Abstract
An essential step in Serial Analysis of Gene Expression (SAGE) is tag mapping, which refers to the unambiguous determination of the gene represented by a SAGE tag. Current resources for tag mapping are incomplete, and thus do not allow assessment of the efficacy of SAGE in transcript identification. A method of tag mapping is described here and applied to the Drosophila melanogaster and Caenorhabditis elegans genomes, which permits detailed SAGE assessment and provides tag-mapping resources that were unavailable previously for these organisms. In our method, a conceptual transcriptome is constructed using genomic sequence and annotation by extending predicted coding regions to include UTRs on the basis of EST and cDNA alignments, UTR length distributions, and polyadenylation signals. Analysis of extracted tags suggests that, using the standard SAGE procedure, expression of 8% of D. melanogaster and 15% of C. elegans genes cannot be detected unambiguously by SAGE due to shared sequence or lack of NlaIII-anchoring enzyme sites. Both increasing tag length by 2–3 bp and using Sau3A instead of NlaIII as the anchoring enzyme increases potential for transcript detection. This work identifies and quantifies genes not amenable to SAGE analysis, in addition to providing tag-to-gene mappings for two model organisms.
Serial Analysis of Gene Expression (SAGE) (Velculescu et al. 1995) is a method of large-scale gene expression analysis that has the potential to determine relative levels of almost all mRNA molecules within a cell population. It has been used for expression analysis in organisms including Saccharomyces cerevisiae (Velculescu et al. 1997), Cryptococcus neoformans (Steen et al. 2002), Caenorhabditis elegans (Jones et al. 2001), Drosophila melanogaster (Jasper et al. 2001; Fujii and Amrein 2002; Gorski et al. 2003), Rattus norvegicus (Madden et al. 1997), Mus musculus (Virlon et al. 1999), and Homo sapiens (Velculescu et al. 1999). It involves sequencing small segments of expressed transcripts (SAGE tags) in such a way that the number of times a SAGE tag sequence is observed is directly proportional to the abundance of the transcript from which it is derived (Fig. 1).
Figure 1.
SAGE procedure. A population of mRNA transcripts is first extracted from a tissue or cell type, and the mRNA is reverse transcribed to produce cDNA. A series of enzyme digestions, ligations, and PCR amplifications are performed as shown. As the 3′ end of the cDNA is anchored during the first digestion, the tags are extracted from the 3′-most anchoring enzyme site, an important fact to consider when producing tag mappings. The procedure produces 14-bp SAGE tags derived from the sequence of the mRNA transcripts, the frequency of which represents the expression level of the transcripts.
An essential step in SAGE is the unambiguous correlation of the 14-bp SAGE tag with the transcript from which it is derived, a process termed tag mapping. This generally involves automated searching for the tag sequence in sequence databases, and the choice of database and methods used have implications for the quality of the tag mapping, and thus, the utility of the gene expression data obtained. Ideally, tag sequences would be mapped to full-length cDNA sequences, as these represent most closely the transcripts from which the SAGE tags are experimentally derived. However, cDNA sequences are not available for all genes in any organism, and despite full-length cDNA sequencing projects focused on Human (Strausberg et al. 1999), D. melanogaster (Stapleton et al. 2002), and other organisms, it is unlikely that full-length transcript sequences will be derived for all genes for some time to come. Therefore, many SAGE tags will not be found in sequenced cDNAs. Not only does this reduce the number of tags that can be mapped to transcripts, but it also introduces the potential for tags to be misassigned. If two transcripts produce the same tag, but only one is represented in cDNA databases, the tag will be assigned unambiguously to the sequenced transcript. Instead, that tag should be given an ambiguous designation, as it could derive from either transcript. Having a complete gene set for tag mapping is thus imperative for accurate attribution of tags to genes and for determining the number of genes that share identical SAGE tags resulting in ambiguous tag mappings.
A valuable and commonly used resource (Nacht et al. 2001; Waghray et al. 2001) for tag mapping is NCBI's SAGEmap (http://www.ncbi.nlm.nih.gov/SAGE/). As described in their most recent publication (Lash et al. 2000), this tag mapping is done on the basis of matching SAGE data to tags extracted from Expressed Sequence Tag (EST) and cDNA sequences found in the NCBI UniGene clusters (Schuler 1997) (http://www.ncbi.nlm.nih.gov/UniGene/). The UniGene database is composed of expressed sequences (ESTs and cDNAs) clustered on the basis of sequence similarity to gene sequences found in GenBank. The 3′-most predicted SAGE tags are extracted from each expressed sequence, and experimentally derived tags can be compared with these predicted tags to determine the UniGene cluster that most likely represents the gene from which the experimental SAGE tag was derived. However, there are several drawbacks to this method of tag mapping, primarily due to the underlying data.
The same gene may be represented in multiple clusters, resulting in a single tag mapping to multiple clusters, and thus being erroneously assigned as ambiguous. The sequences in the clusters are primarily ESTs, which have an error rate estimated at ∼1% (1 in 100 bp), resulting in a tag error rate of ∼10% (Lash et al. 2000). This error rate will result in tags not matching the appropriate cluster and/or matching an incorrect cluster. Because this approach is based primarily on expressed sequences, any genes that are not represented by ESTs or cDNAs in GenBank will not be represented by tags in SAGEmap, resulting in tag-to-gene assignments that are missing or potentially erroneous.
As a result, tag assignments can be ambiguous, incorrect, or unavailable, leading to potentially incomplete or erroneous gene expression interpretation. Also, this resource is only available currently for mammalian and Arabidopsis thaliana sequences, whereas the SAGE technique is increasingly being applied to other organisms (Jasper et al. 2001; Jones et al. 2001; Steen et al. 2002).
Many of the difficulties encountered when tag mapping, based solely on expressed sequences, could be addressed by using genomic sequence and annotation. As genomic sequence is more accurate, with a low estimated error rate of <1/10,000 (C. elegans Sequencing Consortium 1998; Adams et al. 2000), the potential for results to be obfuscated due to sequence errors in the tag mappings is reduced. Also, annotated gene sets are potentially more complete than expressed sequence sets alone due to the use of additional information such as Genefinder predictions and exons detected through conserved protein similarity, and the curation of this data to form the annotation (Stein et al. 2001; FlyBase Consortium 2002). Basing tag mappings entirely on predicted genes from genomic databases is problematic, however, as gene annotations rarely include untranslated regions (UTRs) of expressed transcripts. Because SAGE tags will correlate to the 3′-most anchoring enzyme site, typically that of the four-cutter restriction enzyme NlaIII, which occurs on average every 256 bp, many SAGE tags are expected to be derived from 3′ UTR sequence.
