 and preloaded internally with 1 mm arginine or lysine. The internal substrate pool was labeled with [3H]arginine or [3H]lysine by carrier-mediated exchange equilibration. Then, the proteoliposomes were passed through Sephadex G-75. The efflux of [3H]arginine (A) or [3H]lysine (B) was started by adding buffer A alone (○) or in buffer A (4 mm palmitoylcarnitine (▿), 5 mm arginine (■, in A) or 5 mm lysine (♦, in B), 20 mm pyridoxal 5′-phosphate and 0.1 mm HgCl2 (●), 20 mm pyridoxal 5′-phosphate, 0.1 mm HgCl2, and 4 mm palmitoylcarnitine (▾), and 20 mm pyridoxal 5′-phosphate, 0.1 mm HgCl2, and 5 mm arginine (in A) or lysine (in B) (□)). The data are means ± S.D. of four independent experiments.
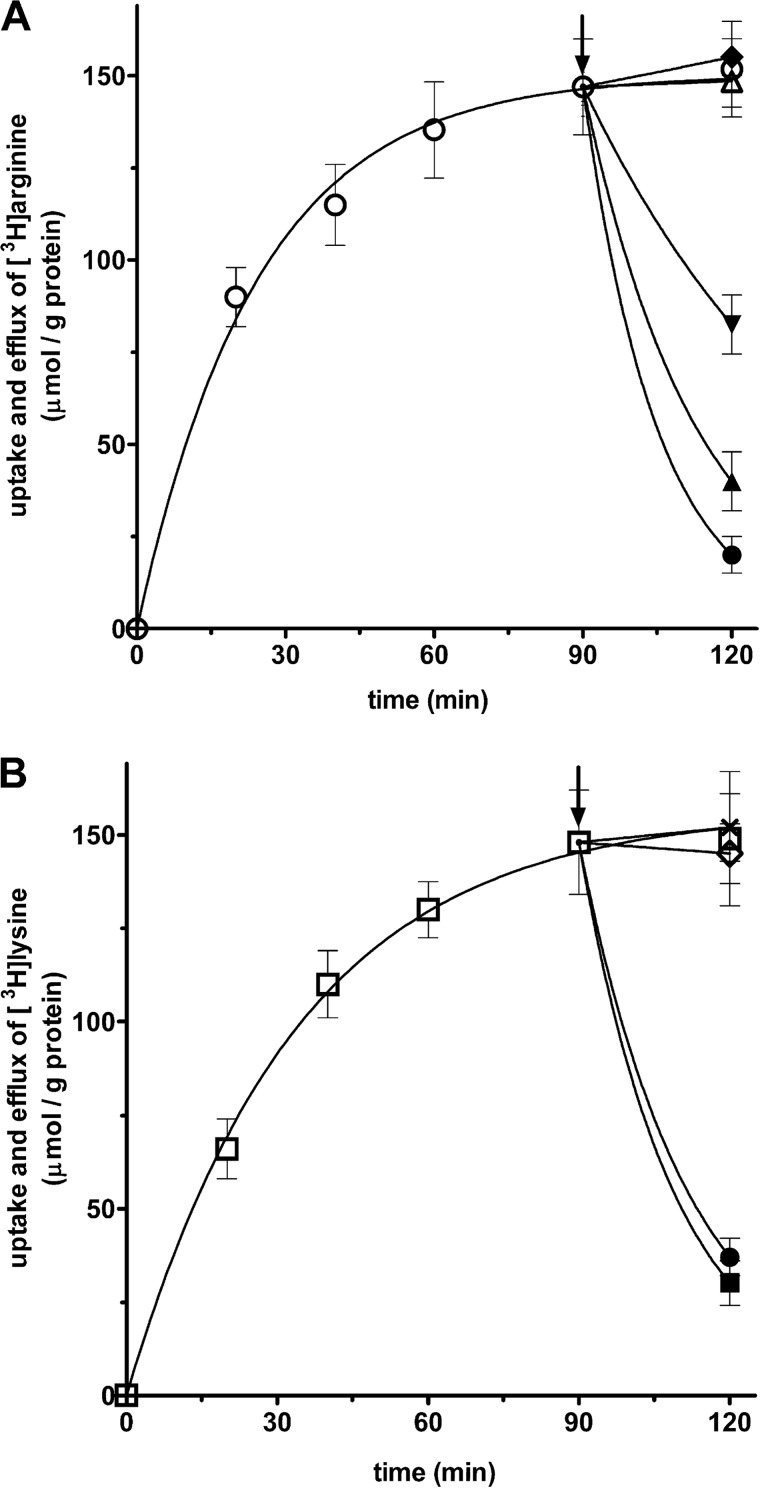
In another set of experiments, the efflux of [3H]arginine or [3H]lysine from SLC25A29-reconstituted liposomes was investigated after their uptake in the absence of internal substrate (Fig. 10). The addition of 2 mm arginine (●), methylarginine (▴), or ornithine (▾) (Fig. 10A) or 2 mm arginine (●) or lysine (■) (Fig. 10B) after 90-min incubation, when radioactive uptake by the proteoliposomes had approached equilibrium, caused an extensive efflux of intraliposomal radioactivity. This efflux indicates that the [3H]arginine or [3H]lysine taken up by uniport via SLC25A29 is released by exchange for the externally added substrates of SLC25A29. By contrast, the addition of palmitoylcarnitine (♦) and octanoylcarnitine (▵) (Fig. 10A), propionylcarnitine (Χ), and miristoylcarnitine (♢) (Fig. 10B) or glutamate, leucine, and agmatine (not shown) up to 4 mm had no effect as compared with controls (○ in A, and □ in B). These results clearly indicate that arginine, lysine, and ornithine are substrates for reconstituted SLC25A29, whereas the carnitine esters, glutamate, leucine, and agmatine are not.
