This work analyzes and elucidates the function of the LHC protein isoform LHCBM9. It provides evidence that the green microalga C. reinhardtii has evolved a LHCII-dependent, medium-term process for photoprotection during environmental stress conditions.
Abstract
Photosynthetic organisms developed multiple strategies for balancing light-harvesting versus intracellular energy utilization to survive ever-changing environmental conditions. The light-harvesting complex (LHC) protein family is of paramount importance for this function and can form light-harvesting pigment protein complexes. In this work, we describe detailed analyses of the photosystem II (PSII) LHC protein LHCBM9 of the microalga Chlamydomonas reinhardtii in terms of expression kinetics, localization, and function. In contrast to most LHC members described before, LHCBM9 expression was determined to be very low during standard cell cultivation but strongly increased as a response to specific stress conditions, e.g., when nutrient availability was limited. LHCBM9 was localized as part of PSII supercomplexes but was not found in association with photosystem I complexes. Knockdown cell lines with 50 to 70% reduced amounts of LHCBM9 showed reduced photosynthetic activity upon illumination and severe perturbation of hydrogen production activity. Functional analysis, performed on isolated PSII supercomplexes and recombinant LHCBM9 proteins, demonstrated that presence of LHCBM9 resulted in faster chlorophyll fluorescence decay and reduced production of singlet oxygen, indicating upgraded photoprotection. We conclude that LHCBM9 has a special role within the family of LHCII proteins and serves an important protective function during stress conditions by promoting efficient light energy dissipation and stabilizing PSII supercomplexes.
INTRODUCTION
The efficient use of sunlight for production of chemical energy by phototrophic organisms relies on the efficiency of light harvesting and energy transfer to the reaction centers of photosystem I (PSI) and photosystem II (PSII). For this purpose, photosynthetic organisms have developed light-harvesting systems composed of pigments and specialized pigment binding proteins. The structure and composition of the light-harvesting systems varies, ranging from extrinsic cyanobacterial phycobilisomes to intrinsic, membrane-spanning light-harvesting pigment protein complexes of plant chloroplasts (Grossman et al., 1995; Elrad and Grossman, 2004; Ballottari et al., 2012). Light-harvesting complex (LHC) proteins of plants and eukaryotic microalgae are located in the thylakoid membrane of the chloroplasts. According to their predominant association with PSI or PSII, the LHC proteins are classified as LHCI or LHCII, respectively, and are encoded by Lhca or Lhcb genes. LHCII proteins are further subdivided into monomeric and trimeric isoforms. Monomeric LHCII proteins are less abundant and are located in close proximity to the PSII core complex. By contrast, the major LHCII proteins are far more abundant and form the outer light antenna system (Dekker and Boekema, 2005). Chlorophyll a and b are the most abundant photoactive pigments associated with LHCII and participate in the energy transfer toward the primary electron donor, P680, of PSII (Barber and Archer, 2001). In addition, lutein, neoxanthin, and xanthophyll cycle carotenoids are associated with LHCII (Croce et al., 1999), being involved in energy transfer and/or dissipation and reactive oxygen species (ROS) scavenging upon excess irradiation (Niyogi et al., 1997; Ballottari et al., 2012). Light harvesting and energy transfer are highly adaptive processes, allowing the photosynthetic cells to react quickly to environmental changes, such as fluctuating light intensity, temperature changes, or nutrient availability. This is reflected by the fact that the expression of LHC proteins is strictly regulated on all major levels of gene expression, from mRNA transcription to protein degradation (Escoubas et al., 1995; Flachmann and Kühlbrandt, 1995; Lindahl et al., 1995; Durnford et al., 2003; Mussgnug et al., 2005). In addition, the size of the functional PSI and PSII LHC antenna can be adjusted via so-called state transitions, involving phosphorylation and lateral migration of LHCII proteins to PSI (Bennett et al., 1980; Kruse, 2001; Wollman, 2001; Finazzi et al., 2002; Depège et al., 2003; Bellafiore et al., 2005). Besides their role as photon energy collectors, certain LHC-type proteins have been reported to play an important alternative role under distinct stress conditions (Li et al., 2000; Peers et al., 2009). Here, LHC proteins participate to dissipate excitation energy, a process observed as nonphotochemical quenching (NPQ) of chlorophyll fluorescence, preventing oxidative damage of the cell caused by ROS produced from reaction of triplet chlorophyll states with O2 (Niyogi et al., 1997; Müller et al., 2001). Trimeric LHCII complexes contain three different carotenoids, lutein, neoxanthin, and violaxanthin, the latter being converted to zeaxanthin in the xanthophyll cycle activated by excess light. Each of these carotenoids play synergistic functions in chlorophyll triplet excited states quenching and ROS scavenging (Elrad et al., 2002; Liu et al., 2004; Dall’Osto et al., 2007a, 2007b, 2012).
It is interesting to note that a variety of homologous LHC genes has evolved in the plant genome. In case of the model green alga Chlamydomonas reinhardtii, the Genome Project (Merchant et al., 2007) has shown at least 20 different LHC isoforms (Elrad and Grossman, 2004). The reasons why such a large number of homologous genes are preserved in the nuclear genome with all of them being functionally expressed (Elrad and Grossman, 2004) are not yet fully understood, but it seemed reasonable to suggest that despite their homology, functional differences do exist and knockout/downregulation of specific members have been shown to affect specific functions such as NPQ or state 1 to state 2 transitions (Elrad et al., 2002; Ferrante et al., 2012).
In previous work, we were able to show that LHCII isoform gene LHCBM9 shows a striking and unique mRNA transcription profile: being hardly expressed under standard growth conditions, and strongly induced when cells are transferred to sulfur-depleted medium (Nguyen et al., 2008), a result that has since been confirmed in several studies (González-Ballester et al., 2010; Aksoy et al., 2013). Strong transcriptional induction was also observed for the gene encoding LHCSR3, which was shown to be involved in protective energy dissipation in C. reinhardtii (Peers et al., 2009; Bonente et al., 2011; Tokutsu and Minagawa, 2013).
It is widely accepted that H2 production in C. reinhardtii acts as a valve system to scavenge stromal protons and electrons that cannot be used for the production of redox equivalents due to a shift from linear to cyclic electron transport under anaerobic conditions (Melis, 2007; Kruse and Hankamer, 2010; Hemschemeier and Happe, 2011). The efficiency of hydrogenase activity very much depends on substrate supply (H+ and e-), on the efficiency of energy capture by LHCII proteins (Melis, 2012; Oey et al., 2013), and on the absence of intracellular O2 (Melis et al., 2000; Kruse et al., 2005; Tolleter et al., 2011). Anaerobiosis, however, represents a potential threat to the survival of phototrophic organisms. As a reaction to nonfunctional oxidative phosphorylation in mitochondria and the resulting perturbed photochemical electron transport in chloroplasts, C. reinhardtii cells can induce acclimation processes like LHC state transitions and NPQ (Wollman and Delepelaire, 1984; Finazzi et al., 2002; Peers et al., 2009; Allorent et al., 2013). Recent transcriptome analysis of the hydrogen production process indicated that perturbation of stress-induced expression of energy quenching proteins affects H2 production capacity due to changes in ROS-induced damage of photosynthetic linear electron transport (Nguyen et al., 2011; Toepel et al., 2013).
In this work, we performed detailed investigation of the LHCII isoform LHCBM9 with respect to its protein expression profile, localization, and functional role during nutrient starvation and hydrogen production.
RESULTS
LHCBM9 Is a Member of the Homologous LHCII Protein Family of C. reinhardtii with Unique Features
Sequencing of the nuclear genome of C. reinhardtii (Merchant et al., 2007) showed that the gene subfamily encoding the major LHCII proteins is composed of nine members, designated LHCBM1-9. The chromosomal loci for seven of the isoforms have been assigned, revealing that four LHCII genes are localized on chromosome 6 (LHCBM4, 6, 8, and 9), two on chromosome 12 (LHCBM2 and 7), and one at chromosome 3 (LHCBM5), while two isoforms (LHCBM1 and 3) could not yet be mapped. For all of these genes, mRNA transcripts could be detected, indicating that none of them represent pseudogenes (Nguyen et al., 2008; González-Ballester et al., 2010). Except for the N-terminal region, all corresponding proteins have a high degree of amino acid sequence identity and contain a sequence region similar or identical to the plant LHCII trimerization motif (WYGPDR) (Hobe et al., 1995) (Supplemental Figure 1A).
According to their amino acid sequence, LHCII proteins can be subdivided into four types (types I to IV; Supplemental Figure 1B and Supplemental Data Set 1; Minagawa and Takahashi, 2004) with LHCBM9 being of type I. The basic features of LHCBM9, which was first mentioned by Elrad et al. (2002), are presented in Supplemental Table 1. In this work, we strictly follow the gene/protein assignments from C. reinhardtii genome sequence (v4.3), which is why the sequence designations and derived alignments presented here do not fully correspond to recent work from Minagawa and coworkers (Supplemental Figure 1B) (Minagawa and Takahashi, 2004; Harris et al., 2009). According to the DNA sequence, the unprocessed LHCBM9 protein comprises 254 amino acids and a corresponding calculated molecular mass of 27.1 kD. It is most likely that the pre-LHCBM9 contains an N-terminal transit sequence of 16 amino acids and in silico modeling indicates a typical transmembrane organization of the mature protein (Supplemental Figure 1C).
The Protein Expression of LHCBM9 Is Strongly Induced by Nutrient Deficiency Stress Conditions
In previous work, we showed that mRNA expression for subunit LHCBM9 was generally low and strongly upregulated specifically during sulfur-deprived anaerobic hydrogen production (up to 3000-fold; for details, see Nguyen et al., 2008).
Since LHCII proteins share a high degree of amino acid sequence identity, functional analysis based on immunoblots or mass spectrometry is not trivial. We generated a specific polyclonal antibody for epitope LQKNGVQF of LHCBM9 and used it to examine whether the increased transcription during hydrogen production is reflected by increased protein levels. A strong increase of LHCBM9 protein expression could be detected in the parental control strain (PCS) CC-406 after cell transfer from aerobic standard growth to anaerobic hydrogen production conditions, while the total amount of LHCII proteins showed a limited decrease (Figure 1A).
