Summary
Investigations into the role of tomato ARF6 and ARF8 reveal that they are critical components in floral and gynoecium development before anthesis.
Keywords: ARF6, ARF8, expression, female sterility, flower development, tomato.
Abstract
Auxin regulates the expression of diverse genes that affect plant growth and development. This regulation requires AUXIN RESPONSE FACTORS (ARFs) that bind to the promoter regions of these genes. ARF6 and ARF8 in Arabidopsis thaliana are required to promote inflorescence stem elongation and late stages of petal, stamen, and gynoecium development. All seed plants studied thus far have ARF6 and ARF8 orthologues as well as the microRNA miR167, which targets ARF6 and ARF8. Whether these genes have broadly conserved roles in flower development is not known. To address this question, the effects of down-regulation of ARF6 and ARF8 were investigated through transgenic expression of Arabidopsis MIR167a in tomato, which diverged from Arabidopsis before the radiation of dicotyledonous plants approximately 90–112 million years ago. The transgenic tomato plants overexpressing MIR167a exhibited reductions in leaf size and internode length as well as shortened petals, stamens, and styles. More significantly, the transgenic plants were female-sterile as a result of failure of wild-type pollen to germinate on the stigma surface and/or to grow through the style. RNA-Seq analysis identified many genes with significantly altered expression patterns, including those encoding products with functions in ‘transcription regulation’, ‘cell wall’ and ‘lipid metabolism’ categories. Putative orthologues of a subset of these genes were also differentially expressed in Arabidopsis arf6 arf8 mutant flowers. These results thus suggest that ARF6 and ARF8 have conserved roles in controlling growth and development of vegetative and flower organs in dicots.
Introduction
Various hormonal signals regulate flower and subsequent fruit development (Kelley and Gasser, 2009; Nemhauser et al., 1998; Nitsch, 1952; Serrani et al., 2008; Vivian-Smith and Koltunow, 1999). Among them, auxin is critical in regulating gene expression within the flower and young fruit (Dharmasiri et al., 2005; Guilfoyle, 1986; Liscum and Reed, 2002). Auxin response factors (ARFs) bind to auxin response elements (5′-tgtctc-3′ or other similar sequences) in the promoter regions of numerous early auxin-inducible genes (Boer et al., 2014; Franco-Zorrilla et al., 2014; Tiwari et al., 2003; Ulmasov et al., 1997). Typical ARF transcription factors contain three domains: (i) a conserved N-terminal DNA-binding domain (DBD) of the B3 family, (ii) a non-conserved middle region, which may either activate or repress gene expression, and (iii) a conserved C-terminal dimerization domain (CTD) including motif III and motif IV which can dimerize with Aux/IAA (auxin/indoleacetic acid protein) repressors and other ARFs (Guilfoyle and Hagen, 2007; Guilfoyle et al., 1998). Among Arabidopsis ARF proteins that mediate auxin-induced gene activation (Tiwari et al., 2003; Ulmasov et al., 1999a), ARF6 and ARF8 regulate growth in both vegetative and reproductive tissues. Arabidopsis arf6 and arf8 single mutants have mild delays in stem elongation and flower organ growth (Nagpal et al., 2005). However, arf6 arf8 double mutants have more severe developmental defects, indicating that ARF6 and ARF8 have partially overlapping functions. Arabidopsis arf6 arf8 flowers arrest as closed buds with short petals, short stamens, and indehiscent anthers, as well as defects in gynoecium growth and support of pollen tube growth (Nagpal et al., 2005). Moreover, ARF6 and ARF8 promote jasmonic acid (JA) production, which in turn induces the expression of MYB21 and MYB24 required for petal, stamen, and gynoecium growth at anthesis (Reeves et al., 2012; Tabata et al., 2010). Hence, in Arabidopsis, ARF6 and ARF8 coordinate the development of petals and both male and female organs at the transition from closed buds to mature fertile flowers, which contributes to efficient fertilization.
All seed plants seem to have ARF6 and ARF8 orthologues (Axtell and Bartel, 2005; Oh et al., 2008; Remington et al., 2004; Xing et al., 2011; Yang et al., 2006), yet their functions have not been studied beyond Arabidopsis. Whether these genes have broadly conserved roles in flower development thus remains an outstanding question. The putative importance of ARF6 and ARF8 in plant development is suggested by the observation that they are probably targets of microRNA167 (miR167) in all analysed plants (Axtell and Bartel, 2005; Oh et al., 2008; Remington et al., 2004; Xing et al., 2011; Yang et al., 2006). It has been experimentally determined in Arabidopsis that miRNA167 regulates expression of ARF6 and ARF8 and that overexpression of the miR167 precursor gene MIR167a phenocopies the arf6 arf8 double mutant (Ru et al., 2006; Wu et al., 2006).
Tomato provides an excellent model to test the potentially conserved role of ARF6 and ARF8 in flower development as well as their regulation by miR167. Tomato and Arabidopsis diverged before the radiation of dicotyledonous plants, approximately 90–112 million years ago as estimated by genome sequence comparison and fossil evidence (Ku et al., 2000; Yang et al., 1999). Cultivated tomato (Solanum lycopersicum) evolved from Solanum pimpinellifolium during the process of crop domestication (Blanca et al., 2012) and they exhibit nearly identical gene sequences (Tomato Genome Consortium, 2012). In our laboratory, we routinely use the S. pimpinellifolium accession LA1589 for analyses of tomato development (e.g. Wu et al., 2011; Xiao et al., 2008; Xiao et al., 2009)). We report here that overexpression of the AtMIR167a gene in the wild tomato accession LA1589 leads to down-regulation of SpARF6 and SpARF8, which in turn alters flower maturation as in Arabidopsis. Our results thus suggest that ARF6 and ARF8 play a critical and highly conserved role in flower development in dicot plants.
Materials and methods
Plant materials
S. pimpinellifolium accession LA1589 was transformed with plasmid pB7WG2-MIR167a (Wu et al., 2006) at the Plant Transformation Facility at University of California Davis, USA. Transgenic and wild-type control LA1589 were grown in a greenhouse under standard conditions including supplemental lighting in 5-l pots. The T0 transgenic lines were named 092026-001 through 092026-006, 092370-001 and 092370-002. Owing to their female sterility, the severe lines were maintained by backcrossing the pollen to wild-type LA1589. A subset of those backcrossed lines were sowed as pedigree 10S222 (two seedlings: MIR167a_222-2 and MIR167a_222-3) and were derived from a backcross with T0 plant 092026-003 as the pollen donor. For each seedling, four cuttings were rooted, and floral buds from the rooted cuttings were taken for the RNA-seq analysis.
Identification of ARF6, ARF8, and MIR167 genomic sequences from tomato
By using the DNA-binding domain of Arabidopsis ARF6 and 8 proteins (defined as amino acids 1 to ~350 by Ulmasov et al., 1999b) as query sequences, four cultivated tomato genes were identified that share a likely common ancestor: SlARF6A (Solyc00g196060), SlARF6B (solyc07g043610/043620), SlARF8A (Solyc03g031970), and SlARF8B (Solyc02g037530). As some of the SlARFs were not correctly annotated in the tomato genome sequence, we used the validated ORF sequences obtained from Mohamed Zouine and Mondher Bouzayen (University of Toulouse, France) for phylogenetic analyses and the RNA seq analyses shown in Supplementary Table S2 available at JXB online. SlARF6B and SpARF6B are probably pseudogenes in both cultivated and wild tomato, and were excluded from most of the analyses. For MIR167, we used the mature microRNA sequence as a query to search for possible MIR167 gene candidates in the tomato genome (http://solgenomics.net) with Tomato WGS Scaffolds (SL2.40) and an expected threshold value of 1e–0 to avoid false positives. The SoMART software (http://somart.ist.berkeley.edu) was employed to validate miR167-mediated regulation of SlARF6A, SlARF8A, and SlARF8B. The SoMART software uses the small RNA (from http://smallrna.udel.edu) and degradome RNA deep sequencing data (Li et al., 2012). We used the Slicer Detector (SLY1-3 libraries), dRNA mapper (D51Wt library), and SMART COMPARE programs in SoMART with default settings. Both small RNA and degradome RNA deep sequencing data were generated from the tomato cultivar VF36, whereas the genome sequence is that of Heinz1706 cultivar.
Construction of the class II ARF phylogenetic tree
ARF proteins have a highly conserved DNA-binding domain at the N-terminal region and motif III-IV (two motifs shared with Aux/IAA proteins) at the C-terminal end, but a divergent middle region that generally cannot be aligned among different ARF proteins. The class II ARFs, to which tomato ARF6 and ARF8 belong, are recognized by a glutamine-rich middle region. To generate the phylogenetic tree, we used the core ARF DNA-binding domain that encompasses a span of ~350 amino acids at the N-terminus but excludes the first ~50 variable amino acids (Ulmasov et al., 1999b). Arabidopsis ARF protein sequences were obtained from TAIR (The Arabidopsis Information Resource, http://www.Arabidopsis.org/). The 8 sequences were aligned using MUSCLE (Edgar, 2004) and a tree was generated using RAxML 7.4.2 (Stamatakis, 2006), both under default settings. To root the tree, the ARF1 DNA binding domain sequence was used as an outgroup. To find the consensus sites for ARF alignment, trimAl (Capella-Gutierrez et al., 2009) was run on the full MUSCLE alignment under strictplus. The trees were visualized using FigTree v 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/).
Plant phenotype analyses
For floral organ measurement, flowers were collected from inflorescences of wild-type and transgenic LA1589 plants. The floral organs were separated and placed on half strength MS solid medium for imaging under a dissection microscope (Leica MZFLIII, Germany). Length measurements were performed using Image J (http://rsb.info.nih.gov/ij/). For leaflet measurements, 10 mature terminal leaflets were collected from each line, scanned on a flat-bed scanner and analysed by Tomato Analyzer application (http://www.oardc.ohio-state.edu/vanderknaap/tomato_analyzer.php). The leaf shape index refers to the ratio of height to width.
