Abstract
In many parts of the developing vertebrate nervous system, axons are pruned to establish mature patterns of connectivity. In this issue of Neuron, Schafer et al. (2012) show that microglia may play a role in developmental axon pruning in the thalamus by engulfing presynaptic retinal ganglion cell terminals via a C3- and CR3-dependent mechanism.
Circuitry in the vertebrate peripheral and central nervous systems is initially established as a rough draft, which is refined through significant axon pruning. This pruning is influenced by synaptic activity, can involve elimination of functional synapses, and is generally complete soon after birth. A particularly well-studied example is in the developing mammalian visual system, where retinal ganglion cells (RGCs) from both eyes establish overlapping projections in the dorsal lateral geniculate nucleus (dLGN). Activity-dependent competitive interactions among RGC inputs drive axon remodeling that results in the adult pattern of nonoverlapping eye-specific projections in the dLGN (Shatz, 1990).
Growing evidence implicates proteins of the immune system—known for their roles in recognizing and removing infected, cancerous, and damaged cells—in axon remodeling in the developing visual system. Proteins of the major histocompatibility complex class I (MHCI) and complement cascade (C1q and C3) are expressed in the developing brain and are necessary for normal pruning of RGC axons in the dLGN (Datwani et al., 2009; Huh et al., 2000; Stevens et al., 2007). PirB, an immunoreceptor for MHCI, is not required for development of either retinogeniculate or thalamocortical visual projections, but limits thalamocortical plasticity in response to visual deprivation (Syken et al., 2006). It is tempting to speculate that proteins involved in identification and removal of unwanted cells and debris by the immune system could use analogous mechanisms to identify and remove unwanted inputs during developmental synapse elimination. In some cases, there are hints that this simple model may not fit. For example, MHCI and PirB have functions in neurons that bear no known resemblance to their functions in the immune response: MHCI limits NMDAR-mediated synaptic transmission (Fourgeaud et al., 2010), while PirB serves as a receptor for myelin-derived axon outgrowth inhibitors (Atwal et al., 2008). For the complement system, however, the final molecular signaling pathways and cellular effectors involved in neuronal and immunological functions may be substantially similar. What may distinguish normal neurodevelopmental and pathological clearance of cellular material by the complement cascade is the factor(s) that trigger their recruitment.
The complement cascade consists of over thirty small proteins and protein fragments, present in inactive forms in blood. Binding of C1q initiates the classical complement cascade, including activation of C3, triggering events that target cellular debris for phagocytosis. Previous studies showed that C1q and C3 localize to developing retinogeniculate synapses and are required for anatomical pruning of RGC inputs (Stevens et al., 2007). The precise role of complement in synapse elimination remained unknown, but was hypothesized to involve microglia, the resident macrophages of the central nervous system, given their expression of the C3 receptor, CR3, and their well-known phagocytic ability.
Microglia engulf neuronal debris following a variety of insults and in degenerative disorders. In addition, microglia can engulf synaptic material in the developing mouse hippocampus, and in mice with defects in microglial migration, hippocampal spine densities are higher (Paolicelli et al., 2011). This study was among the first to provide evidence that microglia, in addition to their role in removing damaged cells, may also help clear neuronal components during normal development.
In this issue of Neuron, Schafer et al. (2012) examined this possibility in the developing visual system, using light- and electron-microscopic (E.M.) imaging to visualize interactions between RGCs and microglia in the early postnatal mouse dLGN. RGC inputs from each eye were labeled with intraocular injections of differently colored anterograde tracers, allowing identification of material that originated from either eye. During the time when RGCs were being pruned, microglia contained RGC material from both eyes within their processes and soma. Some RGC-derived material was found in lysosomes, indicating it was destined to be degraded. EM analysis of microglial lysosomes showed double-membrane-bound structures containing components that resembled neurotransmitter vesicles, as well as immunoreactivity for vGluT2, indicating engulfment of presynaptic RGC terminals. Since there is a brief window between phagocytosis and degradation of lysosomal contents, EM studies may underestimate the synaptic content of microglial lysosomes. Together, these experiments suggest microglia can engulf presynaptic terminals of RGCs, though they do not rule out the engulfment of nonsynaptic or postsynaptic structures, as has been seen in hippocampus (Paolicelli et al., 2011).