When using the SAGE technique, it is essential to be aware of the transcripts that will not be identifiable using this method, due to shared sequence resulting in ambiguity or due to lack of an appropriate anchoring enzyme site, as the expression of such transcripts will not be accurately determined. Identification and quantification of these refractory transcripts would potentially permit the choice of appropriate modifications to the SAGE procedure, which could minimize the number of transcripts that will not be profiled using SAGE and increase the utility of this expression profiling method.
To overcome some of the current limitations in SAGE tag mapping and to assess the utility and restrictions of the SAGE approach in transcript identification, we devised a method for constructing a complete predicted transcript set (conceptual transcripts) and deriving SAGE tags from it. These conceptual transcripts are based on a combination of genomic sequence, annotated predicted genes, and expressed sequences, and thus are sufficiently complete to allow this assessment. We applied our method to the model organisms D. melanogaster and C. elegans, as more mature genomic annotation resources are available for these organisms than for the human genome. Also, although the SAGE technique has been utilized recently in these organisms, there are currently no publicly available tag mappings. In addition to providing tag mappings for these organisms, we are able to determine the number of genes lacking a suitable anchoring enzyme site, and to establish the extent to which SAGE tags will be correlated ambiguously to genes. We also determine the most efficient anchoring enzyme choice for each particular organism. This method thus permits assessment of the efficacy of the SAGE approach in transcript identification by distinguishing genes refractory to this profiling method and facilitates SAGE analysis in both D. melanogaster and C. elegans as well as in other organisms to which this method can potentially be applied.
RESULTS
Tag Mapping Using Genomic Sequence
Mapping SAGE tags directly to genomic sequence could potentially deal with the issues of sequence quality and incomplete transcript sets. However, the genome sizes of complex eukaryotes are large enough that tag sequences may be present more than once by chance. To determine the impact of this issue, we mapped experimental C. elegans and D. melanogaster SAGE tags to the corresponding genomes, which are ∼100 MB and ∼120 MB, respectively (C. elegans Sequencing Consortium 1998; Adams et al. 2000). We found that only 59% of mapped SAGE tags occur unambiguously in the C. elegans genome, and 20% of C. elegans SAGE tags occur three or more times in the genome. A total of 59% of D. melanogaster SAGE tags are also unambiguous, and 18% occur three or more times in the genome. Also, only ∼60% of SAGE tags map to the D. melanogaster or C. elegans genomes at all, due in part to some SAGE tags crossing splice boundaries, and thus not being present in the genome. These results suggest to us that it is necessary to use transcript sequences rather than genomic sequence for SAGE tag-togene mapping.
Transcript Construction and Analysis
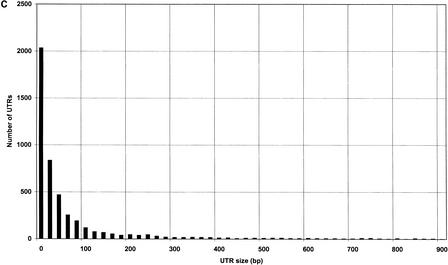
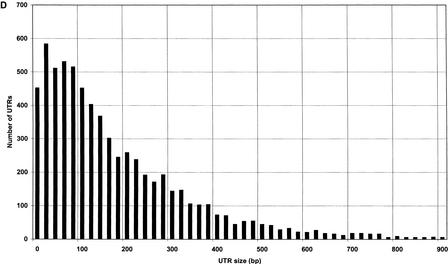
To organize and access the data required to produce the conceptual transcript set, we first constructed a queryable ACEDB (Durbin and Thierry-Mieg 1991) database containing genomic, gene, and expressed sequence information for D. melanogaster (see Methods). A similar database is already available for C. elegans (Stein et al. 2001). Using these databases, conceptual transcripts were constructed as described in the Methods section (Fig. 2A,B). In D. melanogaster, a total of 8213/14,335 (57%) conceptual transcripts have 3′ UTRs constructed based directly on expressed sequence evidence, with an average size of 343 bp and a median of 224 bp. The remaining transcripts have predicted 3′ ends on the basis of empirical size distributions (Fig. 3A,B), indicating that 95% of 3′ UTRs are 1039 bp, or less. In C. elegans, the 6608/20,448 transcripts (32%) found to have 3′ UTRs constructed on the basis of direct sequence evidence were an average of 195 bp and a median of 137 bp, whereas the remaining 3′ UTRs were estimated on the basis of empirical size distributions (Fig. 3C,D) that show 95% of known UTRs to be 574 bp, or less.
Figure 2.
Conceptual transcript construction. Genomic sequence was used to form conceptual transcripts to take advantage of its high-sequence quality. Coding sequences were derived from current genomic annotation. (A) If expressed sequences that extend beyond the predicted coding sequence were available, the alignment position of these sequences was used to determine the extent of the UTR. (B) If no expressed sequences were available, D. melanogaster 5′ and 3′ UTRs were extended to 836 bp and 1039 bp, respectively, and C. elegans UTRs were extended to 388 bp and 574 bp. These UTR size estimates encompass 95% of known UTRs determined in A. If the polyadenylation signal “AATAAA” was found within the estimated 3′ UTR, the UTR was truncated 35 bp downstream of the signal.
Figure 3.
UTR size distributions. Lengths of conceptual transcript UTRs were determined on the basis of expressed sequence alignments (Fig. 2A). (A) D. melanogaster 5′ UTR. (B) D. melanogaster 3′ UTR. C. elegans 5′ UTR. (D) C. elegans 3′ UTR.