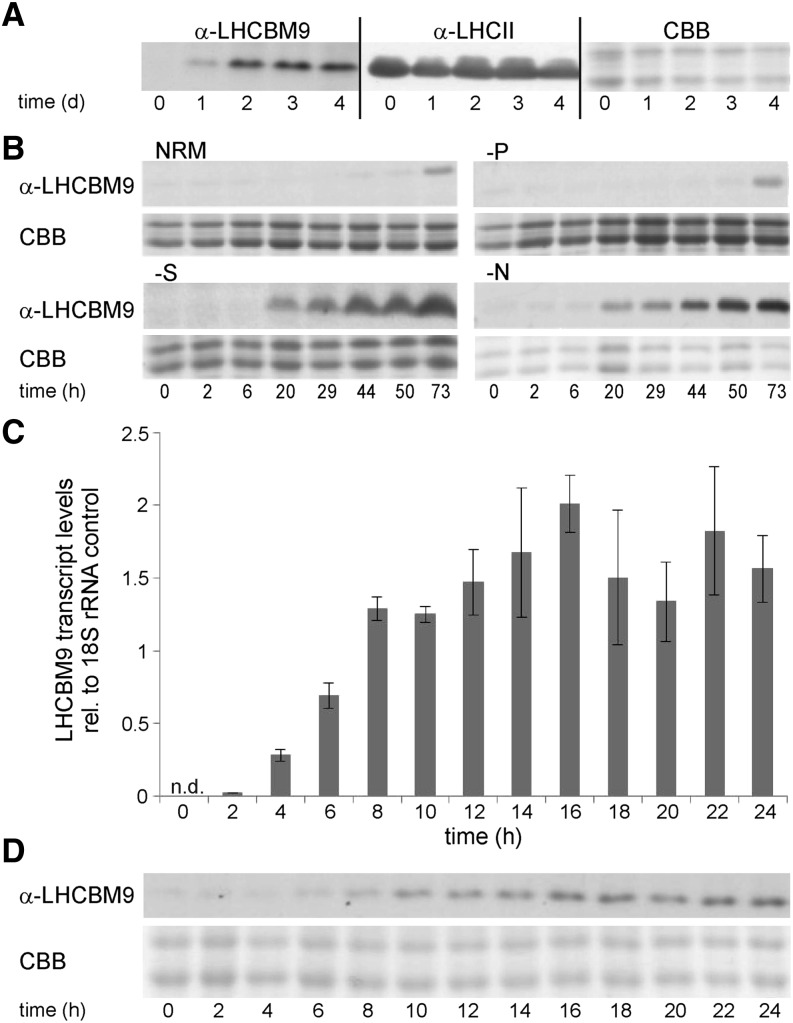
Figure 1.
Time-Resolved Patterns of LHCBM9 Expression during Cultivation in Nutrient-Deficient Media.
(A) Immunoblot analysis of LHCBM9 accumulation under hydrogen production conditions. Samples were harvested before (0 d) and after the initiation of sulfur deprivation at the indicated time points. LHCII, control blot with a LHCII antibody recognizing a conserved LHCII domain (Hippler et al., 2001).
(B) Immunoblot analysis of LHCBM9 accumulation induced by sulfur (-S), nitrogen (-N), and phosphorus (-P) starvation conditions. Nutrient-replete medium (NRM) was used as a control. Samples were tested before (0 h) and after the initiation of each deprivation condition at the indicated time points.
(C) and (D) Detailed time-resolved analyses of LHCBM9 expression during the first 24 h of sulfur deprivation.
(C) RT-Q-PCR analyses of LHCBM9 transcript accumulation, quantified relative to the mRNA level of the housekeeping gene 18S rRNA. n.d., not detectable. Error bars represent se (n = 3).
(D) Concomitant immunoblot analysis of LHCBM9 protein accumulation.
Coomassie Brilliant Blue (CBB)–stained SDS-PAGE gels in (A), (B), and (D) represent the protein loading controls.
In the hydrogen production setup (sulfur depletion of sealed cultures, leading to anaerobiosis followed by H2 production as redox valve system; Melis et al., 2000), cells are subjected to a combination of several factors that could have triggered LHCBM9 biosynthesis. To discriminate if LHCBM9 expression was exclusive to hydrogen production conditions or if it represented a more general stress response, distinct depletion of the major nutrients phosphorus, sulfur, or nitrogen were tested next (Figure 1B).
When cells were transferred into medium without nitrogen or sulfur, accumulation of LHCBM9 protein could be detected already within the first 24 h (Figure 1B, -N and -S), demonstrating that expression was not restricted to hydrogen production conditions. By contrast, LHCBM9 proteins were not detectable for the first 3 d of cultivation when cells were transferred into nutrient replete medium or into medium lacking phosphorus (Figure 1B, NRM and -P). The LHCBM9-specific band appearing after 73 h most likely is a consequence of unspecific nutrient depletion. This could indicate that phosphorus depletion does not trigger LHCBM9 biosynthesis or that the intracellular phosphorus content was still sufficient to prevent stress responses.
More detailed kinetic analyses of LHCBM9 expression during sulfur starvation revealed that considerable mRNA transcript expression started 4 h after cell transfer into nutrient-deprived medium (Figure 1C), with protein accumulation being detectable ∼4 h later (Figure 1D).
Since it is well known that light has a strong effect on LHC expression levels, the effect of high-light treatment was tested next. In the case of LHCBM9, however, light conditions of up to 1500 μmol m−2 s−1, tested in photoautotrophic cultivation conditions, did not provide any evidence that expression of the protein was also controlled by high light stress (Supplemental Figure 2).
In this context, it is important to note that several recent publications described increased LHCBM9 transcription upon different, apparently unrelated stress conditions, e.g., when the cells were subjected to anaerobic conditions induced by N2 bubbling (Nguyen et al., 2008), after prolonged sulfur deprivation (González-Ballester et al., 2010; Aksoy et al., 2013) or in the presence of cationic polyamidoamine dendrimer nanoparticles (Petit et al., 2012). Furthermore, strong LHCBM9 upregulation was also noticed when genes essential for chloroplast transcription or translation were conditionally repressed (Ramundo et al., 2013), clearly supporting the notion that LHCBM9 expression represents a more general stress response.
To gain more detailed insight into the function of LHCBM9, both in vivo as well as in vitro approaches were pursued. First, two LHCBM9 knockdown strains were generated (kd-L9.1 and kd-L9.2, respectively) to study the physiological effects of a reduced amount of LHCBM9 during stress conditions in vivo. Furthermore, recombinant LHCBM9 was expressed in Escherichia coli, reconstituted, and used for subsequent in vitro analyses.
Generation and Characterization of Two LHCBM9 Knockdown Cell Lines
An artificial microRNA approach (Molnar et al., 2009) was applied to generate C. reinhardtii mutants with considerable lower LHCBM9 expression rates. Figure 2A shows the successful isolation of the two knockdown lines kd-L9.1 and kd-L9.2, exhibiting substantial downregulation of LHCBM9 protein accumulation level to ∼30 and 50% of the PCS level for kd-L9.1 and kd-L9.2, respectively. These strains were selected for detailed comparative physiological analysis. RNA interference approaches can sometimes result in cosilencing of related genes. To investigate for cosilencing of other LHCII genes, the PCS and the two knockdown lines were compared under nutrient-replete conditions in terms of individual mRNA amount, determined via quantitative real-time RT-PCR (RT-Q-PCR), LHCII protein amount, determined by immunoblots, and the chlorophyll a/b ratio (Supplemental Figure 3). No evidence for cosilencing was obtained, since LHCBM mRNA, LHCII amount, and chlorophyll a/b ratio were very similar in all strains. However, it should be noted that for a yet unknown reason, the knockdown lines showed an ∼20% decreased average cell length compared with the PCS (3.8 µm versus 4.7 μm), in addition to the significantly reduced LHCBM9 amount.
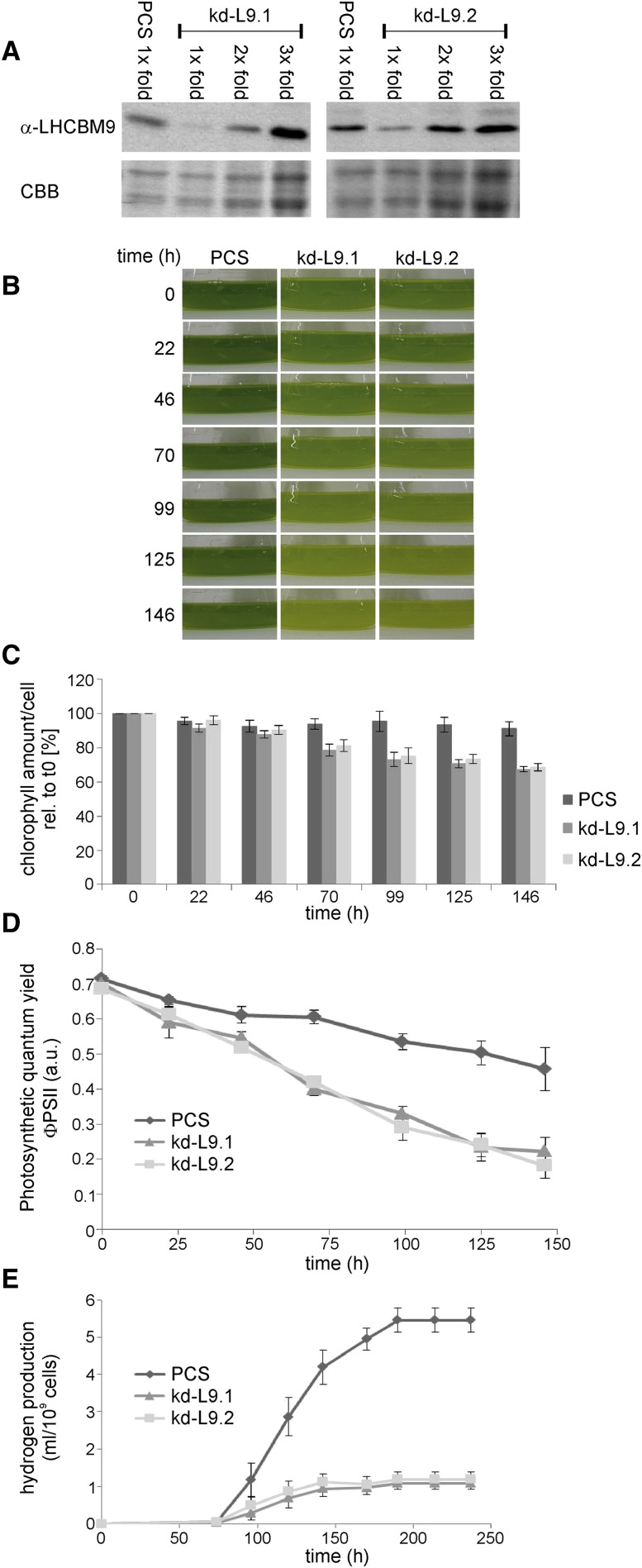
Figure 2.
