This study shows that ACTIN-RELATED PROTEIN6 regulates female meiosis by determining the spatial and temporal patterns of gene expression required for proper meiosis during ovule development, including expression of the recombinase gene DMC1.
Abstract
In flowering plants, meiocytes develop from subepidermal cells in anthers and ovules. The mechanisms that integrate gene-regulatory processes with meiotic programs during reproductive development remain poorly characterized. Here, we show that Arabidopsis thaliana plants deficient in ACTIN-RELATED PROTEIN6 (ARP6), a subunit of the SWR1 ATP-dependent chromatin-remodeling complex, exhibit defects in prophase I of female meiosis. We found that this meiotic defect is likely due to dysregulated expression of meiotic genes, particularly those involved in meiotic recombination, including DMC1 (DISRUPTED MEIOTIC cDNA1). Analysis of DMC1 expression in arp6 mutant plants indicated that ARP6 inhibits expression of DMC1 in the megasporocyte and surrounding nonsporogeneous ovule cells before meiosis. After cells enter meiosis, however, ARP6 activates DMC1 expression specifically in the megasporocyte even as it continues to inhibit DMC1 expression in the nonsporogenous ovule cells. We further show that deposition of the histone variant H2A.Z, mediated by the SWR1 chromatin-remodeling complex at the DMC1 gene body, requires ARP6. Therefore, ARP6 regulates female meiosis by determining the spatial and temporal patterns of gene expression required for proper meiosis during ovule development.
INTRODUCTION
A defining feature of the plant life cycle is the alternation between two multicellular generations, an asexual sporophytic and a sexual gametophytic generation. Male and female meiosis occur in specialized organs within the sporophyte (anthers and ovules, respectively) producing spores. After a distal cell in the ovule primordia is specified and differentiates into the female meiocyte (megaspore mother cell or megasporocyte), it undergoes meiosis to yield four megaspores. Of these, three degenerate, leaving behind one functional megaspore and marking the completion of megasporogenesis. Here, the plant transitions from the sporophyte to the gametophyte generation (Yadegari and Drews, 2004). After three rounds of mitosis, nuclear migration, cellularization, and differentiation, the functional megaspore develops into the mature female gametophyte and completes megagametogenesis (Yadegari and Drews, 2004). Male meiosis occurs during microsporogenesis, in which the microspore mother cell or microsporocyte undergoes meiosis to yield four microspores. After two rounds of mitosis and differentiation, each microspore undergoes microgametogenesis and develops into a mature male gametophyte, the pollen grain (McCormick, 1993). Upon pollination, male and female gametophytes interact to complete double fertilization by facilitating fusion of male and female gametes. Here, the plant transitions from the gametophyte to the sporophyte generation. Coordination of development in the contiguous sexual and asexual cells likely requires intercellular interactions (Feng et al., 2013). However, the gene-regulatory mechanisms critical for plant reproductive cell development, including during meiosis, remain poorly characterized.
Histone modification and ATP-dependent chromatin remodeling regulate chromatin structure to balance chromatin packaging and transcriptional access (Ho and Crabtree, 2010). ATP-dependent chromatin remodeling complexes hydrolyze ATP and use the energy to exchange dimers of canonical histones in nucleosomes for dimers of histone variants (Krogan et al., 2003). Except for H4, all canonical histones have known variants, with H2A having the largest number of variants (March-Díaz and Reyes, 2009). Among H2A variants, H2A.Z is the most conserved (Thatcher and Gorovsky, 1994) and affects multiple biological processes, including regulation of gene expression (Draker and Cheung, 2009).
The Sick With Rat8 (SWR1) complex, a member of the Inositol-requiring 80 (INO80) family of remodelers, mediates deposition of H2A.Z into nucleosomes. The SWR1 complex is unique in swapping H2A-H2B dimers for H2A.Z-H2B dimers (Mizuguchi et al., 2004). Only five of the 14 proteins in the yeast (Saccharomyces cerevisiae) SWR1 complex are essential for viability; the remainder, including the SWR1 ATPase, enhance complex function (Mizuguchi et al., 2004). Homologs of all yeast and 11 human SWR1 subunits have been identified in Arabidopsis thaliana (March-Díaz and Reyes, 2009; Meagher et al., 2009), indicating that Arabidopsis has the SWR1 complex. Additionally, H2A.Z deposition at many target loci is disrupted in Arabidopsis SWR1 subunit-defective mutants (Deal et al., 2007).
ACTIN-RELATED PROTEIN6 (ARP6), one of the nonessential subunits of the yeast SWR1 complex, is required for normal growth of yeast. In the yeast SWR1 complex, ARP6 facilitates binding between other subunits, such as Swc2, and the ATPase domain of SWR1. In Arabidopsis, ARP6 acts in the nucleus (Deal et al., 2005; Meagher et al., 2005) to modulate gene expression in vegetative development and repression of flowering (March-Díaz and Reyes, 2009; Meagher et al., 2009).
The SWR1 complex is a likely candidate to regulate gene expression during meiosis, and its role in this process is just beginning to be understood. Meta-analysis of genome-wide H2A.Z occupancy in Schizosaccharomyces pombe using the Podbat computational tool showed preferential H2A.Z incorporation in the coding regions of genes encoding proteins involved in the regulation of meiosis (Sadeghi et al., 2011). During male meiosis I, colocalization of H2A.Z deposition on chromosomes and meiotic crossover hot spots indicate that H2A.Z may promote the formation or processing of meiotic DNA double-strand breaks (Choi et al., 2013). In Arabidopsis, mutations in SWR1 complex subunits, including ARP6, showed that the SWR1 complex is also involved in female meiosis (Rosa et al., 2013). However, the specific stage of female meiosis when ARP6 acts and the mechanism by which ARP6 regulates female meiosis remain to be identified.
Here, we show that loss of Arabidopsis ARP6 causes defects in prophase I of female meiosis, including aberrant centromere pairing and organization, loss of homologous chromosome pairing, and reduction in normal bivalents. We further show that expression of many meiotic genes, especially those that encode proteins involved in recombination, is dysregulated in arp6 ovules. Among these genes, DMC1 (encoding DISRUPTED MEIOTIC cDNA1) was downregulated in the megasporocyte, while it was simultaneously upregulated in the nonsporogenous cells of the arp6 ovules. We also show that ARP6 is involved in H2A.Z deposition in the DMC1 gene body, providing insights into the mechanism by which ARP6 regulates DMC1 expression. Thus, ARP6 likely determines proper meiosis in ovule primordia by performing a dual function in regulating meiotic gene expression: repression in the nonsporogenous cells and activation in the megasporocyte.
RESULTS
Megasporogenesis Is Defective in arp6 Mutants
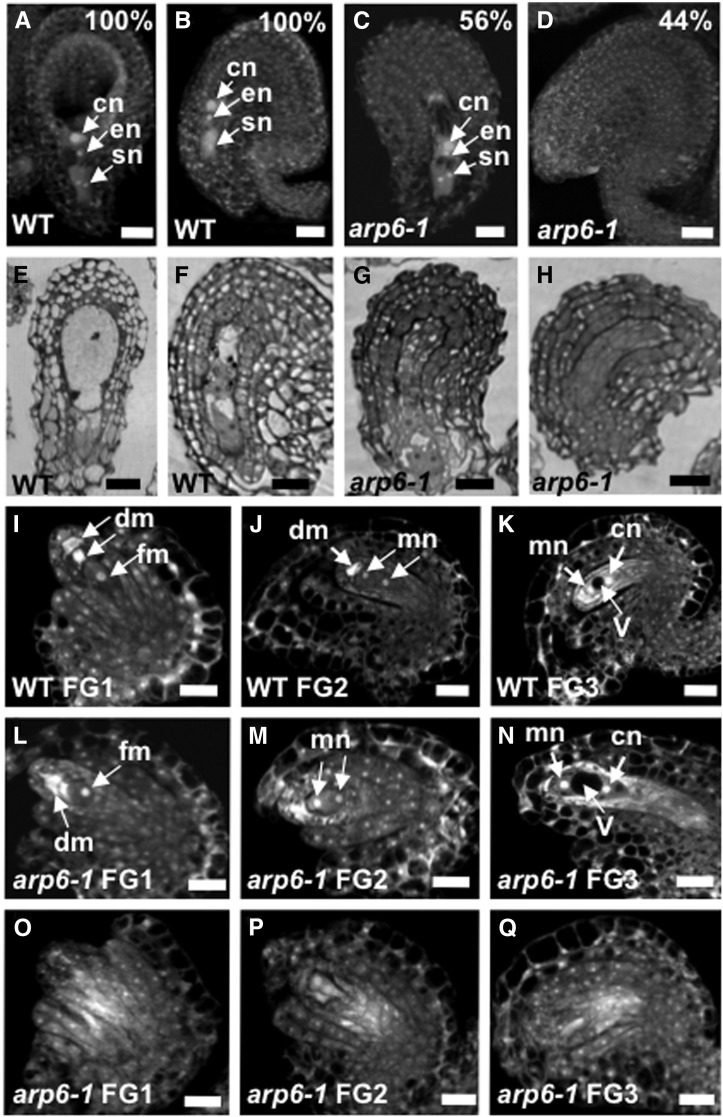
Previous work indicated a maternal role for ARP6 in reproductive development and seed set in Arabidopsis (Deal et al., 2005). In controlled crosses, homozygous arp6-1 plants produced defective ovules (∼46%) that were not targeted properly by wild-type pollen tubes even though pollen tube elongation was comparable in arp6-1 and wild-type pistils (Supplemental Figures 1A to 1C). Semi–in vivo pollen tube targeting assays showed the pollen tube targeting defects originate from arp6 ovules, rather than arp6 stigma or style tissues (Supplemental Figure 1D). Confocal microscopy of ovules from homozygous arp6-1 plants showed ∼44% of ovules lacked a megagametophyte (Figures 1A to 1H; Supplemental Table 1). Analysis of ovules during megagametogenesis revealed defects in arp6-1 ovules with a frequency (Figures 1I to 1Q; Supplemental Table 2) similar to that of defective megagametophytes (Supplemental Table 1). This analysis showed that the Female Gametophyte1 (FG1) stage (Christensen et al., 1997) was abnormal in arp6-1 (Figure 1O), pointing to an earlier defect in the meiotic divisions of megasporogenesis.
Figure 1.
Megagametogenesis Is Defective in arp6-1 Ovules.
(A) to (H) Some ovules in an arp6-1 pistil lack a female gametophyte.
(A) and (B) Confocal laser scanning microscopy (CLSM) images of a wild-type ovule at FG7 stage in two orientations.
(C) CLSM image of an arp6-1 ovule at FG7 stage with a normal female gametophyte.
(D) CLSM image of an abnormal arp6-1 ovule at FG7 stage lacking a female gametophyte. Quantification of CLSM observations of wild-type and arp6-1 ovules ([A] to [D]) at FG7 stage is provided in Supplemental Table 1.
(E) to (H) Light micrographs of thick plastic section of ovules at FG7 stage.
(E) and (F) Light micrographs of wild-type ovule at FG7 stage with a normal female gametophyte.
(G) Light micrograph of an arp6-1 ovule with a normal female gametophyte.
(H) Light micrograph of an abnormal arp6-1 ovule lacking a female gametophyte.
(I) to (Q) CLSM images of first three stages of megagametogenesis (female gametophyte development) in wild-type and arp6-1 ovules. Quantification of CLSM observations of megagametogenesis ([I] to [Q]) is reported in Supplemental Table 2.
(I) to (K) CLSM images of wild-type ovules at female gametophyte (FG) stage 1, 2, and 3 (FG1, FG2, and FG3), respectively.