Tag-to-Gene Mapping
On the basis of this conceptual transcript set, tag-to-gene mappings are derived by extracting all tags from the transcripts, that is, the 10 bp downstream of each NlaIII (CATG) site, for a total of 14 bp for each tag. It is important for this tag-mapping method to consider all anchoring enzyme sites and not just the 3′-most, for two reasons. First, gene prediction programs often have difficulty correctly defining the ends of genes (Rogic et al. 2001), so that the apparent 3′-most tag site may not be correct. Second, estimated UTRs added when no expressed sequence is available are a length that would encompass 95% of known UTRs, and are thus known to be overestimates of real UTR length. In many cases, therefore, the true 3′ end of the transcript and the associated 3′-most SAGE tag will be upstream of the estimated one. SAGE tags are then assigned to the transcript containing the correct tag sequence. In cases in which a SAGE tag is found in more than one transcript, we take advantage of the fact that the SAGE procedure is expected to derive the SAGE tag from the 3′-proximal position (Fig. 1), and resolve this ambiguity where possible by assigning the correct tag as the one closest to the 3′ end of the transcript. If a tag is found in the same relative position in more than one transcript, that tag cannot be resolved and is considered to be ambiguous. In this tag-to-gene mapping, tags derived from alternative transcripts are consolidated and assigned to a single gene locus, so as not to determine a tag that matches two alternative transcripts to be ambiguous, when in fact, it unambiguously represents a single locus. This also means that the same gene may be represented unambiguously by two or more different SAGE tags from different alternative transcripts.
Our tag mappings, as they are derived from accurate (<1/10,000 error rate) (C. elegans Sequencing Consortium 1998; Adams et al. 2000) genomic sequence and incorporate expressed sequence information with the predicted transcriptome, are expected to be more complete than mappings from expressed sequences alone. To assess our tag-mapping method, a test set of 5606 tags were extracted from full-length cDNAs, which are assumed to be accurate representations of true expressed transcripts. The accuracy of each mapping method, including mapping to ESTs or to conceptual transcripts, was determined by its ability to correctly assign tags to genes from this validated test set (Table 1). As relatively few full-length cDNAs are available for C. elegans, this comparison was done for D. melanogaster only, although the results are expected to be similar for both organisms. We found that conceptual transcripts constructed using EST data in conjuction with gene predictions produced significantly more accurate tag mappings (85% correct) compared with EST sequences alone (70% correct). There were also slightly more tags assigned ambiguously when mapping to EST sequences alone (6% of tags compared with 4%), most likely due to sequencing errors in the EST sequences that produce extra, erroneous tags, or due to chimeric ESTs. Simulating the situation in which a set of genes have no associated ESTs, and thus, only predicted protein-coding genes from genomic annotation are available for tag mapping, we found that tag mappings derived from such sequences alone (48% correct) were significantly less accurate than tag mappings from conceptual transcripts constructed with estimated UTRs (81% correct). This is not unexpected, given that 56% of SAGE tags extracted from D. melanogaster cDNAs are derived from the UTR sequence, which is lacking in the predicted protein-coding sequences. The high proportion of tags derived from UTR sequence emphasizes the importance of estimating UTRs when no expressed sequence evidence is available. Overall, between 81% and 85% of tag mappings derived from conceptual transcripts are correct when full-length cDNA data is not incorporated, a significant improvement over tag mapping with ESTs or genomic annotations alone. When full-length cDNA data is incorporated into the conceptual transcripts, as is normally the case where possible, 93% of test tags are mapped correctly (Table 1); the remaining incorrectly mapped tags are due to gene prediction errors.
Table 1.
Tag Mapping Accuracy Using Conceptual Transcripts Compared With Other Available Sequences
| Test tags assigned to genes by mapping to... | Tags mapped unambiguously | Unambiguously mapped tags correctly assigned to genes |
|---|---|---|
| Full-length cDNAs (5606) | 99% | 100% |
| EST sequences (204,380) | 94% | 70% |
| Predicted protein coding sequences from genomic annotation (13,489) | 97% | 48% |
| Conceptual transcripts constructed from predicted protein coding sequences from genomic annotation, UTRs estimated (13,489) | 96% | 81% |
| Conceptual transcripts constructed from predicted protein coding sequences from genomic annotation, UTRs derived from EST data or estimated if ESTs unavailable (13,489) | 96% | 85% |
| Conceptual transcripts constructed from predicted protein coding sequences from genomic annotation, UTRs derived from EST and cDNA data or estimated if ESTs and cDNAs unavailable (13,489) | 96% | 93% |
Analysis of SAGE Transcriptome
As the conceptual transcript sets for D. melanogaster and C. elegans are based on essentially complete genomic sequence representing all known and predicted genes, quantification of the genes that are not amenable to SAGE in these organisms is possible. For this analysis, we considered alternative transcripts to be a single gene, so as not to erroneously assign tags as ambiguous if they are derived from multiple alternative transcripts belonging to the same locus, as described above. Tag mappings are also available (see Methods) that assign tags to individual alternative transcripts.
As described above, genes that produce ambiguous tags, shared with other genes, will not be uniquely identifiable in a SAGE library. A total of 6% of D. melanogaster genes and 12% of C. elegans genes fall in this category, in the common situation of 14-bp SAGE tags extracted using the NlaIII anchoring enzyme. As recent work has yielded a SAGE procedure that produces a 21-bp tag (Saha et al. 2002), it is relevant to ask how SAGE tag length influences the ambiguity of the tag mappings for each organism. We observe (Fig. 4A,B) that increasing SAGE tag length by 2–3 bp decreases ambiguity in tag assignments, after which increasing length has little effect. This information can be used to make an informed decision about the ideal tag length for a particular SAGE experiment, especially as increasing tag size decreases the number of SAGE tags that can be obtained from each sequence read, and thus decreases the efficiency of the SAGE approach.
Figure 4.
SAGE tag ambiguity varies with tag length. Number of ambiguous genes and number of ambiguous tags derived from conceptual transcripts is shown with varying tag length (length includes anchoring enzyme site) and anchoring enzyme. (A) D. melanogaster, total 13,489 gene loci. (B) C. elegans, total 19,432 gene loci.