Characterization of the Two LHCBM9 Knockdown Lines kd-L9.1 and kd-L9.2 with Respect to the PCS.
(A) Efficiency of LHCBM9 downregulation as determined from cell samples after 52 h of cultivation in sulfur-deplete medium by immunoblot analyses. Protein loading for the knockdown lines corresponded to 1×, 2×, or 3× of the protein loading applied for the PCS sample. Coomassie Brilliant Blue (CBB)–stained SDS-PAGE gels serve as loading controls.
(B) to (D) Phenotypic characterization of the LHCBM9 knockdown lines kd-L9.1 and kd-L9.2 in response to sulfur deprivation. Cells were adjusted to equal cell density and cultured on an orbital shaker in standard Erlenmeyer flasks at constant illumination (40 μmol m−2 s−1).
(B) Photographic images of liquid cell cultures in sulfur depleted medium. All cultures were adjusted to the same cell number (7 × 106 cells/mL) at the start of the experiment (0 h).
(C) Total chlorophyll amount per cell, relative to the total chlorophyll content at time point 0 h (set to 100%). Error bars indicate se (n = 10).
(D) Kinetics of photosynthetic quantum yield (ΦPSII). Fluorescence parameters Ft and Fm´ were recorded during illumination of the -S cultures and ΦPSII was calculated according to Maxwell and Johnson (2000). a.u., arbitrary units. The PCS served as a control, and error bars indicate se (n = 4).
(E) Total H2 production (mL/109 cells) of the two knockdown strains compared with the PCS. Here, cultures were sealed after transfer into sulfur-deplete medium to obtain oxygen-free conditions and induce subsequent oxygen-sensitive hydrogen production. Error bars indicate se (n ≥ 7).
Reduced Levels of LHCBM9 Cause a Pale-Green Phenotype and a Decrease in Hydrogen Productivity
Since LHCBM9 expression was observed only under elevated stress conditions, the PCS and the knockdown lines were cultivated for further functional analysis until they reached the log growth phase and subsequently shifted to nutrient-depleted medium. Preliminary measurements of the chlorophyll fluorescence parameter ΦPSII indicated that the impairment of photosynthesis was similar in -S and -N conditions for the first 3 d in the PCS. Sulfur depletion was preferred over nitrogen depletion for subsequent phenotypic analyses of the knockdown lines in order to avoid possibly disturbing effects related to gametogenesis, which is induced by nitrogen deficiency in C. reinhardtii. Furthermore, applying sulfur starvation with sealed cell cultures allowed studying the effects of LHCBM9 knockdown on the oxygen-sensitive hydrogen production.
Due to the smaller average cell size and the concomitant lower chlorophyll content per cell (PCS, 16.45 ± 0.4 μg; kd-L9.1, 10.12 ± 0.22 μg; and kd-L9.2, 9.64 ± 0.37 μg chlorophyll per 107 cells), cultures of the two knockdown cell lines appeared to be light green when adjusted to equal cell densities (Figure 2B). During prolonged sulfur depletion, the chlorophyll amount in both PCS and the knockdown lines declined; interestingly, this paling phenotype was significantly more pronounced in the knockdown strains (Figure 2C). Relative quantification demonstrated that the total chlorophyll content per cell of the PCS dropped to 94% ± 2.9% after 70 h, whereas it reached 78.6% ± 3.5% and 81.4% ± 3.5% in kd-L9.1 and kd-L9.2, respectively. After 125 h, the PCS still exhibited a chlorophyll content of 93.5% ± 4.2%, while the mutants showed a content of 70.6% ± 2.2% and 73.4% ± 2.7%, respectively (Figure 2C). This chlorophyll decrease was accompanied by a severe perturbation of the efficiency of photochemistry (ΦPSII) in all strains; however, this effect again was much more pronounced in the knockdown lines (Figure 2D).
These data indicate that the reduced amount of LHCBM9 proteins during nutrient stress conditions in vivo led to severely increased cell damage and bleaching of the cell culture. We conclude that LHCBM9 is involved in one or more stress response mechanisms in C. reinhardtii that are important to ensure survival of the organism.
As we originally monitored LHCBM9 upregulation under hydrogen production conditions (Nguyen et al., 2008), we tested the effect of downregulation of LHCBM9 gene expression on H2 production activity in C. reinhardtii. As shown in Figure 2E, decreased LHCBM9 levels strongly affected H2 production activity.
In summary, these data suggested that this specific LHC-type protein fulfills an important protective role during prolonged stress conditions, most likely by participating in energy dissipation and ROS scavenging processes.
LHCBM9 Can Be Localized as Part of PSII Supercomplexes
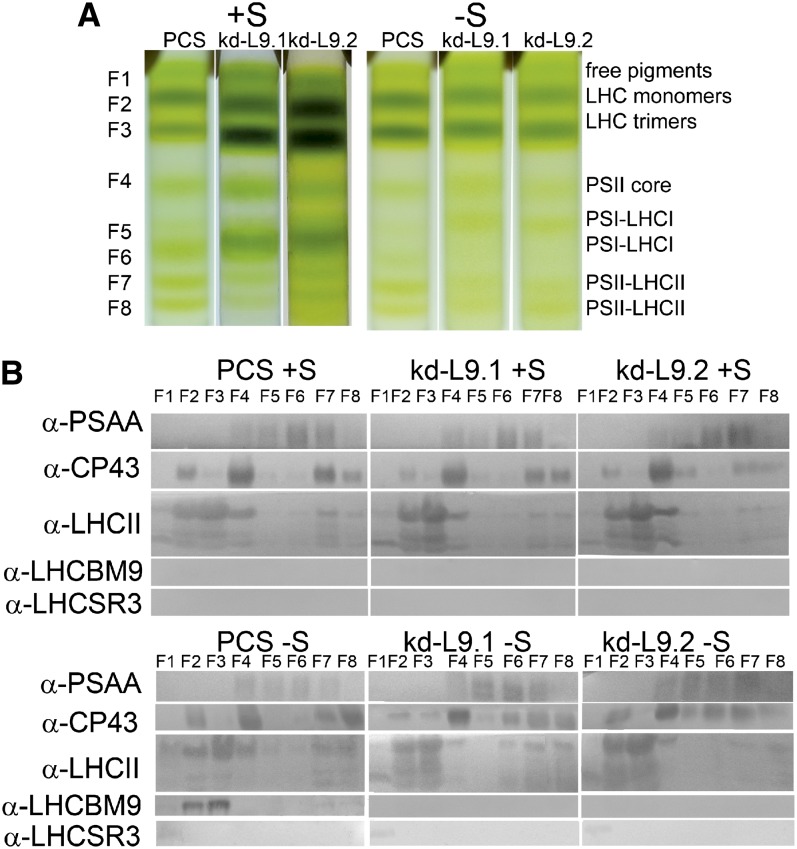
Thylakoid membranes were mildly solubilized and subsequently fractionated by ultracentrifugation in sucrose gradients (Tokutsu et al., 2012) in order to identify the association pattern of LHCBM9 with other photosynthetic complexes. To this aim, thylakoid membranes were isolated from PCS and kd-L9.1/2 cells in control conditions or after sulfur starvation. The chlorophyll-containing bands recovered from sucrose gradients were analyzed by absorption spectroscopy (Supplemental Figure 4) and by denaturing SDS-PAGE plus immunoblotting. Antibodies specific for PSI (PSAA) and PSII (CP43) subunits were selected in order to identify the identity of complexes contained in each band (Figure 3). Eight green bands were resolved in each sample, containing as major components free pigments (F1), LHC monomers (F2), LHCII trimers (F3), PSII core (F4), PSI with small antenna (F5), PSI-LHCI (F6), and two PSII-LHCII supercomplexes with increased chlorophyll b content (F7 and F8). It is worth noting that the PSAA subunit could be found in F4-7, suggesting the presence of PSI complexes of different sizes. Sulfur depletion of PCS led to decreased intensity of the F6 band, while the abundance of bands F7 and F8 was conserved. The chlorophyll distribution in the kd-L9.1/2 lines depleted in LHCBM9 showed that the PSI-LHCI complex (F6) was depleted with a concomitant increase of F5 (PSI with smaller antenna). The abundance of PSII-LHCII supercomplexes with higher molecular density (F7-F8) was reduced in the kd-L9.1/2 mutants in -S conditions, as evidenced by the decrease of CP43 in F8 bands compared with F6 and F7. LHCBM9 was found in PCS -S thylakoid membranes and was located in monomeric and trimeric LHCII (F2 and F3) and in fractions F7 and F8 containing PSII-LHC complexes as detected by the enrichment in CP43 (Supplemental Figure 5A). Only traces of LHCBM9 could be found in any fraction of the knockdown genotypes (Figure 3). These data suggest that LHCBM9 accumulates upon sulfur starvation in monomeric or trimeric LHC antenna complexes (F2 and F3) or as part of the PSII-LHCII supercomplexes (F7 and F8) as detected in the sucrose gradients (Figure 3). In order to further investigate the association of LHCBM9 with PSI and/or PSII supercomplexes, we fractionated thylakoids from PCS +S and -S Clear-Native-PAGE (Supplemental Figure 5B). The presence of LHCBM9, LHCII, as well as PSI and PSII core complex subunits in each green band was then detected by immunoblotting upon analysis by SDS-PAGE in a second dimension (Supplemental Figure 5). The reaction pattern with anti-CP43 showed the presence of PSII as high molecular mass supercomplexes in both -S samples +S samples. PSII supercomplexes in -S conditions contained both, LHCII and LHCBM9, implying that LHCBM9 was a component of PSII antenna irrespective of the PSII/LHCII stoichiometry. In fact, it was previously shown that multiple PSII supercomplex bands with increasing molecular mass correspond to supercomplexes with larger antenna complements (Caffarri et al., 2009).The pattern distribution of PSAA in the high molecular mass range did not match either LHCBM9 or LHCII reactions and was reduced to a single band in correspondence of which no increase of either LHCII or LHCBM9 was evident, suggesting it results from comigration rather than from direct interaction.
Figure 3.
Separation of Chlorophyll Binding Complexes by Ultracentrifugation in Sucrose Gradients and Subsequent Immunoblot Analysis.
(A) Sucrose gradient loaded with solubilized thylakoids purified from PCS and the knockdown lines kd-L9.1 and kd-L9.2 in control conditions (+S) or in sulfur deficiency for 48 h (-S). Fractions 1 to 8 (F1 to F8) and major chlorophyll binding complexes are indicated.
(B) Immunoblot analysis of purified fractions with antibodies specific for PSAA, CP43, LHCII, LHCBM9, and LHCSR3. Fractions were loaded on equal volume basis.
[See online article for color version of this figure.]
