Summary
This work revealed that the Arabidopsis TFIIB-related protein BRP4 is critical for male gametogenesis by modulating mitotic cell-cycle progression.
Key words: Arabidopsis, BRP4, male gametogenesis, mitosis, ORC6, TFIIB-related protein.
Abstract
Male gametogenesis in angiosperms involves two rounds of mitosis that are essential for the generation of two sperm cells to achieve double fertilization, a distinct event in the sexual reproduction of flowering plants. Precise regulation of mitosis during male gametogenesis is critically important for the establishment of the male germline. However, the molecular mechanisms underlying mitotic division during male gametophyte development have not been characterized fully. Here, we report that the Arabidopsis transcription initiation factor TFIIB-related protein BRP4 is involved in the regulation of mitotic cell-cycle progression during male gametogenesis. BRP4 was expressed predominately in developing male gametophytes. Knockdown expression of BRP4 by a native promoter-driven RNA interference construct in Arabidopsis resulted in arrest of the mitotic progression of male gametophytes, leading to a defect in pollen development. Moreover, we showed that the level of expression of a gene encoding a subunit of the origin recognition complex, ORC6, was decreased in BRP4 knockdown plants, and that the ORC6 knockdown transgenic plants phenocopied the male gametophyte defect observed in BRP4 knockdown plants, suggesting that ORC6 acts downstream of BRP4 to mediate male mitotic progression. Taken together, our results reveal that BRP4 plays an important role in the regulation of mitotic cell-cycle progression during male gametogenesis.
Introduction
The formation and development of male gametophytes in flowering plants involve both meiosis and mitosis. These processes generate a pair of functional sperm cells that are required for the success of subsequent double fertilization, an event specific and essential for plant sexual reproduction (McCormick, 1993; Berger and Twell, 2011). Male gametophytes of plants come from the pollen mother cells, which undergo a round of meiosis to produce a tetrad of microspores that are then released within reproductive locules. The uninucleate microspores undergo two rounds of mitotic cell division, which occur either before or after pollination, depending on the plant species (McCormick, 1993). The first mitotic cell division, pollen mitosis I (PMI), occurs asymmetrically to produce two differently sized cells with different fates (McCormick, 1993; Berger and Twell, 2011). The larger vegetative cell arrests at G1 phase and exits from the cell cycle, while the smaller generative cell undergoes a second symmetric mitosis, pollen mitosis II (PMII), and produces two sperm cells that are delivered to the female ovule via pollen tubes for double fertilization (Tanaka, 1997; Twell et al., 1998; Borg et al., 2009). Using both genetic and molecular genetic approaches, some of the key genes involved in microspore formation have been identified in Arabidopsis. Furthermore, the regulatory network underlying early anther development has been characterized (Ma et al., 2012). By contrast, our knowledge of the genetic and molecular regulation of the mitotic cell-cycle progression during male gametogenesis is still limited.
DUO POLLEN1 (DUO1) was the first transcription factor shown to be expressed specifically in generative cells. The microspores of duo1 plants can go through PMI, but the generative cells cannot undergo PMII. This leads to the formation of bicellular-like pollen grains that are incapable of finishing double fertilization (Durbarry et al., 2005; Rotman et al., 2005). DUO1 has been shown to regulate three germline marker genes, Histone H3.3 Variant (MGH3/HRT10), GAMETE EXPRESSED2 (GEX2), and GENERATIVE-CELL SPECIFIC1 (GCS1/HAP2) (Brownfield et al., 2009a). Recently, several additional genes, such as DUO1-ACTIVATED TARGET (DAT), were found possibly to be regulated by DUO1 (Borg et al., 2011). DUO POLLEN3 (DUO3) is another regulator whose function is similar to DUO1. DUO3 coordinates germ-cell division with DUO1, and may share the same regulatory targets as DUO1, as DUO3 is known to be required for the expression of GEX2 and GCS1/HAP2 (Brownfield et al., 2009b). The A-type cyclin-dependent kinase;1 (CDKA;1), a key regulator of both the G1/S and the G2/M phase transitions in the cell-cycle progression, was shown to be required for entry into generative-cell division, as cdka;1 produces bicellular pollen containing a single sperm-like cell and a vegetative cell (Iwakawa et al., 2006). Similarly, mutation of an F-box protein (FBL17) also causes a pollen defect similar to that observed for the cdka;1 mutant. FBL17 forms an SCF-type complex with the SKP1-like protein11 (ASK11) to target Kip-related protein 6 (KRP6) and KRP7 for proteasome-dependent degradation (Kim et al., 2008; Gusti et al., 2009). In addition, disruption of the Aberrant Pollen Development1 (APD1) and APD2 genes, which encode plant-specific RING-finger proteins, increases the percentage of bicellular-like pollen at the mature pollen stage; this phenotype is enhanced by downregulation of APD3 or APD4 (Luo et al., 2012). Recent studies in Arabidopsis have also identified a few genes that participate in the regulation of PMI. For example, simultaneous mutation in RING-H2 group F1a (RHF1a) and RHF2a not only leads to a defect in pollen development mainly due to the arrest of PMI but also blocks mitotic cell division during female gametophyte formation (Liu et al., 2008). This result indicates that these genes function in both male and female gametogenesis. RHF1a was also shown to interact directly with and target a cyclin-dependent kinase inhibitor, ICK4/KRP6, for proteasome-mediated degradation (Liu et al., 2008).
The initiation of eukaryotic gene transcription requires the formation of a pre-initiation complex (PIC) on promoter regions. The PIC is composed of PolII, transcription factor IIB (TFIIB), and TATA box-binding proteins (TBPs) (Roeder, 1996; Kostrewa et al., 2009). TFIIB transcription factors are central components of these complexes, as they not only recruit PolII to the promoter but also bind DNA and TBPs (Nikolov et al., 1995; Kostrewa et al., 2009). The crystal structure of the PolII-B complex shows that a zinc ribbon domain (B-ribbon) at the N terminus of TFIIB proteins can bind PolII, while its C-terminal domain (B-core) binds DNA and TBP (Kostrewa et al., 2009). In Arabidopsis, there are 14 TFIIB-like proteins that have been phylogenetically categorized into the TFIIB, Brf, and Rrn7/TAF1B/MEE12 subfamilies (Knutson, 2013; Niu et al., 2013). The TFIIB subfamily has eight members, including TFIIB1, TFIIB2, and six BRP proteins, among which BRP3 and BRP6 contain only partial TFIIB domains and are predicted to function differently from the other TFIIBs (Knutson, 2013). A recent study revealed that TFIIB1 plays important roles in pollen-tube growth and endosperm development, as mutation in TFIIB1 results in retarded growth of the pollen tube, impaired pollen-tube guidance and reception, and abnormal endosperm development (Zhou et al., 2013). It is interesting that the defective pollen-tube growth and guidance and reception can be restored completely by expression of TFIIB2 driven by the TFIIB1 promoter, and the defective seed development phenotype can be rescued by the expression of TFIIB3/BRP2 driven by the TFIIB1 promoter. These results suggest that these three genes function in a partially redundant and cooperative manner (Zhou et al., 2013). BRP1 is expressed ubiquitously and encodes a plant-specific TFIIB-related protein that is localized both to plastids and to the nucleus (Lagrange et al., 2003). BRP1 has been reported to be involved in RNA polymerase I-dependent rRNA synthesis (Imamura et al., 2008). By contrast, the expression of BRP2 is restricted to reproductive organs and seeds, and BRP2 is involved in the regulation of endosperm growth as brp2 exhibits a slower proliferation rate at the endosperm syncytial stage (Cavel et al., 2011). BRP5/PTF2 also encodes a plant-specific TFIIB-related protein and participates in pollen germination and embryogenesis, and its expression is abundant in developing pollen, embryos, and shoot apical meristems (Niu et al., 2013).