FIGURE 10.
Uptake of [3H]arginine and [3H]lysine into unloaded proteoliposomes and their efflux after the addition of unlabeled substrates. 50 μm [3H]arginine (A) or 80 μm [3H]lysine (B) was added to liposomes reconstituted with SLC25A29 containing 10 mm NaCl and no substrate. The arrows indicate the addition of 2 mm arginine (●), 2 mm methylarginine (▴), 2 mm ornithine (▾), 4 mm palmitoylcarnitine (♦), 4 mm octanoylcarnitine (▵), or none (○) (in A) and 2 mm lysine (■), 2 mm arginine (●), 4 mm propionylcarnitine (Χ), 4 mm miristoylcarnitine (♢), or none (□) (in B). The data are means ± S.D. of at least three independent experiments.
Lack of [3H]Carnitine Efflux from SLC25A29-reconstituted Liposomes
The results of Figs. 2 and 7 indicate that [3H]carnitine, a known substrate along with its esters of the carnitine/acylcarnitine carrier, is poorly or not transported at all by SLC25A29. However, Figs. 3 and 6 show that the amount of [3H]arginine taken up by SLC25A29-reconstituted liposomes containing 10 mm carnitine is little yet significantly more than that in the absence of any internal substrate. This finding can be interpreted to indicate that either SLC25A29 exchanges labeled arginine with internal carnitine very poorly or internal carnitine minimally stimulates the uptake of arginine into SLC25A29-reconstituted liposomes. To further investigate whether carnitine is exchanged with external arginine or carnitine in SLC25A29-reconstituted liposomes, we tested the potential of [3H]carnitine efflux from active proteoliposomes via SLC25A29. To this end, we included [3H]carnitine in the reconstitution mixture of SLC25A29-containing liposomes and measured the efflux of labeled carnitine from [3H]carnitine-preloaded, SLC25A29-reconstituted liposomes in the presence of external carnitine, arginine, or no substrate as compared with the radioactivity entrapped in the proteoliposomes treated immediately after preparation (at time 0) with the SLC25A29 inhibitors pyridoxal 5′-phosphate and HgCl2. After a 90-min incubation period with or without 10 mm external arginine or carnitine, the radioactivity of [3H]carnitine in the proteoliposomes was virtually the same as that measured at time 0 (Table 1), indicating that [3H]carnitine did not efflux from liposomes reconstituted with SLC25A29 under any condition. For controls, we employed liposomes that were reconstituted with recombinant SLC25A20 or SLC25A24 instead of SLC25A29 using the same experimental conditions described for the reconstitution of SLC25A29 into liposomes. SLC25A20 is the mitochondrial carnitine/acylcarnitine carrier (24–26), whereas SLC25A24 is one of the four mitochondrial isoforms of the ATP-Mg/Pi carrier, which catalyzes the exchange of adenine nucleotides for inorganic phosphate and also the physiologically unproductive phosphate/phosphate exchange (12). Contrary to what was observed with liposomes reconstituted with SLC25A29, approximately two-thirds of the intraliposomal [3H]carnitine or [32P]phosphate effluxed from SLC25A20-reconstituted liposomes in the presence of external carnitine (but not arginine) or from SLC25A24-reconstituted liposomes in the presence of phosphate, respectively. Presumably, the remaining radioactivity inside the proteoliposomes after 90-min incubation was that present in the matrix of the liposomes in which the transporter had not been incorporated. These findings show that carnitine preloaded into SLC25A29-reconstituted liposomes does not exchange with external arginine or carnitine; most likely, at high concentrations, intraliposomal carnitine is able to interact with SLC25A29, increasing the uptake of arginine by reconstituted liposomes.
TABLE 1.
Lack of efflux of [3H]carnitine from SLC25A29-reconstituted liposomes
Liposomes were reconstituted with SLC25A29 or SLC25A20 and 0.4 mm [3H]carnitine or with SLC25A24 and 0.4 mm [32P]phosphate. The reaction was initiated by adding 10 mm carnitine or arginine to SLC25A29- or SLC25A20-reconstituted liposomes, and 10 mm phosphate to SLC25A24-reconstituted liposomes or no substrate to each type of liposome. At the indicated times, the reaction was terminated (quenched) by adding the appropriate stop inhibitors (pyridoxal 5′-phosphate and HgCl2 to SLC25A29-reconstituted liposomes, 1.5 mm N-ethylmaleimide to SLC25A20-reconstituted liposomes, or 0.2% tannic acid to SLC25A24-reconstituted liposomes). The values are means ± S.D. of four independent experiments. Differences between the amount of [3H]carnitine or [32P]phosphate inside SLC25A20-reconstituted or SLC25A24-reconstituted liposomes after 90 min in the presence of external carnitine or phosphate, respectively, and the amounts inside the proteoliposomes at time 0 or after 90 min with or without external substrates were significant (p < 0.01, one-way ANOVA). N.D., not determined.