For light microscopy, samples were fixed in a mixture of 3% glutaraldehyde/4% paraformaldehyde/0.05% Triton X-100 in 0.1M potassium phosphate buffer (pH 7.2) for 2h at room temperature and then overnight at 4°C. After three washes with the potassium phosphate buffer, the samples were dehydrated in a graded ethanol series (25%, 50%, 70% and twice 90%), infiltrated with the EMbed 812 resin (Electron Microscopy Services, Hatfield, PA) and 90% ethanol series (1:3, 1:1, 3:1, twice 100% resin), embedded in airtight gelatine capsules (Electron Microscopy Services) and polymerized overnight at 60°C. Five-µm thick sections were collected on glass slides and stained with 0.5% toluidine blue in 0.1% sodium bicarbonate/25% ethanol for light microscopic observation.
For scanning electron microscopy, samples were infiltrated with 3% gluteraldehyde/2% paraformaldehyde in 0.1M potassium phosphate buffer, pH 7.4, for 2h and subsequently stored in 70% ethanol overnight at 4 °C. The samples were then dehydrated in an ethanol series (25%, 50%, 80%, and twice 95%), critical-point dried, sputter-coated with gold, and viewed at 20kV in an scanning electron microscope (Hitachi S-4700, Japan).
For the hormone experiments, selected floral buds were tagged before the petals in the MIR167- transgenic lines turned white, which corresponded to 1–2 days before anthesis in wild-type plants. They were sprayed with 10 µM IAA, 500 µM JA, or 100 µM GA3 (gibberellin A3; in 10% methanol and 0.05% Tween 20) or with buffer alone every 2 d. Fruit development was evaluated one week after the start of hormone treatment.
Pollen tube staining
Flower buds of LA1589 were emasculated one day before anthesis and pollinated using MIR167- transgenic pollen. In parallel, LA1589 pollen was used to pollinate MIR167-transgenic flowers. Pollinated pistils were collected after 16h, and kept in a fixative (3:1 of 95% ethanol: glacial acetic acid) overnight. The pistils were cleared with 5M NaOH softening solution for 24h followed by 6-h staining with aniline blue (ABF: 4′4-[carbonyl bis (benzene-4,1-diyl)-bis (imino)]-bisbenzensulfonic acid) in 0.1M K2HPO4 (pH 10) for callose staining of the pollen tube walls. Slide-mounted pistils were examined using an epi-fluorescence microscope (Leica DM IRB, Germany) equipped with a digital camera. Images were captured at ×50 magnification with QImaging Retiga 2000.
In situ hybridization
To generate RNA probes for RNA in situ hybridization of LA1589 tissues, we amplified linear templates for SpARF6A and SpARF8B from cDNAs using the following specific primer pairs: SpARF6A, 5′-TTTCATGAACCGGAACCATT-3′ and 5′-CAAAATTGCCAACGAGTGTG-3′; SpARF8B, 5′-GGGAAAG GAAGAGGCTGAAT-3′ and 5′-CGAAAGCTAAAGAAGCCAG GT-3′. An antisense or sense RNA probe was prepared by adding a sequence containing the T7 promoter sequence to the 5′ of the reverse or forward primers, respectively. Probes were labelled by in vitro transcription with T7 polymerase using a DIG RNA labeling kit (Roche, Indianapolis, IN, USA). Flower buds at 9 and 4 d before anthesis were fixed with 4% paraformaldehyde in 0.1M phosphate buffer (pH 7.4) and embedded in paraffin. Ten-µm thick sections were obtained with a microtome (American Optical Spencer 820, USA). After dewaxing the sections were washed with 0.2×saline-sodium citrate buffer and incubated with blocking solution (Boehringer, USA) before hybridization with DIG-labelled RNA probes overnight at 55 °C. After buffer washes the DIG-labelled RNA probes were detected by an alkaline-phosphatase-conjugated antibody (Anti-digoxigenin-AP Fab fragments, Roche, USA). After further buffer washes the sections were incubated with the NBT/BCIP solution (Roche, USA) for colour reaction. The mounted slides were observed under the epi-fluorescence microscope (Leica DM IRB) equipped with a digital camera (Q Imaging Retiga 2000, USA).
RNA isolation and RNA seq library construction
Tomato organs and tissues were collected between 9.00h and 10.00h (supplemental lighting was turned on at 6.00h) and immediately frozen in liquid nitrogen. This time point of collection (short duration and 3h after the lights were turned on) reduced the chance to identify differentially expressed genes due to the circadian clock. Sample collections were performed on separate days for the replicates. Total RNA was extracted with Trizol (Invitrogen Inc. Carlsbad, CA, USA) as described by the manufacturer. RNA quantity and quality were assessed using a Qubit 2.0 fluorometer RNA Assay Kit (Invitrogen Inc. USA) and an Agilent 2100 Bioanalyzer RNA 6000 Nano kit (Agilent, Santa Clara, CA, USA).
The expression analysis for SpARF6A, SpARF8A, SpARF8B, and STYLE2.1 in LA1589 included the following samples that were collected from greenhouse-grown plants: newly developing leaves approximately 5mm long, full size green terminal leaflets, flower/inflorescence meristems and flower buds up to 10 d before anthesis, flowers at anthesis, 10 d post-anthesis (DPA) fruit, 20 DPA fruit, and breaker stage ripening fruit. The following organs were collected from 7-day-old seedlings grown in petri dishes in a Conviron incubator (Winnipeg, Manitoba, Canada) (16h light and 8h dark): whole root, hypocotyl from below the cotyledons to above the root zone, cotyledons, and vegetative shoot apex containing the vegetative meristem and leaf primordia. Gene expression analysis of wild-type and MIR167a- overexpressing LA1589 were evaluated in flower/inflorescence meristems and flower buds up to 10 d before anthesis. This corresponds to the ovule initiation step in floral development (Xiao et al., 2009).
Strand-specific single-end RNA-seq libraries with insert size of approximately 250bp were prepared using the protocol described by Zhong et al. (2011) using 10 µg of total RNA. Eight libraries with compatible barcodes were pooled and run on a single lane in a flowcell on the Illumina HiSeq2000 at the Genomics Resources Core Facility at Weill Cornell Medical College (New York, NY, USA) and sequences of 44–51bp length were generated.
Alignment and analysis of Illumina reads
After the Illumina reads were quality checked, demultiplexed and trimmed, they were clustered per library. The reads were aligned to ribosomal RNA sequences using Bowtie2 (Langmead and Salzberg, 2012) with the ‘very-sensitive-local’ preset parameters to identify and remove the ribosomal RNA sequences from the dataset. Owing to the near-identical gene sequence of the S. pimpinellifolium with that of S. lycopersicum, the filtered reads were aligned with TopHat2 (Kim et al., 2013) against the cultivated tomato genome allowing for maximum intron lengths of 5000bp, segment lengths of 22bp, and one mismatch per segment. All other parameters were set to the default values. Reads that mapped to up to 20 genes were counted as one for each match. Reads that mapped to more than 20 genes were not counted. Aligned sequences were then separated into sense and antisense, and the counts of aligned reads for each tomato gene model and from each sample were derived using an in-house Perl script. This script also counted reads that partially mapped to the UTRs. Read counts were used to find differentially expressed genes between LA1589 and the two backcrossed progenies miR167a_222-2, and miR167a_222-3 with DESeq2 (Anders et al., 2013) using both parametric and local dispersion fits. Heatmaps and principle components analyses were used to identify possible outlier datasets. Differentially expressed genes with an adjusted P<0.05 found in both comparisons were considered for further analysis. Reads per kilobase of exon model per million mapped reads (RPKM) were calculated using an in-house script based on the ITAG 2.3 exon lengths and the total number of reads that mapped to the tomato genome. For the expression analyses of selected genes from different tissues, the average RPKM values for each tissue type are shown. All raw reads for the wild type compared with miR167a_222-2, and miR167a_222-3 lines are deposited in the NCBI sequence read archive (SRA) with accession number SRA057458. All raw reads for expression analysis of the different tissues in LA1589 are deposited with accession number SRA061767. The average RPKM values per sample and for all genes are deposited at http://ted.bti.cornell.edu/cgi-bin/TFGD/digital/home.cgi. Functional category enrichment analysis was performed using the ‘phyper’ function in R (http://www.r-project.org/). Genes were classified into 136 categories based on MapMan BINs using Slyc_ITAG2.3 annotation as the reference.
AuxRE promoter analysis
The upstream sequence for each significant differentially expressed tomato gene was obtained using custom perl scripts. We used the first 1kb that was 5′ of the start codon or the full 5′ non-coding sequence if the next gene was within 1kb. The sequence and the start codon locations were obtained from http://solgenomics.net/, ITAG v 2.4 for the gene models and v 2.5 for the chromosome sequences. A custom perl script was written to count the instances of two putative AuxRE elements within 20bp of each other on either strand. Three potential AuxRE sequences were analysed: TGTCGG, TGTCGA, and TGTCTC (Franco-Zorrilla et al., 2014), both for pairs of the same elements and for combinations of two different elements, with 1bp mismatches to the search sequences allowed. To evaluate AuxRE occurrence in randomized promoter sequences, the upstream sequences were shuffled 100 times using shuffleseq (http://emboss.sourceforge.net/) and AuxRE pairs were counted each time. The mean, standard deviation, and fraction of the times the shuffled sequence had fewer hits than the actual sequence was reported. We considered AuxRE elements that occurred more in the actual than reshuffled sequences at 0.95 or higher to be significant.