Here, we report that Arabidopsis transcription initiation factor BRP4, another member of the TFIIB-related protein family, is involved in the regulation of mitotic cell-cycle progression during male gametogenesis. BRP4 was highly expressed in developing male gametophytes, with a peak in expression at the tetrad stage of microspore development. Knockdown of BRP4 expression by a native promoter-driven RNA interference (RNAi) construct in Arabidopsis predominantly aborted male gametophyte development by arrest of the male gametophyte mitotic cell-cycle progression. We have provided evidence that a gene encoding a subunit of the origin recognition complex, ORC6, may act downstream of BRP4, and showed that ORC6 is required for male gametophyte cell-cycle progression. These findings suggest that BRP4-mediated male gametogenesis may function, at least in part, through ORC6-regulated cell division machinery.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana accession Columbia-0 plants were used in this study, with the exception that the small organ 3-D (smo3-D) mutant was initially isolated from a WS2 background. All seeds were sterilized in 0.5% sodium hypochlorite for 15min, and then geminated on 1/2 MS medium in a culture room at 22 °C under a 16h light/8h dark photoperiod with an illumination intensity of 80–90 μmol m–2 s–1. Seven-day-old seedlings were transferred to soil and grown in a growth room at 22±1 °C under the same photoperiod and illumination as those in the culture room (Jing et al., 2009). For morphological characterization of smo3-D, 30-d-old plants were used for examination of the leaf area, and 50-d-old plants were used to determine the plant height and silique length.
Sequence alignment and phylogenetic tree generation
The sequences of TFIIBs and BRP2 were obtained from The Arabidopsis Information Resource (TAIR) database. Alignment analysis was performed with the MUSCLE program (http://www.ebi.ac.uk/Tools/msa/muscle/), and then manually optimized with Genedoc software (Nicholas and Nicholas, 1997; Liu et al., 2013). The full-length amino acid sequences of the BRP homologues in Arabidopsis were used for maximum-likelihood phylogenetic analysis. The phylogenetic tree was reconstructed with PHYML version 2.4 using the GTR+I+G model, and 1000 bootstrap replicates were performed (Guindon and Gascuel, 2003; Liu et al., 2013).
Gene expression analysis
Total RNA was isolated using TRIzol reagent (Invitrogen). RNA was digested with DNase I and reverse transcribed with Superscript III reverse transcriptase (Invitrogen) into cDNA for reverse transcription (RT)-PCR or quantitative (q)RT-PCR analysis. GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE C SUBUNIT (GAPC) or ACTIN2 was used as an internal control in the RT-PCR or qRT-PCR analysis, respectively. qRT-PCR analysis was carried out with SYBR Premix Ex Taq Mix on a Rotor-Gene3000 (Corbett Research) by three biological replicates, according to the manufacturer’s instructions. All primers used in the expression analyses are listed in Supplementary Table S1 at JXB online.
For the RNA in situ hybridization experiments, inflorescences were collected from 30 individual 5-d-old plants and fixed in 4% paraformaldehyde. The inflorescences were then embedded in paraffin and sectioned to an 8 μm thickness. A 157bp cDNA fragment specific for BRP4 was amplified by PCR to generate digoxigenin-labelled sense and antisense probes. RNA in situ hybridization was performed according to a previously described protocol (Chen et al., 2007), and hybridization signals were visualized under a microscope (Olympus BX51).
For the β-glucuronidase (GUS) staining assay, the inflorescences and other organs of homozygous transgenic plants carrying a proBRP4:GUS construct were incubated in a 50mM sodium phosphate solution (pH 7.0) containing 5mM K4Fe(CN)6, 5mM K3Fe(CN)6, 0.1% Triton X-100, and 1mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid at 37 °C for several hours (Feng et al., 2011).
Plasmid construction and Arabidopsis transformation
To generate the pro35S:BRP4, pro35S:antiBRP4, pro35S:BRP2, and pro35S:TFIIBs constructs, the coding sequences of BRP4, TFIIBs, and BRP2 were amplified by RT-PCR and ligated into the pEASY-Blunt vector (TransGen Biotech). The plasmids were sequenced for verification and then digested with appropriate restriction endonuclease(s) and cloned into the pVIP96 or pCAMBIA1302 plasmid (Hu et al., 2003). A BRP4 fragment digested with SpeI and AscI was cloned into the pMDC83 plasmid for generation of pro35S:BRP4-GFP (Curtis and Grossniklaus, 2003). A BRP4 fragment digested with XhoI and SpeI was cloned into the pER10 plasmid to generate the chemically inducible proXVE:BRP4 construct (Zuo et al., 2000). For the RNAi constructs, a 157bp cDNA fragment specific for BRP4 or a 210bp fragment from ORC6 was cloned into the pCAMBIA1300 plasmid containing an RNAi fragment in both the sense and antisense orientations (Qin et al., 2005), and then the appropriate promoter region was cloned into the resulting plasmid to generate proBRP4:BRP4 RNAi, proACT11:BRP4 RNAi, and proBRP4:ORC6 RNAi constructs. For the proBRP4:GUS construct, a 2500bp genomic fragment from the BRP4 promoter was cloned into the pBI101 vector following digestion with BamHI and SalI. All primers used for generation of the constructs are listed in Supplementary Table S2 at JXB online.
All constructs were introduced into Arabidopsis by Agrobacterium tumefaciens-mediated transformation via the floral dip method, as described previously (Clough and Bent, 1998). At least 15 independent lines harbouring a single T-DNA insertion from each construct were generated, and at least three independent lines of their T3 plants were used for detailed characterization.
Alexander staining and 4′,6-diamidine-2-phenylindole dihydrochloride (DAPI) staining
To observe the developmental status of male gametophyte, anthers were dissected from flowers and mounted on 1/2 MS medium and visualized under a microscope (Olympus BX51). To determine the viability of mature pollen, pollen grains were stained in Alexander’s solution (Alexander, 1969). To visualize nuclei of microspores, stamens at different stages were detached and placed into 0.5 µg µl–1 of DAPI staining solution (Park et al., 1998; Liu et al., 2008). The Alexander- and DAPI-stained microspores or pollen grains were photographed, and their developmental status was determined under the microscope.
DAPI fluorescence was used to quantify the relative nuclear DNA content in generative cells. DAPI-stained images were recorded with a fixed exposure and then the DAPI fluorescence values of generative-cell nuclei were determined with ImageJ version 1.4.3.67 software (http://rsb.info.nih.gov/ij/). A net value for each nucleus was calculated after subtraction of the corresponding cytoplasmic background signal. The mean fluorescence value of the generative prophase nuclei from wild-type (WT) bicellular pollen was considered as 2C, and the values obtained were then normalized against this value to calculate the relative C values (Gusti et al., 2009).