| Quenching time (min) | Additions | SCL25A29 liposomes ([3H]carnitine (cpm/mg of phospholipids)) | SLC25A20 liposomes ([3H]carnitine (cpm/mg of phospholipids)) | SLC25A24 liposomes ([3H]phosphate (cpm/mg of phospholipids)) |
|---|---|---|---|---|
| 0 | None | 8844 ± 363 | 8912 ± 289 | 13665 ± 474 |
| 90 | None | 8977 ± 234 | 9134 ± 424 | 13599 ± 463 |
| 90 | Carnitine | 8737 ± 262 | 2798 ± 148 | N.D. |
| 90 | Arginine | 9175 ± 258 | 8698 ± 257 | N.D. |
| 90 | Pi | N.D. | N.D. | 4659 ± 215 |
Phylogenetic Analysis
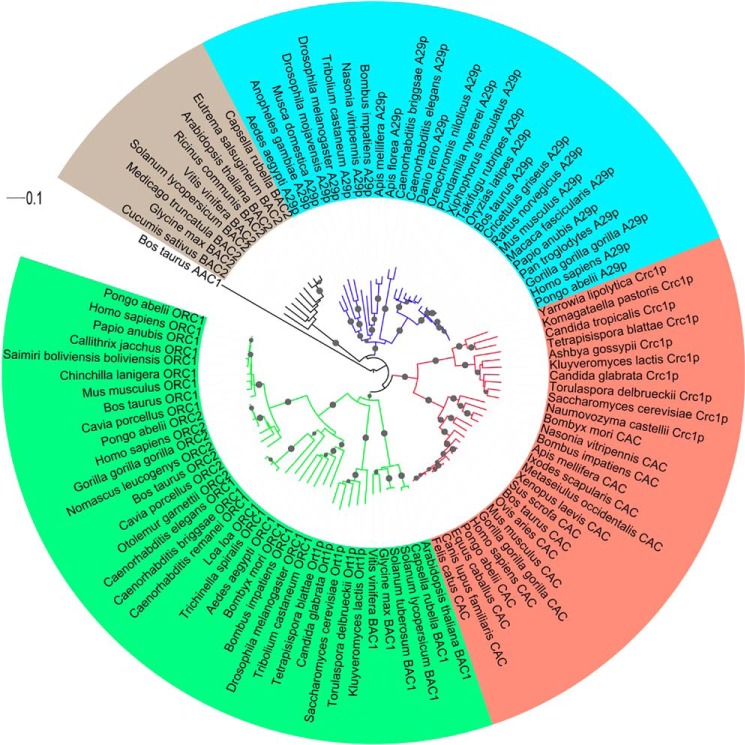
A previous phylogenetic tree of all mitochondrial carriers in Homo sapiens, Saccharomyces cerevisiae, and A. thaliana (5) revealed that SLC25A29 is located in a branch (containing BAC2 and the unidentified SLC25A45 and SLC25A47 besides SLC25A29) of a clade grouping the ornithine carriers (human ORC1–2 and yeast Orc1p) (27, 28), the plant carriers for basic amino acids (BAC1 and BAC2) (29), and the carnitine/acylcarnitine carriers (human CAC or SLC252A20 and yeast Crc1p (10, 30)). The present targeted phylogenetic analysis of 102 mitochondrial carriers from metazoa, fungi, and plants, related to SLC25A29, ORC1–2, Orc1p, BAC1–2, CAC and Crc1p, consistently identified four well separated sequence clades for SLC25A29, ornithine, carnitine/acylcarnitine, and BAC2 carriers (Fig. 11). More specifically, the ornithine and carnitine/acylcarnitine sequences were phylogenetically closer to each other than to the SLC25A29 sequences. Therefore, SLC25A29 also differs from the ornithine and carnitine/acylcarnitine carriers phylogenetically, although all these proteins share a common ancestral sequence. Furthermore, SLC25A29 orthologs based on phylogenetic relationships are found only in metazoan species, whereas the orthologs of ornithine and carnitine/acylcarnitine carriers are also found in fungi and/or plants (Fig. 11). This observation suggests a more recent evolutionary acquisition of SLC25A29 as compared with the other two carriers, although a double loss of SLC25A29 orthologs in fungi and plant lineages cannot be excluded. Finally, to our knowledge, none of the SLC25A29 orthologs reported in Fig. 11 have been characterized biochemically.
FIGURE 11.