Microglia are exquisitely sensitive to injury and inflammation, and the above studies involved intraocular injections, which might cause microglia to target RGCs. To control for this possibility, a genetically encoded marker was used to label RGCs, eliminating the need for injections, and similar microglial engulfment of RGC material was still seen. The idea that microglial engulfment is part of a normal developmental pathway is further supported by the fact that engulfment of RGC components in vivo roughly paralleled the timing of developmental remodeling.
Previous work demonstrated that C3 is present at synapses during the early postnatal period and is required for normal developmental remodeling of retinogeniculate axons (Stevens et al., 2007). Given that microglia are the only known resident brain cells to express the C3 receptor, CR3, Schafer et al. (2012) hypothesized that C3-CR3 interactions might recruit microglia to RGC axons as they remodel. To directly test the requirement for CR3 in remodeling, similar RGC-tracer experiments were performed in transgenic mice lacking functional CR3. Overlap between inputs from the two eyes was increased, and engulfment of RCG material by microglia was reduced, in CR3-deficient mice, effects that were mimicked by pharmacologically inhibiting microglial activity in WT animals. The increase in overlap in CR3-deficient mice was paralleled by an increase in synapse density in adults, as assessed by colocalization of VGlut2 (a marker for RGC presynaptic terminals) and GluR1 (a marker of postsynaptic sites) by array tomography. These and other results show that in the absence of complement C3 or its microglial receptor, CR3, microglia contain less RGC-derived material than in WT, and inappropriate axon projections and synapses are present.
Interestingly, CR3-deficient mice showed increase in both VGlut2-containing synapses and VGlut2 puncta not associated with synapses. Some of the nonsynaptic VGlut2 could represent retracting RGC axons that successfully underwent elimination in the absence of CR3-dependent mechanisms. This is likely, since significant pruning still occurs in CR3 knockouts. However, these recently pruned inputs should also be present in WT dLGNs. In fact, if pruning is impaired in CR3-deficient animals, this might be expected to lead to fewer recently pruned inputs than WT, not more. So what is the source of the extra nonsynaptic VGlut2 puncta in CR3 knockouts? One possibility is that increases in nonsynaptic vGlut2 above WT levels could be evidence of increased presynaptic RGC sprouting in CR3-deficient mice. Such sprouting could contribute to both increased synapse density and ectopic axons in CR3-deficient mice. Indeed, axon sprouting in response to neurotrophic factors delays synapse elimination in the peripheral nervous system (Nguyen et al., 1998). Therefore, it will be important to determine if manipulations of C3/CR3 and microglia influence RGC sprouting as well as RGC phagocytosis. This counterintuitive possibility is particularly worth exploring given that microglia may promote growth and regeneration in some systems (Glezer et al., 2007). Intriguingly, although inputs from both eyes are engulfed by microglia, increased overlap in CR3-deficient mice stems primarily from a larger-than usual ipsilateral projection, suggesting that CR3 may promote engulfment and/or limit sprouting preferentially for ipsilateral inputs.
Retinogeniculate remodeling is thought to be an activity-dependent competitive process. Reducing activity in a subset of inputs (e.g., with TTX) promotes their elimination, while enhancing activity in a subset of inputs (e.g., with forskolin) reduces their elimination. Schafer et al. (2012) used monocular injection of these drugs in one eye to test the prediction that cells with elevated activity would be phagocytosed less than controls, while reduced activity would facilitate removal. Indeed, microglia preferentially engulfed less-active RGC inputs, the same inputs that are known to undergo more extensive synaptic pruning.
Overall, these results convincingly and elegantly demonstrate that microglia can internalize RGC axon material, and that this phagocytosis shows the same timing and activity dependence as the remodeling process as a whole. Furthermore, they show that loss of microglial involvement (either by disrupting CR3 receptors or by pharmacologically inhibiting microglial activation) is associated with reduced phagocytosis of RGC material and an increase in synapses and inappropriate axonal projections. These results raise several exciting questions for future study.
