Abstract
With the completion of the human and mouse genome sequences, the task now turns to identifying their encoded transcripts and assigning gene function. In this study, we have undertaken a computational approach to identify and classify all of the protein kinases and phosphatases present in the mouse gene complement. A nonredundant set of these sequences was produced by mining Ensembl gene predictions and publicly available cDNA sequences with a panel of InterPro domains. This approach identified 561 candidate protein kinases and 162 candidate protein phosphatases. This cohort was then analyzed using TribeMCL protein sequence similarity clustering followed by CLUSTALV alignment and hierarchical tree generation. This approach allowed us to (1) distinguish between true members of the protein kinase and phosphatase families and enzymes of related biochemistry, (2) determine the structure of the families, and (3) suggest functions for previously uncharacterized members. The classifications obtained by this approach were in good agreement with previous schemes and allowed us to demonstrate domain associations with a number of clusters. Finally, we comment on the complementary nature of cDNA and genome-based gene detection and the impact of the FANTOM2 transcriptome project.
Regulation of protein activity by reversible phosphorylation, so-called phosphoregulation, is an important posttranslational control mechanism implicated in many areas of biology. In this work, we use the term phosphoregulators to refer to both the protein kinases and the protein phosphatases. These enzymes regulate the phosphorylation status of the protein complement of a cell, and in turn, regulate the activity of their target phosphoproteins in cellular processes. The protein kinases covalently attach a phosphate group to a target, whereas the protein phosphatases remove these groups. Defining the entire complement of these proteins in the mouse gives us an opportunity to view the system as a whole.
The phosphorylation status of a protein, or more specifically, the pattern of phosphorylation on a given protein can determine its activity. The presence or absence of a phosphate group can change the conformation of the target protein, thereby modifying its activity. Phospho motifs in some cases provide binding sites for interactors, such as the 14-3-3's (Yaffe 2002). Alternatively, phosphorylation provides binding sites for enzymes catalyzing secondary modifications, such as further phosphorylation, dephosphorylation, acetylation (HIPK2; Hofmann et al. 2002), or in the case of ubiquitin ligases, facilitate ubiquitination and targeting of the protein for proteolysis (Ding and Dale 2002).
Phosphorylation events often occur in a cascade, in which activity of one kinase or phosphatase is dependent on the upstream activity of another. One of the best-studied examples of this is the regulation of the mitogen activated protein kinase (MAPK)-signaling cascade. MAPK signaling has no fewer than five levels of kinase regulation, MAP4K, MAP3K, MAP2K, MAPK, and MAPKAPK (Larochelle and Suter 1995; Cobb 1999; Dan et al. 2001) and one level of phosphatase regulation (MKP) (Theodosiou and Ashworth 2002). Furthermore, there is considerable cross talk between signaling cascades involving other phosphoregulators (Lehman and Gomez-Cambronero 2002), resulting in a network of phosphoregulators rather than a linear cascade.
Phosphoregulation is implicated in many areas of biology; these include transcriptional control (HIPK2; Hofmann et al. 2002; Pierantoni et al. 2002), signal transduction (MAPK; Cobb 1999), regulation of the cell cycle (Cyclin dependent kinases, NIMA kinases, cdc25 phosphatases; Nigg 2001), immunoproliferation (CD45 phosphatase; Koretzky et al. 1991), development (wnt signaling,β-catenin; casein kinase 1, glycogen synthase kinase-3; Ding and Dale 2002; Hedgehog signaling, Cubitus interruptus, CK1, GSK-3, Protein kinase A; Price and Kalderon 2002), apoptosis (casein kinase-2; Ahmed et al. 2002), and targeted proteolysis (Li and Blow 2001; Ding and Dale 2002).
Spatially, phosphoregulators play specific roles throughout the cell. Many cell surface markers are receptor kinases or phosphatases. Examples include the ephrin receptor kinases, TGFβ receptor kinases and cd45 receptor phosphatase. There are cytoplasmic kinases with affinity to the cytoskeleton, such as the Microtubule affinity-regulating kinases that are involved in microtubule dynamics (Drewes et al. 1997). There are nuclear kinases such as the Homeodomain-interacting protein kinases that are involved in transcriptional regulation (Hofmann et al. 2002). Interestingly, there are some phosphoregulators that shuttle dynamically between the cytoplasm and the nucleus. The shuttling of cdc25 phosphatases (Davezac et al. 2000) and cyclin/cdk complexes (Toyoshima et al. 1998) is dependent upon the phase of the cell cycle or induction of a checkpoint, and is dependent upon phosphorylation events.