Phylogenetic tree of SLC25A29, ornithine, carnitine/acylcarnitine and plant basic amino acid carriers from some metazoa, fungi, and plants. The tree was constructed using 102 sequences whose accession number IDs are reported as follows: NP_001034444.1, XP_004055738.1, XP_510163.3, XP_002825151.2, XP_005562260.1, XP_003902313.1, XP_003503758.1, NP_001010958.1, NP_001071339.1, NP_851845.1, NP_001025408.1, XP_003962766.1, XP_003440688.1, XP_005801326.1, XP_005723914.1, XP_004082289.1, XP_001659279.1, XP_319886.4, XP_970813.1, XP_001605887.1, XP_003493430.1, XP_005178048.1, XP_001121709.2, XP_003690819.1, XP_002004109.1, NP_609380.1, XP_002637782.1, NP_506621.1, XP_004235715.1, XP_004141775.1, XP_003543644.1, XP_002284619.1, NP_178108.1, XP_006389712.1, XP_006301202.1, XP_002516060.1, XP_003604475.1, NP_180938.2, XP_006294643.1, XP_004248107.1, XP_006361030.1, XP_003528461.1, XP_002280023.1, NP_114153.1, XP_003266564.1, XP_004042747.1, XP_002816047.1, XP_003782159.1, NP_001137558.1, XP_003477287.1, XP_974555.1, XP_003490747.1, XP_004927907.1, NP_001259423.1, XP_001653099.1, XP_003381554.1, XP_002648990.1, NP_498094.1, XP_003092176.1, XP_003140440.1, NP_055067.1, XP_003913850.1, XP_002824250.1, XP_003934443.1, XP_005387690.1, XP_002749041.1, XP_003477487.2, NP_001039791.1, NP_851842.1, NP_014773.4, XP_448670.1, XP_003678868.1, XP_004177581.1, XP_454073.1, NP_001088580.1, XP_002413523.1, XP_003747410.1, XP_004924428.1, XP_001604306.1, XP_003493905.1, XP_624739.1, NP_000378.1, XP_004034154.1, NP_001126542.1, XP_863142.1, XP_003982254.1, XP_001494767.2, NP_001071404.1, NP_001120749.1, XP_003483226.1, NP_065266.1, NP_014743.1, XP_003675139.1, XP_003682160.1, XP_445168.1, NP_983832.1, XP_452799.1, XP_004178505.1, XP_002491830.1, XP_501359.1, and XP_002546098.1. The figure also indicates the new designations of the sequences, based on the evolutionary relationships disclosed in this study. Nodes with bootstrap values >90% for 1,000 replicates are indicated with black dots. The clades in green, red, blue, and gray contain ornithine, carnitine/acylcarnitine, SLC25A29, and plant basic amino acid BAC2 carriers, respectively. The Bos taurus ADP/ATP carrier (AAC1; 1okc.pdb) has also been included in the tree as an outlier. Abbreviations: A29, SLC25A29; BAC1, basic amino acid carrier 1 from plants; BAC2, basic amino acid carrier 2 from plants; ORC1 and ORC2, isoforms 1 and 2 of the ornithine carrier; Crc1p, carnitine/acylcarnitine carrier from fungi; and Ort1p, ornithine carrier from fungi (Crc1p and Ort1p are designations of proteins applied to fungi).
DISCUSSION
This study further demonstrates the utility of overexpressing mitochondrial carriers in E. coli or S. cerevisiae and reconstituting them into liposomes to study their activity (see Refs. 1–3 and 5 for reviews). The transport properties and kinetic characteristics of recombinant SLC25A29, together with its localization to mitochondria reported before (7, 8), demonstrate that this protein is a mitochondrial transporter of basic amino acids with a preference for arginine and lysine. In contrast to the great majority of mitochondrial carriers, which are obligatory exchangers (31–37), SLC25A29 catalyzes a substantial uniport besides exchange of substrates. In this respect, SLC25A29 resembles the carriers for phosphate (38), glutamate (39), and carnitine/acylcarnitine (40), which are also capable of mediating uniport. All these carriers, including SLC25A29, operate exchanges in vitro at a much higher rate than uniports (from 2.8- to 10-fold higher). However, with the exception of the carnitine/acylcarnitine carrier, which catalyzes both uniport and exchange in vivo, the phosphate and glutamate carriers and most likely SLC25A29 mediate only uniport physiologically because the exchanges they may operate are nonproductive. Of note, the specific activity of reconstituted SLC25A29 is comparable with the activities of other SLC25 proteins (19, 23, 41, 42), and the Km values of arginine and lysine for SLC25A29 are lower or not much higher than their cytosolic concentrations (43, 44).
As previously inferred by complementation experiments (8), our results prove that SLC25A29 transports ornithine, although with a much lower affinity than arginine and lysine. Therefore, SLC25A29 is not the human ornithine transporter isoform 3 (ORNT3) (8). In this respect, it is worth mentioning that the percentage of identical amino acids between SLC25A29 and the two human isoforms of the ornithine carrier (31%) is not much higher than the basic homology existing between the different members of the mitochondrial carrier family. Furthermore, the percentage of identical amino acids between known isoforms of the same carrier in humans varies from 65% (glutamate carrier) to 92% (ADP/ATP carrier). The present data are also not consistent with the previous conclusion that SLC25A29 functions as carnitine/acylcarnitine transporter-like (CACL) (7). Several observations demonstrate that none of the tested carnitine/acylcarnitine carrier substrates, including acetylcarnitine, propionylcarnitine, octanoylcarnitine, miristoylcarnitine, and palmitoylcarnitine, are transported by SLC25A29 in exchange for arginine or lysine. Furthermore, [3H]carnitine is neither taken up nor effluxed by or from SLC25A29-reconstituted liposomes in the presence or absence of unlabeled arginine or carnitine as counter-substrate. In addition, no SLC25A29-mediated carnitine/acylcarnitine transport activity was detected when the influence of the carrier environment was investigated by changing the phospholipid composition extensively during proteoliposome reconstitution (data not shown). Instead, both our transport and our bioinformatics data provide evidence for the conclusion that SLC25A29, ornithine and carnitine/acylcarnitine carriers are different subfamilies of the mitochondrial carrier family, although they may share some of their substrates. Indeed, a certain degree of substrate overlapping is a common phenomenon among mitochondrial carriers (45).