(L) to (N) CLSM images of an arp6-1 ovule with a normal female gametophyte at FG1, FG2, and FG3 stage, respectively.
(O) to (Q) CLSM images of an arp6-1 ovule lacking a female gametophyte at FG1, FG2, and FG3 stage, respectively.
en, egg cell nucleus; FG, female gametophyte; cn, central cell nucleus; sn, synergid cell nucleus; fm, functional megaspore; dm, degenerating megaspore; mn, micropylar nucleus; cn, chalazal nucleus; v, vacuole. Bars in (A) to (H) = 20 μm, and bars in (I) to (Q) = 20 μm.
To clearly define the stage of the life cycle affected in arp6 mutants, we performed a series of genetic tests (Drews and Yadegari, 2002; Yadegari and Drews, 2004) using the two null alleles (arp6-1 and arp6-2; Deal et al., 2005) and showed that the reduced fertility of arp6 plants is not likely due to male or female gametophytic defects (Table 1; Supplemental Table 3). The same genetic tests demonstrated that arp6-1 and arp6-2 pollen from homozygous mutant plants is functional (Supplemental Table 3), indicating that these mutants do not affect microsporogenesis. In support of this conclusion, tetrad analysis with arp6 quartet (Preuss et al., 1994) double mutants produced only tetrads with four normal pollen grains (Supplemental Figures 2A to 2D), showing that meiotic divisions of microsporogenesis in arp6-1 and arp6-2 are apparently normal. Together, our data suggest that ARP6 is required for proper megasporogenesis.
Table 1. Transmission Frequency of arp6 Mutations through Male and Female Gametophytes Is Unaffected.
| Female Parent | Male Parent | Expected Ratio (Bastar:Bastas) | Observed No. of Offspringa |
|
|---|---|---|---|---|
| Bastar | Bastas | |||
| arp6-1/+ | arp6-1/+ | 3:1 | 171 | 62 |
| arp6-1/+ | +/+ | 1:1 | 163 | 160 |
| +/+ | arp6-1/+ | 1:1 | 202 | 194 |
| arp6-2/+ | arp6-2/+ | 3:1 | 206 | 65 |
| arp6-2/+ | +/+ | 1:1 | 104 | 94 |
| +/+ | arp6-2/+ | 1:1 | 165 | 141 |
Observed ratio did not deviate from expected ratio (χ2 test)
ARP6 Regulates Meiosis I during Megasporogenesis
To identify the defect in arp6 mutants, we first examined the earliest events in megasporogenesis using a pSPL:GUS line (harboring the SPOROCYTELESS promoter fused to the β-glucuronidase reporter gene; Ito et al., 2004) as a marker for differentiation of the archesporial cell to a megasporocyte (Yang et al., 1999) at the distal end of each ovule primordium (Schneitz et al., 1995). Wild-type and arp6-1 mutant ovules showed similar GUS staining (Supplemental Figure 3), indicating that arp6-1 nucellus differentiates normally and is capable of producing a megasporocyte.
To determine if the megasporocytes in arp6 ovules proceeded through meiotic divisions properly, we stained callose in the cell plates of megasporocytes undergoing meiosis in stage 2-III/IV ovules (Schneitz et al., 1995) with aniline blue (Rodkiewicz, 1970). In the early stages, we found comparable frequencies of aniline blue–stained callose bands in wild-type (95.97%; n = 296 ovules) and arp6-1 (92.22%; n = 180 ovules) or arp6-2 ovules (92.86%; n = 182 ovules), indicating normal initiation of meiosis in mutant megasporocytes.
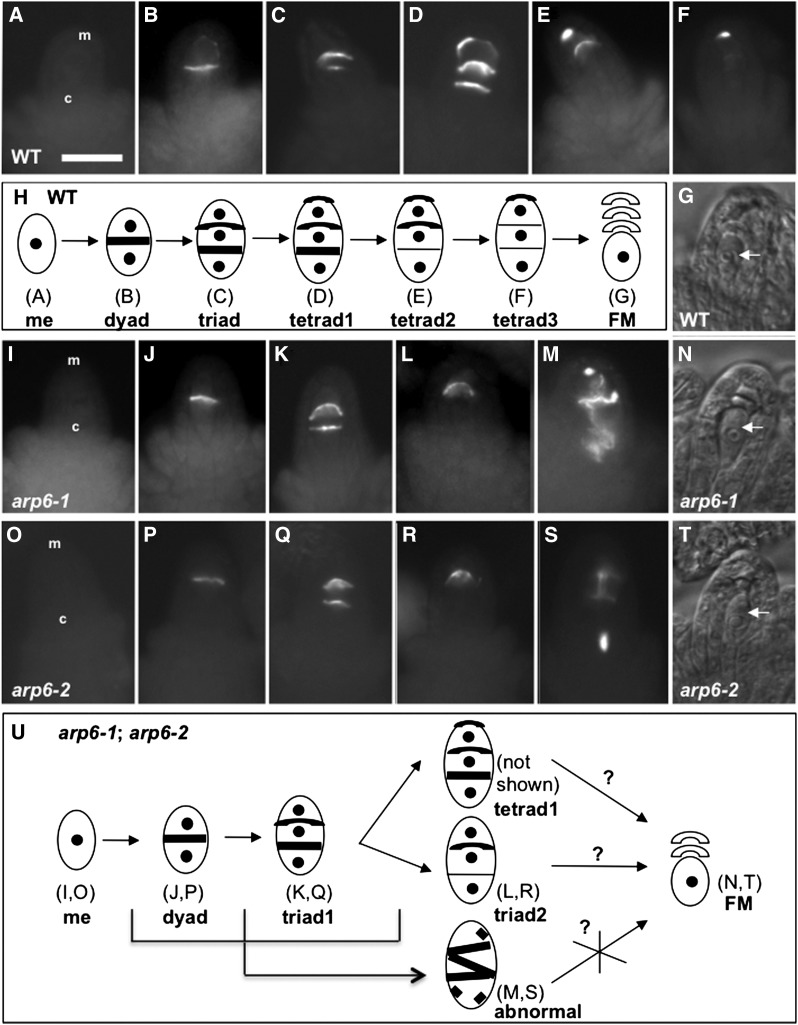
We next tested if the progression of meiotic divisions during megasporogenesis is altered in arp6 mutants by quantifying the frequency of each key stage of meiosis in callose-stained ovule primordia (Siddiqi et al., 2000). We detected a higher occurrence of dyad (Figures 2B, 2H, 2J, 2P, and 2U; Supplemental Table 4) and triad1 stages (Figures 2C, 2H, 2K, 2Q, and 2U; Supplemental Table 4) in the mutant ovule primordia than in the wild type. In contrast to the wild type (Figures 2D and 2H), few of the mutant ovule primordia contained megasporocytes that were at the tetrad1 stage and none were detected at the tetrad2 or tetrad3 stages (Supplemental Table 4). Many more mutant megasporocytes were at the triad2 stage compared with wild-type megasporocytes (Figures 2L and 2R; Supplemental Table 4). Finally, many arp6 megasporocytes exhibited abnormal callose staining (Figures 2M and 2S; Supplemental Table 4). These results identified defects in meiosis progression in arp6-1 and arp6-2 ovule primordia. In some cases, mutant megasporocytes likely formed megaspores (Figures 2N and 2T), but the remaining defective megasporocytes likely did not complete meiosis and failed to produce functional megaspores (Figure 2U).
Figure 2.
Meiotic Divisions of Megasporogenesis Is Defective in arp6 Megasporocytes.
(A) to (F), (I) to (M), and (O) to (S) Epifluorescence microscopy images of aniline-blue stained wild-type, arp6-1, and arp6-2 megasporocytes, respectively.
(A) No callose staining in a wild-type megasporocyte before meiosis.
(B) Callose staining in a transverse cell plate of a wild-type dyad.
(C) Two callose bands in a wild-type triad.
(D) A megasporocyte with three callose bands in a wild-type tetrad, with the fourth band invisible in this orientation (tetrad1).
(E) A wild-type tetrad without a callose-stained wall in the chalazal end (tetrad2).
(F) A wild-type tetrad without callose-stained walls in middle and chalazal ends (tetrad3).
(G) Bright-field image of a wild-type ovule with a functional megaspore (FM; white arrow).
(H) A summary of meiosis stages in wild-type megasporocytes. Thick lines refer to callose bands.
(I) and (O) No callose staining in an arp6-1 and arp6-2 megasporocyte, respectively, before meiosis.
(J) and (P) Callose staining in transverse cell plate of an arp6-1 and arp6-2 dyad, respectively.
(K) and (Q) An arp6-1 and arp6-2 megasporocyte, respectively, with two callose bands (triad1).
(L) and (R) An arp6-1 and arp6-2 megasporocyte, respectively, with one callose band (triad2).
(M) and (S) Abnormal callose staining in an arp6-1 and arp6-2 megasporocyte, respectively.
(N) and (T) Bright-field images of arp6-1 and arp6-2 ovules, respectively, with a functional megaspore (white arrows).
(U) A summary of meiosis stages in arp6-1 and arp6-2 megasporocytes. Possibilities of a tetrad and a triad2 developing into a functional megaspore (question mark) and abnormally stained megasporocytes likely not developing into a functional megaspore (X mark). Thick lines refer to callose bands.
In each ovule, the micropylar end (m) is at the top and the chalazal end (c) is at the bottom (indicated in [A], [I], and [O]). Quantification of stages of meiotic divisions of megasporogenesis is provided in Supplemental Table 4. Bars = 20 μm.
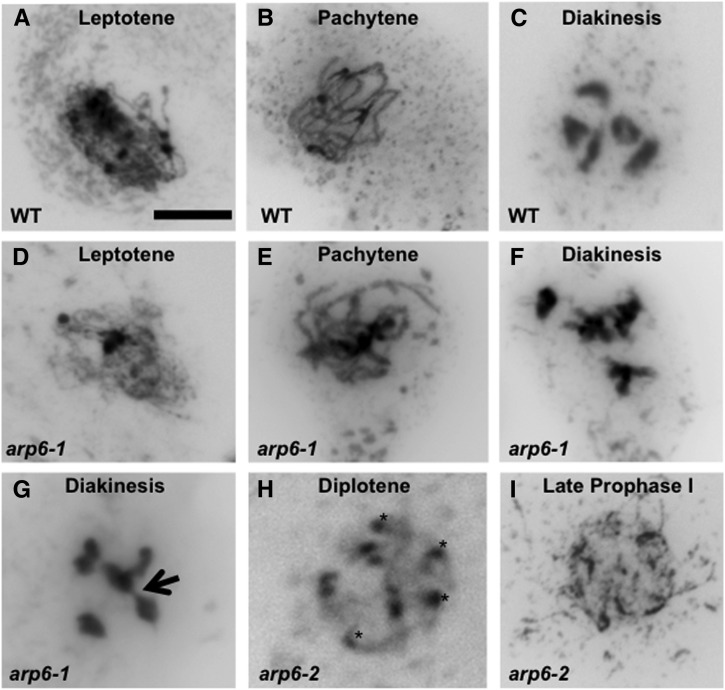
To further characterize defects in arp6 mutants, we examined meiotic spreads of wild-type and arp6 megasporocytes. In arp6-1 megasporocytes, we detected apparently normal-looking leptotene and pachytene of prophase I (Figures 3D and 3E) that were comparable to the wild-type pattern (Figures 3A and 3B) as described for Arabidopsis (Armstrong and Jones, 2003). All wild-type megasporocytes at diakinesis contained five highly condensed bivalents (Figure 3C). However, in arp6-1, we detected two types of megasporocytes at late prophase/diakinesis: one group similar to the wild type with five highly condensed bivalents (Figure 3F) and one containing defective meiotic structures such as those with abnormal linkages between chromosomes (Figure 3G). arp6-2 megasporocytes also contained defective meiotic structures. We detected arp6-2 megasporocytes that contained a mixture of bivalent and univalent (Figure 3H) or those with disorganized and fragmented chromosomes, as evident from the presence of more than five densely staining regions and the presence of discontinuous threads extending from densely stained chromatin (Figure 3I). A similar analysis of meiotic spreads of microsporocytes showed no detectable differences between the wild type and arp6-2 (Supplemental Figures 2E to 2L), indicating that meiotic divisions of microsporogenesis are not affected in arp6-1 or arp6-2 plants, as expected from our genetic evidence (Table 1; Supplemental Table 2). Thus, meiotic chromosome pairing is defective during prophase I in a subset of arp6 megasporocytes but not in arp6 microsporocytes.