In order to further exclude a role of LHCBM9 in PSI activity, we proceeded to measure the cyclic electron flow (CEF) and the level of LHC state transition induction in control (+S) and in sulfur deficiency (-S) conditions (Supplemental Figure 6). PCS and kd-L9.1/2 strains were similar in showing an increase of the CEF rate in -S conditions compared with +S conditions, which was between ∼1.5- and 2.5-fold. Similarly, a clear transition to state II could be observed for PCS and kd.L9.1/2 strains in -S conditions, without major differences. It was recently shown that induction of CEF in C. reinhardtii seems to be independent from the association of LHCII proteins to PSI or PSII (Takahashi et al., 2013). In agreement, our data show that the accumulation of LHCBM9 in thylakoids did not abolish the cells’ ability to increase CEF during nutrient deficiency. Although the relationship between CEF and state transition is under debate (Takahashi et al., 2013), the accumulation of LHCBM9 in thylakoids did not significantly affect the LHC transition to state II in sulfur deficiency, suggesting that LHCBM9 function is exerted in the close vicinity of PSII.
PSII-Bound LHCBM9 Induces Faster Fluorescence Decay Kinetic under Sulfur Deficiency
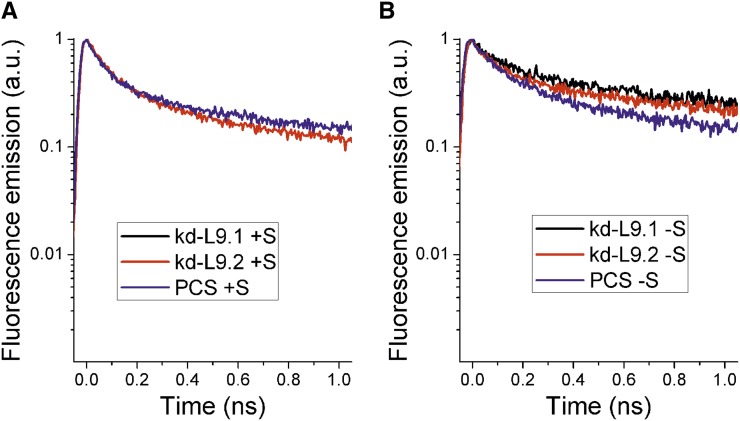
PSII supercomplexes were purified from thylakoids of PCS and kd-L9.1/2 genotypes as described in the previous section and analyzed by time-resolved fluorescence spectroscopy in order to verify whether the accumulation of LHCBM9 could affect the level and distribution of chlorophyll excited states. Fraction 8 was selected for analysis because this sample contained the PSII supercomplexes with the highest chlorophyll b content (Supplemental Figure 4). As shown in Figure 4A, PSII supercomplexes purified from cells grown in +S conditions showed similar fluorescence decay kinetics, irrespective of the genotype. However, when PSII supercomplexes were isolated after -S treatment, the presence of LHCBM9 in the PCS resulted in faster fluorescence decay compared with the kd-L9.1/2 samples (Figure 4B).
Figure 4.
Time-Resolved Fluorescence Emission Decay Analysis of PSII Supercomplexes in the LHCBM9 Knockdown Lines (kd-L9.1/2) and the PCS.
Fluorescence emission decay kinetics were measured for PSII supercomplexes purified from PCS and kd-L9.1/2 cells grown in presence of sulfur (A) or after cultivation under sulfur deficiency for 48 h (B).
Fluorescence decay kinetics could be satisfactorily fitted using three exponential decay components (Table 1): The fastest component had a time constant (τ) below 100 ps, the intermediate component had a time constant in the range of 280 to 590 ps, and the longest component had one of 3.0 to 3.8 ns. These components do not necessarily represent discrete physical processes but comprehensively describe PSII fluorescence decay kinetics. The long component is usually associated with LHCII complexes unquenched by the photochemical activity of PSII. In -S conditions, the relative abundance of this long component was considerable higher in both kd-L9.1 and kd-L9.2 compared with PCS in the same conditions (-S), suggesting a greater number of LHCII complexes poorly connected to the core moiety within PSII supercomplexes in the absence of LHCBM9 (Table 1).
Table 1. Decay Kinetics of PSII Supercomplex Fluorescence Emission.
| Strain and Growth Condition | A1 (%) | τ1 (ps) | A2 (%) | τ2 (ps) | A3 (%) | τ3 (ns) | τAV (ps, τ3 Not Included) | τAV (ps, τ3 Included) |
|---|---|---|---|---|---|---|---|---|
| PCS +S | 54.3 | 75 | 22.5 | 283 | 14.0 | 3.3 | 104 | 562 |
| kd-L9.1 +S | 66.7 | 71 | 18.9 | 362 | 14.4 | 3.2 | 116 | 573 |
| kd-L9.2 +S | 60.2 | 64 | 27.6 | 282 | 12.2 | 3.0 | 116 | 490 |
| PCS -S | 50.5 | 79 | 33.5 | 355 | 17.3 | 3.5 | 189 | 757 |
| kd-L9.1 -S | 44.0 | 74 | 28.0 | 453 | 26.0 | 3.5 | 221 | 1108 |
| kd-L9.2 -S | 43.0 | 90 | 32.0 | 653 | 24.5 | 3.8 | 330 | 1177 |
Decay kinetics were fitted with three exponentials with amplitude (A1 to A3) and time constants (τ1 to τ3) reported in the table. τ AV, average lifetime of PSII supercomplex fluorescence emission calculated by the weighted average of τ1-2 or τ1-3. PSII supercomplexes from PCS and LHCBM9 knockdown lines kd-L9.1 and kd-L9.2 were analyzed after cultivation in sulfur-replete (+S) or sulfur-deficient (-S) conditions. Error bars are below 5% for amplitude and time constants (n = 3); R2 is higher than 0.996 for all samples.
Interestingly, in PSII supercomplexes purified from PCS after sulfur depletion, the presence of LHCBM9 reduced the lifetime of the intermediate component, which can be assigned to fluorescence emission from the LHC proteins well connected to PSII reaction center. The average lifetime calculated for PSII supercomplexes analyzed are reported in Table 1. We also calculated the average lifetime without considering the 3.5- to 3.8-ns component, a procedure that was proposed to yield a more representative lifetime value for the PSII-LHCII supercomplex by avoiding the contribution of ill-connected antennas (Caffarri et al., 2009): In +S conditions, the average lifetime was similar for PCS and kd-L9.1/2, while the presence of LHCBM9 in PCS -S PSII supercomplexes induced a shortening of the average fluorescence lifetime that was not evident in the knockdown lines. This LHCBM9-dependent quenching in PCS -S originates from an increase connection of the outer LHCII to PSII core complexes (reduced A3 in PCS -S, compared with kd-L9.1/2 mutant; Table 1), partially restoring the situation of PSII supercomplexes in +S conditions, and from a reduction of the time constant associated with the energy transfer from the connected antenna proteins to the reaction center (τ2). These results are consistent with the hypothesis of LHCBM9 being a better quencher compared with most of the other LHCBM proteins and being important for stabilization of PSII supercomplex.
Nonphotochemical Quenching Induction
In order to evaluate a possible role of LHCBM9 in NPQ of PSII, PCS and knockdown lines were grown in high-light conditions (400 μmol m−2 s−1) to induce activation of the LHCSR3-dependent stress response (Peers et al., 2009; Bonente et al., 2012). LHCSR3 accumulated to similar levels in all genotypes (Supplemental Figure 7A). Upon 48 h of sulfur deficiency, slightly increased levels of LHCSR3 were detected in all three cell lines. The sulfur deficiency treatment also caused a significant accumulation of LHCBM9 in the parental cell line (PCS -S), while this effect was much lower in the respective knockdown lines (kd-L9.1/2 -S). NPQ traces of PCS and kd-L9.1/2 mutants in +S and -S conditions are shown in Supplemental Figures 7B and 7C. Although the treatment induced similar NPQ activity and kinetics in both PCS and knockdown genotypes, the effect was consistently higher, by 30%, in the knockdown lines. Since similar levels of LHCSR3 accumulated in the three genotypes, the differences in NPQ activity could not be ascribed to differential LHCSR3 accumulation. The PCS strain showed lower NPQ activity than the knockdown lines despite having higher LHCBM9 levels, therefore demonstrating that LHCBM9 is not specifically involved in excess-light-induced NPQ.