One drawback of the SAGE technique is that genes that lack the appropriate anchoring enzyme recognition sequence will not be represented in SAGE libraries. For instance, genes lacking a CATG site will not be represented in SAGE libraries constructed with NlaIII. However, judicious choice of anchoring enzyme could improve the utility of the SAGE approach. On the basis of the conceptual transcript set, there are 261 genes (2% of the annotated transcriptome) lacking NlaIII sites in D. melanogaster and 563 (3%) in C. elegans. However, when tags are extracted with other four-cutter anchoring enzymes, we observe that differing numbers of genes contain no anchoring enzyme site or produce ambiguous SAGE tags (Fig. 5A,B). Sau3A, which has occasionally been used in SAGE library construction (Virlon et al. 1999), allows recovery of more genes unambiguously in D. melanogaster and C. elegans than NlaIII. Interestingly, however, at higher tag length in D. melanogaster, NlaIII has this property (Fig. 4A). It is important to consider, however, that NlaIII is compatible with the BsmFI-tagging enzyme in such a way that an extra base pair of tag length can be obtained, resulting in the potential for a 15-bp rather than 14-bp tag. The AciI (CCGC) enzyme, which has not been used in SAGE library construction, also has the same property and allows more genes to be resolved in D. melanogaster than NlaIII. This knowledge allows the preselection of an anchoring enzyme, that may be organism specific, which produces the best representation of the expressed genes within an mRNA population.
Figure 5.
Number of genes not resolvable by SAGE varies with anchoring enzyme. Number of ambiguous genes with no anchoring enzyme site is shown for various restriction enzymes used as the anchoring enzyme. (A) D. melanogaster, total 13,489 gene loci. (B) C. elegans, total 19,432 gene loci.
Tag-to-Gene Mapping of Experimental SAGE Tags
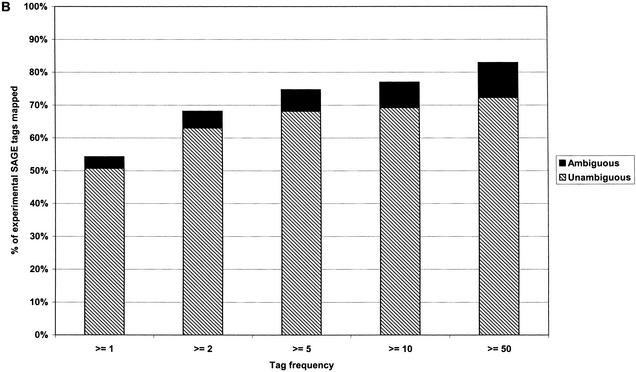
To further analyze our tag-to-gene mappings and compare predicted levels of ambiguity with experimental levels, we mapped C. elegans and D. melanogaster experimental SAGE tags using our method of tag-to-gene mapping (Fig. 6A,B). In this comparison, we cannot compare the number of ambiguous genes, as we do not know how many genes are represented by an ambiguous experimental SAGE tag. Thus, we instead compare the number of SAGE tags that are ambiguous. In both organisms, an even higher proportion of experimental SAGE tags map ambiguously to genes than expected. For C. elegans, 5.0% of 14-bp tags were predicted to be ambiguous (Fig. 4B), and 7.5% of experimental tags were ambiguous; for D. melanogaster, the predicted (Fig. 4A) and experimental ambiguities were 2.5% and 3.6%, respectively. It is also notable that the proportion of tags that are mapped to genes increases with expression level for both organisms.
Figure 6.
Experimental SAGE tags mapped to conceptual transcripts. Subsets of experimental SAGE tags with varying minumum frequency were mapped to conceptual transcripts, and the percentage that mapped ambiguously and unambiguously determined. (A) D. melanogaster. (B) C. elegans.
Online Resources
Both the constructed transcript sets described above for D. melanogaster and C. elegans and their associated tag mappings are available from http://sage.bcgsc.ca/tagmapping/. The D. melanogaster ACEDB database is also available from this site.
DISCUSSION
We have shown that utilizing genomic annotation provides a more accurate strategy for the assignment of SAGE tags to gene transcripts. We have also estimated the total resolving power of the SAGE approach, determining that 93% and 86% of genes can be detected in D. melanogaster and C. elegans, respectively, when optimum anchoring enzymes are utilized. Our findings suggest that the use of NlaIII, most commonly used in SAGE experiments, is potentially suboptimal.
Ambiguity in SAGE
The observed rate of SAGE tag ambiguity is higher than would have been expected on the basis of statistical calculations that predict 13,489 genes would produce tags with a 98.7% probability of being unique (see Methods). The observed increase in ambiguity is likely due to the nonindependent nature of gene sequences, as there is similarity within gene families and even between distantly related genes. In addition, the presence of repetitive elements in the 3′ UTRs of genes can contribute to increased rates of ambiguity. The 6% increase in ambiguity seen in C. elegans compared with D. melanogaster may be due to a combination of increased gene number and expansion of gene families (Friedman and Hughes 2001). The same factors of nonindependent and repetitive sequence are likely to be responsible for the higher than expected rates of >40% ambiguity observed when mapping directly to genomic sequence, which makes such an approach less feasible even for relatively small genomes of ∼100 MB.
The ambiguity predicted on the basis of conceptual transcripts is likely to be an underestimate of the true ambiguity. This is demonstrated by the higher than predicted proportion of ambiguous tags seen in experimental data. There are two factors that contribute to this. First, the genomic annotations remain under constant revision and our current understanding of both transcription and gene prediction suggests that complete transcript predictions will not be available for some time. Thus, the total gene count may be an underestimate, as not all genes have necessarily been identified. EST data in D. melanogaster suggests there may be as many as 10%–20% more genes than currently predicted (Andrews et al. 2000; Gorski et al. 2003), and 5% of full-length cDNAs do not match known or predicted genes (data not shown). Increased gene number increases the chance of shared sequence, and thus shared SAGE tags. Also, increasing numbers of alternative transcripts, which are poorly predicted by genefinders and are constantly being discovered on the basis of expressed sequences, can also increase ambiguity if an alternative transcript produces a tag that is already present in a transcript from a different locus. Second, 43%–68% of 3′ UTRs predicted in this analysis are estimated, and thus are likely to be longer than the actual UTRs, and so will extend into less-conserved intergenic sequence. It is expected that this sequence, as it is more random, will be less likely to be shared between genes, and thus less likely to yield ambiguous tags. As this hypothesis would predict, updating the transcripts with increasing amounts of expressed sequence data has resulted in shorter UTRs and increased ambiguity as more tags are derived from conserved sequence (data not shown).
The increase in ambiguity with expression level that occurs in C. elegans is likely because ambiguous tags have the potential to represent the sum of expression of several genes, thus increasing the observed tag frequency. For instance, the tag “AAAAAAAAAA” which can be derived from the poly(A) tail of many transcripts is seen at a relatively high frequency in most SAGE libraries (data not shown). Such an increase may not be visible for D. melanogaster in Figure 6A because of the lower overall ambiguity, and fewer tags available with high frequencies in the libraries under study (<200 tags with expression >50 tags).