Northern blot analysis
For Northern blots, 20 µg of total RNA per sample was separated on a 1.2% agarose gel in 1× MOPS buffer and transferred to Hybond N membrane (GE Biosciences, Pittsburgh, PA, USA). Primers used for generating template for labelling were: SpARF6A, 5′-AGTGGGTGGCGAGTATCCCG-3′ and 5′-CACCAAGGAGGAGAACATCA-3′; SpARF8A/B, 5′-TTTCTC ACAGACACCACCCT-3′; and 5′-CTGCCGTTGACTCATCCC-3′; SpARF3, 5′-GATTGTTTTGCTCCCTTGGA-3′ and 5′-TGCTCA GCTGCATCTTCTGT-3′; STYLE2.1, 5′-GATTCGCAATCGTCG CTCTA-3′ and 5′-CTGATGATTGCTGCTTCTGG-3′; eIF4a, 5′-CA GCTTTTGCCACCAAAAAT-3′ and 5′-TCTGATCCATGTCTCC GTGA-3′. DNA probes were generated using α32P-dCTP and a three-cycle PCR amplification of the templates obtained from RT-PCR. Hybridization was performed at 65 °C in modified Church buffer (0.5M sodium phosphate pH 7.2, 1mM EDTA, and 7% SDS). Blots were washed twice with 2×SSC, 0.1% SDS at 65 °C and twice with 0.2×SSC, 0.1% SDS at 65 °C. Radioactive bands were visualized using a Phosphor Imager (Model Storm840, Molecular Dynamics, GE Biosciences, Pittsburgh PA, USA).
For low molecular weight RNA, 25 µg of total RNA extracted from anthesis-stage flowers was suspended in 20 µl of RNA loading buffer (95% formamide, 5mM EDTA, 0.025% SDS, 0.025% bromophenol blue and xylene cyanol FF) and separated in 15% denaturing polyacrylamide gel containing 8M urea. Antisense miR167 (5′-TAGATCATGCTGGCAGCTTCA-3′) and miR166 (5′-GGGGAATGAAGCCTGGTCCGA-3′) probes were prepared by end-labelling with T4-polynucleotide kinase (New England Biolabs, Ipswich MA, USA) in the presence of γ32P-ATP. Hybridization was performed at 42 °C in ULTRAhyb ® hybridization buffer (Invitrogen, USA) at 42 °C. Blots were washed with 2×SSC, 0.1% SDS at 42 °C and with 0.2×SSC, 0.1% SDS at 42 °C. Radioactive signals were visualized as mentioned above.
Southern blot analysis
Genomic DNA was extracted from the young leaves of four-week old tomato plants. Ten µg of genomic DNA was digested with EcoRI and EcoRV (New England Biolabs, USA), separated on a 0.8% agarose gel, and blotted onto a Hybond-N+ membrane (GE Biosciences, USA) under alkaline conditions. The coding region of BAR gene amplified with primers (5′-TGCCAGTTCCCGTGCTT-3′ and 5′-CAACTCGATCGAGGGGATC-3′) was used as template for the three-cycle PCR labelling reaction. Hybridization and visualization were performed as described for the northern blot analysis.
Results
Identification of tomato ARF6A/B and ARF8A/B
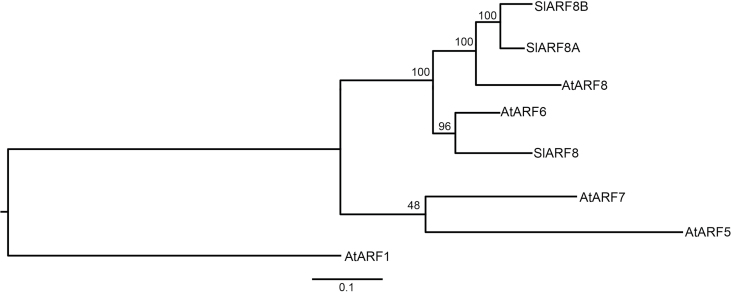
The DNA-binding domains of Arabidopsis ARF6 and ARF8 were used to identify tomato orthologues from the genome sequence (Tomato Genome Consortium, 2012; Ulmasov et al., 1999b). We found two putative orthologues of ARF6 (SlARF6A Solyc00g196060 and SlARF6B Solyc07g043610/043620) and two orthologues of ARF8 (SlARF8A Solyc03g031970 and SlARF8B Solyc02g037530). SlARF6B is probably a pseudogene given its annotation as two genes (Solyc07g043610 and Solyc07g043620) and given the presence of a premature stop codon in both the wild (LA1589) and cultivated tomato (Heinz1706) sequences. The sequences of SlARF6A and SlARF8A/B were validated by Mohamed Zouine and Mondher Bouzayen (University of Toulouse, France). These SlARFs are clustered in the Class II subclade based on phylogenetic analysis (Fig. 1).
Fig. 1.
Phylogeny of SlARF6A, SlARF8A and SlARF8B and closely related Arabidopsis ARF proteins. The DNA binding motif was used to construct the phylogenetic tree using MUSCLE and RAxML version 7.4.2. AtARF1 is used as outgroup.
Expression of SpARF6A, SpARF8A, and SpARF8B in tomato organs and tissues.
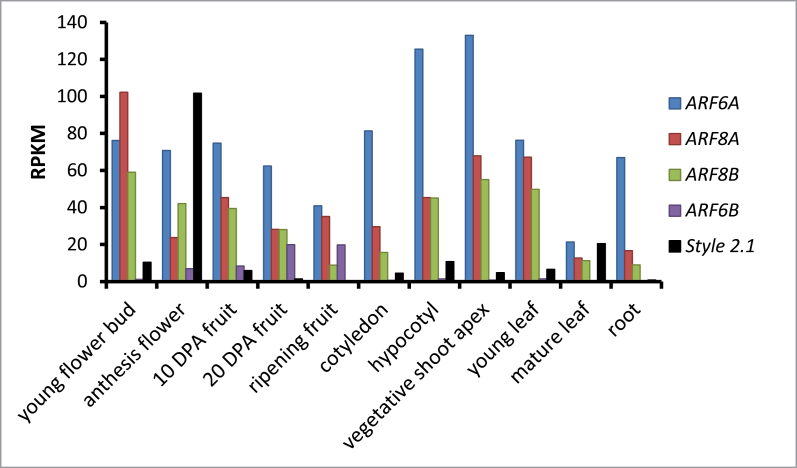
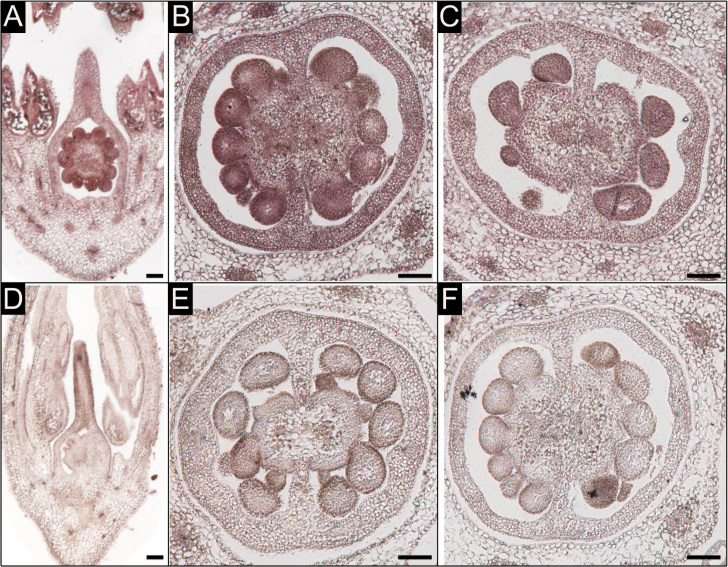
The expression of the three ARF genes in different organs/tissues of LA1589 was investigated from a dataset that was generated previously (Huang et al., 2013). The gene expression levels were averaged over three-to-four biological replicates and expressed in reads per kilobase per million mapped reads (RPKM) (Supplementary Table S1 available at JXB online). In general, SpARF6A was expressed at higher levels than SpARF8A and SpARF8B (Fig. 2). All three genes were expressed in multiple tissues including seedlings, shoot meristems, young leaves, flowers, fruits at multiple stages, and roots. The lowest expression level of the three ARF genes was found in mature terminal leaflets. We also evaluated the expression of the SpARF6B pseudogene by adding the RPKM for Solyc07g043610 and Solyc07g043620 divided by 2. The gene had a very low expression level except in 20 DPA fruits as well as ripening fruits. The significance of the low but detectable expression level of SpARF6B is not clear. In situ hybridization of flower buds at 9 and 4 days before anthesis using SpARF6A and SpARF8B probes demonstrated strong expression levels in the developing ovules and pollen (Fig. 3). This expression pattern suggests potential regulatory roles of these genes in flower development, particularly in the female organs.
Fig. 2.
Expression of SpARF6A, SpARF6B, SpARF8A, SpARF8B and STYLE 2.1 in tomato organs and tissues. The values represent average reads per kilobase of gene and per million reads that map to the annotated genome. (This figure is available in colour at JXB online.)
Fig. 3.
In situ hybridization of SpARF6A and SpARF8B in tomato flowers. (A, B) Wild-type LA1589 flower at 9 and 4 days before anthesis hybridized with the antisense SpARF6A probe. (C) Wild-type ovary at 4 days before anthesis hybridized with the antisense SpARF8B probe. (D, E) MIR167 transgenic flowers at 9 and 4 days before anthesis hybridized with the antisense SpARF6A probe. (F) A 4-days before anthesis wild-type ovary hybridized with the sense SpARF6A probe as control. Bar=100 µm.
Identification of MIR167 in tomato
The Arabidopsis miR167 regulates the expression of AtARF6 and AtARF8. We wanted to know whether this regulatory mechanism is conserved in tomato. Cultivated tomato miR167 was initially identified in a conventional small RNA cloning approach, and its expression pattern was developmentally regulated based on small RNA gel blot (Itaya et al., 2008) and deep sequencing (http://smallrna.udel.edu). Mature Sly-miR167 shares an identical sequence with the Arabidopsis miR167. Previously, tomato miR167-mediated cleavage of SlARF8B was confirmed by 5′-RACE (Moxon et al., 2008). The target site is shown in Supplementary Figure S1A available at JXB online. We employed the newly developed online software tool SoMART and available small RNA sequencing data from VF36 and Microtom (Li et al., 2012) to evaluate whether SlARF6A and SlARF8A were also targets of miR167. miR167-mediated cleavage products were found for SlARF6A, SlARF8A, and SlARF8B (Supplementary Fig. S1A available at JXB online). The results indicated that these tomato genes are targets of miR167, as are the orthologous ARF6 and ARF8 genes in Arabidopsis.