Figure 3.
Prophase I of Meiosis during Megasporogenesis Is Defective in arp6.
(A) and (B) Wild-type megasporocyte in the leptotene and pachytene stages, respectively, of prophase I.
(C) Wild-type megasporocyte in the diakinesis stage of prophase I containing five pairs of bivalents.
(D) and (E) Apparently normal arp6-1 megasporocytes in leptotene and pachytene stages, respectively, of prophase I.
(F) An apparently normal arp6-1 megasporocyte in the diakinesis stage of prophase I.
(G) A defective arp6-1 megasporocyte in the diakinesis stage of prophase I showing disorganized chromosomes with some linked to each other with chromosomal bridges (black arrow).
(H) A defective arp6-2 megasporocyte in the diplotene stage of prophase I showing a mixture of univalent and bivalent; black asterisks point to univalents.
(I) A defective arp6-2 megasporocyte in the late prophase I stage containing disorganized and fragmented chromosomes.
Bar in (A) = 10 μm for (A) to (I).
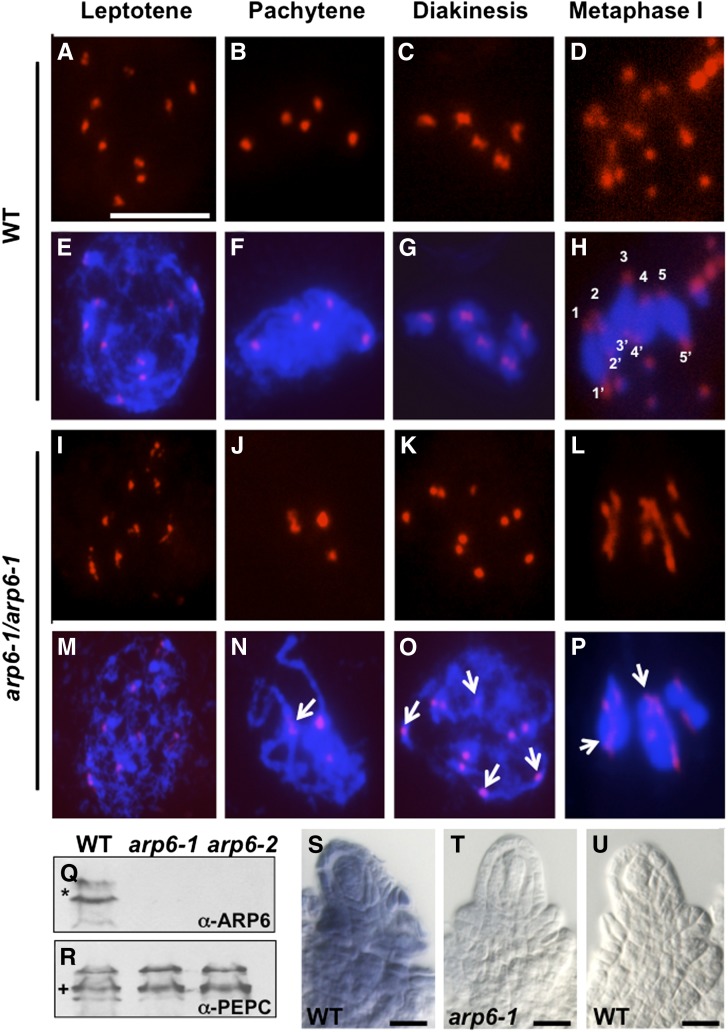
Centromere pairing precedes meiotic chromosome pairing in Arabidopsis (Armstrong et al., 2001). To compare centromere pairing and organization in wild-type (Figures 4A to 4H) and arp6 (Figures 4I to 4P) megasporocytes, we performed fluorescence in situ hybridization (FISH) using a 180-bp centromeric repeat-specific probe. FISH also distinguishes megasporocytes undergoing meiosis from the surrounding nonsporogenous ovule cells undergoing mitosis (Armstrong et al., 2001), enabling more reliable analysis of meiotic progression. In arp6-1, FISH identified two types of megasporocytes at prophase I: those similar to the wild type (56%; n = 41 megasporocytes) and those containing defective centromeres (44%; n = 41 megasporocytes; Figures 4I to 4P). In leptotene, wild-type spreads (Figures 4A and 4E) showed 10 condensed, unpaired centromeres, but arp6-1 spreads showed diffused, unpaired centromeres (Figures 4I and 4M). In arp6-1 megasporocytes that reached pachytene, as evident from the extent of chromosome condensation, some centromeres remained diffused (Figures 4J and 4N), in contrast to the wild type, which contained five pairs of centromeres (Figures 4B and 4F). Unlike in the wild type (Figures 4C and 4G), diakinesis in arp6-1 megasporocytes contained a mixture of paired and unpaired centromeres (Figures 4K and 4O). Additionally, metaphase I was defective, with diffuse and incompletely separated centromeres (Figures 4L and 4P), unlike the punctate and well-separated centromeres in wild-type megasporocytes (Figures 4D and 4H). These results indicated that arp6-1 megasporocytes have defects in centromere pairing and organization during prophase I, and the earliest manifestation of the arp6 defect in megasporocytes was at the leptotene stage of meiosis.
Figure 4.
Centromere Pairing Is Defective during Prophase I of Meiosis in arp6-1 Megasporocytes.
(A) to (D) and (I) to (L) Epifluorescence microscopy images of megasporocyte spreads subjected to FISH (red signal).
(E) to (H) and (M) to (P) Merged images of centromere (red) and DAPI-stained chromosomes (blue) of megasporocyte spreads.
(A) and (E) Wild-type megasporocyte in leptotene showing 10 unpaired and dispersed centromeres.
(B) and (F) Wild-type megasporocyte in pachytene with five pairs of centromeres.
(C) and (G) Wild-type megasporocyte in diakinesis with five pairs of centromeres of the five bivalents.
(D) and (H) Wild-type megasporocyte in metaphase I containing five pairs of condensed chromosomes with separated centromeres in each pair pointing to opposite poles.
(I) and (M) arp6-1 megasporocyte in leptotene showing 10 centromeres, many of which are abnormally diffused.
(J) and (N) arp6-1 megasporocyte in pachytene with at least one abnormally diffused centromere (white arrow in [N]).
(K) and (O) arp6-1 megasporocyte in diakinesis with a mixture of univalents (white arrows) and bivalents.
(L) and (P) arp6-1 megasporocyte in metaphase I containing abnormally diffused centromeres with chromosome bridges (white arrows).
(Q) and (R) Immunoblotting of ARP6 (Q) or PEPC protein (R) in wild-type, arp6-1, and arp6-2 ovaries containing stage 2-III/IV ovules undergoing meiosis. Blots were first probed with ARP6 monoclonal antibody (mAbARP6a; α-ARP6), or the immunoblot was stripped and reprobed with anti-PEPC polyclonal antibody (α-PEPC). Protein sizes in kilodaltons; *, 50 kD; +, 100 kD.
(S) to (U) In situ localization of ARP6 mRNA in Arabidopsis whole-mount ovules.
(S) In situ hybridization of ARP6 antisense RNA to wild-type ovules (89% of stained ovules [n = 106] showed this pattern).
(T) In situ hybridization of ARP6 antisense RNA to arp6-1 ovules (100% of stained ovules [n = 65] showed this pattern).
(U) Control hybridization of wild-type ovules using ARP6 sense RNA did not give any signal (100% of stained ovules [n = 51] showed this pattern).
Bar in (A) = 10 µm for (A) to (P), and bars in (S) to (U) = 10 μm.
We showed that the arp6 mutation is responsible for these meiotic defects by complementing the arp6 reduced-fertility phenotype through transforming a wild-type ARP6 transgene into mutant plants and scoring for restoration of seed set, as previous complementation experiments had focused on other floral and vegetative defects of arp6 (Deal et al., 2005). Seed set in plants transformed with a 5.4-kb genomic DNA fragment containing ARP6 was restored to >96% in 19 of 28 arp6-1 T1 transformants and 21 of 25 arp6-2 T1 transformants (Supplemental Data Set 1). Moreover, we confirmed that the arp6 mutation causes the megasporocyte defect by scoring for the presence of normal-looking megaspores in a subset of these transformants. We detected a megaspore in all ovules of two individual arp6-1 transformants (100%; n = 254 ovules) and two arp6-2 transformants (100%; n = 152 ovules), each carrying an ARP6 transgene.
To characterize ARP6 expression during ovule development and further confirm that the occurrence of the arp6 meiotic defect coincides with its expected expression during megasporogenesis, we performed immunoblotting, RNA in situ hybridization and protein-YFP (yellow fluorescent protein) localization experiments. We detected ARP6 protein (Figures 4Q and 4R) in wild-type ovaries but not in arp6-1 or arp6-2 ovaries from stage 9 flowers corresponding to the period of meiotic divisions in megasporogenesis. Using in situ hybridization of whole-mount ovules with ARP6 probes, we detected ARP6 mRNA in the megasporocyte and nonsporogenous cells of stage 2-III/IV wild-type but not arp6-1 ovules (Figures 4S and 4T). We further confirmed this expression pattern by protein localization experiments using transformants carrying a functional ARP6-YFP fusion protein (Columbia-0 [Col-0] ARP6 fused at its C terminus to YFP and expressed from the ARP6 promoter; Supplemental Figure 4 and Supplemental Data Set 1). Taken together, our data indicate a role for ARP6 in megasporogenesis through the regulation of meiosis I.
Expression of Genes Associated with Meiotic Recombination Is Upregulated in arp6 Ovules
ARP6 is a component of the SWR1 chromatin-remodeling complex, which functions in both gene activation and silencing in eukaryotes (March-Díaz and Reyes, 2009). In arp6 mutants, mRNA levels of repressors of transition to flowering are lower than in the wild type (Deal et al., 2007). In other instances, the arp6 mutants display increased expression of genes related to salicylic acid–dependent plant immunity (March-Díaz et al., 2008), phosphate starvation response (Smith et al., 2010), and heat shock response factor 70 (Kumar and Wigge, 2010). To test whether ARP6 modulates expression of genes involved in meiosis during megasporogenesis, we used reverse transcription coupled with quantitative PCR (RT-qPCR) to compare the mRNA levels of 13 meiosis-related genes in stage 2-III/IV (Schneitz et al., 1995) of wild-type and arp6 ovules. Expression of 7 of the 13 genes was significantly upregulated in ovules in at least one of the two arp6 alleles tested (Table 2). Of these, four genes, including DMC1, ASY1 (ASYNAPTIC1), SWITCH1/DYAD, and MND1 (MEIOTIC NUCLEAR DIVISION PROTEIN1), showed significant levels of upregulation in both alleles. Interestingly, these four genes have been implicated to play a role in the steps leading to pairing and synapsis of homologous chromosomes and double-strand break repair during meiotic recombination (Siddiqi et al., 2000; Armstrong et al., 2002; Mercier et al., 2003; Panoli et al., 2006; Vignard et al., 2007; Da Ines et al., 2012). We also tested the effect of arp6 mutations on the expression of the same genes in microsporocytes (stages 5 to 7; Sanders et al., 1999) and showed that most genes, except for SAP (STERILE APETELA), did not show significant level of upregulation in both alleles (Supplemental Table 5). Therefore, our results indicate that the expression of many meiotic genes in ovules, particularly those associated with meiotic recombination, is regulated by ARP6.