A key question is how active a role microglia play in deciding the outcome of the synapse elimination process. Experiments bidirectionally manipulating activity indicate that microglial pruning follows the same rules of activity dependence as the pruning process itself. This could be because microglia detect activity levels and instruct removal of less-active inputs, or because microglia engulf inputs that have been tagged for elimination through as-yet-unidentified activity-dependent mechanisms. In mice lacking C1q or C3, although retinal ganglion cell axons show incomplete anatomical remodeling, often only one strong input remains (Stevens et al., 2007). This indicates that competitive interactions among neurons have not been halted. Instead, it suggests that C1q and C3 are required to physically remove inputs that have already been functionally weakened through competitive interactions. Consistent with this model, Schafer et al. (2012) found that mitochondria were absent from several engulfed presynaptic terminals, a characteristic of terminals with decreased activity that are destined to undergo elimination. It will be important to examine synaptic strength in C3R KOs, to determine if activity-dependent synaptic weakening is prevented by loss of CR3 in microglia. This will help clarify if microglia destroy otherwise strong and healthy axon branches or arrive on the scene after the competitive damage has been done. In either case, the fact that synapse density is increased in CR3-deficient animals suggests that the ultimate readout of developmental competition in this system—net physical removal of synapses—requires microglia.
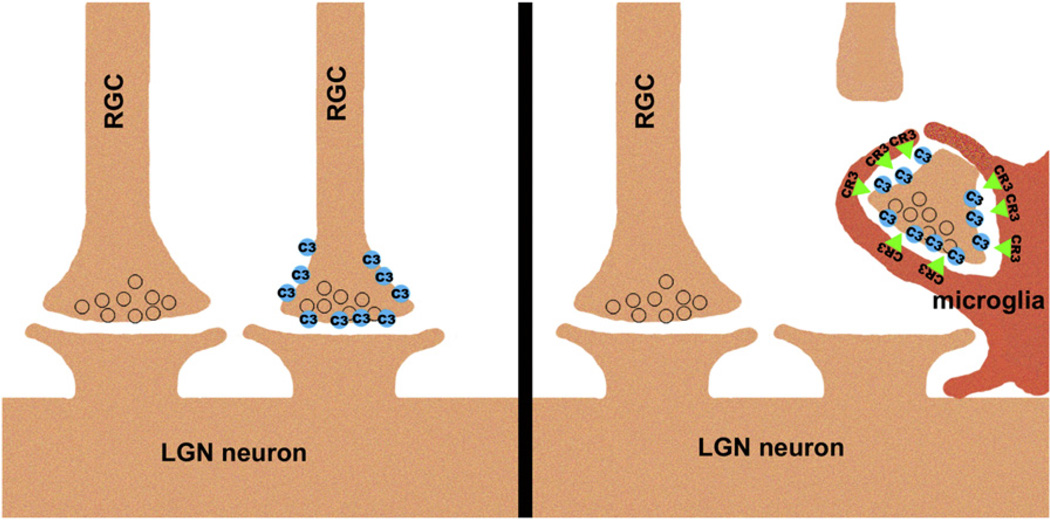
A related question concerns the molecular cascade that precedes microglial engulfment of RGC processes. The current paper suggests that C3 bound to RGCs could interact with microglial CR3 (Figure 1), although direct binding in this context has not been demonstrated. Array tomography shows that C1q is present at a subset of synapses in the developing dLGN (Stevens et al., 2007). However, it remains unknown what leads to deposition of C1q and C3 at some synapses and not others. In other words, what instructs complement to bind to specific connections and mark them for microglial uptake?
Figure 1.