Eukaryotic protein kinases fall into two major classes on the basis of distinct substrate preferences as follows: (1) serine/threonine kinases, and (2) tyrosine kinases. Similarly eukaryotic protein phosphatases are classified as (1) serine/threonine, (2) tyrosine, or (3) dual-specificity phosphatases. Dual-specificity phosphatases are able to dephosphorylate serine, threonine, and tyrosine residues. Each of these classes has well-conserved catalytic domains, and as such, these classes are amenable to domain-based sequence mining. These proteins are well represented within the InterPro database of protein-sequence domains (Apweiler et al. 2001; http://www.ebi.ac.uk/interpro). InterPro contains 12 domain entries implicated in protein-kinase activity and 10 domain entries associated with protein-phosphatase activity (Table 1). In some cases, these domains represent subclasses of more general domain predictions. For example, a protein kinase can contain a eukaryotic protein kinase motif (IPR000719) as well as a tyrosine protein kinase motif (IPR001245). In other cases, the domains represent distinct classes with distinct domain structure. For example, the low molecular weight phosphatase motif IPR000106 is unrelated to the tyrosine specific protein phosphatase motif IPR000242.
Table 1.
InterPro Domains Associated With Protein Kinases and Phosphatases
| Kinases | |
|---|---|
| IPR000719 | Eukaryotic protein kinase |
| IPR002290 | Serine/Threonine protein kinase |
| IPR001245 | Tyrosine protein kinase |
| IPR000961 | Protein kinase C-terminal domain |
| IPR001426 | Receptor tyrosine kinase class V |
| IPR002373 | cAMP-dependent protein kinase |
| IPR001824 | Receptor tyrosine kinase class III |
| IPR002011 | Receptor tyrosine kinase class II |
| IPR002374 | cGMP-dependent protein kinase |
| IPR003527 | MAPK |
| IPR000239 | GPCR kin |
| IPR002291 | Phosphorylase kinase γ catalytic subunit |
| Phosphatases | |
|---|---|
| IPR000387 | Tyrosine specific protein phosphatase and dual specificity protein phosphatase family |
| IPR000340 | Dual specificity protein phosphatase |
| IPR000934 | Serine/threonine specific protein phosphatase |
| IPR001932 | Protein phosphatase 2C domain |
| IPR000242 | Tyrosine specific protein phosphatase |
| IPR003595 | Protein tyrosine phosphatase, catalytic domain |
| IPR000222 | Protein phosphatase 2C subfamily |
| IPR002115 | Mammalian LMW phosphotyrosine protein phosphatase |
| IPR000106 | Low molecular weight phosphotyrosine protein phosphatase |
| IPR000751 | MPI_Phosphatase. |
Using the InterPro domains detailed in Table 1, we set out to identify all potential protein kinases and protein phosphatases of mouse. To this end, we mined all publicly available cDNA sequences (including the FANTOM2 set) and the Ensembl gene predictions. Once defined, the protein phosphatase and protein kinase complements were subjected to clustering, and the resulting clusters examined for functional groups and domain associations. An alternative approach to traditional multiple alignment was adopted for clustering of these two classes.
A recent development has been the release of a new protein sequence-similarity clustering tool known as TribeMCL (Enright et al. 2002; http://www.ebi.ac.uk/research/cgg/tribe). TribeMCL uses Markov clustering (MCL), an algorithm based upon probability and graph flow theory (Van Dongen 2000) to assign proteins to a given class. Distances are calculated on BLASTP output. The BLASTP output is parsed by the Tribe part of the package to produce a matrix of protein similarities (BLASTP e-values), which is then subjected to Markov clustering. TribeMCL has been reported to handle problem sequences such as multidomain proteins and partial sequences (Enright et al. 2002). It is a very fast algorithm, and as such, is amenable to large data sets. It has been applied to the draft human and mouse genomes with great success (Lander et al. 2001; http://www.ensembl.org) and forms the basis of the Ensembl gene family assignments.
Markov clustering (MCL) can be thought of starting with a matrix in which every protein node is interconnected by a probability of transition (moving from one node to another). Nodes that are highly related have a high probability of transition; nodes that are dissimilar have a low probability of transition. Starting at any given node, a random walk to another node has a higher probability of moving within the same natural cluster than between clusters. During iterative rounds of expansion and inflation, terms specific to MCL, the strong connections (those with a high probability of transition) are strengthened, whereas weak connections are weakened further. Inflation effectively severs flow between clusters, whereas expansion dissipates flow within clusters (Enright 2002). Increasing the inflation value can increase the severity of pruning, and hence, lead to higher granularity of clusters. Over several rounds of expansion and inflation, the matrix reaches a steady state, in which further expansion and inflation have no effect, and clusters have effectively been isolated.