Because mitochondria are equipped in their matrix with the entire machinery required for the synthesis of 13 proteins encoded by mtDNA (cytochrome b, 7 subunits of respiratory complex I, 3 subunits of cytochrome c oxidase and 2 subunits of ATP synthase), a primary physiological role of SLC25A29 is to catalyze uptake of the basic amino acids arginine, lysine, and histidine into mitochondria for mitochondrial protein synthesis. This role is consistent with the content of basic amino acids in the mtDNA-encoded proteins, which varies between 2.87% for subunit 6 of complex I and 13.2% for subunit 8 of ATP synthase, with a mean value of 7.25%. It should be noted that SLC25A29 is the seventh transporter for amino acids identified until now among the 53 members of the human mitochondrial carrier family (2, 46). The other known mitochondrial carriers for amino acids are the two isoforms of the glutamate carrier (GC1 and GC2), the two isoforms of the aspartate-glutamate carrier (AGC1 and AGC2), and the two isoforms of the ornithine carrier (ORC1 and ORC2), which are transporters for acidic or basic amino acids (27, 39, 47). However, the presence of structural characteristics typical of the amino acid class of mitochondrial carriers, described by Robinson and Kunji (48), implies that there are additional mitochondrial carriers for amino acids encoded by the human genome: i.e. SLC25A38, SLC25A39, SLC25A40, SLC25A45, SLC25A47, and SLC25A48, whose transported substrates are not yet known.
Another role of SLC25A29 is to provide the common substrate arginine to arginase II and mitochondrial NOS (49–51). In contrast with arginase I, which is the hepatic isoform, arginase II is mainly extrahepatic and is localized in mitochondria (49). Notably, arginase II has been shown to play a critical role in the pathophysiology of atherosclerotic endothelial dysfunction (52). Furthermore, arginase II activity regulates mitochondrial NOS-derived production of NO (53), which exerts important biological functions by reversibly inhibiting cytochrome c oxidase and by reacting with mitochondrial thiol-containing proteins and with superoxide anion to generate peroxynitrite (50, 54). Similarly, intramitochondrial lysine is the substrate of the first enzyme (aminoadipate-semialdehyde synthase) of the main metabolic route of lysine degradation in upper eukaryotes, the saccharopine pathway, which is a mitochondrial pathway leading to the formation of acetyl-CoA (55).
Finally, as SLC25A29 transports lysine with high efficiency, this protein is probably responsible for importing this cationic amino acid into mitochondria in patients with HHH syndrome, which is caused by a defective ornithine carrier isoform 1 (ORC1) (27, 56). In fact, in such patients lacking ornithine in the mitochondria, intramitochondrial ornithine carbamoylase condenses carbamoyl phosphate with lysine to form homocitrulline, leading to homocitrullinuria, one of the hallmarks of HHH syndrome (2, 56). On the other hand, it is highly unlikely that SLC25A29 plays a role in the urea cycle, as suggested previously (8). Indeed, compensation of the defective ORC1-mediated (cytosolic) ornithine/(intramitochondrial) citrulline exchange in HHH syndrome-affected patients by SLC25A29 cannot be accomplished because this carrier does not transport citrulline and has a much lower affinity for ornithine than ORC1. Future studies are warranted to investigate the effects of SLC25A29 knockdown by RNA interference or SLC25A29 knock-out in cellular and animal models and hence provide further insight on the physiological roles of SLC25A29.
This work was supported by grants from the Ministero dell'Università e della Ricerca (MIUR), the National Research Council (CNR), Centro di Eccellenza in Genomica Comparata (CEGBA), Apulia Region, and the Italian Human ProteomeNet Grant RBRN07BMCT_009 (MIUR).
- HHH
- hyperornithinemia-hyperammonemia-homocitrullinuria
- CAC
- carnitine/acylcarnitine carrier
- ANOVA
- analysis of variance
- Car
- carnitine.