Table 2. Expression of Meiosis-Related Genes Increases in arp6 Ovules.
| Gene |
arp6-1/WT |
arp6-2/WT |
||||
|---|---|---|---|---|---|---|
| LSM | sd | P Value | LSM | sd | P Value | |
| DMC1 | 4.04a | 1.01 | 0.01* | 4.55a | 1.45 | 0.01* |
| ASY1 | 3.58ab | 0.94 | 0.01* | 3.43ab | 1.14 | 0.02* |
| DYAD | 3.37ab | 0.90 | 0.01* | 4.75a | 2.02 | 0.03* |
| MND1 | 2.35ab | 0.53 | 0.01* | 2.74ab | 1.01 | 0.03* |
| AML4 | 3.77ab | 1.39 | 0.03* | 3.13ab | 2.17 | 0.14 |
| SPO11 | 2.47ab | 0.45 | 0.01* | 3.82ab | 1.88 | 0.05 |
| DIF1 | 3.77ab | 1.83 | 0.06 | 3.54ab | 1.65 | 0.05* |
| ATM | 3.81ab | 1.89 | 0.06 | 3.97ab | 2.25 | 0.07 |
| SDS | 2.02ab | 0.93 | 0.13 | 2.07ab | 0.98 | 0.08 |
| AML1 | 1.93ab | 0.72 | 0.15 | 1.38ab | 0.90 | 0.25 |
| RAD51 | 1.44ab | 0.39 | 0.16 | 1.07b | 0.64 | 0.33 |
| SAP | 1.35ab | 0.32 | 0.12 | 1.30b | 0.41 | 0.21 |
| SPL | 1.28ab | 0.17 | 0.06 | 1.48ab | 0.74 | 0.14 |
Least-squares mean (LSM) of change in expression of a meiosis-related gene in arp6-1 or arp6-2 compared to that in wild-type (WT) ovules in five technical replicates (three biological replicates) assayed by RT-qPCR. The P value is the probability value associated with a pairwise test between change in the expression of a gene in arp6-1 or arp6-2 compared to that in wild-type ovules and change in the expression of ACTIN2 in arp6-1 or arp6-2 compared to that in wild-type ovules (*P ≤ 0.05). Any pair of least-squares means that do not share the same letter are significantly different (P ≤ 0.05) from each other in pairwise comparisons for change in expression of a meiosis-related gene in arp6-1 or arp6-2 compared to that in wild-type ovules.
ARP6 Specifically Mediates Both Activation of DMC1 Expression in the Megasporocyte and Repression in Nonsporogenous Cells of the Ovule Primordium
The large change in meiotic gene expression program observed in arp6 mutant ovules (Table 2) would be expected to affect megasporocyte function. Among the affected genes, DMC1 showed the highest level of increase in expression in ovules of both arp6 alleles (Table 2). DMC1 is specifically expressed in the megasporocyte (Klimyuk and Jones, 1997; Siddiqi et al., 2000). Moreover, DMC1 facilitates centromere pairing in meiosis during megasporogenesis (Da Ines et al., 2012) and formation and/or stabilization of bivalents (Couteau et al., 1999), features of meiotic divisions that are disrupted in arp6 megasporocytes (Figures 3 and 4). Therefore, we chose DMC1 as a candidate gene regulated by ARP6 and characterized its spatial and temporal transcriptional activity using wild-type and arp6 plants carrying a promoter-reporter (pDMC1:GFP [green fluorescent protein]) transgene.
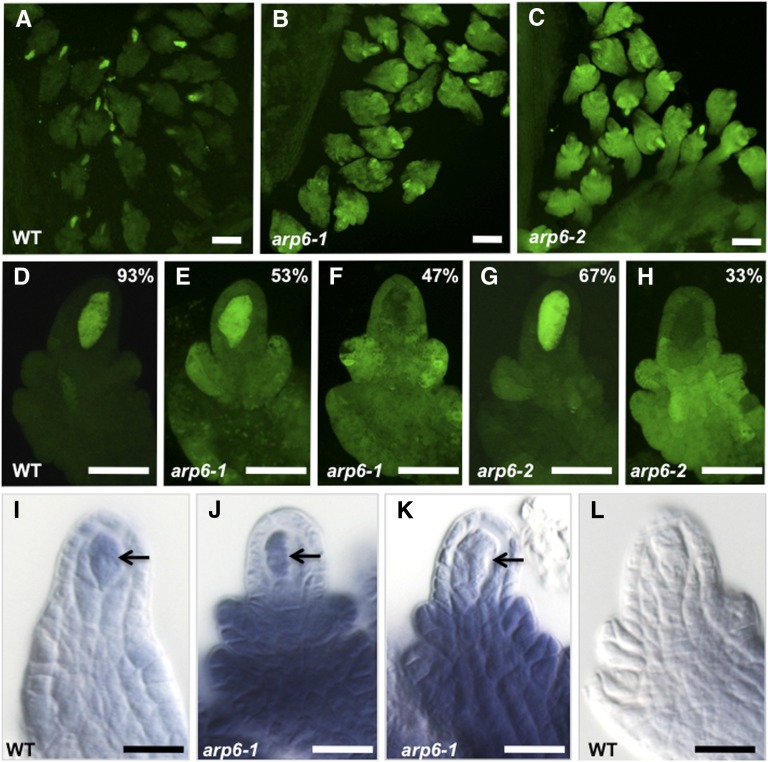
Ovules at stage 2-III/IV contain multiple cell types, including the nurse cells surrounding the megasporocyte (nucellus), integument primordia that develop into inner and outer integuments and the future seed coat, and funiculus primordium that develops into the funiculus, a stalk that subtends the ovule (Schneitz et al., 1995). As reported (Klimyuk and Jones, 1997; Siddiqi et al., 2000), we detected DMC1’s transcriptional activity in wild-type ovules only in megasporocytes (Figures 5A and 5D; Supplemental Table 6). However, in homozygous arp6-1 plants, only 52.9% of megasporocytes showed reporter expression at the same stage of development (Figures 5B and 5E; Supplemental Table 6) with the rest showing no detectable GFP (Figures 5B and 5F; Supplemental Table 6). Interestingly, unlike in ovules from wild-type plants, we detected increased GFP expression in the nonsporogenous cells, including the nucellus, integument, and funiculus primordia in all mutant ovules (compare Figures 5B with 5A and Figures 5E and 5F with 5D). The increase in DMC1’s transcriptional activity in arp6-1 ovules was confirmed by detecting a corresponding increase in GFP mRNA levels in ovules compared with wild-type ovules using RT-qPCR (Supplemental Figure 5). Similar results were obtained with ovules from homozygous arp6-2 plants (Figures 5C, 5G, 5H, and 4I) with the exception that the frequency of GFP-expressing megasporocytes was higher in arp6-2 than arp6-1 (∼67% versus ∼53%; Supplemental Table 6; Figures 5G and 5E). For both alleles, the earliest we could detect ectopic expression of pDMC1:GFP was stage 2-I (Supplemental Figures 6A to 6F), during which the megasporocyte has completed differentiation but has not entered meiosis (Schneitz et al., 1995). No ectopic expression was detected at earlier stages of ovule development (100%; n = 500 ovules at stages 1-I or 1-II) where the ovule primordium consists of a homogeneous group of cells (Schneitz et al., 1995).
Figure 5.
DMC1 Expression in Ovules Undergoing Meiosis Is Altered in arp6 Mutants.
(A) to (H) CLSM images of pDMC1:GFP expression in wild-type ([A] and [D]), arp6-1 ([B], [E], and [F]), and arp6-2 ([C], [G], and [H]) stage 2-III/IV ovules undergoing meiosis during megasporogenesis. Images in (A) to (C) are projections of a Z stack of optical sections spanning 50 μm and were captured using identical image acquisition settings. CLSM images in (D) to (H) are 5-μm single optical sections that were captured using identical image acquisition settings. Percentages refer to number of ovules that displayed the GFP expression pattern shown in each panel; quantification details are provided in Supplemental Table 6.
(I) to (L) In situ localization of DMC1 mRNA in Arabidopsis whole-mount ovules.
(I) In situ hybridization of DMC1 antisense RNA to wild-type ovules of Col-0 ecotype (28% of the stained ovules [n = 181] showed this pattern).
(J) and (K) In situ hybridization of DMC1 antisense RNA to arp6-1 ovules.
(J) A representative arp6-1 ovule that showed DMC1 expression in the megasporocyte and nonsporogenous cells (16% of stained ovules [n = 187] showed this pattern).
(K) A representative arp6-1 ovule that showed DMC1 expression in the nonsporogenous cells but lacked DMC1 expression in the megasporocyte (72% of stained ovules [n = 187] showed this pattern).
(L) Control hybridization of wild-type ovules (Col-0 ecotype) with DMC1 sense RNA did not give any signal (100% of stained ovules [n = 47] showed this pattern).
Arrows in (I) to (K) point to the megasporocyte. Bars = 40 μm in (A) to (C), 20 μm in (D) to (H), and 10 μm in (I) to (L).
To confirm whether the observed ectopic activation of the GFP reporter activity in mutant ovules reflects changes in spatial pattern of expression of the endogenous DMC1 gene, we compared DMC1 mRNA localization in whole-mount ovules from wild-type and arp6-1 plants using in situ hybridization. As with the localization of the GFP reporter activity (Figures 5E and 5F), arp6-1 ovules displayed both DMC1-expressing and DMC1-poorly expressing megasporocytes with the majority showing the latter pattern (Figures 5J and 5K). However, as with the GFP reporter activity (Figures 5E to 5H), we detected a dramatically increased level of DMC1 mRNA accumulation in the nonsporogenous cells of all mutant ovules irrespective of the presence or absence of DMC1 expression in the megasporocyte (Figures 5J and 5K).
To determine whether DMC1 upregulation in the arp6 mutant background is a general plant gene-regulatory response or specific to the developing ovules, we assayed for DMC1 expression broadly in wild-type and mutant plants. First, we detected no ectopic pDMC1:GFP expression in floral organs, including stigma, style, transmitting tissue, or ovary wall in arp6-1 or arp6-2 mutants (100%, n = 10 arp6-1 pistils; 100%, n = 10 arp6-2 pistils). Second, we found no apparent difference in the extent of reporter-gene activity between wild-type and arp6-1 or arp6-2 seedlings (Supplemental Figures 6G to 6L), and RT-qPCR analysis showed that the levels of DMC1 mRNA in the mutant seedlings was comparable to that of the wild type (Supplemental Figure 6M). Third, we found no apparent difference in the extent of reporter-gene activity between wild-type and arp6-1 or arp6-2 trichomes (Supplemental Figures 6N to 6P). Finally, unlike in arp6 ovules, we observed no difference in cell-specific expression of pDMC1:GFP reporter in wild-type, arp6-1, or arp6-2 anthers (Supplemental Figures 7A to 7F and 7J). That is, no ectopic GFP expression was detected in nonsporogenous cells that surround the mutant microsporocytes (100%, n = 18 arp6-1 anther locules; 100%, n = 18 arp6-2 anther locules), and no difference in pDMC1:GFP expression was identified between wild-type tetrads and either arp6-1 or arp6-2 tetrads (Supplemental Figures 7G to 7I). Thus, unlike in ovules undergoing megasporogenesis, DMC1 expression was unaltered in arp6-1 and arp6-2 anthers undergoing microsporogenesis. Together, our data indicate a dual role for ARP6 in the spatial and temporal regulation of DMC1 expression during ovule development: ARP6 represses DMC1 expression in nonsporogenous cells of the ovule primordium during megasporocyte differentiation prior to and after the initiation of meiotic divisions and at the same time partly activates DMC1 expression specifically in the differentiated megasporocyte after the initiation of meiotic divisions.