In Vitro Reconstitution and Functional Characterization of LHCBM9 Holoprotein
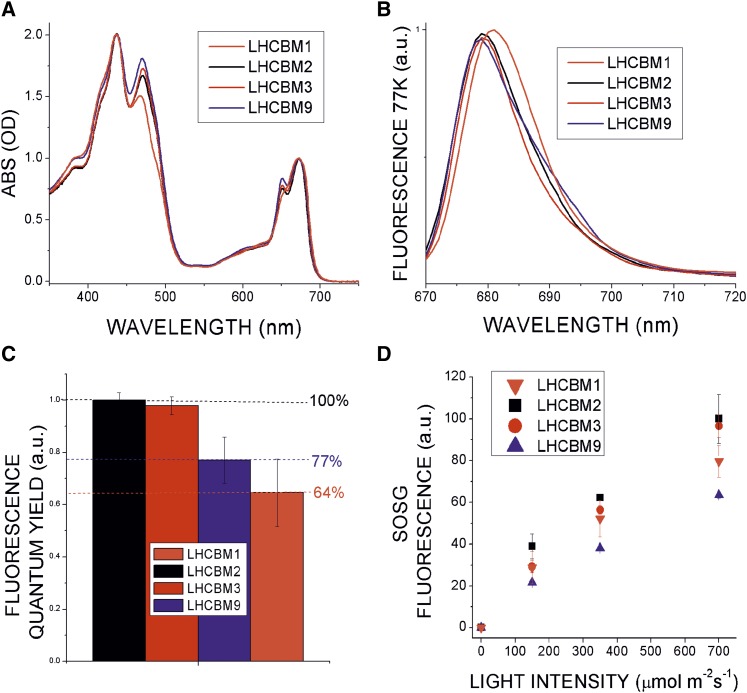
In order to obtain direct information on the functional role of LHCBM9, the mature LHCBM9 apoprotein was expressed in E. coli and the holoprotein was subsequently obtained by in vitro refolding in the presence of C. reinhardtii pigment extracts, as described previously (Giuffra et al., 1996; Bonente et al., 2011). The same procedure was applied to LHCBM1, LHCBM2, and LHCBM3, which were used as controls. All reconstituted complexes were correctly refolded as judged from the establishment of efficient energy transfer from chlorophyll b and xanthophylls to chlorophyll a, measured by fluorescence emission spectra. Amino acid sequence alignments suggested that all chlorophyll binding residues present in LHCII were conserved in LHCBM9 (Supplemental Figure 8). The pigments bound by the recombinant LHCBM proteins were analyzed by HPLC (Supplemental Table 2). All monomeric complexes contained chlorophylls a and b with a similar chlorophyll a/b ratio; however, slightly lower chlorophyll b content was determined for LHCBM1 and 9 compared with LHCBM2 and 3. Neoxanthin, lutein, and violaxanthin were found be bound to all LHCBM proteins yielding three to four carotenoids per each refolded LHCII complex, with roughly 1.4 neoxanthin, 1.7 luteins, and a substoichiometric fraction of violaxanthin per apoprotein. In the case of LHCBM1, the violaxanthin content was slightly reduced compared with other LHCBM proteins. Violaxanthin is mainly bound to LHCII at the peripheral V1 site (Liu et al., 2004), which is less stably bound (Caffarri et al., 2001). Absorption spectra of reconstituted proteins did not reveal major differences (Figure 5A) but for a slightly reduced absorption at 475 and 650 nm in LHCBM1, from reduced chlorophyll b content. The fluorescence emission spectra at 77K of the LHCBM proteins revealed significant differences between the individual LHCII protein isoforms. In particular, the peak of LHCBM1 was the most red-shifted at 681 nm versus 679 nm for LHCBM2, 3, and 9 (Figure 5B). Interestingly, LHCBM9 showed enhanced amplitude of the red-most component located at 695 nm. Such “red” emission forms are associated to chlorophyll ligands with transitions at low energy, potentially acting as quenching sites (Miloslavina et al., 2008). In order to test the possibility of LHCBM9 acting as a quencher, the relative fluorescence quantum yield of reconstituted LHCBM proteins was measured (Figure 5C). Among the investigated LHCBM proteins, LHCBM1 showed the lowest relative fluorescence quantum yield, followed by LHCBM9. By contrast, LHCBM2 and 3 had higher yields. These results suggest a similar function for LHCBM1 and LHCBM9, having the most red-shifted emission bands and the highest quenching properties. This is in agreement with the fluorescence emission decay data. Since quenching of excitation energy prevents the formation of ROS species, we suggest a photoprotective role for LHCBM9. In order to further investigate on this hypothesis, we measured 1O2 production upon light exposure for the different reconstituted LHCBM proteins (Figure 5D) by incubation with Singlet Oxygen Sensor Green (Flors et al., 2006). As a result, the lowest production of 1O2 was detected for LHCBM9, followed by LHCBM1 and LHCBM2/LHCBM3, indicating that the presence of LHCBM9 limits ROS production. Consistent with this, we isolated LHCII trimers from PCS+S/-S, kd-L9.1-S, and kd-L9.2-S thylakoid membranes (Supplemental Figure 9). Significantly reduced fluorescence quantum yield and 1O2 production were observed with LHCII trimers including LHCBM9 when compared with LHCII trimers without LHCBM9. This effect was less pronounced when LHCBM9 expression was repressed in the knockdown lines, pointing to a direct and specific connection of this phenotype to the relative amount of LHCBM9 proteins within the LHCII trimers.
Figure 5.
Spectroscopic Properties of LHCBM9 Compared with LHCBM1, 2, and 3.
(A) Absorption spectra of recombinant reconstituted LHCBM1 (orange), LHCBM2 (black), LHCBM3 (red), and LHCBM9 (blue).
(B) Fluorescence emission spectra measured at 77K, with the same color code used as in (A).
(C) Relative fluorescence quantum yield; the percentage of fluorescence quantum yield for LHCBM1, LHCBM3, and LHCBM9 is reported in relation to LHCBM2, set to 100%. Error bars represent sd (n = 5).
(D) Singlet oxygen production measured as increasing Singlet Oxygen Sensor Green fluorescence at 530 nm upon illumination at different light intensities. Error bars represent sd (n = 5).
LHCBM9 Is a Major Antenna Component in -S Conditions
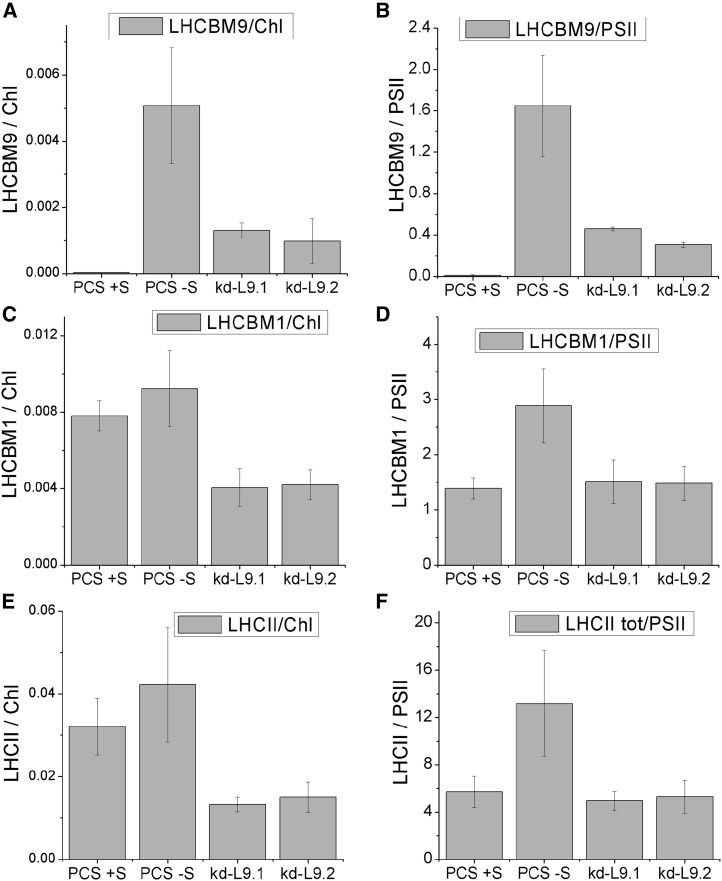
The previous results indicated that sulfur depletion caused major LHC antenna reorganization. We therefore investigated to what extent the LHCBM9 protein could replace other LHCBM proteins (Supplemental Figure 10) and the stoichiometry between LHCBM9 and PSII core complexes (Supplemental Figure 11). We followed the degradation pattern of LHCII complexes during cultivation in sulfur deficiency compared with the accumulation of LHCBM9 in the same conditions via SDS-PAGE. The subsequent immunoblots were performed with an antibody cross-reacting with all LHCBM proteins, typically resulting in a pattern of three distinct bands with apparent molecular masses in the range of ∼20 to ∼30 kD and LHCBM9 being part of the top band (Supplemental Figure 10). During sulfur depletion, the overall LHCII amount strongly decreased in all cell lines. However, the expression of LHCBM9 in the PCS partly alleviated the decrease of the top band, confirming a certain degree of LHCBM protein substitution. The relative amount of LHCBM9 per PSII in thylakoids from PCS and knockdown lines was determined next. For quantification, recombinant LHCBM9 was loaded on a gel together with thylakoid membranes, LHCII trimers, and purified PSII core complexes (Supplemental Figure 11). The relative intensity of the signal obtained with anti-CP43 antibodies served as a reference for the determination of PSII in thylakoids. Also, recombinant LHCBM1 was used as a reference for its abundance in thylakoids from the reaction with LHCII-specific antibodies (Supplemental Figure 11). From the results of this immunotitration, we calculated a stoichiometry of 1.6 LHCBM9 per PSII core complex in PCS after LHCBM9 induction (-S for 48 h; Figure 6). Interestingly, in -S conditions, the LHCII level was significantly higher in PCS thylakoids compared with LHCBM9 knockdown genotypes. These results show that a substantial fraction of LHCII was made up by LHCBM9 upon sulfur depletion and that the presence of this gene product was effective in increasing the abundance of LHCII per chlorophyll upon stress.
Figure 6.
Levels of LHCBM9, LHCBM1, and LHCII per Chlorophyll and per PSII in PCS and LHCBM9 Knockdown Mutants.
The amount of LHCBM9 ([A] and [B]), LHCBM1 ([C] and [D]), and LHCII trimers ([E] and [F]) per chlorophyll ([A], [C], and [E]) and per PSII ([B], [D], and [F]) were determined by immunoblot analysis (Supplemental Figure 11). Error bars represent sd (n = 3).
DISCUSSION
Two general functions can be assigned to LHCII proteins of plants and algae, one being harvesting light energy to fuel the photosynthetic reactions and the other being energy dissipation and ROS scavenging to protect the cells from excess irradiation (Müller et al., 2001; Ballottari et al., 2012). This functional diversity is reflected by the fact that plants and algae have accumulated multiple LHC gene isoforms in the nuclear genome during evolution. The functional characterization of individual LHC isoforms has been assessed for a few of the corresponding proteins. A functional role in thermal energy dissipation could be assigned to LHCBM1 (Elrad et al., 2002), most likely as a partner of LHCSR3 (Peers et al., 2009; Bonente et al., 2011).
Mutant strains unable to accumulate LHCBM1 showed a reduced capacity for NPQ, indicating that this subunit plays a role as an energy quencher to avoid photodamage. LHCBM1 belongs to a subfamily of LHCBM proteins that also include the antenna proteins LHCBM2 and LHCBM7 (Supplemental Figure 1). The homologous proteins LHCBM2, 5, and 7 have been shown to be needed for state 1 to state 2 transitions (Takahashi et al., 2006; Ferrante et al., 2012). Finally, a role in maintaining LHCI structure has been postulated for the LHCA3 protein (Naumann et al., 2005).
In this work, we investigated the features and functional role of LHCBM9, a LHCII-type protein from C. reinhardtii. Our results show that LHCBM9 is a pigment binding protein, with affinity for chlorophyll a/b and carotenoids, similar to other LHCII subunits (Caffarri et al., 2001; Liu et al., 2004) with which it shares high sequence similarity, especially to the LHCII-type one isoforms LHCBM3, 4, 6, and 8 (Supplemental Figure 1). In contrast to LHCBM1-8, expression of LHCBM9 is induced only when cells are subjected to continuous stress conditions over several hours. The finding that LHCBM9 is enriched in trimeric complexes is consistent with the presence of an amino acid motif essential for LHC trimer formation (Supplemental Figure 1) in LHCBM9 sequence (Minagawa and Takahashi, 2004). LHCBM9 is also found in PSII supercomplexes (Figure 3; Supplemental Figure 5).