The tomato genome contains four putative precursor genes that could encode miR167 even though only one, Sly-MIR167, is registered in miRBase (Griffiths-Jones, 2004; Tomato Genome Consortium, 2012). The genome sequences surrounding these four putative genes share sequence homology to Ath-MIR167a and Sly-MIR167. Together, this suggests the presence of four MIR167 genes encoding miR167 precursors (Supplementary Fig. S1B available at JXB online). Thus, we renamed the registered Sly-MIR167 as Sly-MIR167a-1 and the additional three genes as Sly-MIR167a-2, Sly-MIR167a-3, and Sly-MIR167a-4. The predicted precursor structures of these four Sly-MIR167 genes are shown in Supplementary Figure S1B available at JXB online. We also found the corresponding miR167* sequences (from the other strand of the stem-loop precursors) for these four genes in the small RNA deep sequencing database (http://smallrna.udel.edu). The tomato genome has two additional sequences with high similarity to mature miR167. However, we were unable to validate these as bona fide MIR167 genes based on the absence of a strongly supported predicted stem-loop precursor structure using in silico folding analysis (Supplementary Fig. S1C available at JXB online), extremely low reads in small RNA deep sequencing (fewer than 10 reads), and the lack of miRNA* sequences for these candidate genes in deep sequencing data (http://smallrna.udel.edu/). Thus, we concluded that tomato has four MIR167 genes that produce miR167, which regulates SlARF6 and SlARF8 genes. Thus, regulation of ARF6 and ARF8 by the miR167 family is conserved between Solanaceae and Brassicaceae.
Expression of Arabidopsis MIR167a in tomato plants decreases expression of ARF6A and ARF8A/B and causes growth defects.
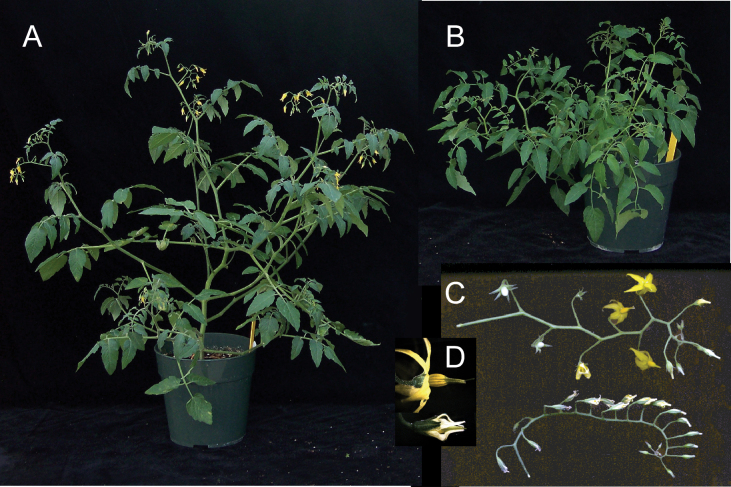
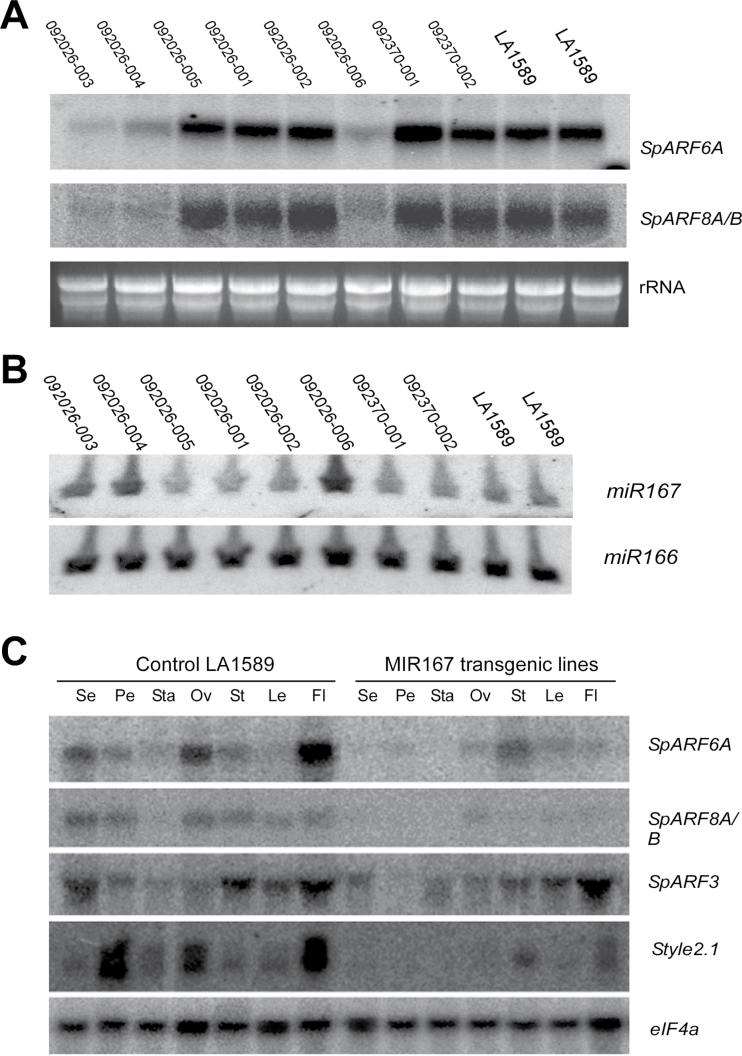
We next investigated whether SpARF6 and SpARF8 might regulate vegetative and flower development similarly in tomato and in Arabidopsis. As miR167 regulates ARF6 and ARF8 genes in both species, we expected that plasmid pB7WG2-MIR167a, which expresses Arabidopsis MIR167a under the control of the CaMV 35S promoter (Wu et al., 2006), would target tomato SpARF6 and SpARF8 genes. We obtained eight independent transgenic LA1589 lines carrying pB7WG2-MIR167a. Compared with control lines, three MIR167a-transgenic lines (092026-003, 092026-004, and 092026-006) exhibited smaller stature, smaller terminal leaflets, and incomplete floral development (Fig. 4, Table 1). The remaining transgenic lines resembled non-transformed control plants. Southern blot analysis showed that most lines harboured two or more copies of the transgene (Supplementary Fig. S2 available at JXB online). However, transgene copy number did not seem to be correlated with the severity of phenotypes. Northern blot analysis of RNA isolated from anthesis-stage flowers showed that the lines with strong phenotypes had a significant reduction in the expression of SpARF6A and SpARF8A/B (Fig. 5A) as well as increased miR167 expression over endogenous levels (Fig. 5B). Moreover, RNA seq analysis of very young flower buds in control and transgenic MIR167 lines showed that SpARF6A and SpARF8B were most significantly down-regulated (3 and 1.7-fold respectively), whereas SpARF8A was not down-regulated in these tissues (Supplementary Table S2 available at JXB online). Expression of the pseudogene SlARF6B was practically undetectable in very young flower buds (Supplementary Table S2 available at JXB online). Therefore, the data demonstrated that increased miR167 expression in the transgenic tomato plants led to reduced expression of ARF6 and ARF8 resulting in altered developmental phenotypes.
Fig. 4.
Phenotypes of the MIR167 transgenic tomato plants. (A) Wild-type LA1589. (B) MIR167 transgenic tomato line 092026-003 showing reduced plant height/size compared with wild-type control tomato. Photos were taken two months after sowing. (C) Wild-type inflorescence with opened flowers and young fruit (top) and MIR167 inflorescence with unopened flowers and no fruit (bottom). (D) Close-up mature flowers of wild type (top) and MIR167 (bottom).
Table 1.
Terminal leaflet characteristics of LA1589 control and MIR167 transgenic plants
| Wild type | MIR167 transgenic lines | ||||||
|---|---|---|---|---|---|---|---|
| LA1589 (n=10) | 092026-001 (n=10) | 092026-002 (n=10) | 092026-003 (n=10) | 092026-004 (n=10) | 092026-005 (n=10) | 092026-006 (n=10) | |
| Perimeter (cm) | 15.2±1.6 | 16.7±1.7 | 14.7±0.5 | 10.5±1.2* | 11.7±2.2* | 14.1±0.6 | 11.6±1.4* |
| Area (cm2) | 7.7±1.2 | 8.4±0.9 | 6.4±1.4 | 3.7±1.1* | 3.3±0.4* | 5.9±0.7 | 4.7±0.7* |
| Maximum width (cm) | 2.4±0.3 | 2.4±0.2 | 2.2±0.3 | 1.6±0.2* | 1.5±0.1* | 2.2±0.2 | 2.0±0.2* |
| Maximum length (cm) | 5.8±0.5 | 6.6±0.9 | 5.6±0.2 | 4.0±0.4* | 4.4±0.4* | 5.4±0.3 | 4.6±0.8* |
| Leaf shape index | 2.4±0.2 | 2.7±0.4 | 2.5±0.4 | 2.5±0.3 | 2.8±0.2 | 2.5±0.2 | 2.3±0.5 |
| Proximal angle | 56.2±4.7 | 52.9±5.4 | 55.0±3.6 | 56.5±8.5 | 58.2±7.4 | 52.5±6.4 | 57.2±6.3 |
The values represent the mean±SD for wild type and primary transformants. *Denotes significant difference by student’s t test (P<0.05) compared with wild type.
Fig. 5.
Gene expression analysis in MIR167 transgenic and wild-type tomato flower and floral organs. (A) Expression of SpARF6A, SpARF8A/B in anthesis-stage flowers of wild-type, and MIR167 transgenic lines. (B) Mature miR167 levels in wild-type and transgenic lines. miR166 expression served as control. (C) Expression analysis of selected SpARF and Style2.1 in floral organs, terminal leaflets, and whole flowers. Se, sepal; Pe, petal; Sta, stamen; Ov, ovary; St, stem; Le, terminal leaflet; Fl, whole flower at anthesis. Northern blots were carried out with 20 µg total RNA.