H2A.Z Deposition at the DMC1 Locus Requires ARP6
We showed that ARP6 both promotes and inhibits DMC1 expression in the megasporocyte and nonsporogenous cells of ovules, respectively. Therefore, we hypothesized that ARP6 carries out this function by modulating deposition of H2A.Z at the DMC1 chromatin, similar to its role in regulating the expression of other genes (March-Díaz and Reyes, 2009). To test this hypothesis, we performed chromatin immunoprecipitation using a H2A.Z antibody coupled with qPCR (ChIP-qPCR) on chromatin isolated from wild-type and arp6-1 floral buds. In wild-type floral buds, we detected a significantly higher level of H2A.Z deposition in the DMC1 gene body similar to the high level of H2A.Z deposition near the transcription start site of the control gene FLC (FLOWERING LOCUS C) (Deal et al., 2007; Table 3; Supplemental Figures 8 and 9). The enrichment in the DMC1 gene body was significantly higher (∼2.5- to 4-fold) than that in the DMC1 promoter, including the region that spans the transcription start site of DMC1 (Table 3; Supplemental Figures 8 and 9), indicating that H2A.Z is preferentially deposited at the DMC1 gene body, a pattern reminiscent of H2A.Z deposition in responsive genes (Coleman-Derr and Zilberman, 2012). In arp6-1 floral buds, H2A.Z was not enriched in any of the locations assayed in DMC1 (Table 3; Supplemental Figures 8 and 9). Moreover, H2A.Z enrichment was significantly lower in arp6-1 compared with wild type in the DMC1 gene body, although the FLC control showed a greater reduction in H2A.Z enrichment compared with DMC1 (Table 3; Supplemental Figures 8 and 9). These results indicate that ARP6 is involved in depositing H2A.Z preferentially at the DMC1 gene body.
Table 3. H2A.Z Deposition at DMC1 Requires ARP6.
| Gene |
ChIP-qPCR Measurement of H2A.Z Enrichment |
Fold Difference in H2A.Z Enrichment (WT/arp6-1) | P Value | |||
|---|---|---|---|---|---|---|
| WT |
arp6-1 |
|||||
| LSM | sd | LSM | sd | |||
| DMC1 | ||||||
| 1 | 1.89e | 0.59 | 1.76ab | 0.88 | 1.07 | 0.377 |
| 2 | 1.19f | 0.25 | 1.02d | 0.13 | 1.17 | 0.033 |
| 3 | 1.31f | 0.44 | 1.56bc | 0.41 | 0.84 | 0.036 |
| 4 | 1.14f | 0.44 | 0.80def | 0.42 | 1.43 | 0.041 |
| 5 | 1.42f | 0.59 | 1.39c | 0.62 | 1.02 | 0.898 |
| 6 | 0.78g | 0.30 | 0.72ef | 0.41 | 1.08 | 0.550 |
| 7 | 2.48d | 0.86 | 0.99d | 0.56 | 2.51 | <0.001 |
| 8 | 3.41b | 1.41 | 0.97de | 0.21 | 3.52 | <0.001 |
| 9 | 3.97a | 1.88 | 1.88a | 1.36 | 2.11 | <0.001 |
| FLC | ||||||
| A | 3.00c | 2.04 | 0.64f | 0.67 | 4.69 | <0.001 |
Numbers in the first column refer to DMC1 genomic region assayed by quantitative PCR for H2A.Z enrichment (as shown in Supplemental Figures 8 and 9). “A” is the genomic region immediately downstream of the transcription start site assayed by quantitative PCR for H2A.Z enrichment in FLC. Least-squares mean (LSM) of H2A.Z enrichment in eight technical replicates (four biological replicates). Within a genotype, any pair of least-squares means that do not share the same letter are significantly different (P ≤ 0.05) from each other in pairwise comparisons for H2A.Z enrichment. The final column is the probability value associated with a pairwise test between LSMs of H2A.Z enrichment at each locus between wild-type (WT) and arp6-1 floral buds. P values ≤ 0.05 indicate significant differences between genotypes.
DISCUSSION
ARP6 Regulates Megasporocyte-Specific Expression of DMC1
Our analysis of DMC1 expression in arp6 mutants revealed an intriguing aspect of DMC1 expression in wild-type ovules: Before entering meiosis, ARP6 represses DMC1 expression in both nonsporogenous cells and the megasporocyte. After initiation of meiosis, ARP6 has dual roles: It inhibits DMC1 expression in nonsporogenous cells even as it promotes DMC1 expression in the megasporocyte undergoing meiosis. Thus, instead of activating DMC1 only in the megasporocyte of ovules at stage 2-III/IV, spatial and temporal regulation of DMC1 expression is complex. BRG1, an ATPase subunit of mouse SWI/SNF chromatin remodeling complex, is involved in repression and activation of CD4 (CLUSTER OF DIFFERENTIATION4) expression during thymocyte development (Chi et al., 2003). Our results show that ARP6 regulates the expression of the same gene differently in ovules: It represses DMC1 expression where it is not required (in nonsporogenous cells of ovules at stage 2-I to 2-IV and differentiating megasporocytes of ovules at stage 2-I/II) and promotes DMC1 expression where it is required for meiosis (in megasporocytes of ovules at stage 2-III/IV). Our results showing that DMC1 expression is reduced in the megasporocyte and causes defects in female meiosis starting from the leptotene stage is also consistent with the reduction in DMC1 foci on leptotene chromosomes in arp6 male meiotic spreads (Choi et al., 2013).
The ectopic activation of DMC1 caused by arp6 mutations is restricted to ovules. These results raise two possibilities. DMC1 expression in vegetative tissues is repressed by an inhibition mechanism and derepressed selectively in certain cell types (trichomes and true leaves of seedlings; Supplemental Figure 6) or is selectively regulated by a cell-specific activation mechanism of DMC1 expression, akin to the regulation of DMC1 expression in ovules. Expression of DMC1 in megasporocytes of ovules undergoing meiosis (stage 2-III/IV) could result from the removal of inhibition of expression that exists prior to meiosis or from promotion of DMC1 expression in the megasporocyte after it enters meiosis. Our results indicate that the latter possibility is more likely. Relief of ARP6-dependent inhibition in the megasporocyte would be expected to produce expression of pDMC1:GFP in all arp6 megasporocytes at stage 2-III/IV. Instead, we found that pDMC1:GFP expression was lost in a substantial portion of arp6 megasporocytes in ovules at stage 2-III/IV, even though all mutant megasporocytes in ovules at stage 2-I expressed pDMC1:GFP (Figure 6A). Therefore, we propose that an ARP6-dependent, cell-specific mechanism promotes DMC1 expression in the megasporocyte undergoing meiosis.
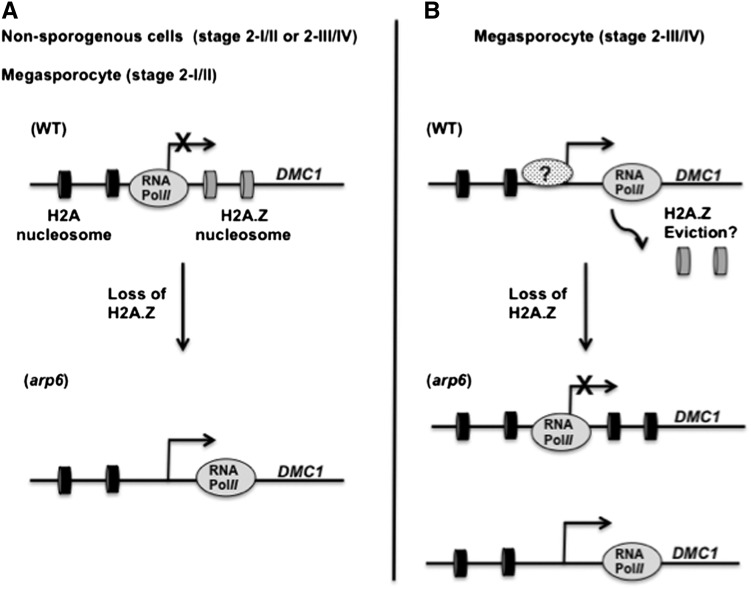
Figure 6.
Model for ARP6 Function in Meiosis during Megasporogenesis.
Proposed model of ARP6 regulation of DMC1 expression in nonsporogenous cell and megasporocyte of wild-type or arp6 ovules by modulating H2A.Z deposition at the DMC1 locus. Black and gray disks mark H2A- and H2A.Z-containing nucleosomes, respectively.
(A) According to this model, in a wild-type nonsporogenous ovule cell, DMC1 is not expressed because H2A.Z prevents RNA polymerase II (RNA pol II; gray oval) progression. This block is nonexistent in an arp6 nonsporogenous ovule cell, leading to DMC1 expression.
(B) In a wild-type megasporocyte of a stage 2-III/IV ovule, DMC1 expression is promoted by eviction of H2A.Z nucleosomes and/or binding of an ARP6-dependent activator (spotted oval with a question mark). Significant decrease in H2A.Z deposition in the DMC1 gene body in an arp6 megasporocyte causes loss of DMC1 expression due to the lack of ARP6-dependent activator (top). However, partial substitution for ARP6 function by another chromatin remodeling complex and/or other nuclear-localized members of the ARP family (ARP4 and ARP7) may activate DMC1 expression in a subset of arp6 megasporocytes (bottom).
Other proteins in the megasporocyte, such as other nuclear-localized members of ARP family (ARP4 and ARP7; Meagher et al., 2005), may function redundantly with ARP6 in activating DMC1 expression (Figure 6B). This may explain why many arp6 megasporocytes of stage 2-III/IV ovules show pDMC1:GFP expression (Figures 5E, 5G, 5J, and 5K). Alternatively, incomplete penetrance of DMC1 expression in arp6 megasporocytes could reflect the effect of ARP6 on the stochastic nature of DMC1 expression, as chromatin modification influences stochastic gene expression in eukaryotes (Viñuelas et al., 2013). This partial role of ARP6 in activating DMC1 expression in the megasporocyte may be the basis of the incompletely penetrant phenotype of arp6, which shows partial defects in megaspore formation and thus also shows reproductive defects in megagametogenesis, pollen tube targeting of ovules, and reduced seed set (Figure 1, Table 1; Supplemental Figure 1 and Supplemental Tables 1 to 3). Unlike the partial role of ARP6 in promoting DMC1 expression in the megasporocyte, ARP6 would be uniquely required for inhibition of DMC1 expression in the nonsporogenous ovule cells, as its loss results in ectopic DMC1 expression in all mutant ovules (Figures 5E to 5H, 5J, and 5K).
ARP6 Regulates Female Meiosis
ARP6 plays a role in meiosis by regulating the expression of many meiosis-related genes during megasporogenesis. We demonstrated that during ovule development, ARP6 regulates the expression of DMC1, which plays an important role in meiosis. Consistent with these findings, the meiosis I defects we identified in arp6 mutants resemble those reported in dmc1 mutants (Couteau et al., 1999; Siddiqi et al., 2000; Da Ines et al., 2012). Besides DMC1, expression of three other meiosis-related genes is also significantly higher in both arp6-1 and arp6-2 ovules compared with the wild type (Table 2). These genes are also likely ectopically expressed in the nonsporogenous cells of arp6 ovules, similar to DMC1. Cell-specific expression analysis can test this prediction and determine if the expression of these genes is abolished in arp6 megasporocytes, similar to DMC1. We predict that this might be the case, as some of the meiotic defects in arp6 resemble those reported for mutants deficient in these four meiosis-related genes.