The expression and accumulation of LHCBM9 to substantial amounts in thylakoid membranes (Figure 3) has a strong photoprotection effect under stress conditions, as clearly indicated by the fact that the PCS has a higher chlorophyll content per cell compared with LHCBM9 knockdown cell lines (Figure 2C). Since the cell number in the culture also decreased under sulfur starvation, this implies that the accumulation of LHCBM9 substitutes for preexisting LHCII gene products and increases light energy dissipation and prevention of cell damage by ROS.
The photoprotective effect of LHCBM9 is exerted through multiple mechanisms: First, both native LHCII trimers containing LHCBM9 and recombinant LHCBM9 refolded in vitro have a lower fluorescence yield and lower production of 1O2 upon illumination (Figure 5; Supplemental Figure 9). This effect appears to be well suited to nutrient deficiency conditions, when PSII core complexes are partially degraded and a fraction of LHCII trimers remains in the thylakoid membrane in the absence of photochemical quenching of the energy they absorb. It could be asked whether the lower yield of 1O2 derives from the lower level of 1Chl* excited states and, thus, from an increased probability of 3Chl* formation. We observed that the decrease of 1O2 formation in LHCBM9, as compared with LHCBM2/3, is stronger than the decrease of fluorescence yield, suggesting that additional mechanisms are also involved such as improved 3Chl* quenching and/or 1O2 scavenging by bound carotenoids (Ballottari et al., 2013). Second, in sulfur deficiency, PSII supercomplexes containing LHCBM9 are found in a more dissipative state compared with PSII supercomplexes purified from kd-L9.1/2, thus decreasing overexcitation of PSII reaction centers and photoinhibition (Figure 4, Table 1). It is interesting that the energy dissipation effect elicited by LHCBM9 is substantially different with respect to that catalyzed by LHCBM1 and LHCSR3 upon exposure to excess light conditions. In fact, while quenching in LHCSR3-containing PSII-LHCII supercomplexes has been reported to occur in a pH-dependent manner (Tokutsu and Minagawa, 2013), in our experiments (Figure 4), quenching occurred at pH 7.0, implying that it was active in nonacidic conditions. However, the LHCBM9-dependent quenching mechanism observed in PCS -S supercomplexes could also derive from a LHCBM9-mediated more stable connection of outer LHCBM proteins to PSII core complex. Third, the abundance of PSII-LHCII supercomplexes is increased by the presence of LHCBM9, suggesting a stabilization of this supramolecular organization that is preferentially disrupted upon the photoinhibitory effect of sulfur deprivation. Fourth, the replacement of other LHCBMs by LHCBM9 in sulfur deficiency resulted in a reduced capacity of LHCII trimers to produce ROS, suggesting a specific role of LHCBM9 in reducing photooxidative damage in stress conditions.
It has been suggested that LHC downregulation could be a viable strategy to increase hydrogen production with C. reinhardtii. Very recently it was shown that the simultaneous silencing of LHCBM1,2 and 3 resulted in a remarkable boost of hydrogen production efficiency (Oey et al., 2013). In this work, we show that, by contrast, downregulation of the LHCBM9 isoform via artificial microRNA leads to a decreased potential for hydrogen production (Figure 2E), most likely because of the specific role of this LHC subunit upon stress induction. The inability of expressing considerable amounts of LHCBM9 under H2 production conditions most likely causes an increase in photoinhibition and, therefore, a reduced provision of electrons for the hydrogenase enzymes. As the efficiency of substrate supply is the major bottleneck in H2 production with microalgae (Kruse and Hankamer, 2010), reduced availability of protons and electrons derived from the water-splitting reaction at PSII should result in a perturbation of hydrogen production activity. Recently, Volgusheva et al. (2013) showed that the amount of active PSII during the hydrogen production phase is directly correlated to the amount of H2 generation. This could mean that in the absence of LHCBM9 and the concomitant higher production of ROS, photoinhibitory damage of PSII is more pronounced, therefore leading to lower overall amounts of H2 production. This indicates that a biotechnological strategy aiming at improving light penetration and hydrogen production rates by downregulation of all LHCBM proteins may not be fruitful. Combined approaches including differential up- and downregulation of individual LHCBM proteins could be more suitable for maintaining the integrity of photosynthetic apparatus during H2 production.
In this work, we demonstrate that certain nutrient starvation triggers LHCBM9 accumulation and that this protein is important in preventing stress-dependent reduction of LHCII content. No indication could be found for a light-dependent regulation of the LHCBM9 expression. The fact that LHCBM9 mRNA and proteins are virtually undetectable when cells are not subjected to stress conditions implies that transcription and translation have to take place after the environmental stress condition has set in. In this work, LHCBM9 mRNA and subsequent protein accumulation were detected in the timeframe of hours (Figure 1). Therefore, it is most likely that the protection function of LHCBM9 in vivo is most important when stress conditions prevail for several hours or longer. The signal transduction pathway triggering LHCBM9 expression is not known. It could therefore be possible that ROS do not directly lead to LHCBM9 accumulation but that indirect factors, e.g., resulting from photoinhibition of PSII complexes, are responsible. This notion would be in agreement with results from Ramundo et al. (2013) where LHCBM9 upregulation was detected when genes for chloroplast transcription or translation were repressed for several days.
In conclusion, the data presented in this work provide evidence that the green microalgae C. reinhardtii has evolved a LHCII-dependent, medium-term acclimation process for nutrient deficiency stress conditions that involves the remodeling of PSII-LHCII supercomplexes, leading to reduced ROS formation and, therefore, to enhanced photoprotection of the cell. Although proteins very similar to LHCBM9 are found in a wide range of algae and plants, the high degree of isoform similarity makes it difficult to assign putative functional analogs. Proteins could be identified in Volvox carteri (NCBI/GenBank XP_002951894.1) and in Chlamydomonas incerta (NCBI/GenBank ABD37913) for which the mature forms showed 90 or 97% sequence identity with C. reinhardtii LHCBM9, respectively, indicating that functional analogs likely exist at least within these phyla.
METHODS
Strains and Culture Conditions
Liquid cultures of Chlamydomonas reinhardtii cell wall–deficient PCS CC-406 and the two LHCBM9 knockdown transformants (kd-L9.1 and kd-L9.2) were cultivated mixotrophically in TAP (Tris acetate phosphate) medium (Harris et al., 2009) under continuous illumination (40 μmol m−2 s−1) and agitation. For experiments in nutrient-deprivation medium, cells were grown mixotrophically until they reached the early logarithmic growth phase, washed three times (2500g, 3 min, 20°C) with TAP without sulfur (-S), nitrogen (-N), or phosphorus (-P), and were adjusted to equal optical (OD750 = 0.8) or cell densities (7 × 106 cells/mL).
Artificial MicroRNA Construct and Screening for Knockdown Transformants
The pChlamiRNA3int vector (Molnar et al., 2009) was modified to contain a LHCBM9-specific artificial microRNA and then transformed into the PCS. The amiRNA sequence chosen was (5′-TTCGGGAGTAACAATGCGCAG-3′), targeting exon 3 of the LHCBM9 coding region. Transformants were selected on TAP agar plates containing 10 µg/mL paromomycin. For screening, the transformants were transferred into sulfur-depleted medium to induce LHCBM9 expression.
Chlorophyll and Chlorophyll Fluorescence Measurements
Chlorophyll contents were determined spectroscopically after extraction with 80% acetone according to Arnon (1949). Photosynthetic quantum yield ΦPSII was determined by illuminated cell suspensions with a MINI-PAM (Waltz). Fluorescence parameters were recorded and ΦPSII calculated [ΦPSII = (Fm’ − Ft)/Fm’] according to Maxwell and Johnson (2000).
SDS-PAGE, Immunoblotting, and Native Electrophoresis
Proteins were separated by Tris-glycine or Tris-Tricine-SDS-PAGE (Schägger and von Jagow, 1987; Bonente et al., 2011) and detected by immunoblotting using enhanced chemiluminescence (ECL; Pierce) or by alkaline phosphatase detection (Sigma-Aldrich). The specific polyclonal antibody for epitope LQKNGVQF of LHCBM9 was produced by Agrisera. Native electrophoresis was performed using Deriphat-PAGE gel as described (Ferrante et al., 2012) or using a Clear-Native-PAGE system (Järvi et al., 2011). The LHCII antibodies used were raised against the synthetically produced 25 N-terminal amino acids of the LHCBM6 precursor protein as described (Hippler et al., 2001) and the antibody raised against LHCBM5 (Agrisera).
Hydrogen Production
Hydrogen production experiments were performed as described earlier (Doebbe et al., 2007) with the following modifications. The centrifugation steps were performed at 2500g for 3 min, the cultures were adjusted to ∼23 µg of chlorophyll per mL, and the experiment was performed at 450 μmol m−2 s−1 without the addition of glucose.
Singlet Oxygen Production
Singlet oxygen production in isolated LHCBM proteins was measured using Singlet Oxygen Sensor Green dye (Flors et al., 2006) as described (Betterle et al., 2010), illuminating the samples with variable light intensities as described.
RNA Isolation and Quantification
RNA was isolated according to Chomczynski and Sacchi (1987) and treated with RNase free DNaseI (Promega). RT-Q-PCR was performed using the SensiMix One-Step kit (Bioline). The 18S rRNA gene was used as a control gene (Nguyen et al., 2011). The primer pairs used for RT-Q-PCR were: LHCBM9, 5′-AGGCCTTCTGGATGTACCAC-3′ and 5′-ATGGTTCTGGACACAACTGC-3′; 18S rRNA, 5′-CCTGCGGCTTAATTTGACTC-3′ and 5′-ACCGGAATCAACCTGACAAG-3′; and Gpxh, 5′-ACGCCACAATACAAGGGTCT-3′ and 5′-TCACGTCCGACTTCTCCATA-3′.
Thylakoid Preparation and Native Chlorophyll Binding Protein Isolation
Thylakoids from C. reinhardtii were prepared as described (Ferrante et al., 2012). In order to isolate the different chlorophyll proteins, thylakoid membranes were solubilized with 1% β-dodecyl-maltoside, loaded on sucrose gradients, and ultracentrifuged overnight as described (Tokutsu et al., 2012). Fractions F1 to F8, isolated from the sucrose gradients, were loaded on SDS-PAGE on a volumetric base (Figure 3), maintaining the chlorophyll distribution observed in the sucrose gradients.
Overexpression and in Vitro Refolding of Recombinant LHCBM9 Protein
The mature LHCBM1, 2, 3, and 9 coding sequences were cloned into the expression vector as described (Flors et al., 2006). Chloroplast transit peptides were identified using ChloroP prediction software (http://www.cbs.dtu.dk/services/ChloroP/). Recombinant apoprotein was refolded in vitro with pigments extracted from C. reinhardtii as previously described (Giuffra et al., 1996).