Microglia Phagocytose RGC Axon Material in a C3- and CR3-Dependent Manner
One candidate for this instructive signal comes from recent studies in cultured mouse neurons. In these studies, neurites bound C1q, and were taken up by cocultured microglial cells in a CR3-dependent manner, but only after enzymatic removal of sialic acid residues from the neuronal glycocalyx (Linnartz et al., 2012). Several neuronal cell surface proteins are sialated, including the neural cell adhesion molecule (NCAM), and sialation is developmentally regulated, disappearing from most brain regions in the adult (Mühlenhoff et al., 1998). Understanding the distribution of sialation at individual, competing inputs, as well as its dynamics in response to changes in activity, will help clarify if this molecular mark could play an instructive role in the deposition of complement and subsequent recruitment of microglia during development.
Another open question is the relationship, if any, among the similar axon remodeling phenotypes seen in MHCI-deficient (β2 m−/−TAP−/−, Kb−/−Db−/−) and complement cascade-deficient (C3−/−, Cd11b−/−, and C1q−/−) mice (Huh et al., 2000; Datwani et al., 2009; Stevens et al., 2007; Schafer et al., 2012). MHCI molecules and C1q are closely associated at retinogeniculate synapses by array tomography (Datwani et al., 2009), indicating they could function together in developmental remodeling.
In addition to their new role in neurodevelopmental remodeling, microglia have been implicated in neurodevelopmental disease pathology. Microglial activation is increased in the brains of patients with autism (Vargas et al., 2005), and WT microglia arrest the progression of neuropathology in Mecp2-null mice (Derecki et al., 2012), suggesting that microglial defects may be important in the pathogenesis of Rett syndrome. Thus, understanding the nature of the signals that recruit microglia to developing axons may help identify the factors that target synapses for elimination in the CNS, either during development or in disease states.
REFERENCES
- Atwal JK, Pinkston-Gosse J, Syken J, Stawicki S, Wu Y, Shatz C, Tessier-Lavigne M. Science. 2008;322:967–970. doi: 10.1126/science.1161151. [DOI] [PubMed] [Google Scholar]
- Datwani A, McConnell MJ, Kanold PO, Micheva KD, Busse B, Shamloo M, Smith SJ, Shatz CJ. Neuron. 2009;64:463–470. doi: 10.1016/j.neuron.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derecki NC, Cronk JC, Lu Z, Xu E, Abbott SB, Guyenet PG, Kipnis J. Nature. 2012;484:105–109. doi: 10.1038/nature10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourgeaud L, Davenport CM, Tyler CM, Cheng TT, Spencer MB, Boulanger LM. Proc. Natl. Acad. Sci. USA. 2010;107:22278–22283. doi: 10.1073/pnas.0914064107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glezer I, Simard AR, Rivest S. Neuroscience. 2007;147:867–883. doi: 10.1016/j.neuroscience.2007.02.055. [DOI] [PubMed] [Google Scholar]
- Huh GS, Boulanger LM, Du H, Riquelme PA, Brotz TM, Shatz CJ. Science. 2000;290:2155–2159. doi: 10.1126/science.290.5499.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnartz B, Kopatz J, Tenner AJ, Neumann H. J. Neurosci. 2012;32:946–952. doi: 10.1523/JNEUROSCI.3830-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlenhoff M, Eckhardt M, Gerardy-Schahn R. Curr. Opin. Struct. Biol. 1998;8:558–564. doi: 10.1016/s0959-440x(98)80144-9. [DOI] [PubMed] [Google Scholar]
- Nguyen QT, Parsadanian AS, Snider WD, Lichtman JW. Science. 1998;279:1725–1729. doi: 10.1126/science.279.5357.1725. [DOI] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, et al. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, Stevens B. Neuron. 2012;74:691–705. doi: 10.1016/j.neuron.2012.03.026. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatz CJ. J. Neurobiol. 1990;21:197–211. doi: 10.1002/neu.480210113. [DOI] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, et al. Cell. 2007;131:1164–1178. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- Syken J, Grandpre T, Kanold PO, Shatz CJ. Science. 2006;313:1795–1800. doi: 10.1126/science.1128232. [DOI] [PubMed] [Google Scholar]
- Vargas DL, Nascimbene C, Krishnan C, Zimmerman AW, Pardo CA. Ann. Neurol. 2005;57:67–81. doi: 10.1002/ana.20315. [DOI] [PubMed] [Google Scholar]