Previous attempts to classify protein kinases have relied on multiple sequence alignment followed by hierarchical tree generation (Kostich et al. 2002). The assumption behind such an approach is that there is conserved sequence between all members to be classified. In the case of the protein kinases, this is the catalytic region. The boundaries of the conserved catalytic region must be determined and an optimal alignment produced. Decisions must be made on how much of the alignment to use, how partial sequences will be handled, and whether to include sequences that do not align well. Sequences that do not share conserved sequence cannot be classified in this way. For example, the protein phosphatases represent multiple classes with distinct evolutionary origins. The tyrosine phosphatases use a different catalytic mechanism to the serine threonine phosphatases (Kerk et al. 2002). TribeMCL does not require or assume conserved sequence, and as such, is amenable to this multiple class problem.
In this work, we report on the use of TribeMCL clustering to classify the murine complement of phosphoregulators. We also report on domains associated with each of the clusters. Finally, we discuss the impact of the FANTOM2 transcriptome data (FANTOM Consortium and the RIKEN GSC Genome Exploration Group 2002) on these two important classes of proteins and the use of TribeMCL as a useful tool for dissecting out distinct classes of proteins.
RESULTS AND DISCUSSION
Identification of Protein Kinases and Protein Phosphatases in Mouse
We used the Sequence Retrieval System (Etzold and Argos 1993; http://srs.ebi.ac.uk) to query two public protein databases, SWALL and IPI (International Protein Index). By use of this tool, mouse sequences annotated as containing the kinase and phosphatase InterPro domains detailed in Table 1 were extracted. The RIKEN FANTOM2 group produced a nonredundant protein set (RPS), which combines public sequences with high-quality representative sequences from the FANTOM2 data set. We again used InterPro domain annotations to extract sequences from RPS3 (nonredundant representative protein set 3, available from RIKEN, http://fantom2.gsc.riken.go.jp). Additional partial RIKEN sequences earmarked as novel kinases or phosphatases during the MATRICS curation phase, which failed to make it through to RPS3, were also included.
Structure-based superfamily predictions (http://scop.mrc-lmb.cam.ac.uk/scop, Structural classification of proteins home page) are also available for the FANTOM2 data, however, the SCOP predictions for the kinases and phosphatases are broader and less well defined than the InterPro predictions. The SCOP PK-like (protein kinase-like) prediction encompasses many protein kinases, but it also includes many related sequences that are not protein kinases. In this respect, the SCOP predictions represent a superset of those identified by InterPro. For this reason, the SCOP-based predictions were not used for identifying the phosphoregulators.
Mapping of the Sequences to a Common Identifier
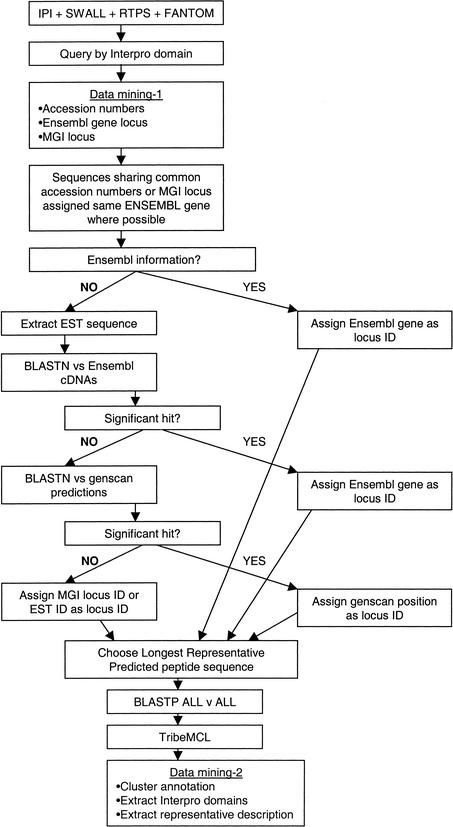
To identify transcripts from the same gene, the sequences were mapped to a common identifier by use of a combination of approaches (Fig. 1). The majority of sequence entries within IPI contain a reference to an Ensembl gene identifier. They also contain cross-references to the original peptide sequences within SWISS-PROT, TrEMBL, and REFSEQ. Similarly, for the RPS and SWALL entries, there are references to the original peptide entries. Mappings to an MGD locus (Blake et al. 2002, http://www.informatics.jax.org/) are also provided for some RPS and IPI entries. In cases in which entries from the different data sources shared original sequence entries, they inherited the Ensembl gene identifier and MGD locus. Sequences that could not be directly assigned to an Ensembl gene had their respective cDNA sequences extracted. These cDNA sequences were then compared with the Ensembl gene cDNA sequences (available for download at http://www.ensembl.org/Mus_musculus) using BLASTN. These alignments were inspected manually. Those with significant hits were assigned the respective Ensembl gene identifier. The remaining sequences were compared with the Ensembl Genscan predictions (available in the same directory). Significant hits were assigned the Genscan locus. Finally, any remaining sequences were assigned the MGD locus (if available) or the EST accession number from which they came. The gold standard for mapping such a group of sequences would be to map them to a genomic location with a given orientation. We provide genomic positions for the majority of the sequences, however, eight of the kinase-related sequences could not be mapped.