Results
Ectopic expression of BRP4 has a pleiotropic effect on organ development
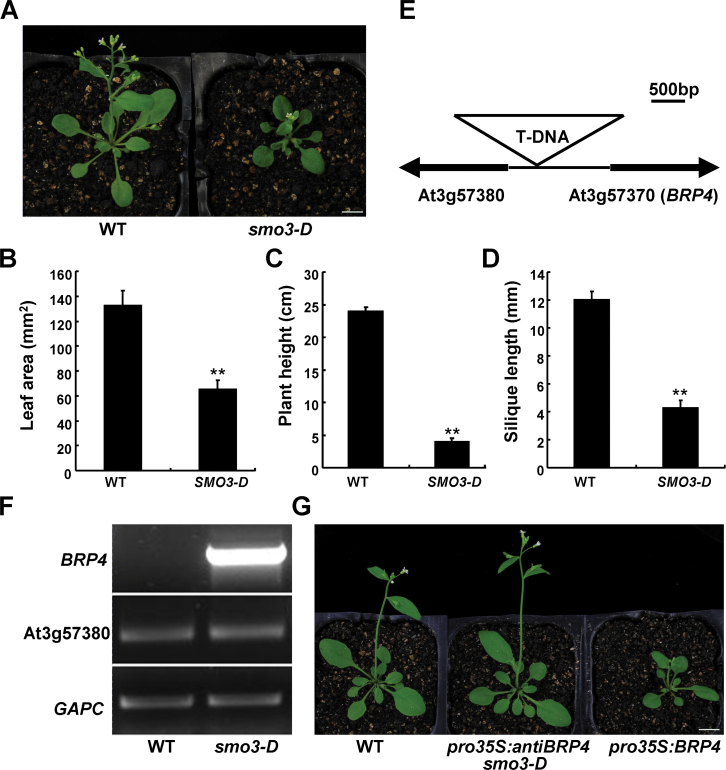
We initially isolated a dominant mutant, smo3-D, from a transgenic Arabidopsis population harbouring the T-DNA activation-tagging plasmid pSKI015 (Weigel et al., 2000). The most striking phenotype of smo3-D plants was the small size of their aerial organs (Fig. 1A). Compared with that of WT, the size of leaves, inflorescences, and siliques in the smo3-D plants was obviously reduced (Fig. 1B–D). Genetic analysis of a backcrossed F2 progeny indicated that the segregation ratio of mutant:WT was 83:28 (3:1, P=0.96, χ2 test), in which all of the mutant phenotypes co-segregated with a T-DNA insertion event. These results suggested that smo3-D is a single-gene dominant mutant that may be caused by a T-DNA insertion. To identify the gene responsible for the smo3-D phenotype, we amplified the genomic sequence flanking the T-DNA by thermal asymmetric interlaced-PCR (Liu et al., 1995), and found that a T-DNA segment was inserted between At3g57380 and At3g57370 (BRP4), 1014bp upstream of the start codon of BRP4 (Fig. 1E). RT-PCR analysis showed that the expression of BRP4 was highly upregulated in smo3-D plants (Fig. 1F). To examine whether the ectopic expression of BRP4 was responsible for the small aerial organ phenotype in smo3-D, we introduced a pro35S:BRP4 construct into WT plants and introduced a pro35S:antiBRP4 construct into smo3-D plants. As shown in Fig. 1G, transgenic plants overexpressing BRP4 also showed the small aerial organ phenotype observed in smo3-D, while introduction of the pro35S:antiBRP4 construct fully restored smo3-D to WT morphology. These results indicated that the small aerial organ phenotype in smo3-D results from the elevated expression of BRP4, and that the ectopic expression of BRP4 has a pleiotropic effect on organ development.
Fig. 1.
Ectopic expression of BRP4 affects aerial organ development. (A) Morphology of 23-d-old WT, left) and smo3-D plants (right). Bar, 1cm. (B) The area of fully expanded fifth leaves of WT and smo3-D plants. At least six leaves from each genotype were used to determine areas, and the data are shown as means±standard deviation (SD) (Student’s t-test, **P<0.01). (C, D) Plant height (C) and silique length (D) of WT and smo3-D plants. Fifty-day-old plants were used for determination of plant height and silique length; the data were from 15 plants of each genotype and data are presented as means±SD (Student’s t-test, **P<0.01). (E) Schematic illustration of the genomic region at the BRP4 locus in smo3-D. The coding regions of both At3g57370 (BRP4) and At3g57380 are indicated as black arrows, and the intermediate genomic sequence is indicated as a black line. T-DNA was inserted in the promoter region of BRP4, 1014bp upstream of ATG. (F) RT-PCR analysis of BRP4 and At3g57380 expression in WT and smo3-D plants. (G) Morphology of 3-week-old transgenic pro35S:BRP4 plants and smo3-D carrying a pro35S:antiBRP4 construct. Bar, 1cm.
BRP4 is a TFIIB-related protein
BLAST searching in the TAIR database showed that the amino acid sequence of BRP4 was highly similar to that of BRP2 (At3g29380), TFIIB1 (At2g41630), and TFIIB2 (At3g10330), all of which belong to the TFIIB family. Alignment indicated that these four proteins shared a conserved zinc ribbon domain and a C-terminal domain with two cyclin folds (Supplementary Fig. S1A at JXB online). However, compared with the other three TFIIB members, BRP4 had longer B-finger and B-linker domains (Supplementary Fig. S1A). In Arabidopsis, there are 14 TFIIB-like proteins. Phylogenetic analysis showed that TFIIB1, TFIIB2, BRP2, BRP4, BRP6, and BRP3 were clustered into a clade, although BRP4 appeared to be far from the other five members (Knutson, 2013; Niu et al., 2013) (Supplementary Fig. S1B). To test for the possibility of functional redundancy among these TFIIB family members, we overexpressed TFIIB1, TFIIB2, and BRP2 in Arabidopsis. Compared with the small aerial organs observed in the transgenic plants overexpressing BRP4, no morphological phenotypes were observed in transgenic plants overexpressing TFIIB1, TFIIB2, or BRP2 (Supplementary Fig. S1C, D), suggesting that BRP4 might have a unique function in plant development.