Our genetic evidence supports a role for ARP6 in meiosis during megasporogenesis, pinpointing the origin of fertility defects in arp6-1 and arp6-2 to the sporophyte, rather than the female gametophyte. Importantly, our results showed that seed set defects in arp6-1 and arp6-2 are primarily due to defects in meiosis during megasporogenesis. However, meiosis during both microsporogenesis and megasporogenesis is disrupted in arp6-3, and it was shown that in this mutant meiosis progressed normally until anaphase II during microsporogenesis (Rosa et al., 2013). All three T-DNA alleles lack full-length transcript: arp6-1 is in the first exon, arp6-2 is in the 4th exon, and arp6-3 is in the 4th intron. The intron insertion in arp6-3 may be one reason for these allele-specific effects. Clarifying the basis for this discrepancy awaits further experiments. Nevertheless, based on likely null phenotype of arp6-3 and our extensive characterizations (including molecular complementation of all phenotypes), arp6-1 and arp6-2 provide an important tool to identify the mechanisms that regulate meiosis during megasporogenesis.
ARP6-Dependent Deposition of H2A.Z at DMC1
As part of the SWR1 complex, ARP6 may swap H2A.Z for H2A at the DMC1 locus, remodel DMC1 chromatin, and facilitate access to the transcriptional repression or activation machinery. In support of this model for ARP6 function in regulating DMC1 expression, we showed that in floral buds, H2A.Z is preferentially deposited in the DMC1 gene body and that this preferential deposition of H2A.Z requires ARP6.
Gene expression profiling studies in yeast, animals, and plants showed a strong association between H2A.Z and expression of developmental response genes and inducible genes (Coleman-Derr and Zilberman, 2012). Here, we found a similar relationship between H2A.Z deposition and developmental expression of DMC1. ARP6-dependent high occupancy of H2A.Z in the DMC1 gene body and low H2A.Z occupancy in the promoter region of DMC1 is similar to the ARP6-dependent H2A.Z enrichment in Heat Shock Protein70, a thermosensory gene (Kumar and Wigge, 2010). In Arabidopsis, H2A.Z enrichment in the gene body negatively correlates with expression levels and positively correlates with higher responsiveness (Coleman-Derr and Zilberman, 2012). Consistent with these observations, enrichment of H2A.Z at the DMC1 gene body may influence its responsiveness to developmental stimuli by poising it for rapid activation in the megasporocyte after it enters meiosis, as proposed for inactive yeast genes (Guillemette et al., 2005) and human genes expressed in CD4+ T cells (Barski et al., 2009).
Our ChIP analysis showed that H2A.Z deposition in the DMC1 gene body is ARP6 dependent. Interestingly, our promoter-reporter transgene analysis recapitulated the endogenous DMC1 expression both in the wild-type and arp6 ovules (Figure 5), suggesting that the upstream regulatory sequences of DMC1 are sufficient to program ARP6-dependent regulation of DMC1. However, the tissues used for ChIP analysis contained multiple cell types; therefore, our results did not allow us to distinguish between a model where H2A.Z deposition is required to activate DMC1 in the megasporocyte versus one in which H2A.Z deposition represses DMC1 in the nonsporogeneous cells. Nevertheless, given that ARP6 is expressed in both the megasporocyte and nonsporogenous cells, we propose that the dual roles of ARP6 in regulation of DMC1 expression likely involve (Figures 6A and 6B): (1) H2A.Z enrichment at DMC1 gene body inhibiting its expression in nonsporogenous ovule cells (stage 2-I/II and 2-III/IV) and the megasporocyte (stage 2-I/II) and (2) H2A.Z eviction from DMC1 gene body promoting its expression in the megasporocyte undergoing meiosis (stage 2-III/IV).
H2A.Z deposition in the DMC1 gene body could inhibit its transcription in the nonsporogenous cells of stage 2-I to 2-IV or megasporocyte of stage 2-I/II ovules by blocking elongation of RNA polymerase II (Kumar and Wigge, 2010; Santisteban et al., 2011) (Figure 6A). This block may be exacerbated if the stalled RNA polymerase II prevents interaction between activator proteins and their cognate DNA binding sites (Marques et al., 2010). By contrast, in megasporocytes of stage 2-III/IV ovules, activation of DMC1 expression may depend on H2A.Z eviction (Figure 6B), where the block to RNA polymerase II transcription might be relieved in the megasporocyte and expose DMC1 to a transcriptional activator (Gallant-Behm and Mustoe, 2010), which is megasporocyte specific and ARP6 dependent (Figure 6B). In this model, another chromatin-remodeling complex or other nuclear-localized members of ARP family (ARP4 and ARP7; Meagher et al., 2005) may substitute for ARP6 in activating DMC1 expression in a subset of arp6 megasporocytes. Alternatively, ARP6 may indirectly regulate DMC1 via H2A.Z deposition at other positive or negative regulatory proteins that in turn regulate the expression of DMC1, as has been proposed for regulation of other genes (Deal et al., 2007).
Coordinated Development of the Megasporocyte and Nonsporogenous Ovule Cells
The coordinated, differential regulation of DMC1 expression in ovules before and during meiotic divisions of megasporogenesis is consistent with proposed crosstalk between megasporocyte and nonsporogenous ovule cells during megasporocyte development (Feng et al., 2013). Inhibition of DMC1 expression in ovules at stage 2-I/II prior to activation specifically in the megasporocyte could also point to ovules being on a holding pattern until megasporocyte differentiation is completed. This may allow nonsporogenous ovule cells to maintain epigenomic plasticity to differentiate a new megasporocyte in case megasporocyte differentiation fails. Analogous to the mammalian SWI complex (BAF), remodeling of chromatin to induce sequential developmental transitions of T cells in response to external signals (Ho and Crabtree, 2010), the SWR1 complex may function as a developmental sensor of crosstalk between the megasporocyte and nonsporogenous ovule cells to ensure initiation of meiosis occurs only in a fully differentiated megasporocyte.
METHODS
Plant Materials and Growth Conditions
Wild-type Arabidopsis thaliana (Col-0 ecotype), arp6-1 (Garlic_599_G03), and arp6-2 (Garlic_236_C07) were grown as described (Qin et al., 2009). arp6 mutants were selected on plates with 10 μg/mL Basta or in soil by spraying with 240 mg/L Basta.
Analysis of Pollen Tube Growth and Guidance
Pollen tube targeting of ovules was examined as described (Johnson et al., 2004) and semi–in vivo assay of pollen tube–ovule interactions was performed as described (Palanivelu and Preuss, 2006). Ovule targeting by pollen tube (ovule targeting efficiency) values were analyzed as a randomized complete block design with dates of observation as blocks and the pistil-ovule combinations as treatments.
Statistical Analysis of Pollen Tube Growth and Guidance Using a Semi–in Vivo Assay
Ovule targeting by pollen tube (ovule targeting efficiency) values were analyzed as a randomized complete block design with dates of observation as blocks and the pistil-ovule combinations as treatments. Ovule targeting efficiency values were arcsine transformed before analysis and were subjected to mixed-model ANOVA with date considered a random effect and treatment a fixed effect. Untransformed least-squares means from this analysis are reported in Supplemental Figure 1D. Analysis was done using PROC MIXED in SAS/STAT version 9.3 of the SAS System for Windows (SAS Institute). Pairwise comparisons of least-squares means for each pistil-ovule combination were accomplished using the PDIFF option in PROC MIXED. Statistical significance was assigned at P ≤ 0.05 in two-sided tests.
Thick Plastic Sections and Light Microscopy
Thick plastic sections of pistils from flower buds (Smyth et al., 1990) were prepared as described (Sandaklie-Nikolova et al., 2007) and observed under bright-field optics using a Zeiss Axiovert 200M microscope (Carl Zeiss).
Confocal Microscopy of Megagametogenesis
Pistils from stage 12a-c flowers were prepared for microscopy to score megagametogenesis as described (Christensen et al., 1997) and imaged on a Zeiss LSM 510 confocal microscope.
pSPL:GUS Expression Analysis
Transgenic plants heterozygous for SPL promoter driving GUS (pSPL:GUS) transgene (Ito et al., 2004) were crossed with arp6-1 plants. F1 plants with positive GUS activity were selfed, and arp6-1/arp6-1 and pSPL:GUS/pSPL:GUS lines were identified among the F2 segregants. Pistils from stage 10/11 flowers were stained for GUS expression as described (Johnson et al., 2004) and imaged using differential interference contrast optics on a Zeiss Axiophot microscope (Carl Zeiss).
Callose Detection
Aniline blue staining and observation for callose detection in the megaspores was as described (Siddiqi et al., 2000) with minor modifications, which include fixation of inflorescences in FAA for 16 h and 8 to 12 h of incubation in 0.1% aniline blue in 100 mM Tris, pH 8.5. Stained pistils were mounted in 30% glycerol and observed on a Zeiss Axiophot microscope (Carl Zeiss) using 365-nm excitation and a 420-nm long-pass emission filter. Staging of ovules was as described (Schneitz et al., 1995).
Meiotic Chromosome Spreads and FISH
Analysis of meiotic chromosome spreads of female and male megasporocytes was performed as described (Agashe et al., 2002) with minor modifications. The enzyme digestion mixture contained 0.3% cellulase/pectinase/driselase. Stock solutions of cellulase (C-9422; Sigma-Aldrich), pectinase (P-4716; Sigma-Aldrich), and driselase (D-9515; Sigma-Aldrich) were prepared in a solution containing 10 mM citrate, pH 4.5, and 45% glycerol and stored at –20°C. Staining of chromosomes was done using 1 μM 4′,6-diamidino-2-phenylindole (DAPI) in PBS/50% glycerol. Chromosomes were observed on a Leica DM6000 microscope using a 365-nm excitation, 420-nm long-pass emission filter, digitally photographed using a Leica DFC360 camera, and edited using Adobe Photoshop.
FISH analysis was conducted according to a method described (Han et al., 2009) with minor modifications. Slides of meiotic chromosome spreads were prepared as described above, UV cross-linked for 2 min, washed in 2× SSC (3 × 5 min), and then rinsed in 70, 95, and 100% ethanol for 5 min each and air-dried for 30 min. After application of 6 μL centromere probe solution (4 ng/μL of probe in 2× SSC and 1× TE buffer, denatured for 5 min in 95°C and then placed on ice), the slides were heated for 5 min at 95°C and then incubated at 37°C for 6 h in a humid chamber. After hybridization, the slides were washed in 2× SSC and mounted in 1 μM DAPI in PBS/50% glycerol. The FISH images were captured using a Leica DM6000 microscope on a Leica DFC360 camera and processed with Adobe Photoshop.