Pigment Extraction and HPLC Analysis
Pigment extractions from C. reinhardtii cells and chlorophyll binding complexes as well as HPLC analysis were performed as described (Ferrante et al., 2012).
Absorption and Steady State Fluorescence Measurements
Absorption and fluorescence emission spectra of isolated LHC proteins were measured as described (Bonente et al., 2011). Low-temperature (77K) fluorescence was measured in a cryostat with 80% glycerol added to each mixture, and 77K fluorescence emission spectra were measured from whole cells in light-adapted samples as described by Bonente et al. (2012).
Cyclic Electron Flow Measurement
Cyclic electron flow was measured transferring cell cultures into state I and state II, respectively, as described (Iwai et al., 2010), with the variation that ChlP700 activity was estimated as described (Ferrante et al., 2012).
Time-Resolved Fluorescence
Time-resolved fluorescence measurements were performed using a femtosecond laser source and streak camera detection system (Hamamatsu C5680). An unamplified Ti:Sapphire laser (Coherent Chameleon Ultra II) operating at 80 MHz was tuned to provide pulses with central wavelengths of 880 nm, energies of ∼50 nJ, and temporal and spectral bandwidths of ∼140 fs and ∼5 nm, respectively. A β-barium borate crystal was used for type I phase-matched second harmonic generation, leading to pulses with central wavelengths of 440 nm. Measurements of fast decays (t < 1 ns) used a synchroscan voltage sweep module, yielding a maximum temporal resolution of ∼3.6 ps. Slower decays (t > 1 ns) used a linear voltage sweep module under 2 MHz excitation, achieved through the use of an acousto-optical modulating pulse picker (APE Pulse Select), yielding a maximum temporal resolution of ∼50 ps.
Accession Numbers
We follow the nomenclature of the C. reinhardtii genome sequence v4.3 (http://www.phytozome.net/; Joint Genome Institute, Department of Energy, and Center for Integrative Genomics, University of California). Sequence data from this article can be found in the Phytozome and NCBI GenBank databases. The accession numbers (NCBI-GenBank accession numbers indicated in parentheses) are: LHCBM1, Cre23.g766250 (AAM18057); LHCBM2, Cre12.g548400 (XP_001693987); LHCBM3, Cre32.g781300 (XP_001703699); LHCBM4, Cre06.g283950 (XP_001695344); LHCBM5, Cre03.g156900 (XP_001697526); LHCBM6, Cre06.g285250 (XP_001695353); LHCBM7, Cre12.g548950 (XP_001694115); LHCBM8, Cre06.g284250 (XP_001695467); and LHCBM9, Cre06.g284200 (XP_001695466).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Alignment, Phylogenic Tree, and I-Tasser Model of LHCBM9.
Supplemental Figure 2. Expression Pattern of LHCSR3 and LHCBM9 after High-Light Cultivation.
Supplemental Figure 3. Phenotypic Characterization of the LHCBM9 Knockdown Lines kd-L9.1 and kd-L9.2 with Respect to the Parental Control Strain in Nutrient-Replete Conditions.
Supplemental Figure 4. Absorption Spectra of Sucrose Gradient Fractions.
Supplemental Figure 5. Analysis of Interactions between LHCBM9 and Reaction Center Complexes by 2D Electrophoresis and Immunoblotting.
Supplemental Figure 6. Cyclic Electron Flow around PSI and LHC State Transition Measurements.
Supplemental Figure 7. LHCBM9 Accumulation and NPQ Characteristics in High-Light Conditions.
Supplemental Figure 8. Protein Sequence Alignment of LHCBM9 with LHCBM1 and LHCII from Arabidopsis thaliana (Lhcb1 Subunit).
Supplemental Figure 9. Fluorescence Quantum Yield and Singlet Oxygen Production of Native LHCII Trimers.
Supplemental Figure 10. Immunoblot Analysis of LHCBM Protein Degradation during Sulfur Deficiency.
Supplemental Figure 11. Immunoblot Analysis of Thylakoids and LHCII Fractions Purified from PCS, kd-L9.1 and kd-L9.2 Mutants, and Recombinant LHCBM9 and LHCBM1.
Supplemental Table 1. Gene and Protein Characteristics of LHCBM9 as Determined by in Silico Analyses.
Supplemental Table 2. Pigment Analysis of Reconstituted LHCBM Proteins.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis in Supplemental Figure 1B.
Supplementary Material
Acknowledgments
We thank A. Alboresi for his assistance in cloning of the LHCBM coding sequences, K. Rogge for her assistance in mRNA preparations, and M. Hippler for the LHCII antibody and LHCBM9 antibody specificity tests. This work was supported by the EU-FP7 project SOLAR-H2/Contract 212508 and the German Federal Ministry of Science (BMBF) project HydroMicPro/Contract 03SF0361G (to O.K.), the EU-FP7 project 316427 ACCLIPHOT, and the Italian Ministry of Agriculture, Food, and Forestry (MIPAAF) project HYDROBIO (to R.B.).
AUTHOR CONTRIBUTIONS
S.G., M.B., J.H.M., B.H., R.B., and O.K. designed research. S.G., M.B., M.A., C.D., and O.B. performed research. S.G., M.B., J.H.M., M.A., C.D., O.B., B.H., R.B., and O.K. analyzed data. S.G., M.B., J.H.M., R.B., and O.K. wrote the article. The authors declare no conflict of interest.
Glossary
- PSII
photosystem II
- PSI
photosystem I
- LHC
light-harvesting complex
- ROS
reactive oxygen species
- NPQ
nonphotochemical quenching
- PCS
parental control strain
- RT-Q-PCR
quantitative real-time RT-PCR
- CEF
cyclic electron flow
- TAP
Tris acetate phosphate
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Aksoy M., Pootakham W., Pollock S.V., Moseley J.L., González-Ballester D., Grossman A.R. (2013). Tiered regulation of sulfur deprivation responses in Chlamydomonas reinhardtii and identification of an associated regulatory factor. Plant Physiol. 162: 195–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allorent G., et al. (2013). A dual strategy to cope with high light in Chlamydomonas reinhardtii. Plant Cell 25: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon D.I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballottari M., Girardon J., Dall’osto L., Bassi R. (2012). Evolution and functional properties of photosystem II light harvesting complexes in eukaryotes. Biochim. Biophys. Acta 1817: 143–157 [DOI] [PubMed] [Google Scholar]
- Ballottari M., Mozzo M., Girardon J., Hienerwadel R., Bassi R. (2013). Chlorophyll triplet quenching and photoprotection in the higher plant monomeric antenna protein Lhcb5. J. Phys. Chem. B 117: 11337–11348 [DOI] [PubMed] [Google Scholar]
- Barber J., Archer M.D. (2001). P680, the primary electron donor of photosystem II. J. Photochem. Photobiol. 142: 97–106 [Google Scholar]
- Bellafiore S., Barneche F., Peltier G., Rochaix J.D. (2005). State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433: 892–895 [DOI] [PubMed] [Google Scholar]
- Bennett J., Steinback K.E., Arntzen C.J. (1980). Chloroplast phosphoproteins: regulation of excitation energy transfer by phosphorylation of thylakoid membrane polypeptides. Proc. Natl. Acad. Sci. USA 77: 5253–5257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betterle N., Ballottari M., Hienerwadel R., Dall’Osto L., Bassi R. (2010). Dynamics of zeaxanthin binding to the photosystem II monomeric antenna protein Lhcb6 (CP24) and modulation of its photoprotection properties. Arch. Biochem. Biophys. 504: 67–77 [DOI] [PubMed] [Google Scholar]
- Bonente G., Ballottari M., Truong T.B., Morosinotto T., Ahn T.K., Fleming G.R., Niyogi K.K., Bassi R. (2011). Analysis of LhcSR3, a protein essential for feedback de-excitation in the green alga Chlamydomonas reinhardtii. PLoS Biol. 9: e1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonente G., Pippa S., Castellano S., Bassi R., Ballottari M. (2012). Acclimation of Chlamydomonas reinhardtii to different growth irradiances. J. Biol. Chem. 287: 5833–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarri S., Croce R., Breton J., Bassi R. (2001). The major antenna complex of photosystem II has a xanthophyll binding site not involved in light harvesting. J. Biol. Chem. 276: 35924–35933 [DOI] [PubMed] [Google Scholar]
- Caffarri S., Kouril R., Kereïche S., Boekema E.J., Croce R. (2009). Functional architecture of higher plant photosystem II supercomplexes. EMBO J. 28: 3052–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Croce R., Weiss S., Bassi R. (1999). Carotenoid-binding sites of the major light-harvesting complex II of higher plants. J. Biol. Chem. 274: 29613–29623 [DOI] [PubMed] [Google Scholar]
- Dall’Osto L., Cazzaniga S., North H., Marion-Poll A., Bassi R. (2007a). The Arabidopsis aba4-1 mutant reveals a specific function for neoxanthin in protection against photooxidative stress. Plant Cell 19: 1048–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Osto L., Fiore A., Cazzaniga S., Giuliano G., Bassi R. (2007b). Different roles of alpha- and beta-branch xanthophylls in photosystem assembly and photoprotection. J. Biol. Chem. 282: 35056–35068 [DOI] [PubMed] [Google Scholar]
- Dall’Osto L., Holt N.E., Kaligotla S., Fuciman M., Cazzaniga S., Carbonera D., Frank H.A., Alric J., Bassi R. (2012). Zeaxanthin protects plant photosynthesis by modulating chlorophyll triplet yield in specific light-harvesting antenna subunits. J. Biol. Chem. 287: 41820–41834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J.P., Boekema E.J. (2005). Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 1706: 12–39 [DOI] [PubMed] [Google Scholar]
- Depège N., Bellafiore S., Rochaix J.D. (2003). Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299: 1572–1575 [DOI] [PubMed] [Google Scholar]
- Doebbe A., Rupprecht J., Beckmann J., Mussgnug J.H., Hallmann A., Hankamer B., Kruse O. (2007). Functional integration of the HUP1 hexose symporter gene into the genome of C. reinhardtii: Impacts on biological H(2) production. J. Biotechnol. 131: 27–33 [DOI] [PubMed] [Google Scholar]
- Durnford D.G., Price J.A., McKim S.M., Sarchfield M.L. (2003). Light-harvesting complex gene expression is controlled by both transcriptional and post-transcriptional mechanisms during photoacclimation in Chlamydomonas reinhardtii. Physiol. Plant. 118: 193–205 [Google Scholar]
- Elrad D., Grossman A.R. (2004). A genome’s-eye view of the light-harvesting polypeptides of Chlamydomonas reinhardtii. Curr. Genet. 45: 61–75 [DOI] [PubMed] [Google Scholar]