BRP4 is predominately expressed during the development of male gametophytes
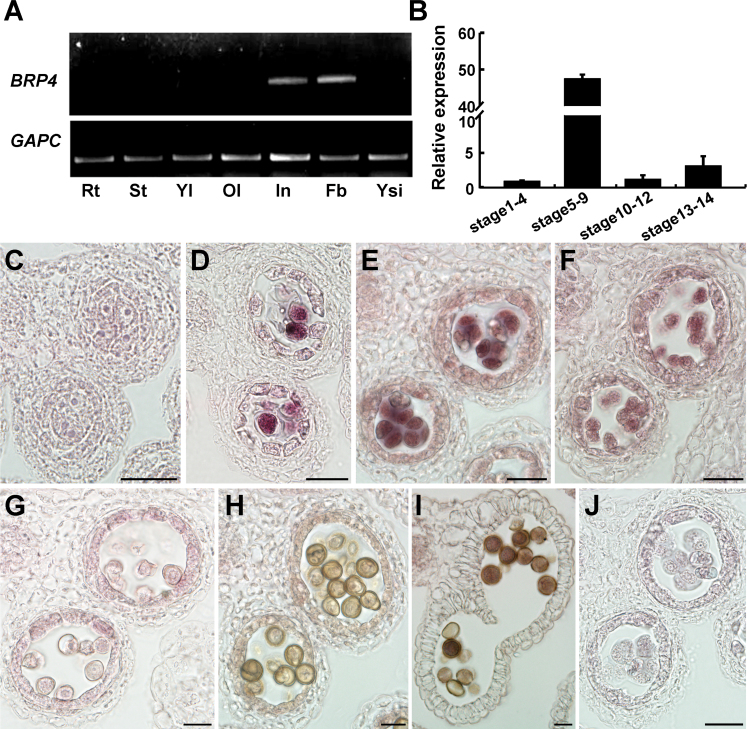
To explore the role of BRP4 in plant development, we initially examined the organ-specific expression of BRP4 with RT-PCR analysis and found that BRP4 transcripts were detected only in the inflorescences and flower buds (Fig. 2A). We then generated transgenic plants carrying a proBRP4:GUS construct, and GUS staining assays showed that BRP4 expression was expressed predominately in developing anthers and mature pollen (Supplementary Fig. S2A, B, at JXB online). These observations were consistent with the expression profiles of BRP4 revealed by the microarray data in the TAIR database (Supplementary Fig. S2C, D). To define the stage-specific expression of BRP4, we carried out qRT-PCR analysis with anthers at various developmental stages, and found that BRP4 was expressed primarily from anther development stages 5–9 (Fig. 2B), during which time the pollen mother cells are undergoing meiosis to produce tetrads and the microspores are then released (Sanders et al., 1999; Ma et al., 2012). Further RNA in situ hybridization verified that mRNA of BRP4 was initially detected at anther development stage 6, when pollen mother cells were undergoing meiotic division (Fig. 2C, D), and that BRP4 mRNA accumulation reached a peak level in the tetrads at anther development stage 7. BRP4 mRNA was also detectable in microspores at stage 8 (Fig. 2E, F). However, no BRP4 mRNA was detected in anthers after stage 9 (Fig. 2G–J). BRP4 mRNA was also detected in tapetum from anther development stages 5–8 (Fig. 2D–F). These findings strongly suggested that BRP4 may function during male gametophyte development. Additionally, we examined the cellular localization of BRP4 with pro35S:BRP4-GFP transgenic plants, which again showed the small aerial organ phenotype of pro35S:BRP4 plants (Supplementary Fig. S2E). Green fluorescent protein (GFP) fluorescence of the BRP4–GFP fusion protein in both root and petal epidermal cells showed that BRP4 was localized to the nucleus (Supplementary Fig. S2F, G).
Fig. 2.
Expression of BRP4. (A) RT-PCR analysis of BRP4 expression in various organs. Rt, root; St, stem; Yl, young leaf; Ol, old leaf; In, inflorescence; Fb, flower bud; Ysi, young silique. (B) Expression of BRP4 in anthers at the anther different developmental stages assayed by qRT-PCR. The data are from three biological replicates and are presented as means±SD. (C–J) BRP4 mRNA accumulation assayed by RNA in situ hybridization. RNA in situ hybridization was performed with anthers at stage 5 (C), stage 6 (D), stage 7 (E), stage 8 (F), stage 9 (G), stage 11 (H), and stage 12 (I). The anther at stage 7 hybridized with a BRP4 sense probe is shown as an example negative control (J). Bars, 20 µm.
Knockdown of BRP4 expression results in pollen developmental defects
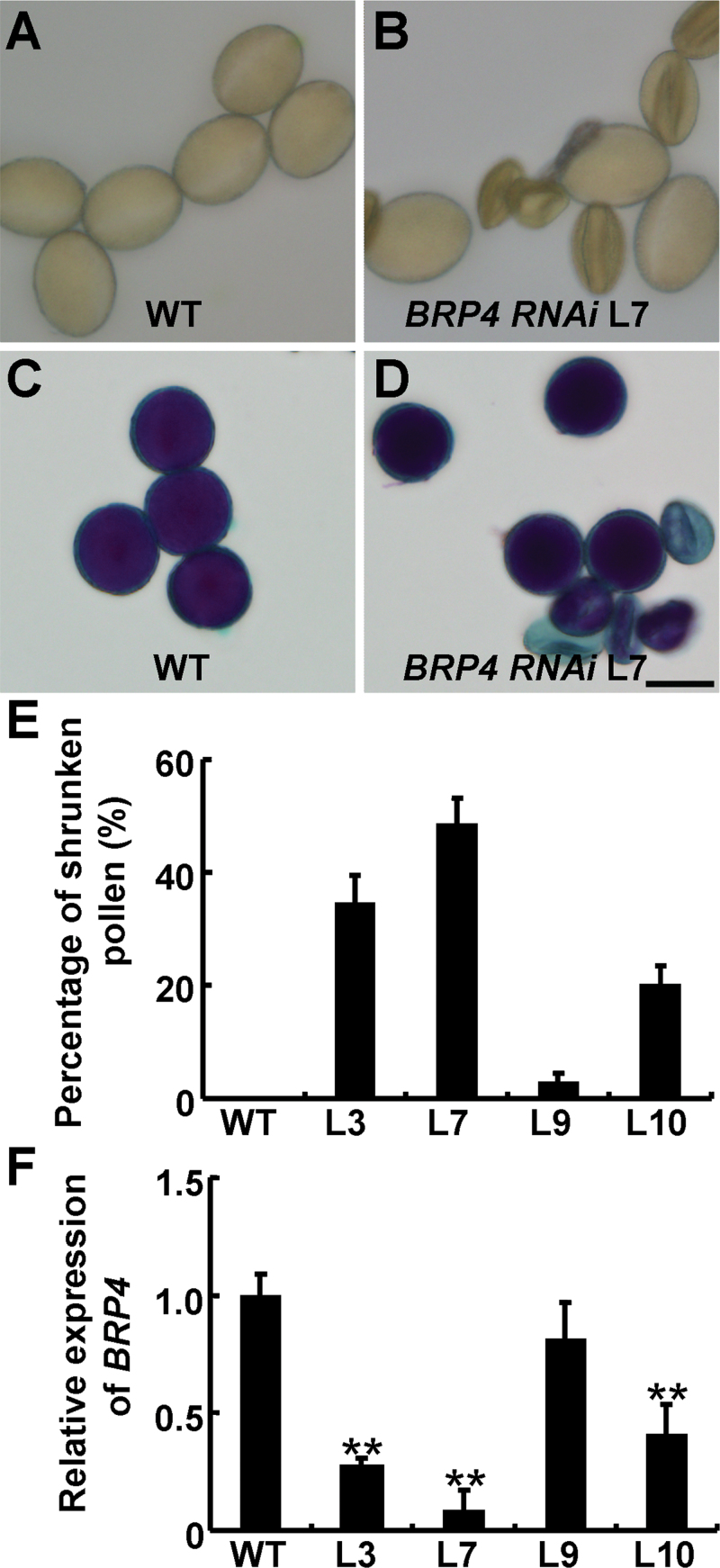
As no knockdown or knockout mutant of BRP4 in Arabidopsis is publically available, we used an RNAi approach to investigate the biological function of BRP4. A native promoter-driven BRP4 RNAi construct (proBRP4:BRP4 RNAi) was generated with a 157bp cDNA fragment specific for BRP4 and introduced into Arabidopsis (Supplementary Fig. S3A at JXB online). All of the transgenic plants from the 34 independent proBRP4:BRP4 RNAi lines we obtained developed normally in their organ morphology. However, 18 lines displayed a pollen abortion phenotype, with varied percentages of shrunken pollen (Fig. 3A, B). Further analysis of mature pollen in the T3 plants using Alexander staining confirmed the different percentages of abnormal pollen (Fig. 3C, D). Of the four independent lines that were examined closely, L7 produced about 48.64% shrunken pollen, while the percentages of shrunken pollen in L3, L10, and L9 were 34.75, 20.39, and 2.94%, respectively (Fig. 3C–E). qRT-PCR analysis showed that the variations of the aborted pollen phenotype were closely correlated to the reduced BRP4 expression levels in these lines (Fig. 3E, F). Expression of other TFIIB genes closely homologous to BRP4 was not obviously altered in these proBRP4:BRP4 RNAi lines (Supplementary Fig. S3B). These results suggested that the pollen development phenotype in these transgenic plants resulted from the knockdown of BRP4 expression. In addition, these transgenic plants also exhibited reproductive abnormalities. Compared with WT plants, which produced only 1.04% undeveloped ovules or seeds in siliques, the transgenic lines L7, L3, L10, and L9 had 40.62, 10.06, 3.87, and 1.22% aborted ovules or seeds, respectively (Supplementary Fig. S3C), demonstrating that knockdown of BRP4 expression also affects plant fertility.