Complementation of arp6 Phenotypes
The ARP6 complementation plasmid was constructed by amplifying a 5.4-kb genomic DNA fragment containing 3.0 kb of 5′ flanking region, 1.7-kb coding sequence, and 0.5-kb 3′ untranslated region using Phusion polymerase (New England Biolabs) with T4P3 BAC DNA as a template and primers ARP6 5′ Kpn2 (5′-ACGTGGTACCCACACTCATTTGGATTGACACC-3′) and ARP6 3′ Sac1 (5-ACGTGAGCTCGTGCAACAGCCTGATACAGTAGC-3′). It was then restriction digested with KpnI/SacI and cloned into a customized pBI101 binary vector (with the GUS-nos3′ cassette removed as BamHI/EcoRI and replaced with pUC19 MCS (Wang et al., 2006). An Agrobacterium tumefaciens strain (GV3101 pMP90) carrying a DNA construct containing the 5.4-kb ARP6 genomic DNA fragment was introduced into arp6-1 and arp6-2 plants by the floral dip method (Clough and Bent, 1998), and transformants were selected on kanamycin. Megaspores in developing ovules of T1 transformants were scored by fixing pistils from stage 11 flowers in FAA (10% formalin, 5% acetic acid, and 50% ethanol) and observing under a light microscope using differential interference contrast optics. Seed set in siliques of T1 transformants were scored using a Zeiss Stemi 2000-C stereomicroscope.
Immunoblotting
Protein extraction and immunoblotting was performed as described (Deal et al., 2005). Ovaries were from stage 9 flowers (Smyth et al., 1990) that contain ovules undergoing meiosis. Protein blots were probed with ARP6 monoclonal primary antibody (1:100 dilution) and horseradish peroxidase–conjugated secondary antibody (Amersham Biosciences; 1:8000 dilution). Equal loading and transfer of proteins were confirmed by staining a duplicate gel with Coomassie Brilliant Blue. PEP carboxylase (PEPC) polyclonal antibody was used at 1:10,000 dilution.
In Situ Hybridization of Whole-Mount Ovules
Ovules from wild-type and arp6-1 Col-0 ecotype plants were dissected from stage 11 pistils, fixed, and processed as described previously (Hejátko et al., 2006). For the DMC1 probe, a 328-bp cDNA fragment was cloned into pTA2 vector (Toyobo). The PCR primers used are listed in Supplemental Table 7. This plasmid was digested with NotI and transcribed with T7 RNA polymerase to generate the antisense probe or digested with HindIII and transcribed with T3 RNA polymerase to generate the sense probe. For the ARP6 probe, a 335-bp cDNA fragment was cloned into pTA2 vector (Toyobo). This plasmid was digested with BamHI and transcribed with T7 RNA polymerase to generate the antisense probe or digested with HindIII and transcribed with T3 RNA polymerase to generate the sense probe.
ARP6-YFP Fusion Protein Localization
The ARP6-YFP reporter plasmid was constructed by amplifying a 4.8-kb genomic fragment containing 3.1 kb of ARP6 promoter, 5′ flanking region, and the coding region of ARP6 by Phusion polymerase (New England Biolabs) using T4P3 BAC DNA as a template and primers ARP6 5′ Kpn2 (5′-ACGTGGTACCCACACTCATTTGGATTGACACC-3′) and ARP6 3′ Xho (5′-ACGTCTCGAGATGAAAGAATCGTCTACGACACC-3′). It was then restriction digested with KpnI/XhoI and cloned into the pBN-YFP vector (Wang et al., 2006). Plants were transformed using Agrobacterium GV3101 pMP90.
RT-qPCR
Total RNA from wild-type, arp6-1, and arp6-2 stage 2-III/IV ovules or wild-type, arp6-1, and arp6-2 stage 5-7 anthers was isolated using the Qiagen RNeasy kit followed by treatment with DNase I (Thermo Scientific) prior to first-strand cDNA synthesis by reverse transcription using a ThermoScript RT-PCR kit (Life Technologies). Quantitative PCR was performed using FastStart DNA Master SYBR Green I master mix (Roche) in a LightCycler system (Roche). Primers used in RT-qPCR are listed in Supplemental Table 7. The PCR cycle conditions used for quantitative PCR were as follows: 95°C for 5 min followed by 45 cycles of 95°C for 10 s, 60°C for 15 s, and 72°C for 15 s. For each meiosis-related gene analyzed in Table 2 and Supplemental Table 5, five reactions were performed (two technical replicates for first two biological replicates and one technical replicate for the third biological replicate), and ACTIN2/8 expression (threshold cycle [CT] values of 19) was used to normalize mRNA levels in wild-type, arp6-1, and arp6-2 stage 2-III/IV ovules or wild-type, arp6-1, and arp6-2 stage 5-7 anthers. ACTIN2/8 was used to normalize mRNA levels in RT-qPCR reactions reported in Table 2, Supplemental Figure 5, and Supplemental Table 5, and HK2 was used to normalize mRNA levels in Supplemental Figure 6M (Czechowski et al., 2005). The comparative threshold cycle (CT) method (Livak and Schmittgen, 2001) was used to calculate the relative expression ratio of the meiosis-related genes in arp6 and wild-type tissues (ovules or anthers).
Statistical Analysis of RT-qPCR
PROC MIXED of the SAS System for Windows (SAS Institute) was used to calculate least-squares means and standard deviations of relative expression ratio of the meiosis-related genes in arp6 and wild-type tissues (ovules or anthers) (Table 3; Supplemental Table 5). Assuming a randomized complete block design, biological and technical replicates (nested within biological replicates) were considered random effects in these analyses and genotype/gene combinations were fixed effects. Sequential pairwise comparisons of least-squares means for the wild type or arp6-1 or arp6-2 genotypes and ACTIN2 (1.00) for each locus were accomplished using the PDIFF option in PROC MIXED. Statistical significance was assigned at P ≤ 0.05 and all tests of significance were two sided.
pDMC1:GFP Expression Analysis
The pDMC1:GFP reporter plasmid was constructed by amplifying a 2.7-kb genomic fragment containing the 5′ flanking region and the first three amino acids using PfuTurbo polymerase (Stratagene) with Landsberg erecta genomic DNA as template and primers DMC1 2.7 5′ Sal (5′-ACGTGTCGACACACCTAATCGGTGATTGCC-3′) and DMC1 3′ BglII (5′-GCAGATCTAGCCATCATTTTCTCGCTCTAAGAC-3′), restriction digested with SalI/BglII, and cloned into the SalI and BamHI sites of the binary vector pBI-GFP (S65T) (Yadegari et al., 2000). Landsberg erecta was used as template because Col-0 ecotype has an 1874-bp transposon located within the promoter region (Klimyuk and Jones, 1997). An Agrobacterium strain (GV3101 pMP90) containing the pDMC1:GFP reporter construct was produced. T1 plants showed expression in the megaspore mother cell, immature flower cluster, and anthers (Supplemental Table 8). T2 lines were screened for single T-DNA insertions by germination on kanamycin. A single T2 plant with multiple T-DNA insertions (>75% Kanr) showing visible expression was crossed to arp6-1 and arp6-2 plants (Supplemental Table 8), and arp6 mutants carrying the pDMC1:GFP transgene were identified among F2 segregants. Ovules and anthers were mounted in 50% glycerol and observed on a TCS SP5 confocal microscope (Leica Microsystems). Staging of ovules and anthers were as described by Schneitz et al. (1995) and Sanders et al. (1999), respectively.
ChIP-qPCR Analysis of H2A.Z Deposition
ChIP was performed as described (Gendrel et al., 2005) in replicates, four each for the wild type and arp6-1. For each ChIP experiment, from 1 g of floral buds stage 12 or younger (Smyth et al., 1990), chromatin was isolated as follows and split into two aliquots: The suspended chromatin was sonicated for five cycles with each cycle lasting for 5 min during which a sonication pulse was applied for 15 s at a power setting of 5 on the sonicator (Bioruptor UCD-200) and paused for 15 s before the next 15-s pulse. The size of fragmented chromatin DNA was estimated to be between 100 and 300 bp using gel electrophoresis. Floral buds isolated included buds containing ovules at stages 1-II and stages 2-I to 2-IV. One aliquot of chromatin was immunoprecipitated with polyclonal H2A.Z antibody (Deal et al., 2007) and the other served as no antibody control. The H2A.Z antibody was used at a dilution of 1:100. Relative enrichment of H2A.Z-associated DNA fragments was analyzed by qPCR in duplicates with the ACTIN2/8 as the endogenous control in both qPCR reactions as described above in the RT-qPCR methods section. Primers used in ChIP-qPCR are listed in Supplemental Table 7. The relative quantity value from qPCR was calculated by the comparative CT method and is reported as fold enrichment of Table 3 and Supplemental Figure 9.
Statistical Analysis of H2A.Z deposition by ChIP-qPCR
PROC MIXED of the SAS System for Windows (SAS Institute) was used to calculate least-squares means and standard deviations for the relative quantity values calculated in ChIP-qPCR experiments for each locus and genotype (Table 3; Supplemental Figure 9). Assuming a randomized complete block design, biological replicates were considered random effects in these analyses and genotype/locus combinations were fixed effects. Sequential pairwise comparisons of least-squares means for wild-type or arp6-1 genotypes for each locus were accomplished using the PDIFF option in PROC MIXED. Similarly, pairwise comparisons between the wild type and arp6-1 least-squares means for each locus were done using the PDIFF option. Statistical significance was assigned at P ≤ 0.05 and all tests of significance were two sided.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: ARP6, At3g33520; DMC1, At3g22880; SAP, At5g35770; SPL, At4g27330; AML1, At5g61960; AML4, At5g07290; RAD51, At5g20850; SPO11, At1g63990; DYAD, At5g51330; ATM, At3g48190; SDS, At1g14750; ASY1, At1g67370; MND1, At4g29170; DIF1, At5g05490; and HK2, At4g26410.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Reproductive Defects in arp6-1 Pistils.
Supplemental Figure 2. Analysis of Meiotic Divisions of Microsporogenesis.
Supplemental Figure 3. Differentiation of an Archesporial Cell into a Megasporocyte in arp6-1 Ovules Is Comparable to Wild Type.
Supplemental Figure 4. Confocal Laser Scanning Microscopy Analysis of ARP6-YFP Fusion Protein Localization in Ovules.
Supplemental Figure 5. RT-qPCR Analysis of GFP Expression Levels in Wild-Type, arp6-1, and arp6-2 Stage 2-III/IV Ovules Carrying pDMC1:GFP.
Supplemental Figure 6. Analysis of pDMC1:GFP Expression during Ovule and Seedling Development in Wild-Type and arp6 Plants.
Supplemental Figure 7. pDMC1:GFP Expression during Meiosis of Microsporogenesis Remains Apparently Unaltered in arp6 Mutants.
Supplemental Figure 8. DMC1 Gene Structure in Arabidopsis (Col-0 Ecotype).
Supplemental Figure 9. H2A.Z Deposition at DMC1 Is Partially Dependent on ARP6.
Supplemental Table 1. Confocal Laser Scanning Microscopy Observations of Wild-Type and arp6-1 Ovules.
Supplemental Table 2. Confocal Laser Scanning Microscopy Observations of Megagametogenesis.
Supplemental Table 3. Reduced Fertility in arp6-1 and arp6-2 Mutants Is Caused by Defects in Sporophytic Tissues in Pistils.
Supplemental Table 4. Meiosis during Megasporogenesis Is Abnormal in arp6 Megasporocytes.
Supplemental Table 5. Expression of Meiosis-Related Genes in arp6 Anthers.
Supplemental Table 6. DMC1 Expression Is Affected in arp6 Megasporocytes.
Supplemental Table 7. List of Primers Used in This Study.
Supplemental Table 8. Identification of Transgenic Plants Expressing pDMC1:GFP.
Supplemental Data Set 1. Complementation of Reduced Fertility Defects Caused by arp6-1 and arp6-2 Mutations Using pARP6:gARP6 and ARP6-YFP Fusion Protein Expressed from ARP6 Promoter.