The three MIR167a-transgenic tomato lines that showed reduced ARF6 and ARF8 expression and severe phenotypes were further investigated. The shorter stature of MIR167a-plants was caused by reduced internode growth (Table 2). After transition to flowering, control LA1589 carried inflorescences after every third internode as is customary for indeterminate tomato. The internode that carried the first inflorescence was the 7th or 8th internode in both wild type and transgenic lines. In contrast, five of the MIR167a-plants carried the second inflorescence after four internodes whereas the other five carried the second inflorescence after three internodes (Table 2), suggesting that down-regulation of SpARF6 and SpARF8 delayed inflorescence meristem termination in the sympodial shoot. In addition, floral development in the MIR167a-plants differed from wild-type development starting just before anthesis when the flowers failed to open completely as compared with the wild-type controls (Fig. 4C, D). In mature post-anthesis flowers, petals, stamens, and style were all shorter in MIR167a plants than in wild-type plants, with the largest defects in petals (Table 3). This reduced organ length was not rescued by exogenous application of MeJA (methyl jasmonic acid), and was partially rescued by application of GA (gibberellic acid; for the style and stamen) (Table 3 and Supplementary Table S3A available at JXB online).
Table 2.
Internode lengths in wild-type LA1589 and MIR167 tomato plants
| Internode number | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7+8a | 9 | 10 | 11+12b | |
| Wild type (mm) n=10 | 21.4±2.8 | 21.5±2.5 | 23.4±2.4 | 35.9±8.4 | 46.9±9.6 | 54.8±11 | 107.8±9.4 | 83.9±13 | 67.7±8.6 | 91.2±12.1 |
| MIR167 (mm) n=10 | 16±2.4* | 18.6±2.3* | 17.8±1.8* | 23.5±2.6* | 20.5±3.2* | 18.5±2.4* | 39.0±4.1* | 35.5±4.8* | 31.2±4.0* | 45.6±6.8* |
The values represent the mean±SD. Asterisk denotes significant difference by student’s t-test (P<0.05) compared with wild type. aTermination of the vegetative meristem in the inflorescence meristem occurs after the 7th or 8th leaf. Values are the sum of the lengths of internodes just above and below the first inflorescence. bIn wild type, sympodial growth continues for exactly three internodes until terminating again in an inflorescence meristem. In MIR167, the second inflorescence forms either after 3 or 4 internodes from the first inflorescence. Values are the sum of the lengths of internodes just above and below the second inflorescence.
Table 3.
Floral organ lengths two days post-anthesis
| LA1589 (n=16) | MIR167 (n=26) | MIR167 + MeJA (n=21) | |
|---|---|---|---|
| Sepal (mm) | 4.9±0.3 | 4.9±0.2 | 4.7±0.2 |
| Petal (mm) | 12.8±1.6 | 7.9±0.6* | 7.9±0.6* |
| Stamen cone (mm) | 8.7±0.4 | 7.4±0.5* | 7.3±0.4* |
| Ovary (mm) | 1.2±0.1 | 1.1±0.1 | 1.0±0.2 |
| Style (mm) | 8.8±0.2 | 6.6±0.9* | 6.6±1.3* |
The values represent the mean±SD. Asterisk denotes significant difference by student’s t-test (P<0.05) compared with wild type. No significant differences were found in MIR167 transgenics treated with or without methyl jasmonate.
Analyses of multiple flower buds from different positions along the inflorescence showed that the styles of transgenic plants had grown less than those of wild-type plants as early as 5–6 days before anthesis, whereas petal and stamen growth defects occurred just before and after anthesis (Table 3 and Supplementary Table S3B available at JXB online). Thus, ectopic expression of MIR167a affected petal, stamen, and style growth, with the earliest effects on style elongation and later effects on petal and stamen elongation. Petals also seemed not to senesce after flower opening. In control LA1589, lack of fertilization normally leads to flower drop. In the lines overexpressing MIR167a, flowers and floral organs remained on the peduncle well after the normal time of flower opening (Fig. 4C).
We also examined the accumulation of SpARF6A and SpARF8A/B transcripts in different floral organs of wild-type and transgenic lines. Northern blot analysis showed that in wild type the highest levels of SpARF6A and SpARF8A/B were found in the ovary, sepal, and petal, but these genes were barely expressed in stamens (Fig. 5C). In MIR167a-transgenic plants, the transcripts of SpARF6A and SpARF8A/B were barely detected in any floral organs. As a control, SpARF3 was expressed at similar levels in wild-type and transgenic plants (Fig. 5C). We also evaluated the expression of STYLE2.1. This gene controls stigma exsertion in tomato and its reduced expression leads to shorter styles, thereby facilitating inbreeding and selfing (Chen et al., 2007). In control plants, expression of STYLE2.1 was robust in flowers at anthesis, and low or undetectable in other organs and tissue types (Fig. 2). STYLE2.1 was highly expressed in petal, ovary, and entire flowers in wild-type plants. However, in the MIR167a transgenic lines, expression of STYLE2.1 was markedly reduced in floral organs. These results suggest that STYLE2.1 may function downstream of ARF6 and ARF8 and its reduced expression in MIR167a-plants contributed to the reduced style growth.
Defects in female organs in MIR167a-overexpressing tomato plants cause sterility
The MIR167a plants did not produce fruit, suggesting that they might be sterile. To investigate whether the lack of fecundity arose from male or female sterility, reciprocal crosses were conducted between MIR167a-overexpressing and control tomato plants. Pollination of wild-type pistils with MIR167a transgenic pollen resulted in high fruit set (n=19 pollinations and 80–85% fruit set for T0 lines 092026-003 and 092026-004) and viable seed production. In contrast, pistils from the severe MIR167a lines 092026-003 and 092026-004 pollinated with wild-type pollen did not yield any fruit (n=15 and 17 pollinations for lines 003 and 004, respectively). Thus, the MIR167a-transgenic tomato plants were female sterile but male fertile.
To determine the basis for the female sterility of MIR167a plants, we examined pollen tube growth in pistils after manual pollination. Whereas MIR167a pollen tubes grew well in wild-type pistils, wild-type pollen was unable to grow in the styles or stigmas of the severe MIR167a plants (Supplementary Fig. S3A–C available at JXB online). To ensure that pollen would stick to the stigmas in these situations, we applied lanolin onto the stigmas before pollination. Regardless of lanolin application, no pollen tubes were found in styles of the severe MIR167a-transgenic lines (data not shown). These results suggested that female sterility was due to defects in pollen recognition on the stigma surface, pollen germination and/or pollen tube growth in the transmitting tracts of MIR167a plants.
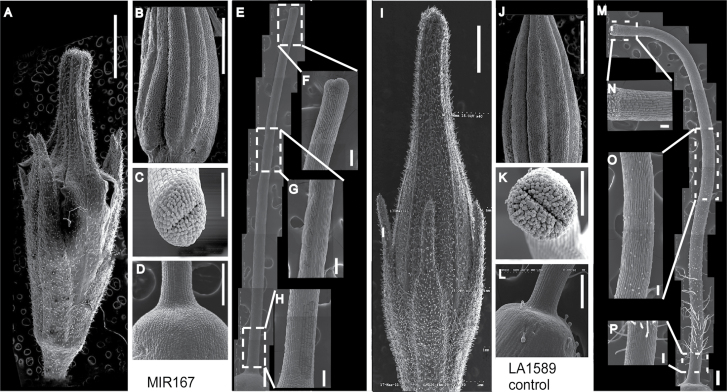
To characterize pistil growth in greater detail, we examined wild-type and MIR167a tomato floral organs by scanning electron microscopy (SEM). As shown in Fig. 6, overall morphology of the sepals, petals, stamen, and stigma seemed similar in wild-type and MIR167a lines although petals, stamens and style were shorter in the transgenic lines as noted above. Notably, trichomes were abundant near the bases of the styles in wild-type plants but were completely absent from the styles of MIR167a plants. Jasmonate-insensitive mutants of tomato also lack stylar trichomes (Li et al., 2004). However, application of exogenous MeJA did not restore the trichome defect of MIR167 styles, and application of IAA, MeJA, or GA did not restore female fertility (data not shown). Cross-sections of style and ovary showed similar cellular organization in wild type and transgenic lines, except possibly for the placenta, which seemed smaller in the MIR167 plants (Fig. S3D–M). In summary, our results suggest the defect in female fertility probably arose as a result of arrested development at the stigma surface and/or in the transmitting tract. More generally, the stigma or style defects and the lack of trichomes together indicate that the pistils of MIR167a plants failed to mature.
Fig. 6.
SEM of floral organs from MIR167 and control tomato plants. Flowers were collected 1 day before anthesis. (A–H) MIR167 transgenic plants. (I–P) Wild-type LA1589. Whole flower buds just before opening (A and I); stamen (B and J); stigma (C and K); top of ovary and base of style (D and L). Style morphology of the MIR167 transgenic (E–H) and wild-type (M–P) plants. Close-up of the corresponding style regions boxed in white are shown in (F–H) for MIR167 and (N–P) for control. Size bar in A, B, I, J =1mm; size bar in C, K=100 µm; size bar in D, E, G, M = 200 µm; size bar in F–H and N–P=500 µm.
To determine the cellular mechanism by which reduced ARF6 and ARF8 expression led to reduced style length, the cell number and length of mature styles were measured one day before anthesis in both transgenic and wild-type flowers. Close to the ovary, cell length varied less as compared with that in the distal regions of the style where cell length was three-to-four times longer in wild-type than in MIR167a styles. On the other hand, total cell number along the style was higher in MIR167a transgenic lines than in wild-type control lines (Table 4). Thus, MIR167a overexpression and the concomitant reduction in SpARF6 and SpARF8 expression led to shorter styles through a greatly reduced cell length even though cell number increased.
Table 4.