REFERENCES
- 1. Palmieri F. (2004) The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch. - Eur. J. Physiol. 447, 689–709 [DOI] [PubMed] [Google Scholar]
- 2. Palmieri F. (2013) The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol. Aspects Med. 34, 465–484 [DOI] [PubMed] [Google Scholar]
- 3. Palmieri F., Agrimi G., Blanco E., Castegna A., Di Noia M. A., Iacobazzi V., Lasorsa F. M., Marobbio C. M. T., Palmieri L., Scarcia P., Todisco S., Vozza A., Walker J. (2006) Identification of mitochondrial carriers in Saccharomyces cerevisiae by transport assay of reconstituted recombinant proteins. Biochim. Biophys. Acta 1757, 1249–1262 [DOI] [PubMed] [Google Scholar]
- 4. Palmieri F. (2008) Diseases caused by defects of mitochondrial carriers: a review. Biochim. Biophys. Acta 1777, 564–578 [DOI] [PubMed] [Google Scholar]
- 5. Palmieri F., Pierri C. L., De Grassi A., Nunes-Nesi A., Fernie A. R. (2011) Evolution, structure and function of mitochondrial carriers: a review with new insights. Plant J. 66, 161–181 [DOI] [PubMed] [Google Scholar]
- 6. Palmieri L., Runswick M. J., Fiermonte G., Walker J. E., Palmieri F. (2000) Yeast mitochondrial carriers: bacterial expression, biochemical identification and metabolic significance. J. Bioenerg. Biomembr. 32, 67–77 [DOI] [PubMed] [Google Scholar]
- 7. Sekoguchi E., Sato N., Yasui A., Fukada S., Nimura Y., Aburatani H., Ikeda K., Matsuura A. (2003) A novel mitochondrial carnitine-acylcarnitine translocase induced by partial hepatectomy and fasting. J. Biol. Chem. 278, 38796–38802 [DOI] [PubMed] [Google Scholar]
- 8. Camacho J. A., Rioseco-Camacho N. (2009) The human and mouse SLC25A29 mitochondrial transporters rescue the deficient ornithine metabolism in fibroblasts of patients with the hyperornithinemia-hyperammonemia-homocitrullinuria (HHH) syndrome. Pediatr. Res. 66, 35–41 [DOI] [PubMed] [Google Scholar]
- 9. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huizing M., Iacobazzi V., Ijlst L., Savelkoul P., Ruitenbeek W., van den Heuvel L. P., Indiveri C., Smeitink J., Trijbels F. J. M., Wanders R. J. A., Palmieri F. (1997) Cloning of the human carnitine-acylcarnitine carrier cDNA, and identification of the molecular defect in a patient. Am. J. Hum. Genet. 61, 1239–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Giangregorio N., Tonazzi A., Console L., Indiveri C., Palmieri F. (2010) Site-directed mutagenesis of charged amino acids of the human mitochondrial carnitine/acylcarnitine carrier: insight into the molecular mechanism of transport. Biochim. Biophys. Acta 1797, 839–845 [DOI] [PubMed] [Google Scholar]
- 12. Fiermonte G., De Leonardis F., Todisco S., Palmieri L., Lasorsa F. M., Palmieri F. (2004) Identification of the mitochondrial ATP-Mg/Pi transporter: bacterial expression, reconstitution, functional characterization and tissue distribution. J. Biol. Chem. 279, 30722–30730 [DOI] [PubMed] [Google Scholar]
- 13. Fiermonte G., Walker J. E., Palmieri F. (1993) Abundant bacterial expression and reconstitution of an intrinsic membrane transport protein from bovine mitochondria. Biochem. J. 294, 293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fiermonte G., Palmieri L., Dolce V., Lasorsa F. M., Palmieri F., Runswick M. J., Walker J. E. (1998) The sequence, bacterial expression and functional reconstitution of the rat mitochondrial dicarboxylate transporter cloned via distant homologs in yeast and Caenorhabditis elegans. J. Biol. Chem. 273, 24754–24759 [DOI] [PubMed] [Google Scholar]
- 15. Todisco S., Di Noia M. A., Castegna A., Lasorsa F. M., Paradies E., Palmieri F. (2014) The Saccharomyces cerevisiae gene YPR011c encodes a mitochondrial transporter of adenosine 5′-phosphosulfate and 3′-phospho-adenosine 5′-phosphosulfate. Biochim. Biophys. Acta 1837, 326–334 [DOI] [PubMed] [Google Scholar]
- 16. Palmieri F., Indiveri C., Bisaccia F., Iacobazzi V. (1995) Mitochondrial metabolite carrier proteins: purification, reconstitution and transport studies. Methods Enzymol. 260, 349–369 [DOI] [PubMed] [Google Scholar]
- 17. Bisaccia F., Indiveri C., Palmieri F. (1985) Purification of reconstitutively active α-oxoglutarate carrier from pig heart mitochondria. Biochim. Biophys. Acta 810, 362–369 [DOI] [PubMed] [Google Scholar]
- 18. Palmieri F., Klingenberg M. (1979) Direct methods for measuring metabolite transport and distribution in mitochondria. Methods Enzymol. 56, 279–301 [DOI] [PubMed] [Google Scholar]
- 19. Agrimi G., Di Noia M. A., Marobbio C. M. T., Fiermonte G., Lasorsa F. M., Palmieri F. (2004) Identification of the human mitochondrial S-adenosylmethionine transporter: bacterial expression, reconstitution, functional characterization and tissue distribution. Biochem. J. 379, 183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Palmieri L., Agrimi G., Runswick M. J., Fearnley I. M., Palmieri F., Walker J. E. (2001) Identification in Saccharomyces cerevisiae of two isoforms of a novel mitochondrial transporter for 2-oxoadipate and 2-oxoglutarate. J. Biol. Chem. 276, 1916–1922 [DOI] [PubMed] [Google Scholar]