Figure 1.
Pipeline for identifying protein kinase and protein phosphatase containing sequences.
By use of our mock locus approach with a preference for Ensembl gene identifiers, it was possible to consider all sequences. This made it simple to compare genomic predictions with transcript data. In Supplementary Tables 1 and 2, we provide extensive cross-referencing for all genes identified. Where possible, we provide representative IPI, RPS3 (and RPS6.3), SWALL, RIKEN, Ensembl, and MGD identifiers. We also provide a representative accession number and genomic position.
Nonredundant Sets of Protein Kinases and Protein Phosphatases
By use of the mock locus assigned in the previous step, the data sets were cross-referenced. For cluster analysis, the combined data set was sorted by mock locus, and the longest representative sequence taken. Intermeshing the predictions from the various protein sets resulted in an estimated 561 candidate protein kinases and 162 candidate protein phosphatases. It is worth noting that only 11 kinase-related sequences and 1 phosphatase sequence failed to map to an Ensembl locus or Genscan prediction.
A total of 541 of the candidate protein kinase sequences mapped to an Ensembl gene, 77 of these are predictions by Ensembl with no supporting transcript evidence in mouse. Of the remaining sequences, nine mapped to Genscan predictions, six were assigned to MGD loci, and the remaining five were labeled with their respective accession numbers.
Similarly, 158 of the candidate protein phosphatase sequences mapped to an Ensembl gene, 19 of these are predictions by Ensembl with no supporting evidence in mouse. A further three mapped to Genscan predictions, and the remaining one was assigned to an MGD locus.
Considering only those sequences with transcript evidence, 104 of the 484 candidate kinases were only supported by RIKEN evidence. This represents a significant proportion of kinases confirmed by transcript (21.5%). Similarly, for the candidate protein phosphatases, 27 of the 143 were only represented by RIKEN (18.9%). These sequences are novel transcripts in the respect that they are the only publicly available transcript evidence for these genes. However, the term novel may be inappropriate, as some of these sequences have been public since the first phase of the FANTOM project (The RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium 2001). To clarify the terminology, we will use the term novel transcripts to refer to genes in which there is no obvious homolog and for which the FANTOM sequences are the only source of transcript evidence. The term homolog is used to identify sequences in which there is an easily identifiable homolog, and for which FANTOM sequences are the only source of transcript evidence. Additionally, there are uncharacterized sequences from other sources, but for the purpose of this analysis, these are considered known.
Clustering of the Nonredundant Kinase and Phosphatase Sets
TribeMCL-based clustering tended to separate the data sets into a few large clusters and many smaller ones. The number and size of clusters (i.e., the granularity of the clustering) could be adjusted by changing the inflation value used within TribeMCL or by using different expectation value (e-value) cut off values during the BLASTP step. The inflation value (a value between 1 and 5, default of 3) is used during the iterative expansion and inflation steps of TribeMCL, the higher the value the more granular the result. Adjustment of the e-value is not recommended by the authors of the TribeMCL package, however, adjusting the inflation value alone did not provide sufficient granularity. Decreasing the granularity led to smaller distinct clusters being split and merged with multiple large clusters rather than all members of a given smaller cluster going to a given larger cluster. We experimented with different combinations of e-value cutoff and inflation value (data not shown), and in the final data presented here, we use an expectation value of e-10 and an inflation value of 5. Using an e-value restriction of e-10 effectively sets the probability of transition to zero for any pair of nodes in which the e-value was higher than e-10 (i.e., those sequence comparisons with low BLAST scores). These settings tended to generate a large number of smaller clusters, which may indicate fragmentation of larger clusters. It was decided it was better for these to be assigned their own class rather than risk splitting them inappropriately between the larger clusters. These settings also separated smaller classes with distinct biology such as the guanylyl cyclases, TGF-β-type receptor kinases, and myotubularins (Tables 2 and 3).
Table 2.