Supplementary Material
Acknowledgments
We thank R. Meagher (University of Georgia) for the arp6-1 and arp6-2 seeds and H2A.Z, ARP6, and PEPC antibodies; E. Meyerowitz (Caltech, CA) for pSPL:GUS seeds; H. Ma and Z. Cheng (Fudan University, China) for assistance with FISH; J. Wang (SIPPE, China) for help with Bioruptor; and J. Mach for language editing of the article. This work was supported by grants from Ministry of Science and Technology of China (2011CB944603) and National Natural Science Foundation of China (31170290) to Y.Q. and by grants from the U.S. National Science Foundation to R.Y. (IOS-0520008) and R.P. (IOS-0723421).
AUTHOR CONTRIBUTIONS
Y.Q., I.S., Z.Y., R.Y., and R.P. conceived and designed the experiments. Y.Q., L.Z., M.I.S., S.A., T.T., A.P., and K.N.W. performed the experiments. S.S. conducted statistical analysis. Y.Q., R.Y., and R.P. wrote the article. R.Y. and R.P. were joint senior authors of this study. R.P. coordinated the study and was the liaison among the authors.
Glossary
- FISH
fluorescence in situ hybridization
- Col-0
Columbia-0
- RT-qPCR
reverse transcription coupled with quantitative PCR
- ChIP-qPCR
chromatin immunoprecipitation coupled with qPCR
- DAPI
4′,6-diamidino-2-phenylindole
- PEPC
PEP carboxylase
- CT
threshold cycle
- CLSM
confocal laser scanning microscopy
Footnotes
Online version contains Web-only data.
References
- Agashe B., Prasad C.K., Siddiqi I. (2002). Identification and analysis of DYAD: a gene required for meiotic chromosome organisation and female meiotic progression in Arabidopsis. Development 129: 3935–3943 [DOI] [PubMed] [Google Scholar]
- Armstrong S.J., Jones G.H. (2003). Meiotic cytology and chromosome behaviour in wild-type Arabidopsis thaliana. J. Exp. Bot. 54: 1–10 [DOI] [PubMed] [Google Scholar]
- Armstrong S.J., Caryl A.P., Jones G.H., Franklin F.C. (2002). Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 115: 3645–3655 [DOI] [PubMed] [Google Scholar]
- Armstrong S.J., Franklin F.C., Jones G.H. (2001). Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J. Cell Sci. 114: 4207–4217 [DOI] [PubMed] [Google Scholar]
- Barski A., Jothi R., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Zhao K. (2009). Chromatin poises miRNA- and protein-coding genes for expression. Genome Res. 19: 1742–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi T.H., Wan M., Lee P.P., Akashi K., Metzger D., Chambon P., Wilson C.B., Crabtree G.R. (2003). Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity 19: 169–182 [DOI] [PubMed] [Google Scholar]
- Choi K., Zhao X., Kelly K.A., Venn O., Higgins J.D., Yelina N.E., Hardcastle T.J., Ziolkowski P.A., Copenhaver G.P., Franklin F.C., McVean G., Henderson I.R. (2013). Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat. Genet. 45: 1327–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen C.A., King E.J., Jordan J.R., Drews G.N. (1997). Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod. 10: 49–64 [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coleman-Derr D., Zilberman D. (2012). Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 8: e1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F., Belzile F., Horlow C., Grandjean O., Vezon D., Doutriaux M.P. (1999). Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell 11: 1623–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Ines O., Abe K., Goubely C., Gallego M.E., White C.I. (2012). Differing requirements for RAD51 and DMC1 in meiotic pairing of centromeres and chromosome arms in Arabidopsis thaliana. PLoS Genet. 8: e1002636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal R.B., Kandasamy M.K., McKinney E.C., Meagher R.B. (2005). The nuclear actin-related protein ARP6 is a pleiotropic developmental regulator required for the maintenance of FLOWERING LOCUS C expression and repression of flowering in Arabidopsis. Plant Cell 17: 2633–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal R.B., Topp C.N., McKinney E.C., Meagher R.B. (2007). Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 19: 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draker R., Cheung P. (2009). Transcriptional and epigenetic functions of histone variant H2A.Z. Biochem. Cell Biol. 87: 19–25 [DOI] [PubMed] [Google Scholar]
- Drews G.N., Yadegari R. (2002). Development and function of the angiosperm female gametophyte. Annu. Rev. Genet. 36: 99–124 [DOI] [PubMed] [Google Scholar]
- Feng X., Zilberman D., Dickinson H. (2013). A conversation across generations: soma-germ cell crosstalk in plants. Dev. Cell 24: 215–225 [DOI] [PubMed] [Google Scholar]
- Gallant-Behm C.L., Mustoe T.A. (2010). Occlusion regulates epidermal cytokine production and inhibits scar formation. Wound Repair Regen. 18: 235–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Martienssen R., Colot V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2: 213–218 [DOI] [PubMed] [Google Scholar]
- Guillemette B., Bataille A.R., Gévry N., Adam M., Blanchette M., Robert F., Gaudreau L. (2005). Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3: e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F., Gao Z., Birchler J.A. (2009). Reactivation of an inactive centromere reveals epigenetic and structural components for centromere specification in maize. Plant Cell 21: 1929–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejátko J., Blilou I., Brewer P.B., Friml J., Scheres B., Benková E. (2006). In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat. Protoc. 1: 1939–1946 [DOI] [PubMed] [Google Scholar]
- Ho L., Crabtree G.R. (2010). Chromatin remodelling during development. Nature 463: 474–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Wellmer F., Yu H., Das P., Ito N., Alves-Ferreira M., Riechmann J.L., Meyerowitz E.M. (2004). The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 430: 356–360 [DOI] [PubMed] [Google Scholar]
- Johnson M.A., von Besser K., Zhou Q., Smith E., Aux G., Patton D., Levin J.Z., Preuss D. (2004). Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168: 971–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimyuk V.I., Jones J.D. (1997). AtDMC1, the Arabidopsis homologue of the yeast DMC1 gene: characterization, transposon-induced allelic variation and meiosis-associated expression. Plant J. 11: 1–14 [DOI] [PubMed] [Google Scholar]
- Krogan N.J., et al. (2003). A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12: 1565–1576 [DOI] [PubMed] [Google Scholar]
- Kumar S.V., Wigge P.A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- March-Díaz R., Reyes J.C. (2009). The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Mol. Plant 2: 565–577 [DOI] [PubMed] [Google Scholar]
- March-Díaz R., García-Domínguez M., Lozano-Juste J., León J., Florencio F.J., Reyes J.C. (2008). Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 53: 475–487 [DOI] [PubMed] [Google Scholar]
- Marques M., Laflamme L., Gervais A.L., Gaudreau L. (2010). Reconciling the positive and negative roles of histone H2A.Z in gene transcription. Epigenetics 5: 267–272 [DOI] [PubMed] [Google Scholar]
- McCormick S. (1993). Male gametophyte development. Plant Cell 5: 1265–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher R.B., Deal R.B., Kandasamy M.K., McKinney E.C. (2005). Nuclear actin-related proteins as epigenetic regulators of development. Plant Physiol. 139: 1576–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher, R.B., Kandasamy, M.K., McKinney, E.C., and Roy, E. (2009). Nuclear actin-related proteins in epigenetic control. In International Review of Cell and Molecular Biology, W.J. Kwang, ed (Waltham, MA: Academic Press), pp. 157–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R., Armstrong S.J., Horlow C., Jackson N.P., Makaroff C.A., Vezon D., Pelletier G., Jones G.H., Franklin F.C. (2003). The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis. Development 130: 3309–3318 [DOI] [PubMed] [Google Scholar]
- Mizuguchi G., Shen X., Landry J., Wu W.H., Sen S., Wu C. (2004). ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348 [DOI] [PubMed] [Google Scholar]
- Palanivelu R., Preuss D. (2006). Distinct short-range ovule signals attract or repel Arabidopsis thaliana pollen tubes in vitro. BMC Plant Biol. 6: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panoli A.P., Ravi M., Sebastian J., Nishal B., Reddy T.V., Marimuthu M.P., Subbiah V., Vijaybhaskar V., Siddiqi I. (2006). AtMND1 is required for homologous pairing during meiosis in Arabidopsis. BMC Mol. Biol. 7: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D., Rhee S.Y., Davis R.W. (1994). Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264: 1458–1460 [DOI] [PubMed] [Google Scholar]
- Qin Y., Leydon A.R., Manziello A., Pandey R., Mount D., Denic S., Vasic B., Johnson M.A., Palanivelu R. (2009). Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet. 5: e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodkiewicz B. (1970). Callose in cell walls during megasporogenesis in angiosperms. Planta 93: 39–47 [DOI] [PubMed] [Google Scholar]
- Rosa M., Von Harder M., Cigliano R.A., Schlögelhofer P., Mittelsten Scheid O. (2013). The Arabidopsis SWR1 chromatin-remodeling complex is important for DNA repair, somatic recombination, and meiosis. Plant Cell 25: 1990–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi L., Bonilla C., Strålfors A., Ekwall K., Svensson J.P. (2011). Podbat: a novel genomic tool reveals Swr1-independent H2A.Z incorporation at gene coding sequences through epigenetic meta-analysis. PLOS Comput. Biol. 7: e1002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandaklie-Nikolova L., Palanivelu R., King E.J., Copenhaver G.P., Drews G.N. (2007). Synergid cell death in Arabidopsis is triggered following direct interaction with the pollen tube. Plant Physiol. 144: 1753–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders P.M., Bui A.Q., Weterings K., McIntire K.N., Hsu Y.C., Lee Y.R., Troung M.T., Beals T.P., Goldberg R.B. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11: 297–322 [Google Scholar]
- Santisteban M.S., Hang M., Smith M.M. (2011). Histone variant H2A.Z and RNA polymerase II transcription elongation. Mol. Cell. Biol. 31: 1848–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz K., Hülskamp M., Pruitt R.E. (1995). Wild-type ovule development in Arabidopsis thaliana: a light microscope study of cleared whole-mount tissue. Plant J. 7: 731–749 [Google Scholar]
- Siddiqi I., Ganesh G., Grossniklaus U., Subbiah V. (2000). The dyad gene is required for progression through female meiosis in Arabidopsis. Development 127: 197–207 [DOI] [PubMed] [Google Scholar]
- Smith A.P., Jain A., Deal R.B., Nagarajan V.K., Poling M.D., Raghothama K.G., Meagher R.B. (2010). Histone H2A.Z regulates the expression of several classes of phosphate starvation response genes but not as a transcriptional activator. Plant Physiol. 152: 217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher T.H., Gorovsky M.A. (1994). Phylogenetic analysis of the core histones H2A, H2B, H3, and H4. Nucleic Acids Res. 22: 174–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignard J., Siwiec T., Chelysheva L., Vrielynck N., Gonord F., Armstrong S.J., Schlögelhofer P., Mercier R. (2007). The interplay of RecA-related proteins and the MND1-HOP2 complex during meiosis in Arabidopsis thaliana. PLoS Genet. 3: 1894–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viñuelas J., Kaneko G., Coulon A., Vallin E., Morin V., Mejia-Pous C., Kupiec J.J., Beslon G., Gandrillon O. (2013). Quantifying the contribution of chromatin dynamics to stochastic gene expression reveals long, locus-dependent periods between transcriptional bursts. BMC Biol. 11: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Tyson M.D., Jackson S.S., Yadegari R. (2006). Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 13244–13249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadegari R., Drews G.N. (2004). Female gametophyte development. Plant Cell 16 (Suppl): S133–S141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadegari R., Kinoshita T., Lotan O., Cohen G., Katz A., Choi Y., Katz A., Nakashima K., Harada J.J., Goldberg R.B., Fischer R.L., Ohad N. (2000). Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell 12: 2367–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W.C., Ye D., Xu J., Sundaresan V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13: 2108–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.