Choice of Tag Length and Anchoring Enzyme
On the basis of Figure 4, the ideal tag lengths are ∼16 bp for D. melanogaster and 17 bp for C. elegans. It is at these points that the curves of ambiguity versus tag length level off and little is gained from further tag length. However, currently, only SAGE procedures that produce tags of 14 and 21 bp are available. An altered SAGE procedure that produces 18-bp tags has been designed, but it has the added limitation that it does not allow the identification of duplicate ditags that may arise from PCR bias during SAGE library construction (Ryo et al. 2000). It is important also to consider that increasing tag length decreases the efficiency of sequencing SAGE tags due to both longer SAGE tag length, and thus fewer tags per sequence read, and also an increased per-tag error rate (at 1% sequence error, 13% of 14-bp tags are expected to contain sequence errors, whereas 21-bp tags will be erroneous in 19% of cases). Thus, for these two organisms, a tag length of 14 bp is probably an efficient and cost-effective choice, unless a more complete snapshot of the transcriptome is desired or a particular gene or set of genes of interest is identified as ambiguous with a 14-bp tag and not with 21 bp. Alternatively, if a tag of particular interest is found to be ambiguous, it can be resolved with further experimental procedures such as GLGI (Chen et al. 2000), which derive a larger portion of the expressed sequence represented by a SAGE tag, allowing more likelihood of unambiguous gene assignment. The human genome, however, contains both more genes and further expansion to gene families, and thus it is expected that ambiguity would be even higher. A recent analysis of SAGEmap tag mappings (Lee et al. 2002) showed that ∼1/3 of tags from human genes map to more than one UniGene cluster. Although this may not be a completely accurate estimate of SAGE tag ambiguity, as a single gene may be represented in multiple UniGene clusters and some genes may not be represented in UniGene, it emphasizes the importance of ambiguity considerations in SAGE analysis.
As shown in Figure 5A, 5B, there can be a decrease in the number of genes that cannot be accurately analyzed using SAGE, depending on the anchoring enzyme chosen. The number of genes that are not unambiguously identifiable with a 14-bp tag can be decreased by 12% (883 vs. 1001) in D. melanogaster and 5% (2823 vs. 2962) in C. elegans by using Sau3A instead of NlaIII. However, making use of the extra base pair of tag length provided by NlaIII by using a 15-bp tag in analysis would increase the effectiveness of NlaIII. This would not involve a change in the SAGE procedure, only a change in the way in which SAGE tags are extracted from raw sequence derived from serially ligated tags. The tradeoff of such an analysis would be the loss of the small proportion of tags which, due to the slight variation in cutting position of BsmFI, would only be 14 bp even with NlaIII. Our analysis of the effects of the choice of anchoring enzyme allows the potential for tailoring of the SAGE procedure to produce the most comprehensive results for each organism individually.
Tags With No Gene Mapping
It is notable that the proportion of experimental SAGE tags that cannot be mapped to genes or genomic sequence is significant (Fig. 6). This is due, in part, to the presence of sequencing errors in experimental SAGE data. In Figure 6A, 55% of all experimental SAGE tags are mapped to genes, whereas 68% of tags that occur two or more times are mapped. This suggests that a significant proportion of the unmapped tags are singletons (occur only once) in the SAGE library under consideration. A similar trend occurs in Figure 6B. Singleton tags are more likely to represent sequencing errors, which generally produce tags that do not match known sequences. In rare cases, a sequencing error may produce a tag that, by chance, matches another gene, which can confound analysis. Using more sequences for tag-to-gene mapping, such as a large set of EST sequences or clusters, increases the chance of this occurrence, because there is a larger body of sequence to which a match may occur. Using the conceptual transcripts for mapping makes this unlikely, and thus a significant proportion of infrequently occurring, unmapped tags may be due to sequencing errors. Many of these errors can be removed by using only SAGE tags derived from sequence of high quality as determined, for instance, by Phred (Ewing et al. 1998). Techniques have also been proposed (Velculescu et al. 1999) to remove tags that are likely to represent sequence errors of more highly expressed tags. These methods can help to limit the effect of sequence errors in SAGE analysis.
The general increase in proportion of tags mapped to genes with increasing expression level may also be related to the quality of genome annotation. On average, highly expressed genes are easier to study and survey and so are more likely to be known and thus included in the predicted gene set, whereas rarely expressed genes may not be annotated. Quality of annotation may in part explain why fewer tags are assigned to genes in D. melanogaster than C. elegans in Figure 6, as the C. elegans genome has been available for a longer period and, therefore, may have more correct annotations. It is somewhat surprising that the proportion of non-singleton tags mapped to genomic sequence (61%) is even lower than the proportion mapped to genes (68% in C. elegans). We expect that this is because SAGE tags that cross splice boundaries will not exist in genomic sequence.
Another reason that experimental SAGE tags may not be mapped to transcripts or genomic sequence is the presence of polymorphisms. The D. melanogaster and C. elegans strains used to produce the SAGE data described here are slightly different or derived from those used for genomic sequencing, and so there are expected to be base-pair changes in a fraction of SAGE tags that prevent exact matches to genomically derived sequence. The same polymorphism effect can be expected in humans. Large bodies of EST sequence from multiple strains or multiple individuals are likely to represent these polymorphisms; however, it is very difficult to separate true polymorphisms from EST sequence and clonal errors that can cause erroneous mappings as discussed above, and thus this issue is not simplified by the use of EST sequences for tag mapping. Fortunately, the polymorphism rate in most experimental situations is relatively low, estimated to be below 1/1000 bp in human sequences (Lander et al. 2001; Venter et al. 2001). Even a relatively high polymorphism rate of 1/500 bp would be expected to affect <3% of SAGE tags, and thus does not account for the failure to map a significant proportion of SAGE tags.
We find that even after comprehensive mapping to the known transcriptome, 15%–30% of highly expressed (>50 copies) SAGE tags are not identified. This emphasizes the current incompleteness of genomic annotation and underscores the role that SAGE can play in novel gene discovery.