Summary of Protein Kinase Domain Containing Clusters Identified by TribeMCL
| cluster ID | riken | total | branch | cluster or branch description | Hanks' classification | domain associations | |
|---|---|---|---|---|---|---|---|
| Serine/Threonine Kinases (289) | 0.0 | 10 | 66 | SGK | serum glucocorticoid kinase | AGC-OTHER | |
| Ribos6 | Ribosomal S6 kinase | AGC-VI | |||||
| RAC | RAC | AGC-III | Pleckstrin like | ||||
| PKC | Protein kinase-C | AGC-II | PKC, C2 domain, PKN | ||||
| cNMP | cAMP/cGMP dependent PKs | AGC-I | cNMP binding | ||||
| Rho/myotonin | Rho/myotonin | AGC-VII | PKC, Pleckstrin, citron | ||||
| unknown1k | AGC-OTHER | PDZ | |||||
| aurora | aurora-related | OPK-I | |||||
| GPCR | G protein coupled receptor kinase | AGC-IV | regulator of G protein, GPCR kin | ||||
| 0.1 | 25 | 94 | death assoc | death associated kinases | CAMK-I | ||
| CAMKII | calcium/calmodulin-dependent kinase type II | CAMK-I | |||||
| CAMKI | calcium/calmodulin-independent kinase type I | CAMK-I | |||||
| unknown2k | snf1/ELKL/ink76/st22 | CAMK-II | UBQ assoc, kinassoc c-term | ||||
| unknown3k | pim | OPK-OTHER | UBQ assoc | ||||
| MAPKAPK | mitogen activated protein kinase activated protein kinase | CAMK-OTHER | |||||
| PKD/ULK | AGC-II/OPK-V | ||||||
| 0.2 | 13 | 72 | unknown4k | tousled, misc | OPK-VII/CAMK-1 | ||
| MAPK | mitogen activated protein kinase | CMGC-II | mapk | ||||
| CDK | cyclin dependent kinases | CMGC-I | |||||
| GSK, CKII | glycogen synthase kinase, casein kinase II | CMGC-OTHER/III/IV | |||||
| apop MAP3K | apoptosis regulating MAP3 kinase | OPK-IV | |||||
| 0.3 | 1 | 5 | unknown5k | OPK-OTHER | |||
| 0.4 | 13 | 52 | polo | polo like kinases | OPK_I | polo box | |
| MAP3K | mitogen activated protein kinase kinase kinase | OPK-IV | |||||
| NEK | NIMA expressed related kinases | OPK-V | |||||
| DUSP/MAP2K | dual specificity mitogen activated protein kinase kinases | OPK_II | |||||
| PAK/GCK | p21 activated kinases, Germinal center kinases | OPK-III | PAK, citron | ||||
| PAK | p21 activated kinases | OPK-III | PAK | ||||
| MAP4K | mitogen activated protein 4K | OPK-III | citron | ||||
| unknown6k | ste20 related? | OPK-III | |||||
| unknown7k | ste20 related? | OPK-III | |||||
| unknown8k | ste20 related? | OPK-III | |||||
| Tyrosine Kinases (123) | 1.0 | 0 | 8 | EGF/UFO/c-met | EGF/UFO/c-met receptor kinases | PTK-OTHER/XXI/X/XII | Ig, Fibronectin III |
| 1.1 | 7 | 50 | unknown9k | CAMK-I | |||
| RIPK | Receptor interacting PK | Ankyrin | |||||
| MAP3K/MLK | mitogen activated protein 3K, Mixed lineage kinase | OPK-XI | SH3 | ||||
| unknown10k | |||||||
| TIE | tunica interna endothelial cell kinase | PTK-XIII/XVIII | Fibronectin III, EGF like | ||||
| LIMK | lim motif containing kinases | ||||||
| RAF | RAF | OPK-VIII | PKC | ||||
| GFR | VEGFR, FGFR, PDGF | PTK-OTHER/XIV | IG various, RTKII | ||||
| 1.2 | 0 | 27 | unknown11k | SRC related | PTK-III/IV | SH2, SH3 | |
| SRC | SRC proto-onco | PTK-I | SH2, SH3 | ||||
| TEC/BTK | Bruton's tyrosine kinase | PTK-II/V | SH2, SH3, pleckstrin like, Tec/BTK | ||||
| leukocyte | PTK-XVII | RTKII | |||||
| insulin recep | insulin receptor and related kinases | PTK-XVI | RTKII, EGFR-L, furin, fibIII | ||||
| 1.3 | 3 | 28 | eph like | PTK-VIII | eph, RTK-V | ||
| eph receptors | ephrin-type a and B receptors | PTK-XI | eph, SAM, FIII, RTK-V | ||||
| JAK | Janus kinase | PTK-VII/VI | SH2, Band4.1 | ||||
| FAK | focal adhesion kinase | PTK-IX | Band4.1, focaladhesion | ||||
| 1.4 | 1 | 10 | unknown12k | PTK-XIX/XX | Ig, Frizzled, RTK-II | ||
| Other small classes | 2.0 | 1 | 13 | TGF-B recep | TGF-B, activin, BMP | OPK-IX | TGF-B R, TGF-b GS motif, Activin type II |
| 3.0 | 3 | 12 | CK1 | casein kinase 1 | OPK-XII | ||
| 4.0 | 1 | 12 | CLK | cdc-like kinase | CMGC-V | ||
| 5.0 | 2 | 8 | GC | guanylyl cyclase | GC, extracellual ligand binding recep | ||
| 6.0 | 1 | 7 | cdk4/6 | cyclin dep kin 4 and 6 | CMGC-I | ||
| 7.0 | 1 | 5 | cdc2-like | cdc2-like | CMGC-I | ||
| 8.0 | 3 | 5 | IRAK | IL-1 receptor associated | OPK-X | ||
| 9.0 | 1 | 4 | WNK | PK, lysine deficient | OPK-IV | ||
| 10 | 1 | 4 | cdk-like | cdk-like kinases | CMGC-I | ||
| 11 | 1 | 3 | GAK | cyclin G associated kinase | |||
| undefined | 16 | 76 | undefined | cluster <3 members | |||
| TOTAL | 104 | 561 |
Cluster ID refers to clusters identified by TribeMCL and CLUSTALV branches. Hanks' kinase class is provided to demonstrate overall agreement between the clustering (Hanks and Quinn 1991). Clusters labeled as unknown represent clusters in which a common underlying biology could not be identified readily for a given cluster
Table 3.