- Elrad D., Niyogi K.K., Grossman A.R. (2002). A major light-harvesting polypeptide of photosystem II functions in thermal dissipation. Plant Cell 14: 1801–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escoubas J.M., Lomas M., LaRoche J., Falkowski P.G. (1995). Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc. Natl. Acad. Sci. USA 92: 10237–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante P., Ballottari M., Bonente G., Giuliano G., Bassi R. (2012). LHCBM1 and LHCBM2/7 polypeptides, components of major LHCII complex, have distinct functional roles in photosynthetic antenna system of Chlamydomonas reinhardtii. J. Biol. Chem. 287: 16276–16288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finazzi G., Rappaport F., Furia A., Fleischmann M., Rochaix J.D., Zito F., Forti G. (2002). Involvement of state transitions in the switch between linear and cyclic electron flow in Chlamydomonas reinhardtii. EMBO Rep. 3: 280–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flachmann R., Kühlbrandt W. (1995). Accumulation of plant antenna complexes is regulated by post-transcriptional mechanisms in tobacco. Plant Cell 7: 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flors C., Fryer M.J., Waring J., Reeder B., Bechtold U., Mullineaux P.M., Nonell S., Wilson M.T., Baker N.R. (2006). Imaging the production of singlet oxygen in vivo using a new fluorescent sensor, Singlet Oxygen Sensor Green. J. Exp. Bot. 57: 1725–1734 [DOI] [PubMed] [Google Scholar]
- Giuffra E., Cugini D., Croce R., Bassi R. (1996). Reconstitution and pigment-binding properties of recombinant CP29. Eur. J. Biochem. 238: 112–120 [DOI] [PubMed] [Google Scholar]
- González-Ballester D., Casero D., Cokus S., Pellegrini M., Merchant S.S., Grossman A.R. (2010). RNA-seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell 22: 2058–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman A.R., Bhaya D., Apt K.E., Kehoe D.M. (1995). Light-harvesting complexes in oxygenic photosynthesis: diversity, control, and evolution. Annu. Rev. Genet. 29: 231–288 [DOI] [PubMed] [Google Scholar]
- Harris, E.H., Stern, D.B., Witman, G., and Harris, E.H. (2009). The Chlamydomonas Sourcebook. (Amsterdam; London: Elsevier). [Google Scholar]
- Hemschemeier A., Happe T. (2011). Alternative photosynthetic electron transport pathways during anaerobiosis in the green alga Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1807: 919–926 [DOI] [PubMed] [Google Scholar]
- Hippler M., Klein J., Fink A., Allinger T., Hoerth P. (2001). Towards functional proteomics of membrane protein complexes: analysis of thylakoid membranes from Chlamydomonas reinhardtii. Plant J. 28: 595–606 [DOI] [PubMed] [Google Scholar]
- Hobe S., Förster R., Klingler J., Paulsen H. (1995). N-proximal sequence motif in light-harvesting chlorophyll a/b-binding protein is essential for the trimerization of light-harvesting chlorophyll a/b complex. Biochemistry 34: 10224–10228 [DOI] [PubMed] [Google Scholar]
- Iwai M., Takizawa K., Tokutsu R., Okamuro A., Takahashi Y., Minagawa J. (2010). Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature 464: 1210–1213 [DOI] [PubMed] [Google Scholar]
- Järvi S., Suorsa M., Paakkarinen V., Aro E.M. (2011). Optimized native gel systems for separation of thylakoid protein complexes: novel super- and mega-complexes. Biochem. J. 439: 207–214 [DOI] [PubMed] [Google Scholar]
- Kruse O. (2001). Light-induced short-term adaptation mechanisms under redox control in the PS II-LHCII supercomplex: LHC II state transitions and PS II repair cycle. Naturwissenschaften 88: 284–292 [DOI] [PubMed] [Google Scholar]
- Kruse O., Hankamer B. (2010). Microalgal hydrogen production. Curr. Opin. Biotechnol. 21: 238–243 [DOI] [PubMed] [Google Scholar]
- Kruse O., Rupprecht J., Bader K.P., Thomas-Hall S., Schenk P.M., Finazzi G., Hankamer B. (2005). Improved photobiological H2 production in engineered green algal cells. J. Biol. Chem. 280: 34170–34177 [DOI] [PubMed] [Google Scholar]
- Li X.P., Björkman O., Shih C., Grossman A.R., Rosenquist M., Jansson S., Niyogi K.K. (2000). A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403: 391–395 [DOI] [PubMed] [Google Scholar]
- Lindahl M., Yang D.H., Andersson B. (1995). Regulatory proteolysis of the major light-harvesting chlorophyll a/b protein of photosystem II by a light-induced membrane-associated enzymic system. Eur. J. Biochem. 231: 503–509 [DOI] [PubMed] [Google Scholar]
- Liu Z.F., Yan H.C., Wang K.B., Kuang T.Y., Zhang J.P., Gui L.L., An X.M., Chang W.R. (2004). Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature 428: 287–292 [DOI] [PubMed] [Google Scholar]
- Maxwell K., Johnson G.N. (2000). Chlorophyll fluorescence—a practical guide. J. Exp. Bot. 51: 659–668 [DOI] [PubMed] [Google Scholar]
- Melis A. (2007). Photosynthetic H2 metabolism in Chlamydomonas reinhardtii (unicellular green algae). Planta 226: 1075–1086 [DOI] [PubMed] [Google Scholar]
- Melis A. (2012). Photosynthesis-to-fuels: from sunlight to hydrogen, isoprene, and botryococcene production. Energ. Environ. Sci. 5: 5531–5539 [Google Scholar]
- Melis A., Zhang L., Forestier M., Ghirardi M.L., Seibert M. (2000). Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Physiol. 122: 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miloslavina Y., Wehner A., Lambrev P.H., Wientjes E., Reus M., Garab G., Croce R., Holzwarth A.R. (2008). Far-red fluorescence: a direct spectroscopic marker for LHCII oligomer formation in non-photochemical quenching. FEBS Lett. 582: 3625–3631 [DOI] [PubMed] [Google Scholar]
- Minagawa J., Takahashi Y. (2004). Structure, function and assembly of photosystem II and its light-harvesting proteins. Photosynth. Res. 82: 241–263 [DOI] [PubMed] [Google Scholar]
- Molnar A., Bassett A., Thuenemann E., Schwach F., Karkare S., Ossowski S., Weigel D., Baulcombe D. (2009). Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 58: 165–174 [DOI] [PubMed] [Google Scholar]
- Müller P., Li X.P., Niyogi K.K. (2001). Non-photochemical quenching. A response to excess light energy. Plant Physiol. 125: 1558–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussgnug J.H., et al. (2005). NAB1 is an RNA binding protein involved in the light-regulated differential expression of the light-harvesting antenna of Chlamydomonas reinhardtii. Plant Cell 17: 3409–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann B., Stauber E.J., Busch A., Sommer F., Hippler M. (2005). N-terminal processing of Lhca3 is a key step in remodeling of the photosystem I-light-harvesting complex under iron deficiency in Chlamydomonas reinhardtii. J. Biol. Chem. 280: 20431–20441 [DOI] [PubMed] [Google Scholar]
- Nguyen A.V., Thomas-Hall S.R., Malnoë A., Timmins M., Mussgnug J.H., Rupprecht J., Kruse O., Hankamer B., Schenk P.M. (2008). Transcriptome for photobiological hydrogen production induced by sulfur deprivation in the green alga Chlamydomonas reinhardtii. Eukaryot. Cell 7: 1965–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A.V., Toepel J., Burgess S., Uhmeyer A., Blifernez O., Doebbe A., Hankamer B., Nixon P., Wobbe L., Kruse O. (2011). Time-course global expression profiles of Chlamydomonas reinhardtii during photo-biological H₂ production. PLoS ONE 6: e29364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi K.K., Bjorkman O., Grossman A.R. (1997). Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell 9: 1369–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oey M., Ross I.L., Stephens E., Steinbeck J., Wolf J., Radzun K.A., Kügler J., Ringsmuth A.K., Kruse O., Hankamer B. (2013). RNAi knock-down of LHCBM1, 2 and 3 increases photosynthetic H2 production efficiency of the green alga Chlamydomonas reinhardtii. PLoS ONE 8: e61375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peers G., Truong T.B., Ostendorf E., Busch A., Elrad D., Grossman A.R., Hippler M., Niyogi K.K. (2009). An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462: 518–521 [DOI] [PubMed] [Google Scholar]
- Petit A.N., Debenest T., Eullaffroy P., Gagné F. (2012). Effects of a cationic PAMAM dendrimer on photosynthesis and ROS production of Chlamydomonas reinhardtii. Nanotoxicology 6: 315–326 [DOI] [PubMed] [Google Scholar]
- Ramundo S., Rahire M., Schaad O., Rochaix J.D. (2013). Repression of essential chloroplast genes reveals new signaling pathways and regulatory feedback loops in Chlamydomonas. Plant Cell 25: 167–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166: 368–379 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Clowez S., Wollman F.A., Vallon O., Rappaport F. (2013). Cyclic electron flow is redox-controlled but independent of state transition. Nat. Commun. 4: 1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Iwai M., Takahashi Y., Minagawa J. (2006). Identification of the mobile light-harvesting complex II polypeptides for state transitions in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 103: 477–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toepel J., Illmer-Kephalides M., Jaenicke S., Straube J., May P., Goesmann A., Kruse O. (2013). New insights into Chlamydomonas reinhardtii hydrogen production processes by combined microarray/RNA-seq transcriptomics. Plant Biotechnol. J. 11: 717–733 [DOI] [PubMed] [Google Scholar]
- Tokutsu R., Minagawa J. (2013). Energy-dissipative supercomplex of photosystem II associated with LHCSR3 in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 110: 10016–10021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokutsu R., Kato N., Bui K.H., Ishikawa T., Minagawa J. (2012). Revisiting the supramolecular organization of photosystem II in Chlamydomonas reinhardtii. J. Biol. Chem. 287: 31574–31581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolleter D., et al. (2011). Control of hydrogen photoproduction by the proton gradient generated by cyclic electron flow in Chlamydomonas reinhardtii. Plant Cell 23: 2619–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volgusheva A., Styring S., Mamedov F. (2013). Increased photosystem II stability promotes H2 production in sulfur-deprived Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 110: 7223–7228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman F.A. (2001). State transitions reveal the dynamics and flexibility of the photosynthetic apparatus. EMBO J. 20: 3623–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman F.A., Delepelaire P. (1984). Correlation between changes in light energy distribution and changes in thylakoid membrane polypeptide phosphorylation in Chlamydomonas reinhardtii. J. Cell Biol. 98: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.