Limitations of the Conceptual Transcript Set
As the conceptual transcript sets rely heavily on curated sets of known and predicted genes, errors and omissions in gene predictions result in inaccuracies and incompleteness in the tag mappings. For instance, 7% of tags from known D. melanogaster cDNAs in the test set were not mapped correctly, due to gene prediction errors. We presume this is due to the latency in the process of incorporating experimental data into the curated genomic databases. It is anticipated that as gene predictions are updated, refined, and consolidated with more expression data, the constructed transcript sets will become a more accurate reflection of the true transcriptome, and thus yield more accurate tag mappings. For this purpose, the full-length cDNA project currently being carried out for D. melanogaster has positive implications for SAGE analysis in this organism.
Conclusion
The conceptual transcript sets and tag-mapping procedure described here provide tag-to-gene mappings for the D. melanogaster and C. elegans genomes, thus facilitating SAGE analysis in these organisms. They also permit analysis of the limitations of SAGE for transcript identification. As the method used is flexible, it can be applied to other organisms with sequenced genomes. Construction of a complete conceptual transcript set for the human genome, which will soon be possible due to intense genomic annotation efforts, will allow the production of a similar set of tag mappings and a SAGE procedure that is tailored to the human genome. This will provide both a more complete analysis of the genes expressed, and a more thorough knowledge of the limitations of SAGE for the study of human gene expression.
METHODS
ACEDB
An ACEDB (Durbin and Thierry-Mieg 1991) database is available for C. elegans (Stein et al. 2001) and contains a large variety of data including genomic, gene, expression, and sequence similarity information. This database is vital in the construction of the conceptual transcript set for tag mapping, as it can be automatically queried using existing software. It was accessed through a remote connection to WormBase (http://www.wormbase.org) version WS78 (May 2002). This database includes EST alignments determined by BLAT (Kent 2002).
The D. melanogaster databases GadFly (http://www.bdgp.org/annot/index.html) and FlyBase (Flybase Consortium 2002) contain similar information, but do not allow the direct automated manipulation of data in the manner necessary for the integration of sequence information into conceptual transcripts. Accordingly, we constructed an ACEDB database for D. melanogaster, on the basis of the GadFly resources, which contain data on the genomic sequence as well as predicted and known genes, and EST and cDNA alignments. This data-base is directly queryable in an automated manner. Genomic sequence and predicted gene-coding sequence positions were obtained from the GadFly Release 2 mySQL database gadb2c on April 8 2002, accessible from headcase.lbl.gov. EST and cDNA sequences were obtained from the BDGP at ftp.fruitfly.org/pub/genomic/fasta/ on April 11 2002. ESTs and cDNAs were aligned to genomic segments identified as having similarity by BLASTN (Altschul et al. 1990) (v. 2.0.14) (E value <1e-100, word size 32) using the dynamic, intron-aware EST_GENOME (Mott 1997) alignment program (min. score 100). EST_GENOME output was converted into ACEDB format using specialized Perl scripts. In addition, the gene represented by each of the ESTs was determined by BLASTN similarity (E value <1e-3, word size 16) processed by MSPcrunch (Sonnhammer and Durbin 1994) (v. 2.3), requiring that an EST match no more than one gene locus. All other input/output processing was performed using specialized Perl scripts. This D. melanogaster ACEDB database is available for download from http://sage.bcgsc.ca/tagmapping/. Beside its utility in constructing the conceptual transcript set, the D. melanogaster ACEDB database is useful for viewing gene structure, expression, and similarity information. In these respects, it is similar to FlyBase's (Flybase Consortium 2002) GeneSeen (http://www.flybase.org/annot/geneseen-launch-static.html). However, it has the added advantages that it can incorporate user-specific data and be queried directly or through the AcePerl (Stein and Thierry-Mieg 1998) interface to extract sequences and associated information, allowing further whole-genome analysis.
Transcript Construction
Transcripts were constructed using specialized Perl scripts for each organism, which interact with the ACEDB database described above through the AcePerl modules (Stein and Thierry-Mieg 1998). UTRs were added to predicted genes using the genomic sequence as determined by alignments with ESTs and cDNAs for genes in which such data is available. If expressed sequences were unavailable, UTRs were estimated to be a length that would encompass 95% of known UTR sequences on the basis of empirical UTR size distributions and polyadenylation signals.
In C. elegans, UTRs were added on the basis of EST evidence if the EST was assigned to the gene (in the Matching_cDNA field of the gene in ACEDB) and was in the correct orientation (e.g., only 3′ ESTs used to construct 3′ UTRs) and started within 1 kb of the end of the gene (for reasons of efficiency; UTRs in C. elegans are not expected to extend this far). UTRs were extended to encompass the furthest EST from the gene.
In D. melanogaster, UTRs were added on the basis of EST/cDNA evidence if the EST/cDNA both overlapped with the gene's coding region (by EST_GENOME) and was more similar to that gene than any other (by BLAST). As D. melanogaster UTRs are occasionally spliced, breaks in the EST alignments corresponding to introns were excluded from the final transcript sequence. Only ESTs or cDNAs starting within 20 kb of genomic sequence from the end of the gene were considered. As in C. elegans, ESTs were required to be in the correct orientation.
For genes that did not have expressed sequence corresponding to their UTRs, 5′ UTRs were extended to 836 bp in D. melanogaster and 388 bp in C. elegans, and 3′ UTRs were extended to 1039 and 574 bp, respectively. This corresponds to a length >95% of the known UTRs as determined by EST/cDNA alignments. If the most common polyadenylation signal AATAAA (Riddle et al. 1997; Graber et al. 1999) was found in the estimated 3′ UTR, the UTR was truncated 35 bp downstream of the end of this signal. Also, if there was a gene nearby, the estimated UTRs were truncated so as not to overlap ESTs associated with another gene nor extend over one-half the distance to the next nearest coding region, so as to prevent adjacent UTRs from overlapping.
A 30-bp poly(A) sequence was added to the 3′ end of all constructed transcripts to represent the poly(A) tail present on mRNAs, as occasionally SAGE tags contain part of this poly(A) sequence. A 30 bp was chosen because it extends further than the longest SAGE tags (25 bp) used in this analysis, thus permitting SAGE tags to extend their full length into the poly(A) sequence, if necessary. SAGE tags derived from poly(A) sequence will complicate SAGE analysis in any case. All such tags will end in a varying number of As, and thus will be more likely to be ambiguous. Also, the variance in poly-adenylation cleavage sites will result in multiple tags derived from the same gene (Pauws et al. 2001).