- 21. Capobianco L., Bisaccia F., Mazzeo M., Palmieri F. (1996) The mitochondrial oxoglutarate carrier: sulphydryl reagents bind to cysteine 184 and this interaction is enhanced by substrate binding. Biochemistry 35, 8974–8980 [DOI] [PubMed] [Google Scholar]
- 22. Wibom R., Lasorsa F. M., Töhönen V., Barbaro M., Sterky F. H., Kucinski T., Naess K., Jonsson M., Pierri C. L., Palmieri F., Wedell A. (2009) AGC1 deficiency associated with global cerebral hypomyelination. N. Engl. J. Med. 361, 489–495 [DOI] [PubMed] [Google Scholar]
- 23. Marobbio C. M. T., Agrimi G., Lasorsa F. M., Palmieri F. (2003) Identification and functional reconstitution of yeast mitochondrial carrier for S-adenosylmethionine. EMBO J. 22, 5975–5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Indiveri C., Tonazzi A., Palmieri F. (1990) Identification and purification of the carnitine carrier from rat liver mitochondria. Biochim. Biophys. Acta 1020, 81–86 [DOI] [PubMed] [Google Scholar]
- 25. Indiveri C., Iacobazzi V., Giangregorio N., Palmieri F. (1997) The mitochondrial carnitine carrier protein: cDNA cloning, primary structure, and comparison with other mitochondrial transport proteins. Biochem. J. 321, 713–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Indiveri C., Iacobazzi V., Giangregorio N., Palmieri F. (1998) Bacterial overexpression, purification and reconstitution of the carnitine/acylcarnitine carrier from rat liver mitochondria. Biochem. Biophys. Res. Commun. 249, 589–594 [DOI] [PubMed] [Google Scholar]
- 27. Fiermonte G., Dolce V., David L., Santorelli F. M., Dionisi-Vici C., Palmieri F., Walker J. E. (2003) The mitochondrial ornithine transporter: bacterial expression, reconstitution, functional characterization, and tissue distribution of two human isoforms. J. Biol. Chem. 278, 32778–32783 [DOI] [PubMed] [Google Scholar]
- 28. Palmieri L., De Marco V., Iacobazzi V., Palmieri F., Runswick M. J., Walker J. E. (1997) Identification of the yeast ARG-11 gene as a mitochondrial ornithine carrier involved in arginine biosynthesis. FEBS Lett. 410, 447–451 [DOI] [PubMed] [Google Scholar]
- 29. Hoyos M. E., Palmieri L., Wertin T., Arrigoni R., Polacco J. C., Palmieri F. (2003) Identification of a mitochondrial transporter for basic amino acids in Arabidopsis thaliana by functional reconstitution into liposomes and complementation in yeast. Plant J. 33, 1027–1035 [DOI] [PubMed] [Google Scholar]
- 30. Palmieri L., Lasorsa F. M., Iacobazzi V., Runswick M. J., Palmieri F., Walker J. E. (1999) Identification of the mitochondrial carnitine carrier in Saccharomyces cerevisiae. FEBS Lett. 462, 472–476 [DOI] [PubMed] [Google Scholar]
- 31. Klingenberg M. (2008) The ADP and ATP transport in mitochondria and its carrier. Biochim. Biophys. Acta 1778, 1978–2021 [DOI] [PubMed] [Google Scholar]
- 32. Marobbio C. M. T., Di Noia M. A., Palmieri F. (2006) Identification of the mitochondrial transporter for pyrimidine nucleotides in Saccharomyces cerevisiae: bacterial expression, reconstitution and functional characterization. Biochem. J. 393, 441–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Floyd S., Favre C., Lasorsa F. M., Leahy M., Trigiante G., Stroebel P., Marx A., Loughran G., O'Callaghan K., Marobbio C. M. T., Slotboom D. J., Kunji E. R. S., Palmieri F., O'Connor R. (2007) The IGF-I-mTOR signaling pathway induces the mitochondrial pyrimidine nucleotide carrier to promote cell growth. Mol. Biol. Cell 18, 3545–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Palmieri F., Rieder B., Ventrella A., Blanco E., Do P. T., Nunes-Nesi A., Trauth A. U., Fiermonte G., Tjaden J., Agrimi G., Kirchberger S., Paradies E., Fernie A. R., Neuhaus H. E. (2009) Molecular identification and functional characterisation of Arabidopsis thaliana mitochondrial and chloroplastic NAD+ carrier proteins. J. Biol. Chem. 284, 31249–31259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fiermonte G., Dolce V., Palmieri L., Ventura M., Runswick M. J., Palmieri F., Walker J. E. (2001) Identification of the human mitochondrial oxodicarboxylate carrier: bacterial expression, reconstitution, functional characterization, tissue distribution, and chromosomal location. J. Biol. Chem. 276, 8225–8230 [DOI] [PubMed] [Google Scholar]
- 36. Palmieri L., Lasorsa F. M., De Palma A., Palmieri F., Runswick M. J., Walker J. E. (1997) Identification of the yeast ACR1 gene product as a succinate-fumarate transporter essential for growth on ethanol or acetate. FEBS Lett. 417, 114–118 [DOI] [PubMed] [Google Scholar]
- 37. Picault N., Palmieri L., Pisano I., Hodges M., Palmieri F. (2002) Identification of a novel transporter for dicarboxylates and tricarboxylates in plant mitochondria: bacterial expression, reconstitution, functional characterization, and tissue distribution. J. Biol. Chem. 277, 24204–24211 [DOI] [PubMed] [Google Scholar]