Fig. 3.
Knockdown of BRP4 expression causes an aborted pollen phenotype. (A, B) Morphology of mature pollen from WT (A) and transgenic proBRP4:BRP4 RNAi L7 plants (B). (C, D) Alexander staining of pollen from WT (C) and L7 (D) plants. Bar, 20 µm. (E) Percentage of the shrunken pollen in WT and four independent T3 transgenic proBRP4:BRP4 RNAi lines (L3, L7, L9, and L10). (F) qRT-PCR analysis of the expression of BRP4 in WT and proBRP4:BRP4 RNAi lines as described in (E). The data are from three biological replicates and are presented as means±SD (Student’s t-test, **P<0.01).
To further verify whether or not the stage-specific knockdown of BRP4 expression was responsible for the pollen defects in the proBRP4:BRP4 RNAi transgenic plants, we also generated transgenic plants carrying a proACT11:BRP4 RNAi construct (Supplementary Fig. S3A at JXB online). ACT11 was expressed in young pistils from anther development stages 9–12, and was also strongly expressed in mature pollen (Huang et al., 1997). Among the 18 transgenic lines we obtained, no pollen developmental defects were observed, although a decrease in BRP4 expression was detected in the inflorescences of these transgenic plants (Supplementary Fig. S3D–F). This observation further supported the assertion that the expression of BRP4 in developing anthers is required for proper pollen development.
BRP4 affects the progression of male gametophyte mitosis
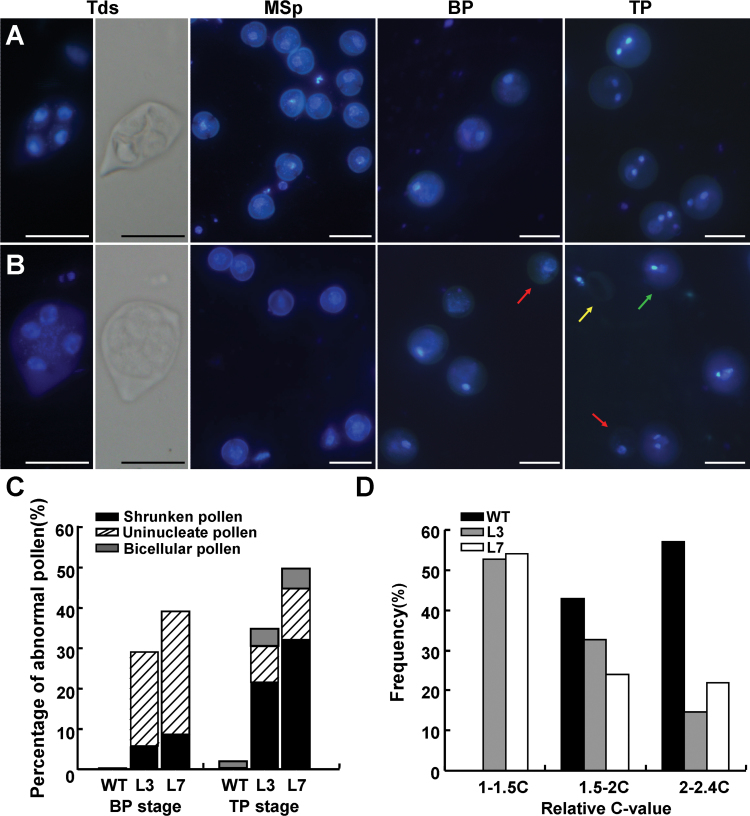
To explore the role of BRP4 during male gametogenesis, we carefully compared the microspore formation and development of WT and T3 proBRP4:BRP4 RNAi transgenic plants. At the tetrad and microspore stages, the male gametophytes appeared to be morphologically normal in both genotypes (Fig. 4A, B). However, a striking difference between the two genotypes was observed starting from the bicellular stage. At this stage, 99.56% of the microspores in the WT had two cells, in which the small generative cell had an intense fluorescent signal but the large vegetative cell had a faint signal (Fig. 4A, C). However, 39.15 and 29.29% of the microspores had only one cell with faint fluorescence in the proBRP4:BRP4 RNAi L7 and L3 plants, respectively (Fig. 4B, C), demonstrating that these microspores failed to go through PMI. At the tricellular stage, the percentages of aberrant microspores or bicellular-like pollen in the proBRP4:BRP4 RNAi L7 and L3 plants reached 49.74 and 35.07%, respectively. This implies that there are an additional 10.59 and 5.78% of bicellular-like pollen that is incapable of undergoing PMII in the L7 and L3 plants, respectively (Fig. 4A–C). These observations demonstrated that knockdown of BRP4 expression had no effect on microspore formation but impaired the progression of both PMI and PMII during male gametogenesis.
Fig. 4.
Knockdown of BRP4 expression arrests the mitotic cell-cycle progression in male gametophytes. (A, B) DAPI staining of tetrads, microspores, and bicellular and tricellular pollen from WT (A) and transgenic proBRP4:BRP4 RNAi L7 (B) plants. Note that the abnormal uninucleate pollen (red arrows) and bicellular-like pollen (green arrow) are still present at the bicellular and tricellular development stages in transgenic proBRP4:BRP4 RNAi plants. The yellow arrow indicates shrunken pollen. Bar, 20 µm. Tds, tetrads; MSp, microspores; BP, bicellular pollen; TP, tricellular pollen. (C) Quantification of different types of abnormally developed pollen from WT and transgenic proBRP4:BRP4 RNAi plants (L3 and L7). BP, bicellular pollen, TP, tricellular pollen. At the BP stage, about 1000 pollen grains were examined, and approximately 2500 pollen grains were examined at the TP stage. (D) Distribution of the DNA content of generative-cell nuclei at prophase from WT bicellular pollen (n=42) and generative-like cell nuclei at anthesis in proBRP4:BRP4 RNAi L3 (n=55) and L7 (n=50). The relative C-value of each nucleus was determined by the DAPI fluorescence value normalized against the mean fluorescence of WT nuclei (DNA=2C).
It was reported previously that Arabidopsis generative cells at prophase contain about 2C of the mean DNA content (Friedman, 1999). We conducted DNA content analysis of the generative-like cells of proBRP4:BRP4 RNAi L7 and L3 plants at anthesis. In WT generative cells, the distribution of the DNA content ranged from 1.5 to 2.4C, which reflects technical variations in the measurement of fluorescence (Brownfield et al., 2009b). By contrast, more than 50% of the generative-like cells from L7 and L3 had a DNA content of 1–1.5C (Fig. 4D), suggesting that these generative-like cells may be arrested at the G1/S phase during the mitotic cell-cycle progression.