Conceptual transcript sequences and corresponding tag mappings, to both gene loci and individual alternative transcripts, is available at http://sage.bcgsc.ca/tagmapping/. Perl code for the construction of transcripts is available upon request from the authors.
Tag-to-Gene Mapping Accuracy
To evaluate the accuracy of tag-to-gene mappings derived from our conceptual transcripts, we used a set of test SAGE tags extracted from the 3′-most position of 6614 full-length D. melanogaster cDNA sequences (Stapleton et al. 2002) obtained from BDGP (http://www.bdgp.org/EST/index.shtml; entire available set as of April 11 2002). We calculated the accuracy of mapping as the percentage of tags that could be correctly and unambiguously assigned to genes, as this is the goal of tag-to-gene mapping. We first attempted to assign each of the cDNA and 252,362 EST sequences (from BDGP sequencing project, http://www.bdgp.org/EST/index.shtml) to one of the 13,489 D. melanogaster predicted genes by BLASTN (E value <e-50); conceptual transcripts, as they are constructed on the basis of predicted gene sequences, are already associated with genes. To correctly compare the accuracy of EST mapping approaches, we only considered in our analysis cDNAs and genes to which we could map EST sequences. We also removed from our analysis ESTs that did not match predicted genes from genomic annotation. Thus, we used for our analysis 5606 SAGE tags extracted from cDNA sequences, 13,489 conceptual transcripts, and 204,380 EST sequences, each assigned to a gene.
We then used each of the sequence sets described in Table 1 to map the test SAGE tags to genes. We constructed sets of conceptual transcripts with and without using EST or cDNA sequences to determine UTRs; if no ESTs or cDNAs were used, the UTRs were estimated as described above. In mapping to EST sequences, tags were assigned to ESTs that were grouped on the basis of gene assignments, so that clustering ESTs on the basis of sequence similarity was unnecessary. Ambiguous mappings would only occur in cases in which two ESTs from different genes contained the same SAGE tag. The number of tags assigned to a single gene using these various sequence sets and the number of those assignments that were correct were determined.
Tag Mapping, Ambiguity, Tag Size, and Enzyme Site Analysis
All tags of a given size for a given enzyme site were extracted from all conceptual transcripts for all loci in the genomes using Perl scripts. A locus was considered not to contain an enzyme site if none of its alternative transcripts contained that site, and a locus was considered ambiguous if any of its transcripts shared a tag at the 3′-most enzyme site with a transcript from a different locus. If, for instance, a particular tag was found in the 3′-most site in one transcript and a site closer to the 5′ end in another transcript, that was not considered to be an ambiguity, and that tag would be assigned to the gene in which it was found at the 3′-most site. Probability of uniqueness was calculated from the formula used in Saha et al. (2002), assuming 13,489 tags of 14 bp each.
Tag Mapping of Experimental SAGE Tags
A total of 4007 different experimentally derived D. melanogaster tags (Gorski et al. 2003) and 9159 different C. elegans tags (G. Vatcher, unpubl.), of at least 99% quality (1% chance of sequence error) and occurring more than once, were derived using software under development in our laboratory (S. Zuyderduyn, unpubl.). All tags of 14 bp were extracted using Perl scripts from NlaIII enzyme sites at every position in the D. melanogaster and C. elegans genomes derived from ACEDB. Experimental tags were compared with genomic tags, and the number of occurrences of each tag determined. The same libraries of experimental tags, with varying frequency cutoffs, were mapped to conceptual transcripts, and the percentage mapped unambiguously and ambiguously determined.
Acknowledgments
We thank BDGP, GadFly, and WormBase for help accessing and processing the genomic databases they make available. We also thank the Genome Sciences Centre Bioinformatics SAGE Group for invaluable help and advice, Greg Vatcher for sharing unpublished C. elegans SAGE data, and Sharon Gorski for sharing unpublished D. melanogaster work and for critically reading the manuscript. E.P. is a Michael Smith Foundation for Health Research Trainee and is supported by the Natural Sciences and Engineering Research Council of Canada. M.M. is a Michael Smith Foundation for Health Research Scholar. We gratefully acknowledge the support of the BC Cancer Agency and the BC Cancer Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
[The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: S. Gorski, G. Vatcher, and S. Zuyderduyn.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.873003. Article published online before print in May 2003.
References
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Andrews, J., Bouffard, G.G., Cheadle, C., Lu, J., Becker, K.G., and Oliver, B. 2000. Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Genome Res. 10: 2030–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282: 2012–2018. [DOI] [PubMed] [Google Scholar]
- Chen, J.J., Rowley, J.D., and Wang, S.M. 2000. Generation of longer cDNA fragments from serial analysis of gene expression tags for gene identification. Proc. Natl. Acad. Sci. 97: 349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin, R. and Thierry-Mieg, J. 1991. A C. elegans Database.