Summary of Protein Phosphatase Domain Containing Clusters Identified by TribeMCL
| Cluster ID | rik | all | branch | branch or cluster description | domain associations | |
|---|---|---|---|---|---|---|
| Tyrosine phosphatase (40) | 0.0 | 0 | 15 | recep 1 | Receptor tyrosine phosphatases—group 1 | Ig, F-III, MAM, tyro catalytic region |
| 0.1 | 1 | 5 | recep 2 | Receptor tyrosine phosphatases—group 2 | ||
| 0.2 | 1 | 7 | recep 3 | Receptor tyrosine phosphatases—group 3 | F-III, tyro catalytic region | |
| 0.3 | 0 | 7 | non-recep 1 | Non-receptor tyrosine phosphatases—group 1 | band4.1 | |
| 0.4 | 0 | 8 | non-recep 2 | Non-receptor tyrosine phosphatases—group 2 | ||
| DUSP (25) | 1.0 | 2 | 7 | DUSP | DUSP | DUSP |
| 1.1 | 3 | 13 | MKP | MAPK phosphatase | DUSP, Rhodanese like | |
| 1.2 | 0 | 5 | T-DSP | DUSP | ||
| PIP3 (9) | 2.0 | 0 | 9 | myotubularin | myotubularin PIP3 phosphatase | tyro/dusp/gram |
| Ser/Thr (9) | 3.0 | 0 | 9 | ser/thr | Ser/thr phos | Ser/thr phos, metallo-phosphoesterase |
| PP2c (9) | 4.0 | 2 | 9 | PP2c | Protein phosphatase 2c | PP2c |
| 5.0 | 0 | 5 | 4a/MPRL | phos 4a and prenylated | prenyl group binding site | |
| 6.0 | 1 | 4 | PTEN like | PTEN PIP3 phosphatase | ||
| 7.0 | 3 | 4 | cdc14 | cdc14 tyrosine phosphatases | ||
| 8.0 | 0 | 3 | sphingo | Acid Sphingomyelinase-like phosphodiesterase | Ser/thr phos, metallo-phosphoesterase | |
| 9.0 | 0 | 3 | mRNA capping | mRNA capping enzyme | Tyr phos and DUSP | |
| 10 | 0 | 3 | ser/thr 2B | ser/thr 2B phosphatase | Ser/thr phos, metallo-phosphoesterase | |
| 11 | 2 | 3 | unknown pp2c | pp2c-like | ||
| 12 | 0 | 3 | cdc25/MPI | cdc25 phosphatase/M-phase inducer phosphatase | M-phase inducer, rhodanese like | |
| undefined | 12 | 40 | undefined | clusters <3 members | ||
| TOTAL | 27 | 162 |
Cluster ID refers to clusters identified by TribeMCL and CLUSTALV branches
Cluster 11 is labeled as unknown pp2C, members contain a pp2C domain but no common underlying biology could be identified for this cluster
Cluster Analysis of Protein Kinases
Two major kinase clusters were identified, a serine/threonine kinase cluster containing 289 members, with 63 previously uncharacterized transcripts, and a tyrosine kinase cluster containing 123 members, with 11 previously uncharacterized transcripts. The remaining 149 sequences, of which 37 were uncharacterized previously, were split into 59 smaller clusters, including 30 singletons (Table 2). A number of these smaller clusters represent kinases and kinase-like proteins with specialized biology and domain architecture; these include a TGF-B receptor kinase cluster containing 13 members, 2 of which were uncharacterized previously (cluster 2.0), a casein kinase-1 cluster with 12 members and 4 previously uncharacterized transcripts (cluster 3.0), and a guanylyl cyclase cluster with 8 members and 2 previously uncharacterized transcripts (cluster 5.0).
To further segment the large serine/threonine kinase and tyrosine kinase clusters, we used CLUSTALV (Higgins et al. 1992) to produce multiple alignments, and then used the alignments to create neighbor-joining trees with 1000 bootstraps. The larger branches from these trees were then used to subclass these two major kinase classes. (To view where subclasses were derived from, please refer to the trees in Supplementary Figs. 1–4). The serine/threonine group split into five major subclusters, as did the tyrosine kinases. These subclusters could be split further into subfamilies that share a conserved set of domains and represent known kinase families (Table 2).