ORC6 acts downstream of BRP4 during male gametogenesis
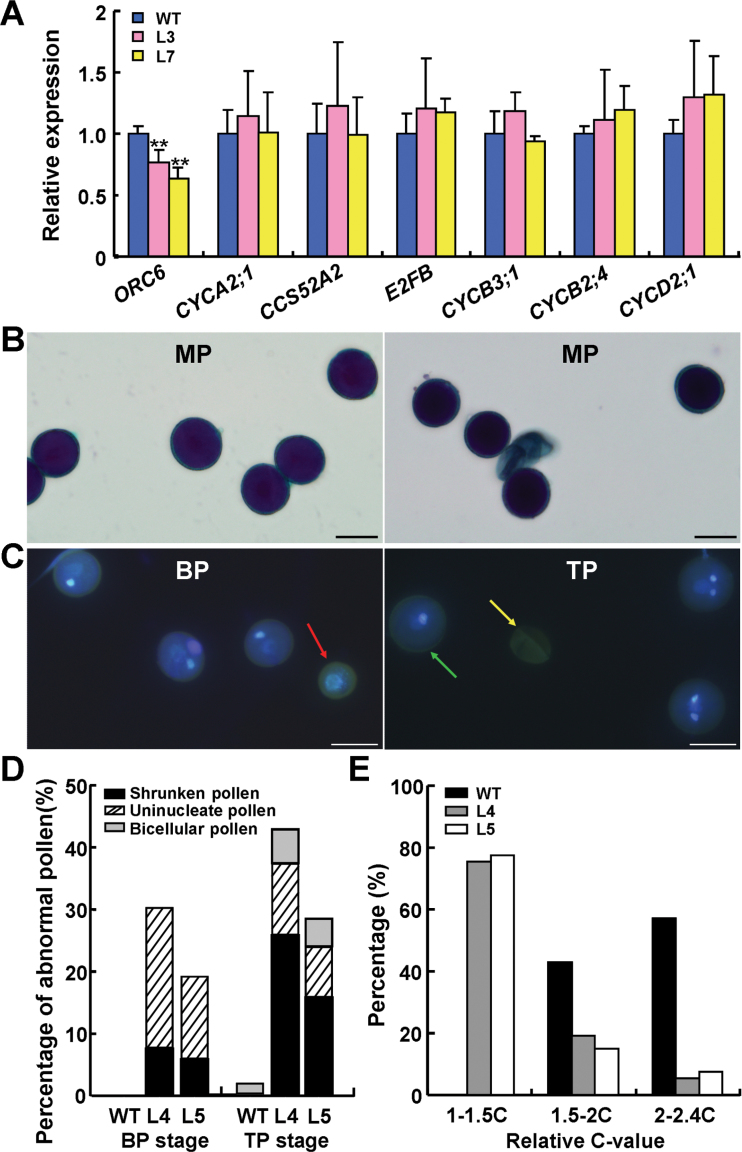
As BRP4 affects the mitotic cell-cycle progression of male gametophytes, we attempted to identify genes that may function downstream of BRP4. We examined expression of genes reported previously to be involved in the regulation of male gametogenesis, DUO1, DUO3, RHF1a, RHF2a, and CDKA;1, in both WT and proBRP4:BRP4 RNAi inflorescences. The expression levels of these genes were similar between the two genotypes (Supplementary Fig. S4A at JXB online), implying that these genes might not be downstream of BRP4. We then selected seven cell-cycle-related genes whose expression levels were reported to be high during male gametophyte development (Pina et al., 2005), and examined their expression in the inflorescences of both WT and proBRP4:BRP4 RNAi plants. As shown in Fig. 5A, the expression of a gene encoding a subunit of the origin recognition complex (ORC), ORC6, was obviously decreased in proBRP4:BRP4 RNAi inflorescences. The expression levels of the other six genes, CYCA2;1, CCS52A2, E2FB, CYCB3;1, CYCB2;4, and CYCD2;1, were similar between WT and proBRP4:BRP4 RNAi plants (Fig. 5A). To validate the possible regulation of ORC6 by BRP4, we generated transgenic plants carrying an oestradiol-inducible BRP4 construct, proXVE:BRP4. After transgenic seedlings were treated with the inducer, BRP4 was dramatically induced within 4h, and ORC6 was obviously upregulated after 8h, supporting the notion that ORC6 functions downstream of BRP4 (Supplementary Fig. S4B, C).
Fig. 5.
ORC6 acts downstream of BRP4 during male gametogenesis. (A) qRT-PCR analysis of cell division-related genes in WT and transgenic proBRP4:BRP4 RNAi inflorescences. The data are from three biological replicates and are presented as means±SD (Student’s t-test, **P<0.01). Note the decreased expression of ORC6 in the L3 and L7 plants. (B) Alexander staining of mature pollen (MP) from WT and transgenic proBRP4:ORC6 RNAi plants. Bars, 20 µm. (C) DAPI staining of pollen at the bicellular and tricellular stages from transgenic proBRP4:ORC6 RNAi anthers. The red arrow shows abnormal uninucleate pollen, the green arrow shows bicellular-like pollen, and the yellow arrow shows shrunken pollen with no DAPI staining. Bars, 20 µm. (D) Percentage of abnormal pollen in transgenic proBRP4:ORC6 RNAi L4 and L5 anthers. BP, bicellular pollen; TP, tricellular pollen. At the BP stage, about 1500 pollen grains were examined, and approximately 2500 pollen grains were examined at the TP stage. (E) DNA content analysis of WT generative-cell nuclei at prophase (n=42) and undivided generative-like cell nuclei at anthesis in proBRP4:ORC6 RNAi L4 (n=57) and L5 (n=53).
To investigate the involvement of ORC6 in BRP4-mediated male gametogenesis, we generated transgenic plants harbouring a BRP4 promoter-driven ORC6 RNAi construct (proBRP4:ORC6 RNAi) (Supplementary Fig. S5A at JXB online). Of the 28 independent transgenic lines examined, 12 displayed pollen developmental defects (Fig. 5B). Detailed examination of the T3 plants of three independent lines (L2, L4, and L5) showed that the percentages of shrunken pollen grains were 36.53, 42.15, and 26.56%, respectively. In contrast, the WT and transgenic plants carrying an empty vector (containing only the BRP4 promoter fragment) produced 0.67 and 2.2% abnormal pollen, respectively (Supplementary Fig. S5B). Expression analysis showed that the reduced expression of ORC6 correlated to the percentages of defective pollen in L2, L4, and L5 plants. The expression of other ORC genes in these plants did not appear to differ from their expression levels observed in the WT (Supplementary Fig. S5C, D). Further DAPI staining in L4 and L5 revealed that 30.45 and 19.4% of the microspores failed to go through PMI, while another 12.62 and 9.29% were arrested at PMII, respectively (Fig. 5C, D). Moreover, the DNA content analysis of generative-like cells from proBRP4:ORC6 RNAi L4 and L5 plants showed that about 75.44 and 77.36% of the aborted generative cells had a DNA content in the range of 1–1.5C (Fig. 4E), suggesting that these generative-like cells are also impaired at the G1/S phase. These observations suggested that ORC6 acts downstream of BRP4, and that it is, at least in part, involved in the BRP4-mediated mitotic cell-cycle progression.