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185. [DOI] [PubMed] [Google Scholar]
- FlyBase Consortium. 2002. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 30: 106–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, R. and Hughes, A.L. 2001. Gene duplication and the structure of eukaryotic genomes. Genome Res. 11: 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, S. and Amrein, H. 2002. Genes expressed in the Drosophila head reveal a role for fat cells in sex-specific physiology. EMBO J. 21: 5353–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski, S.M., Chittaranjan, S., Pleasance, E.D., Freeman, J.D., Anderson, C.L., Varhol, R.J., Coughlin, S.M., Zuyderduyn, S.D., Jones, S.J.M., and Marra, M.A. 2003. A SAGE approach to discovery of genes involved in autophagic cell death. Curr. Biol. 13: 358–363. [DOI] [PubMed] [Google Scholar]
- Graber, J.H., Cantor, C.R., Mohr, S.C., and Smith, T.F. 1999. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl. Acad. Sci. 96: 14055–14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasper, H., Benes, V., Schwager, C., Sauer, S., Clauder-Munster, S., Ansorge, W., and Bohmann, D. 2001. The genomic response of the Drosophila embryo to JNK signaling. Dev. Cell 1: 579–586. [DOI] [PubMed] [Google Scholar]
- Jones, S.J., Riddle, D.L., Pouzyrev, A.T., Velculescu, V.E., Hillier, L., Eddy, S.R., Stricklin, S.L., Baillie, D.L., Waterston, R., and Marra, M.A. 2001. Changes in gene expression associated with developmental arrest and longevity in Caenorhabditis elegans. Genome Res. 11: 1346–1352. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Lash, A.E., Tolstoshev, C.M., Wagner, L., Schuler, G.D., Strausberg, R.L., Riggins, G.J., and Altschul, S.F. 2000. SAGEmap: A public gene expression resource. Genome Res. 10: 1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Clark, T., Chen, J., Zhou, G., Scott, L.R., Rowley, J.D., and Wang, S.M. 2002. Correct identification of genes from serial analysis of gene expression tag sequences. Genomics 79: 598–602. [DOI] [PubMed] [Google Scholar]
- Madden, S.L., Galella, E.A., Zhu, J., Bertelsen, A.H., and Beaudry, G.A. 1997. SAGE transcript profiles for p53-dependent growth regulation. Oncogene 15: 1079–1085. [DOI] [PubMed] [Google Scholar]
- Mott, R. 1997. EST_GENOME: A program to align spliced DNA sequences to unspliced genomic DNA. Comput. Appl. Biosci. 13: 477–478. [DOI] [PubMed] [Google Scholar]
- Nacht, M., Dracheva, T., Gao, Y., Fujii, T., Chen, Y., Player, A., Akmaev, V., Cook, B., Dufault, M., Zhang, M., et al. 2001. Molecular characteristics of non-small cell lung cancer. Proc. Natl. Acad. Sci. 98: 15203–15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauws, E., van Kampen, A.H., van de Graaf, S.A., de Vijlder, J.J., and Ris-Stalpers, C. 2001. Heterogeneity in polyadenylation cleavage sites in mammalian mRNA sequences: Implications for SAGE analysis. Nucleic Acids Res. 29: 1690–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle, D.L., Blumenthal, T., Meyer, B.J., and Preiss, J.R. 1997. C. Elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [PubMed]
- Rogic, S., Mackworth, A.K., and Ouellette, F.B. 2001. Evaluation of gene-finding programs on mammalian sequences. Genome Res. 11: 817–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryo, A., Kondoh, N., Wakatsuki, T., Hada, A., Yamamoto, N., and Yamamoto, M. 2000. A modified serial analysis of gene expression that generates longer sequence tags by nonpalindromic cohesive linker ligation. Anal. Biochem. 277: 160–162. [DOI] [PubMed] [Google Scholar]
- Saha, S., Sparks, A.B., Rago, C., Akmaev, V., Wang, C.J., Vogelstein, B., Kinzler, K.W., and Velculescu, V.E. 2002. Using the transcriptome to annotate the genome. Nat. Biotechnol. 20: 508–512. [DOI] [PubMed] [Google Scholar]
- Schuler, G.D. 1997. Pieces of the puzzle: Expressed sequence tags and the catalog of human genes. J. Mol. Med. 75: 694–698. [DOI] [PubMed] [Google Scholar]
- Sonnhammer, E.L. and Durbin, R. 1994. A workbench for large-scale sequence homology analysis. Comput. Appl. Biosci. 10: 301–307. [DOI] [PubMed] [Google Scholar]
- Stapleton, M., Liao, G., Brokstein, P., Hong, L., Carninci, P., Shiraki, T., Hayashizaki, Y., Champe, M., Pacleb, J., Wan, K., et al. 2002. The Drosophila gene collection: Identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 12: 1294–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen, B.R., Lian, T., Zuyderduyn, S., MacDonald, W.K., Marra, M., Jones, S.J., and Kronstad, J.W. 2002. Temperature-regulated transcription in the pathogenic fungus Cryptococcus neoformans. Genome Res. 12: 1386–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, L., Sternberg, P., Durbin, R., Thierry-Mieg, J., and Spieth, J. 2001. WormBase: Network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res. 29: 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, L.D. and Thierry-Mieg, J. 1998. Scriptable access to the Caenorhabditis elegans genome sequence and other ACEDB databases. Genome Res. 8: 1308–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg, R.L., Feingold, E.A., Klausner, R.D., and Collins, F.S. 1999. The mammalian gene collection. Science 286: 455–457. [DOI] [PubMed] [Google Scholar]
- Velculescu, V.E., Zhang, L., Vogelstein, B., and Kinzler, K.W. 1995. Serial analysis of gene expression. Science 270: 484–487. [DOI] [PubMed] [Google Scholar]
- Velculescu, V.E., Zhang, L., Zhou, W., Vogelstein, J., Basrai, M.A., Bassett Jr., D.E., Hieter, P., Vogelstein, B., and Kinzler, K.W. 1997. Characterization of the yeast transcriptome. Cell 88: 243–251. [DOI] [PubMed] [Google Scholar]
- Velculescu, V.E., Madden, S.L., Zhang, L., Lash, A.E., Yu, J., Rago, C., Lal, A., Wang, C.J., Beaudry, G.A., Ciriello, K.M., et al. 1999. Analysis of human transcriptomes. Nat. Genet. 23: 387–388. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304–1351. [DOI] [PubMed] [Google Scholar]
- Virlon, B., Cheval, L., Buhler, J.M., Billon, E., Doucet, A., and Elalouf, J.M. 1999. Serial microanalysis of renal transcriptomes. Proc. Natl. Acad. Sci. 96: 15286–15291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waghray, A., Schober, M., Feroze, F., Yao, F., Virgin, J., and Chen, Y.Q. 2001. Identification of differentially expressed genes by serial analysis of gene expression in human prostate cancer. Cancer Res. 61: 4283–4286. [PubMed] [Google Scholar]
WEB SITE REFERENCES
- ftp.fruitfly.org/pub/genomic/fasta/; GadFly sequence downloads.
- http://sage.bcgsc.bc.ca/tagmapping/; SAGE tag mapping resources.
- http://www.bdgp.org/annot/index.html; GadFly homepage.
- http://www.bdgp.org/EST/index.shtml; BDGP cDNA and EST sequencing.
- http://www.flybase.org/annot/geneseen-launch-static.html; GadFly GeneSeen Launcher.
- http://www.ncbi.nlm.nih.gov/SAGE/; SAGEmap Home page.
- http://www.ncbi.nlm.nih.gov/UniGene/; UniGene Home page.
- http://www.wormbase.org/; WormBase Home page.