Cell length and number in the styles of control and MIR167 flowers
| Distance from the ovary | Control LA1589 | MIR167 |
|---|---|---|
| Proximal, region 1 (µm) | 27.9±5.0 | 19.5±1.8 |
| Region 2 (µm) | 107.4±25.5 | 58.9±6.5 |
| Region 3 (µm) | 247.0±44.1 | 61.7±6.6 |
| Distal, region 4 (µm) | 79.0±19.4 | 30.6±2.6 |
| Total cell number | 51±3.9 | 84.3±6.0 |
Region 1 is closest to the ovary. Region 2 and 3 are in the middle, whereas region 4 is furthest away from the ovary, proximal to the stigma. Epidermal cell length and number of the styles were evaluated using SEM.
RNA-seq analysis of floral tissues of wild-type and MIR167a-transgenic lines
To gain molecular insights into the impacts of miR167a-regulated ARF6 and ARF8 expression on flower development, we used RNA-seq to profile the global expression patterns of tomato genes in very young flower buds of MIR167a plants in comparison with those in non-transgenic control plants. The samples were collected at flower developmental stages corresponding to ovule initiation and earlier, so that the primary effects of decreased SpARF6 and SpARF8 expression on flower development could be assessed before general growth defects occurred. Statistical evaluation using the DESeq2 package revealed a total of 1094 and 1737 genes that were differentially expressed in miR167a_222-2 and miR167a_222-3 progenies, respectively, as compared with those in wild-type plants. These lines were derived from a backcross of 092026-003 pollen with LA1589. Of these, 687 differentially expressed genes were shared in the two data sets (Supplementary Table S4 available at JXB online). Gene ontology (GO) analyses of these 687 genes showed that most of the bin categories containing four or more members were collectively down-regulated in MIR167a plants compared with the control, whereas other bins show a mixture of up- and down-regulated genes. No bin category with four or more members was up-regulated in MIR167a plants. Many genes involved in transcription regulation were differentially expressed. Of these, MADS box, AP2/EREBP, and MYB-domain/MYB-related genes were represented with the highest numbers. The MADS box genes were mostly up-regulated whereas AP2/EREBP genes were mostly down-regulated in MIR167a plants compared with those in wild-type plants (Supplementary Table S4 available at JXB online). Many genes encoding proteins in cell wall metabolism were under-expressed in the MIR167a flower buds. These included UDP-glucosyl and -glucoronyl transferases, gluco-, manno- and galactosidases, invertases and pectin methyl transferase inhibitors, and beta-1,3-glucan hydrolases. In the hormone bin, genes mostly involved in auxin and ethylene metabolism were differentially expressed, where others were up- or down-regulated. Many genes involved in protein degradation and post-translational modifications were differentially expressed, as were genes encoding receptor kinases and factors in calcium signalling. In the category ‘Development’, we found that genes with similarity to LFY, CUCs, and UFO were up-regulated in MIR167a plants, in addition to many others whose roles in development are less clear. Lastly, many genes involved in transport were differentially expressed, in particular those that transport sugars and peptides as well as ABC transporters (Supplementary Table S4 available at JXB online). Promoter analyses of the differentially expressed tomato genes showed that 109 out of 686 genes contained pairs of putative AuxRE elements that occurred more than random 95% of the time or more (Supplementary Table S5 available at JXB online). The list included two SAUR genes, an expansin gene, and an ARF19-like gene. Arabidopsis homologues of these genes are auxin-inducible, suggesting that a subset of all the differentially expressed genes were directly downstream of ARF6 and ARF8.
To determine whether certain functional categories were overrepresented in the list of differentially expressed genes, we performed hypergeometric tests with the genes in each MapMan BIN (Thimm et al., 2004; Table 5). At the same time, we also compared the differentially expressed tomato genes with those identified in a previous Arabidopsis microarray study of wild-type and arf6 arf8 mutant stage 12 flower buds (just before anthesis) (Reeves et al., 2012). This comparison led to the identification of 185 common differentially expressed genes corresponding to the same Phytozome family in tomato and Arabidopsis. Of these, 142 were down-regulated and 43 were up-regulated as a result of overexpression of MIR167a (Supplementary Table S6 available at JXB online). Thus, decreased ARF6 and ARF8 activity affected the expression of a conserved set of genes in tomato and Arabidopsis. For example, in the category ‘Regulation of transcription’, ARFs and bZIP transcription factors were overrepresented in the common set, as expected, whereas general transcription factor genes (all were members of the GRF family) were overrepresented in the tomato-specific set only. Some other common genes included those in the ‘Cell wall’, ‘Transport’, and ‘Hormone’ categories. Common genes in the ‘Other’ category included those encoding MAP kinases, O-methyl transferases, TCA cycle components, biodegradation machinery, GDSL-motif lipases, tetrapyrrole synthesis enzymes, and UDP-glucosyl and -glucoronyl transferases.
Table 5.
Functional category enrichment of differentially expressed genes between wild-type LA1589 and MIR167 transgenic lines
| MapMan BinCode | Category | Alla | Commonb | Uniquec | Genomed | P-valuef | P-value (common) | P-value (unique) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nume | Freq | num | freq | num | freq | num | freq | ||||||
| Regulation of transcription | 27.3.50 | General transcription (GRF family) | 4 (0) | 0.57% | 0 | 0 | 4 (0) | 0.78% | 31 | 0.09% | 0.011 | 0.263 | 0.010 |
| 27.3.24 | MADS box transcription factor family | 7 (6) | 0.99% | 2 (1) | 1.05% | 5 (5) | 0.97% | 101 | 0.28% | 0.016 | 0.060 | 0.039 | |
| 27.3.4 | Auxin response factor family | 3 (2) | 0.43% | 2 (2) | 1.05% | 1 (0) | 0.19% | 26 | 0.07% | 0.016 | 6.02E–03 | 0.138 | |
| 27.3.40 | Aux/IAA family | 3 (3) | 0.43% | 1 (1) | 0.52% | 2 (2) | 0.39% | 25 | 0.07% | 0.016 | 0.037 | 0.039 | |
| 27.3.26 | MYB-related transcription factor family | 3 (1) | 0.43% | 0 | 0 | 3 (1) | 0.58% | 49 | 0.14% | 0.059 | 0.332 | 0.039 | |
| 27.3.21 | GRAS transcription factor family | 3 (0) | 0.43% | 0 | 0 | 3 (0) | 0.58% | 53 | 0.15% | 0.069 | 0.340 | 0.047 | |
| 27.3.35 | bZIP transcription factor family | 4 (3) | 0.57% | 2 (1) | 1.05% | 2 (2) | 0.39% | 82 | 0.23% | 0.071 | 0.042 | 0.232 | |
| Cell wall | 10.6 | Cell wall degradation | 10 (9) | 1.42% | 3 (3) | 1.57% | 7 (6) | 1.36% | 181 | 0.51% | 0.016 | 0.060 | 0.039 |
| 10.8.1 | Pectinesterases | 6 (6) | 0.85% | 3 (3) | 1.57% | 3 (3) | 0.58% | 87 | 0.24% | 0.016 | 0.012 | 0.111 | |
| 10.1 | Cell wall precursor synthesis | 5 (4) | 0.71% | 2 (2) | 1.05% | 3 (2) | 0.58% | 68 | 0.19% | 0.019 | 0.030 | 0.070 | |
| 10.7 | Cell wall modification | 4 (4) | 0.57% | 3 (3) | 1.57% | 1 (1) | 0.19% | 81 | 0.23% | 0.071 | 0.010 | 0.437 | |
| 10.2 | Cellulose synthesis | 3 (3) | 0.43% | 2 (2) | 1.05% | 1 (1) | 0.19% | 66 | 0.19% | 0.100 | 0.028 | 0.354 | |
| Transport | 34.2 | Transport of sugars | 5 (2) | 0.71% | 2 (1) | 1.05% | 3 (1) | 0.58% | 80 | 0.23% | 0.029 | 0.042 | 0.091 |
| 34.1 | p- and v-ATPases | 3 (3) | 0.43% | 2 (2) | 1.05% | 1 (1) | 0.19% | 62 | 0.17% | 0.084 | 0.027 | 0.334 | |
| 34.15 | Potassium transport | 3 (2) | 0.43% | 2 (1) | 1.05% | 1 (1) | 0.19% | 62 | 0.17% | 0.084 | 0.027 | 0.334 | |
| Lipid metabolism | 11.1 | Fatty acid synthesis and elongation | 7 (6) | 0.99% | 2 (2) | 1.05% | 5 (4) | 0.97% | 132 | 0.37% | 0.029 | 0.100 | 0.060 |
| 11.8 | Lipid metabolism (steroids, squalene etc) | 6 (5) | 0.85% | 0 | 0 | 6 (5) | 1.17% | 114 | 0.32% | 0.039 | 0.503 | 0.023 | |
| Hormone metabolism | 17.6 | Gibberellin | 3 (1) | 0.43% | 2 (0) | 1.05% | 1 (1) | 0.19% | 53 | 0.15% | 0.069 | 0.021 | 0.289 |
| 17.2.3 | Auxin-induced-regulated- responsive-activated | 7 (4) | 0.99% | 4 (2) | 2.09% | 3 (2) | 0.58% | 218 | 0.61% | 0.140 | 0.032 | 0.479 | |
| Others | 29.7 | Protein glycosylation | 6 (6) | 0.85% | 2 (2) | 1.05% | 4 (4) | 0.78% | 49 | 0.14% | 6.6E–03 | 0.021 | 0.016 |
| 26.22 | Short chain dehydrogenase/ reductase | 7 (6) | 0.99% | 1 (1) | 0.52% | 6 (5) | 1.17% | 86 | 0.24% | 0.011 | 0.169 | 0.011 | |
| 30.6 | MAP kinases | 3 (3) | 0.43% | 3 (3) | 1.57% | 0 | 0 | 17 | 0.05% | 0.011 | 1.24E–04 | 0.331 | |
| 16.2 | Phenylpropanoids metabolism. | 10 (3) | 1.42% | 1 (0) | 0.52% | 9 (3) | 1.75% | 179 | 0.50% | 0.016 | 0.340 | 0.011 | |
| 26.6 | O-methyl transferases | 3 (3) | 0.43% | 3 (3) | 1.57% | 0 | 0 | 20 | 0.06% | 0.016 | 1.66E–04 | 0.354 | |
| 8 | TCA | 6 (6) | 0.85% | 6 (6) | 3.14% | 0 | 0 | 87 | 0.24% | 0.016 | 6.39E–05 | 0.778 | |