Discussion
BRP4 participates in mitotic cell-cycle progression during male gametogenesis
In flowering plants, the two rounds of mitotic division during male gametogenesis are not only involved in cell fate determination but are also required for double fertilization. Several genes that participate in the regulation of male gametophyte development in Arabidopsis have been identified recently; these include DUO1, DUO3, RHF1a, RHF2a, FBL17, CDKA;1, and APD1-4. These discoveries have improved our understanding of male gametogenesis, yet the molecular basis of this process has not been fully characterized. Here, we showed that the BRP4, a member of the TFIIB-related protein family, is involved in regulation of the mitotic cell-cycle progression during male gametogenesis. Although the gain-of-function mutant of BRP4, smo3-D, was initially isolated due to its small aerial organ phenotype, our subsequent characterization studies clearly demonstrated that BRP4 performs a critically important function in mitotic cell-cycle progression during male gametogenesis. BRP4 is predominantly expressed in male gametophytes at anther development stages 6–8, when microspores are forming and then being released. Transgenic plants harbouring a native promoter-driven BRP4 RNAi construct displayed a pollen developmental defect. Moreover, our investigations suggested that this pollen defect resulted primarily from the impaired cell-cycle progression before and after PMI, which led to the production of underdeveloped pollen that subsequently aborted. Therefore, our findings show that BRP4 plays an important role in the mitotic cell-cycle progression during male gametogenesis.
BRP4 may function distinctly from other TFIIB-related members
In Arabidopsis, the TFIIB subfamily has eight members: TFIIB1, TFIIB2, and six BRP proteins (Knutson, 2013). However, BRP4 appears to be unique member among the TFIIB subfamily proteins of Arabidopsis. BRP4 has a long B-finger and B-linker domain at its N terminus, which may be important for the selection of transcription start sites and promoter opening (Kostrewa et al., 2009). Our observation that only transgenic plants overexpressing BRP4 exhibited the small aerial organ phenotype also supports the notion that BRP4 may mediate a biological process distinct from the other TFIIB subfamily proteins of Arabidopsis. Moreover, unlike the expression of the other TFIIB genes, which are ubiquitous or restricted to the reproductive organs and seeds, BRP4 is expressed predominately in developing male gametophytes and in the tapetum from anther development stages 6–8 and in mature pollen. The abundant BRP4 expression in developing pollen was reported previously in an expression profile analysis (Honys and Twell, 2004). Our observations strongly suggest that BRP4 plays important roles in male mitotic cell-cycle progression. Additionally, previous transcriptome analysis of anther development stages 4–7 showed that both BRP4 and TFIIB1 are expressed at these stages, but only BRP4 was obviously downregulated in the sporocyteless (spl) and excess male sporocytes1 (ems1) mutants. BRP4 was designated previously as a reproductive preferential gene, implying that BRP4 may be involved in EMS-mediated anther development (Ma et al., 2012).
BRP4 affects male gametogenesis, possibly by influencing the cell-cycle progression
In Arabidopsis, most of the identified factors involved in male gametogenesis affect the cell division progression of either microspore or generative cells. Our work demonstrated that BRP4 participates in the regulation of cell-cycle progression in both microspores and generative cells. Surprisingly, the expression levels of genes known to be involved in the mitotic cell-cycle progression of male gametophytes were comparable in the inflorescences of WT and proBRP4:BRP4 RNAi plants, suggesting that the role of BRP4 in regulation of mitotic cell-cycle progression is not dependent on the expression of these genes. Consistent with this, our further analysis indicated that ORC6 might be downstream of BRP4, and that the knockdown of ORC6 expression in Arabidopsis phenocopied the mitotic defects observed in BRP4 RNAi male gametophytes. This implied that ORC6 acts downstream of BRP4, and, at least in part, participates in BRP4-mediated male gametophyte development. ORC6 encodes a subunit of the ORC, which is required for the initiation of eukaryotic DNA replication, alterations to chromatin structure, and transcriptional silencing (Bell, 2002; Collinge et al., 2004). ORC6 expression peaks at the G1/S phase (Diaz-Trivino et al., 2005). Consistently, our DNA content analyses of generative-like cells in both proBRP4:BRP4 RNAi and proBRP4:ORC6 RNAi plants support the notion that DNA synthesis is impaired in these cells. Therefore, it is likely that BRP4 participates in the regulation of mitotic cell-cycle progression of male gametophytes, at least in part, through the ORC6-mediated cell-cycle regulatory machinery.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Alignment and phylogenetic analysis of BRP4 and TFIIB-related proteins.
Supplementary Fig. S2. Expression pattern and subcellular localization of BRP4.
Supplementary Fig. S3. Characterization of transgenic proBRP4:BRP4 RNAi and proACT11:BRP4 RNAi plants.
Supplementary Fig. S4. Identification of genes downstream of BRP4.
Supplementary Fig. S5. Generation and characterization of transgenic proBRP4:ORC6 RNAi plants.
Supplementary Table S1. The primers used for expression analysis in this study.
Supplementary Table S2. The primers used for the generation of DNA constructs in this study.
Acknowledgements
We thank Dr Kezhen Yang and Dr Huaqin Gong for technical help with the DAPI staining assay, and Mr Xuelei Lin for help with the phylogenetic analysis. This work was supported by grants from the National Natural Science Foundation of China (30800599 and 31121065) and the Ministry of Science and Technology of China (2009CB941500).
Glossary
Abbreviations:
- DAPI
4′,6-diamidine-2-phenylindole dihydrochloride
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- ORC
origin recognition complex
- PM
pollen mitosis
- PIC
pre-initiation complex
- RNAi
RNA interference
- RT-PCR
reverse transcription-PCR
- qRT-PCR
quantitative RT-PCR
- SD
standard deviation
- TBP
TATA box-binding protein
- TAIR
The Arabidopsis Information Resource
- WT
wild type.