We took two of these subclusters as examples to demonstrate the clusters and their domain associations. We considered cluster 0.0, the first subcluster of the serine threonine group and cluster 1.3 of the tyrosine kinases (Table 2).
Within cluster 0.0, there are nine subfamilies, six of which appear to have domains that define them. The nine subfamilies are the ribosomal s6 kinases, the protein kinase C's, a rho/myotonin group, an unknown group, the G protein coupled receptor kinases, the serum glucocorticoid kinases, the RAC/AKT kinases, the cyclic nucleotide dependent kinases (cNMP; cAMP, and cGMP dependent), and the aurora-related kinases. The PKC sequences share a PKC domain (IPR002219), a C2 domain (IPR000008), and a PKN domain (IPR000861). The Rho/myotonin group is defined by a PKC domain, Pleckstrin-like domain (IPR001849), and citron-like domain (IPR001180), a subset of the unknown 1k group contains PDZ domains. The RAC group contains a pleckstrin-like domain, and finally, the cNMP-dependent group contains a cNMP-binding domain (IPR000595). For domain predictions on individual sequences, please refer to Supplementary Table 4, which shows the clusters and all of the InterPro domains found within each of the sequences.
Within the 1.3 cluster of the tyrosine kinases, there are four subfamiles, the ephrin receptors, the Janus kinases (JAK), the focal adhesion kinases (FAK), and a less-well defined group (EPH-like). The ephrin receptors are characterized by four InterPro motifs, these are ephrin (IPR001090), SAM— Sterile Alpha Male (IPR001660), FIII—Fibronectin type III repeats (IPR003961, IPR003962), and RTK-V—receptor-type kinase-V motifs (IPR001426). The EPH-like group consists of six members, two of which share the ephrin and RTK-V motifs of the ephrins, but lack the FIII or SAM domains. The Janus kinase group is defined by an SH2 (IPR000980) motif and a Band4.1 motif (IPR000299), and finally, the focal adhesion group contains a Band4.1 motif and a focal-adhesion targeting region (IPR005189) (Table 2).
Cluster Analysis of Phosphatases
As with the kinases, the phosphatase sequences produced 2 major clusters and 40 smaller clusters that included 19 singletons. The largest was a Tyrosine phosphatase cluster containing 42 members, 2 of which were novel transcripts; the next largest was a Dual specificity phosphatase cluster containing 25 members with 7 novel transcripts. The remaining 64 phosphatases contained 19 novel transcripts. The smaller classes include nine myotubularins, nine serine/threonine phosphatases, nine protein phosphatase 2Cs (two novel transcripts), and five protein tyrosine phosphatase 4a/MPRL (prenylated phosphatases) (Table 3).
Domain Associations Within the Kinases and Phosphatases
All sequences in this study had their corresponding InterPro domain annotations extracted. Supplementary Tables 3 and 4 provide extensive domain information and cluster ID for every sequence in this study. Tables 2 and 3 summarize the most common domain associations for each of the clusters.
A number of InterPro domains were associated with the eukaryotic kinases. Excluding the kinase domains used to identify these sequences, the next most common InterPro domains were SH2 (IPR000980), Immunoglobulin/MHC (IPR001452), SH3 (IPR001452), and Fibronectin, type-III repeats (IPR003961). These fall into two major categories, receptor-type domains and nonreceptor-type domains. Additional receptor-type domains observed and associated with specific groups were the various classes of immunoglobulin-like domains, receptor-type kinase domains, and the ephrin receptor-associated domains. The nonreceptor group appears to be enriched for interaction domains, most notably the SH2, SH3, PDZ (IPR001478), and pleckstrin-like domains (IPR001849), as well as citron (IPR001180) and ubiquitin-associated domains (IPR000449).
Similarly, within the phosphatases, two major groups could be identified. The most common domains associated with the phosphatases were the Fibronectin type-III repeats (IPR003961/IPR003962), Rhodanese-like domain (IPR001763), immunoglobulin-type domains (IPR003006/IPR003599/IPR003598), Band 4.1 (IPR000299), MAM (IPR000998), and GRAM domains (IPR004182). The FIII, Ig, and MAM domains were all associated with Receptor phosphatases. The Rhodanese-like domain is found in cdc25 phosphatases (m-phase inducing phosphatases) and a subset of the Dual specificity phosphatases, the MAPK phosphatases. The Band4.1 motif is associated with cytoskeletal interactors and the GRAM domain is known to occur in myotubularins (InterPro at EBI; http://www.ebi.ac.uk/InterPro/).
Misclassifications and Related Gene Families Detected by TribeMCL
Perhaps the greatest issue when using domain-based prediction is the identification of false positives. We encountered a number of nonphosphatase and nonkinase proteins that contained kinase or phosphatase InterPro domains. Within TribeMCL, these sequences clustered separately as small distinct clusters.