| 26.3 | Gluco-, galacto- and mannosidases | 5 (4) | 0.71% | 1 (1) | 0.52% | 4 (3) | 0.78% | 71 | 0.20% | 0.022 | 0.131 | 0.039 | |
| 5 | Fermentation | 3 (3) | 0.43% | 0 | 0 | 3 (3) | 0.58% | 32 | 0.09% | 0.025 | 0.266 | 0.022 | |
| 31.3 | Cell cycle | 7 (4) | 0.99% | 1 (1) | 0.52% | 6 (3) | 1.17% | 130 | 0.37% | 0.028 | 0.263 | 0.039 | |
| na | SUN-like proteins | 3 (2) | 0.44% | 1 (1) | 0.54% | 2 (1) | 0.40% | 34 | 0.10% | 0.028 | 0.055 | 0.060 | |
| 21.2 | Ascorbate and glutathione | 4 (2) | 0.57% | 2 (1) | 1.05% | 2 (1) | 0.39% | 61 | 0.17% | 0.038 | 0.027 | 0.144 | |
| 24 | Biodegradation of xenobiotics | 4 (2) | 0.57% | 3 (1) | 1.57% | 1 (1) | 0.19% | 64 | 0.18% | 0.042 | 6.02E-03 | 0.346 | |
| 21.1 | Thioredoxin | 4 (4) | 0.57% | 0 | 0 | 4 (4) | 0.78% | 75 | 0.21% | 0.060 | 0.423 | 0.039 | |
| 26.28 | GDSL-motif lipase | 5 (5) | 0.71% | 4 (4) | 2.09% | 1 (1) | 0.19% | 112 | 0.32% | 0.072 | 6.02E–03 | 0.571 | |
| 19 | Tetrapyrrole synthesis | 3 (2) | 0.43% | 3 (2) | 1.57% | 0 | 0 | 58 | 0.16% | 0.075 | 6.02E–03 | 0.641 | |
| 26.2 | UDP glucosyl and glucoronyl transferases | 10 (10) | 1.42% | 7 (7) | 3.66% | 3 (3) | 0.58% | 338 | 0.95% | 0.149 | 7.03E–03 | 0.778 | |
P-values in bold indicate significant enrichment. a, All 687 differentially expressed genes between wild-type tomato and MIR167 transgenic lines. b, The 185 genes that were differentially expressed both owing to overexpression of MIR167 in tomato and in the Arabidopsis arf6 arf8 double mutant. c, The 502 differentially expressed genes unique in tomato. d, Number of genes in the GO category in the entire tomato genome. e, Numbers in the parentheses are the numbers of down-regulated genes. f, Hypergeometric tests were performed in R using ‘phyper’. FDR method was used for P-value correction.
Discussion
ARF6 and ARF8 orthologues are found in diverse angiosperms, including monocots and dicots (Remington et al., 2004), but their roles in plant development have only been tested in Arabidopsis (Nagpal et al., 2005; Reeves et al., 2012; Ru et al., 2006; Tabata et al., 2010). In this study, the functions of the tomato ARF6 and ARF8 genes were evaluated by transgenically overexpressing the Arabidopsis MIR167a in this Solanaceous species. Although this approach does not distinguish between the functions of ARF6 and ARF8, these genes have overlapping expression patterns and it seems likely that they act, at least partially, redundantly in tomato as in Arabidopsis.
Tomato plants overexpressing MIR167a had significantly decreased expression of SpARF6A and SpARF8B, and the flowers were arrested at the time of flower opening. The petals, stamens and styles were each shorter in mature post-anthesis flowers of MIR167a plants compared with wild type. Moreover, the style lacked trichomes that are normally present, and did not support wild-type pollen tube germination and/or growth resulting in female sterility. These phenotypes are similar to those described for flowers of Arabidopsis arf6 arf8 mutants, which also arrest at the time of flower opening, with defects in petal, stamen, ovary, style, and stigma growth, and severely decreased ability of the gynoecium to support pollen tube growth (Crawford and Yanofsky, 2011; Nagpal et al., 2005; Reeves et al., 2012; Ru et al., 2006; Tabata et al., 2010). In each plant species, the organs that grow most rapidly at the time of flower opening are most strongly affected by decreasing ARF6 and ARF8: petals and the style in tomato, and petals and stamens in Arabidopsis.
MIR167a transgenic tomato plants also had shorter internodes and smaller leaves than did wild-type plants, indicating that the products of SpARF6A and SpARF8B genes play a role in promoting stem elongation and leaf expansion. Arabidopsis arf6 arf8 mutants similarly have very short inflorescence stems, whereas they have only a subtle leaf phenotype. In Arabidopsis, ARF6 and ARF8 contribute to leaf expansion together with NPH4/ARF7 and ARF19 (Wilmoth et al., 2005; JWR, unpublished results). In our RNA seq data set, overexpression of MIR167a led to increased expression of tomato SpARF7/ARF19, suggesting that it may compensate for the reduction in SpARF6 and SpARF8 function and perhaps have a similar role in regulating leaf expansion as in Arabidopsis.
Although our data demonstrate that ARF6 and ARF8 regulate flower maturation in Arabidopsis and tomato, differences between MIR167a transgenic tomato and Arabidopsis arf6 arf8 phenotypes may reflect developmental or regulatory differences between the two species. In particular, the anthers of MIR167a tomato flowers still produced viable pollen, whereas the Arabidopsis mutants did not and were therefore male-sterile. This suggests either that the threshold for ARF6 and ARF8 action in tomato anthers is lower than was achieved by the MIR167a transgene expression, or that ARF6 and ARF8 do not regulate anther dehiscence in tomato.
The regulation of ARF6 and ARF8 by miR167 is likely to be highly conserved. miR167 is found in seed-producing plants from gymnosperms to flowering plants (Axtell and Bartel, 2005). In addition to targeting ARF6 and ARF8, it was recently shown that miR167 also guides cleavage of IAA-Ala Resistant 3 (IAR3) transcripts in Arabidopsis (Kinoshita et al., 2012). By using the SoMART degradome RNA library analysis, we demonstrate that miR167 guides the cleavage of SlARF6A, SlARF8A, and SlARF8B transcripts in cultivated tomato, consistent with a previous report (Moxon et al., 2008). We attempted to find and validate additional targets of miR167 in tomato, which are listed in the miSolRNA database (www.misolrna.org). In addition to SlARF6A and SlARF8A/B, three putative candidate genes (Solyc04g077220, Solyc04g073990, and Solyc11g011980) could be targeted by miR167. However, none of them was validated as a likely target of miR167 in the SoMART degradome RNA library database. Similarly, we could not identify breakdown products of the tomato orthologue of IAR3 (Solyc03g121270) in the degradome RNA library database. It should be noted, however, that the SoMART degradome RNA library and other tomato degradome RNA libraries used leaf and fruit samples, but not root samples (Karlova et al., 2013). IAR3 is important for lateral root growth and thus, further studies on tomato root samples may help clarify whether miR167 could mediate regulation of tomato IAR3. Thus far, there is no evidence that miR167 in tomato regulates guided cleavage of any gene other than ARF6A, ARF8A, and ARF8B, consistent with findings from another study (Karlova et al., 2013).
The phenotypes described above indicate that orthologous ARF6 and ARF8 genes have similar developmental functions in tomato and Arabidopsis. It will be interesting in future work to determine whether this similarity extends to targets of ARF regulation. The 187 putative orthologues whose expression is similarly altered in arf6 arf8 Arabidopsis mutants and MIR167a tomato (even though we examined distinct developmental stages using different experimental platforms) suggest that some targets may be conserved. Among these is Style2.1 (which promotes style elongation) in tomato MIR167a plants, and the related AtPRE1 gene in Arabidopsis arf6 arf8 plants, which are underexpressed in MIR167a or arf6 arf8 flowers (Chen et al., 2007; Reeves et al., 2012). Moreover, in both cases, some of the phenotypes could be attributed to reduced jasmonate production or signalling. In Arabidopsis, jasmonate is required for petal growth and anther dehiscence, and decreased jasmonate production accounts for a subset of arf6 arf8 mutant phenotypes (Nagpal et al., 2005; Reeves et al., 2012; Tabata et al., 2010). Analogously, tomato jasmonate-insensitive plants have defective stylar trichomes (Li et al., 2004). Although we could not rescue the stylar development defect in tomato MIR167a plants with exogenous methyl jasmonate, decreased jasmonate production or response might nevertheless contribute to this aspect of the MIR167a overexpression phenotype. Similarly, a family of closely related MYB genes regulates various aspects of flower growth in Arabidopsis, ornamental tobacco, and Petunia (Cheng et al., 2009; Colquhoun et al., 2011; Liu et al., 2009; Liu and Thornburg, 2012; Mandaokar et al., 2006; Reeves et al., 2012; Spitzer-Rimon et al., 2010). These are underexpressed in Arabidopsis arf6 arf8 flowers, and it will be interesting to determine whether members of this gene family in tomato similarly regulate flower growth downstream of ARF6 and ARF8.
Supplementary data
Supplementary data are available at JXB online
Figure S1. Mapped Sly-miR167-mediated cleaved SlARF6A/SlARF8A/SlARF8B RNAs and the predicted folding of the Sly-miR167 genes.
Figure S2. Southern blot analysis of MIR167 transgenic lines.
Figure S3. Pollen tube growth and gynoecium structure in MIR167 and the wild-type LA1589.
Table S1. RPKM value for each gene in each replicate in 11 LA1589 tissues.