References
- Alexander MP. 1969. Differential staining of aborted and nonaborted pollen. Stain Technology 44, 117–122 [DOI] [PubMed] [Google Scholar]
- Bell SP. 2002. The origin recognition complex: from simple origins to complex functions. Genes & Development 16, 659–672 [DOI] [PubMed] [Google Scholar]
- Berger F, Twell D. 2011. Germline specification and function in plants. Annual Reviews Plant Biology 62, 461–484 [DOI] [PubMed] [Google Scholar]
- Borg M, Brownfield L, Khatab H, Sidorova A, Lingaya M, Twell D. 2011. The R2R3 MYB transcription factor DUO1 activates a male germline-specific regulon essential for sperm cell differentiation in Arabidopsis . Plant Cell 23, 534–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg M, Brownfield L, Twell D. 2009. Male gametophyte development: a molecular perspective. Journal of Experimental Botany 60, 1465–1478 [DOI] [PubMed] [Google Scholar]
- Brownfield L, Hafidh S, Borg M, Sidorova A, Mori T, Twell D. 2009a. A plant germline-specific integrator of sperm specification and cell cycle progression. PLoS Genetics 5, e1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownfield L, Hafidh S, Durbarry A, Khatab H, Sidorova A, Doerner P, Twell D. 2009b. Arabidopsis DUO POLLEN3 is a key regulator of male germline development and embryogenesis. Plant Cell 21, 1940–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavel E, Pillot M, Pontier D, Lahmy S, Bies-Etheve N, Vega D, Grimanelli D, Lagrange T. 2011. A plant-specific transcription factor IIB-related protein, pBRP2, is involved in endosperm growth control. PLoS One 6, e17216, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Li HJ, Shi DQ, Yuan L, Liu J, Sreenivasan R, Baskar R, Grossniklaus U, Yang WC. 2007. The central cell plays a critical role in pollen tube guidance in Arabidopsis . Plant Cell 19, 3563–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743 [DOI] [PubMed] [Google Scholar]
- Collinge MA, Spillane C, Köhler C, Gheyselinck J, Grossniklaus U. 2004. Genetic interaction of an origin recognition complex subunit and the Polycomb group gene MEDEA during seed development. Plant Cell 16, 1035–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. 2003. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology 133, 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Trivino S, del Mar Castellano M, de la Paz Sanchez M, Ramirez-Parra E, Desvoyes B, Gutierrez C. 2005. The genes encoding Arabidopsis ORC subunits are E2F targets and the two ORC1 genes are differently expressed in proliferating and endoreplicating cells. Nucleic Acids Research 33, 5404–5414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbarry A, Vizir I, Twell D. 2005. Male germ line development in Arabidopsis. duo pollen mutants reveal gametophytic regulators of generative cell cycle progression. Plant Physiology 137, 297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Qin Z, Yan J, Zhang X, Hu Y. 2011. Arabidopsis ORGAN SIZE RELATED1 regulates organ growth and final organ size in orchestration with ARGOS and ARL. New Phytologist 191, 635–646 [DOI] [PubMed] [Google Scholar]
- Friedman WE. 1999. Expression of the cell cycle in sperm of Arabidopsis: implications for understanding patterns of gametogenesis and fertilization in plants and other eukaryotes. Development 126, 1065–1075 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52, 696–704 [DOI] [PubMed] [Google Scholar]
- Gusti A, Baumberger N, Nowack M, Pusch S, Eisler H, Potuschak T, De Veylder L, Schnittger A, Genschik P. 2009. The Arabidopsis thaliana F-box protein FBL17 is essential for progression through the second mitosis during pollen development. PLoS One 4, e4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D. 2004. Transcriptome analysis of haploid male gametophyte development in Arabidopsis . Genome Biology 5, R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Xie Q, Chua NH. 2003. The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 15, 1951–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SR, An YQ, McDowell JM, McKinney EC, Meagher RB. 1997. The Arabidopsis ACT11 actin gene is strongly expressed in tissues of the emerging inflorescence, pollen, and developing ovules. Plant Molecular Biology 33, 125–139 [DOI] [PubMed] [Google Scholar]
- Imamura S, Hanaoka M, Tanaka K. 2008. The plant-specific TFIIB-related protein, pBrp, is a general transcription factor for RNA polymerase I. EMBO Journal 27, 2317–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa H, Shinmyo A, Sekine M. 2006. Arabidopsis CDKA;1, a cdc2 homologue, controls proliferation of generative cells in male gametogenesis. The Plant Journal 45, 819–831 [DOI] [PubMed] [Google Scholar]
- Jing Y, Cui D, Bao F, Hu Z, Qin Z, Hu Y. 2009. Tryptophan deficiency affects organ growth by retarding cell expansion in Arabidopsis . The Plant Journal 57, 511–521 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Oh SA, Brownfield L, Hong SH, Ryu H, Hwang I, Twell D, Nam HG. 2008. Control of plant germline proliferation by SCFFBL17 degradation of cell cycle inhibitors. Nature 455, 1134–1137 [DOI] [PubMed] [Google Scholar]
- Knutson BA. 2013. Emergence and expansion of TFIIB-like factors in the plant kingdom. Gene 526, 30–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrewa D, Zeller ME, Armache KJ, Seizl M, Leike K, Thomm M, Cramer P. 2009. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature 462, 323–330 [DOI] [PubMed] [Google Scholar]
- Lagrange T, Hakimi MA, Pontier D, Courtois F, Alcaraz JP, Grunwald D, Lam E, Lerbs-Mache S. 2003. Transcription factor IIB (TFIIB)-related protein (pBrp), a plant-specific member of the TFIIB-related protein family. Molecular and Cellular Biology 23, 3274–3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhang Y, Qin G, et al. , 2008. Targeted degradation of the cyclin-dependent kinase inhibitor ICK4/KRP6 by RING-type E3 ligases is essential for mitotic cell cycle progression during Arabidopsis gametogenesis. Plant Cell 20, 1538–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Cui S, Wu F, Yan S, Lin X, Du X, Chong K, Schilling S, Theissen G, Meng Z. 2013. Functional conservation of MIKC*-type MADS box genes in Arabidopsis and rice pollen maturation. Plant Cell 25, 1288–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. 1995. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. The Plant Journal 8, 457–463 [DOI] [PubMed] [Google Scholar]
- Luo G, Gu H, Liu J, Qu LJ. 2012. Four closely-related RING-type E3 ligases, APD1–4, are involved in pollen mitosis II regulation in Arabidopsis . Journal of Integrative Plant Biology 54, 814–827 [DOI] [PubMed] [Google Scholar]
- Ma X, Feng B, Ma H. 2012. AMS-dependent and independent regulation of anther transcriptome and comparison with those affected by other Arabidopsis anther genes. BMC Plant Biology 12, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S. 1993. Male gametophyte development. Plant Cell 5, 1265–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HBJ. 1997. GeneDoc: a tool for editing and annotating multiple sequence alignments. EMBNEW.NEWS 4, 14 [Google Scholar]
- Nikolov DB, Chen H, Halay ED, Usheva AA, Hisatake K, Lee DK, Roeder RG, Burley SK. 1995. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature 377, 119–128 [DOI] [PubMed] [Google Scholar]
- Niu QK, Liang Y, Zhou JJ, Dou XY, Gao SC, Chen LQ, Zhang XQ, Ye D. 2013. Pollen-expressed transcription factor 2 encodes a novel plant-specific TFIIB-related protein that is required for pollen germination and embryogenesis in Arabidopsis . Molecular Plant 6, 1091–1108 [DOI] [PubMed] [Google Scholar]
- Park SK, Howden R, Twell D. 1998. The Arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development 125, 3789–3799 [DOI] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijo JA, Becker JD. 2005. Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiology 138, 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G, Gu H, Zhao Y, et al. 2005. An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 17, 2693–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends in Biochemical Sciences 21, 327–335 [PubMed] [Google Scholar]
- Rotman N, Durbarry A, Wardle A, Yang WC, Chaboud A, Faure JE, Berger F, Twell D. 2005. A novel class of MYB factors controls sperm-cell formation in plants. Current Biology 15, 244–248 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB. 1999. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sexual Plant Reproduction 11, 297–322 [Google Scholar]
- Tanaka I. 1997. Differentiation of generative and vegetative cells in angiosperm pollen. Sexual Plant Reproduction 10, 1–7 [Google Scholar]
- Twell D, Park SK, Lalanne E. 1998. Asymmetric division and cell-fate determination in developing pollen. Trends in Plant Science 3, 305–310 [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, et al. 2000. Activation tagging in Arabidopsis . Plant Physiology 122, 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JJ, Liang Y, Niu QK, Chen LQ, Zhang XQ, Ye D. 2013. The Arabidopsis general transcription factor TFIIB1 (AtTFIIB1) is required for pollen tube growth and endosperm development. Journal of Experimental Botany 64, 2205–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH. 2000. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. The Plant Journal 24, 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.