- 38. Fiermonte G., Dolce V., Palmieri F. (1998) Expression in Escherichia coli, functional characterization, and tissue distribution of isoforms A and B of the phosphate carrier from bovine mitochondria. J. Biol. Chem. 273, 22782–22787 [DOI] [PubMed] [Google Scholar]
- 39. Fiermonte G., Palmieri L., Todisco S., Agrimi G., Palmieri F., Walker J. E. (2002) Identification of the mitochondrial glutamate transporter: bacterial expression, reconstitution, functional characterization, and tissue distribution of two human isoforms. J. Biol. Chem. 277, 19289–19994 [DOI] [PubMed] [Google Scholar]
- 40. Indiveri C., Iacobazzi V., Tonazzi A., Giangregorio N., Infantino V., Convertini P., Console L., Palmieri F. (2011) The mitochondrial carnitine/acylcarnitine carrier: function, structure and physiopathology. Mol. Aspects Med. 32, 223–233 [DOI] [PubMed] [Google Scholar]
- 41. Marobbio C. M. T., Vozza A., Harding M., Bisaccia F., Palmieri F., Walker J. E. (2002) Identification and reconstitution of the yeast mitochondrial transporter for thiamine pyrophosphate. EMBO J. 21, 5653–5661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fiermonte G., Paradies E., Todisco S., Marobbio C. M. T., Palmieri F. (2009) A novel member of solute carrier family 25 (SLC25A42) is a transporter of coenzyme a and adenosine 3′,5′-diphosphate in human mitochondria. J. Biol. Chem. 284, 18152–18159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vallance P., Leiper J. (2004) Cardiovascular biology of the asymmetric dimethylarginine: dimethylarginine dimethylamino-hydrolase pathway. Arterioscler. Thromb. Vasc. Biol. 24, 1023–1030 [DOI] [PubMed] [Google Scholar]
- 44. Zinnanti W. J., Lazovic J., Housman C., LaNoue K., O'Callaghan J. P., Simpson I., Woontner M., Goodman S. I., Connor J. R., Jacobs R. E., Cheng K. C. (2007) Mechanism of age-dependent susceptibility and novel treatment strategy in glutaric acidemia type I. J. Clin. Invest. 117, 3258–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Palmieri F., Pierri C. L. (2010) Structure and function of mitochondrial carriers: role of the transmembrane helix P and G residues in the gating and transport mechanism. FEBS Lett. 584, 1931–1939 [DOI] [PubMed] [Google Scholar]
- 46. Palmieri F., Pierri C. L. (2010) Mitochondrial metabolite transport. Essays Biochem. 47, 37–52 [DOI] [PubMed] [Google Scholar]
- 47. Palmieri L., Pardo B., Lasorsa F. M., del Arco A., Kobayashi K., Iijima M., Runswick M. J., Walker J. E., Saheki T., Satrústegui J., Palmieri F. (2001) Citrin and aralar1 are Ca2+-stimulated aspartate/glutamate transporters in mitochondria. EMBO J. 20, 5060–5069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Robinson A. J., Kunji E. R. S. (2006) Mitochondrial carriers in the cytoplasmic state have a common substrate binding site. Proc. Natl. Acad. Sci. U.S.A. 103, 2617–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu G., Morris S. M., Jr. (1998) Review article. Arginine metabolism: nitric oxide and beyond. Biochem. J. 336, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ghafourifar P., Cadenas E. (2005) Mitochondrial nitric oxide synthase. Trends Pharmacol. Sci. 26, 190–195 [DOI] [PubMed] [Google Scholar]
- 51. Persichini T., Mazzone V., Polticelli F., Moreno S., Venturini G., Clementi E., Colasanti M. (2005) Mitochondrial type I nitric oxide synthase physically interacts with cytochrome c oxidase. Neurosci. Lett. 384, 254–259 [DOI] [PubMed] [Google Scholar]
- 52. Ryoo S., Gupta G., Benjo A., Lim H. K., Camara A., Sikka G., Lim H. K., Sohi J., Santhanam L., Soucy K., Tuday E., Baraban E., Ilies M., Gerstenblith G., Nyhan D., Shoukas A., Christianson D. W., Alp N. J., Champion H. C., Huso D., Berkowitz D. E. (2008) Endothelial arginase II: a novel target for the treatment of atherosclerosis. Circ. Res. 102, 923–932 [DOI] [PubMed] [Google Scholar]
- 53. Lim H. K., Lim H. K., Ryoo S., Benjo A., Shuleri K., Miriel V., Baraban E., Camara A., Soucy K., Nyhan D., Shoukas A., Berkowitz D. E. (2007) Mitochondrial arginase II constrains endothelial NOS-3 activity. Am. J. Physiol. Heart Circ. Physiol. 293, H3317–H3324 [DOI] [PubMed] [Google Scholar]
- 54. Ghafourifar P., Asbury M. L., Joshi S. S., Kincaid E. D. (2005) Determination of mitochondrial nitric oxide synthase activity. Methods Enzymol. 396, 424–444 [DOI] [PubMed] [Google Scholar]
- 55. Blemings K. P., Crenshaw T. D., Swick R. W., Benevenga N. J. (1994) Lysine-α-ketoglutarate reductase and saccharopine dehydrogenase are located only in the mitochondrial matrix in rat liver. J. Nutr. 124, 1215–1221 [DOI] [PubMed] [Google Scholar]
- 56. Camacho J. A., Obie C., Biery B., Goodman B. K., Hu C. A., Almashanu S., Steel G., Casey R., Lambert M., Mitchell G. A., Valle D. (1999) Hyperornithinaemia-hyperammonaemia-homocitrullinuria syndrome is caused by mutations in a gene encoding a mitochondrial ornithine transporter. Nat. Genet. 22, 151–158 [DOI] [PubMed] [Google Scholar]