Table S2. Read count for SpARF6A, SpARF6B, SpARF8A, SpARF8B using the confirmed gene sequences.
Table S3. Floral organ lengths of MIR167 and LA1589 control flowers.
Table S4. Differentially expressed genes in MIR167 lines compared with LA1589 control in two comparisons.
Table S5. AuxRE Promoter analysis of the differentially expressed tomato genes
Table S6. Common differentially expressed genes in tomato and Arabidopsis.
Acknowledgments
This work was supported by USDA-AFRI agreement 2007-35304-18295 to JWR and EvdK. We thank Mondher Bouzayen and Mohamed Zouine from the University of Toulouse for help with the tomato ARF6 and ARF8 annotations. We also thank the Molecular and Cellular Imaging Center, at The Ohio State University, Wooster for the microscopy analyses.
Glossary
Abbreviations:
- ARF
auxin response factor
- Aux/IAA
auxin/indoleacetic acid protein
- CTD
C-terminal dimerization domain
- DBD
DNA-binding domain
- DPA
days post-anthesis
- GA
gibberellic acid
- JA
jasmonic acid
- MeJA
methyl jasmonic acid
- MIR
microRNA precursor gene.
References
- Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD. 2013. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nature Protocols 8, 1765–1786 [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Bartel DP. 2005. Antiquity of microRNAs and their targets in land plants. Plant Cell 17, 1658–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanca J, Canizares J, Cordero L, Pascual L, Diez MJ, Nuez F. 2012. Variation revealed by SNP genotyping and morphology provides insight into the origin of the tomato. PLoS One 7, e48198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer DR, Freire-Rios A, van den Berg WAM, Saaki T, Manfield IW, Kepinski S, López-Vidrieo I, Franco-Zorrilla JM, de Vries SC, Solano R, Weijers D, Coll M. 2014. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156, 577–589 [DOI] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KY, Cong B, Wing R, Vrebalov J, Tanksley SD. 2007. Changes in regulation of a transcription factor lead to autogamy in cultivated tomatoes. Science 318, 643–645 [DOI] [PubMed] [Google Scholar]
- Cheng H, Song S, Xiao L, Soo HM, Cheng Z, Xie D, Peng J. 2009. Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis . PLoS genetics 5, e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun TA, Schwieterman ML, Wedde AE, et al. 2011. EOBII controls flower opening by functioning as a general transcriptomic switch. Plant Physiologyogy 156, 974–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford BC, Yanofsky MF. 2011. HALF FILLED promotes reproductive tract development and fertilization efficiency in Arabidopsis thaliana . Development 138, 2999–3009 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. 2005. The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445 [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Researchearch 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R. 2014. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proceedings of the National Academy of Sciences, USA 111, 2367–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. 2004. The microRNA registry. Nucleic Acids Research 32, D109–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle TJ. 1986. Auxin-regulated gene-expression in higher-plants. Crc Critical Reviews in Plant Sciences 4, 247–276 [Google Scholar]
- Guilfoyle TJ, Hagen G. 2007. Auxin response factors. Current Opinion in Plant Biology 10, 453–460 [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Ulmasov T, Hagen G. 1998. The ARF family of transcription factors and their role in plant hormone-responsive transcription. Cellular and Molecular Life Sciences 54, 619–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Van Houten J, Gonzalez G, Xiao H, van der Knaap E. 2013. Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Molecular genetics and genomics 288, 111–129 [DOI] [PubMed] [Google Scholar]
- Itaya A, Bundschuh R, Archual AJ, Joung JG, Fei Z, Dai X, Zhao PX, Tang Y, Nelson RS, Ding B. 2008. Small RNAs in tomato fruit and leaf development. Biochimica et Biophysica Acta 1779, 99–107 [DOI] [PubMed] [Google Scholar]
- Karlova R, van Haarst JC, Maliepaard C, van de Geest H, Bovy AG, Lammers M, Angenent GC, de Maagd RA. 2013. Identification of microRNA targets in tomato fruit development using high-throughput sequencing and degradome analysis. Journal of Experimental Botany 64, 1863–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Gasser CS. 2009. Ovule development: genetic trends and evolutionary considerations. Sexual Plant Reproduction 22, 229–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita N, Wang H, Kasahara H, Liu J, Macpherson C, Machida Y, Kamiya Y, Hannah MA, Chua NH. 2012. IAA-Ala Resistant3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress. Plant Cell 24, 3590–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku HM, Vision T, Liu J, Tanksley SD. 2000. Comparing sequenced segments of the tomato and Arabidopsis genomes: large-scale duplication followed by selective gene loss creates a network of synteny. Proceedings of the National Academy of Sciences, USA 97, 9121–9126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Orban R, Baker B. 2012. SoMART: a web server for plant miRNA, tasiRNA and target gene analysis. The Plant Journal 70, 891–901 [DOI] [PubMed] [Google Scholar]
- Li L, Zhao Y, McCaig BC, Wingerd BA, Wang J, Whalon ME, Pichersky E, Howe GA. 2004. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16, 126–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Reed JW. 2002. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Molecular Biology 49, 387–400 [PubMed] [Google Scholar]
- Liu G, Ren G, Guirgis A, Thornburg RW. 2009. The MYB305 transcription factor regulates expression of nectarin genes in the ornamental tobacco floral nectary. Plant Cell 21, 2672–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Thornburg RW. 2012. Knockdown of MYB305 disrupts nectary starch metabolism and floral nectar production. The Plant Journal 70, 377–388 [DOI] [PubMed] [Google Scholar]
- Mandaokar A, Thines B, Shin B, Lange BM, Choi G, Koo YJ, Yoo YJ, Choi YD, Browse J. 2006. Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. The Plant Journal 46, 984–1008 [DOI] [PubMed] [Google Scholar]
- Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V, Dalmay T. 2008. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Research 18, 1602–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Ellis CM, Weber H, et al. 2005. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132, 4107–4118 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Zambryski PC, Roe JL. 1998. Auxin signaling in Arabidopsis flower development? Current Opinion in Plant Biology 1, 531–535 [DOI] [PubMed] [Google Scholar]
- Nitsch JP. 1952. Plant hormones in the development of fruits. The Quarterly Review of Biology 27, 33–57 [DOI] [PubMed] [Google Scholar]
- Oh TJ, Wartell RM, Cairney J, Pullman GS. 2008. Evidence for stage-specific modulation of specific microRNAs (miRNAs) and miRNA processing components in zygotic embryo and female gametophyte of loblolly pine (Pinus taeda). New Phytologist 179, 67–80 [DOI] [PubMed] [Google Scholar]
- Reeves PH, Ellis CM, Ploense SE, et al. 2012. A regulatory network for coordinated flower maturation. PLoS genetics 8, e1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington DL, Vision TJ, Guilfoyle TJ, Reed JW. 2004. Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiology 135, 1738–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru P, Xu L, Ma H, Huang H. 2006. Plant fertility defects induced by the enhanced expression of microRNA167. Cell Research 16, 457–465 [DOI] [PubMed] [Google Scholar]
- Serrani JC, Ruiz-Rivero O, Fos M, Garcia-Martinez JL. 2008. Auxin-induced fruit-set in tomato is mediated in part by gibberellins. The Plant Journal 56, 922–934 [DOI] [PubMed] [Google Scholar]
- Spitzer-Rimon B, Marhevka E, Barkai O, Marton I, Edelbaum O, Masci T, Prathapani NK, Shklarman E, Ovadis M, Vainstein A. 2010. EOBII, a gene encoding a flower-specific regulator of phenylpropanoid volatiles’ biosynthesis in Petunia. Plant Cell 22, 1961–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 [DOI] [PubMed] [Google Scholar]
- Tabata R, Ikezaki M, Fujibe T, Aida M, Tian CE, Ueno Y, Yamamoto KT, Machida Y, Nakamura K, Ishiguro S. 2010. Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol 51, 164–175 [DOI] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M. 2004. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal 37, 914–939 [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T. 2003. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15, 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium. 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. 1997. ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. 1999a. Activation and repression of transcription by auxin-response factors. Proceedings of the National Academy of Sciences, USA 96, 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. 1999b. Dimerization and DNA binding of auxin response factors. The Plant Journal 19, 309–319 [DOI] [PubMed] [Google Scholar]
- Vivian-Smith A, Koltunow AM. 1999. Genetic analysis of growth-regulator-induced parthenocarpy in Arabidopsis . Plant Physiology 121, 437–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, Alonso JM, Ecker JR, Reed JW. 2005. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. The Plant Journal 43, 118–130 [DOI] [PubMed] [Google Scholar]
- Wu MF, Tian Q, Reed JW. 2006. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133, 4211–4218 [DOI] [PubMed] [Google Scholar]
- Wu S, Xiao H, Cabrera A, Meulia T, van der Knaap E. 2011. SUN regulates vegetative and reproductive organ shape by changing cell division patterns. Plant Physiology 157, 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Jiang N, Schaffner EK, Stockinger EJ, Van der Knaap E. 2008. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319, 1527–1530 [DOI] [PubMed] [Google Scholar]
- Xiao H, Radovich C, Welty N, Hsu J, Li D, Meulia T, van der Knaap E. 2009. Integration of tomato reproductive developmental landmarks and expression profiles, and the effect of SUN on fruit shape. BMC Plant Biology 9, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H, Pudake RN, Guo G, Xing G, Hu Z, Zhang Y, Sun Q, Ni Z. 2011. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genomics 12, 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Han SJ, Yoon EK, Lee WS. 2006. Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Research 34, 1892–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YW, Lai KN, Tai PY, Li WH. 1999. Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other angiosperm lineages. Journal of Molecular Evolution 48, 597–604 [DOI] [PubMed] [Google Scholar]
- Zhong S, Joung J-G, Zheng Y, Chen Y-r, Liu B, Shao Y, Xiang JZ, Fei Z, Giovannoni JJ. 2011. High-throughput illumina strand-specific RNA sequencing library preparation. Cold Spring Harbor Protocols 2011, 940–949 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.