Proteins that catalyze very similar reactions to protein phosphatases, such as the PIP3 phosphatases, the myotubularins (cluster 2), and the PTEN-related genes (cluster 6) (Taylor et al. 2000, Maehama et al. 2001), represent one class of misclassification. Similarly, more distantly related enzymes, such as the mRNA-capping enzymes, mRNA 5′-triphosphatases (cluster 9) (Changela et al. 2001), acid sphingomyelinase-like phosphodiesterases (cluster 8) (Testi 1996), RNA lariat debranching enzyme (Kim et al. 2000), CD73-5' nucleotidase (Airas et al. 1997), and MRE11A (Hopfner et al. 2001) were all identified as containing protein phosphatase domains. Within the protein kinases, the guanylyl cyclases were also identified (cluster 5) (Lucas et al. 2000).
With the recognition that the data sets contained protein kinase-like and protein phosphatase-like sequences, we divided the data sets into two classes of trust. Clusters are labeled as protein kinase/phosphatase or protein kinase/phosphatase like. Clusters containing member sequences with direct evidence of protein kinase or protein phosphatase activity within the literature and no conflicting reports (as with the myotubularins) were labeled as protein-kinase or protein-phosphatase sequences. Clusters containing sequences in which the literature suggested that they had another role were labeled as protein kinase/phosphatase like. Clusters in which there was no evidence were considered kinase/phosphatase like for downstream analysis purposes. The trust assignments for each sequence cluster are available in supplementary Tables 1–4.
With these new definitions in place, the data sets split into 109 protein phosphatase, 53 protein phosphatase-like, 502 protein kinase, and 59 kinase-like sequences. When considering sequences for which transcripts have been detected, 13 of the 96 protein-phosphatase sequences and 87 of the 435 protein-kinase sequences are only supported by RIKEN transcripts; these represent 13.5% of all protein phosphatases and 20% of all protein kinases detected in the mouse transcriptome. Also, within the protein-kinase-like and protein-phosphatase-like groups, RIKEN transcripts were responsible for 34.7% and 29.8% of these sequences, respectively.
Evaluation of TribeMCL
Examining the clusters obtained by TribeMCL, we observed a number of smaller clusters that seem to have been split from the larger clusters. The cyclin-dependent kinases CDK4 and CDK6 (cluster 6.0) cluster away from the remaining CDKs (subcluster 0.2); whether this represents the true situation is debatable. Similarly, a number of the smaller phosphatase clusters (clusters 10.0 and 11.0) seem to represent proteins with similar domains to larger clusters. There are two Protein phosphatase 2c-type clusters (4.0 and 11.0) and two serine/threonine-type clusters (3.0 and 10.0) (Table 3).
This suggests that the granularity settings used in our analysis may have been set too fine. However, this level of granularity has been important in separating misclassifications from genuine hits (see previous section). Sequences placed within the smaller clusters or classified as singletons represent small classes with distinct biology.
Comparison to Ensembl
As mentioned previously, we identified 561 protein kinase and 162 protein phosphatase, related sequences. A total of 541 of the protein kinase-related sequences and 158 of the phosphatase-related sequences mapped to Ensembl genes. A total of 77 of the Ensembl kinase-related genes are only represented by a genomic prediction. Conversely, 20 kinase-related sequences did not map to Ensembl genes. This indicates an advantage in using a transcriptome-based screen, it is not dependent upon gene predictions. As to whether the 77 Ensembl predictions represent true genes, we will have to wait for transcriptome data to confirm their expression. A list of the sequences that failed to map to an Ensembl gene is available as Supplementary Table 5.
Comparison to Previously Published Classification Schemes
The clustering used in this work, on the basis of whole protein homology, presents an alternative to traditional domain-based homology assignments. A recent global analysis of the human members of the protein-kinase superfamily identified 510 candidate kinase sequences (Kostich et al. 2002), we identified a similar number of sequences, 502 protein kinases, and 59 kinase-like sequences. The study by Kostich et al. (2002), as does most other analyses, uses phenograms constructed using the catalytic domain of the kinase for the alignment and tree assignments. This approach captures relations dependent on the catalytic domain, however, it ignores the effect of other domains that may be important for the biology of the whole protein. Perhaps surprisingly, we obtained similar classifications to those obtained by Kostich et al. (2002) and the earlier Hanks' classification scheme (Hanks and Quinn 1991; Protein Kinase Resource http://pkr.sdsc.edu/html/pk_classification/pk_catalytic/pk_hanks_class.html).
The most notable difference was the splitting of the cyclin-dependent kinases. Both the Hanks' scheme and that of Kostich et al. (2002) place the CDKs together, however, the TribeMCL clustering separated the major CDK group (placed within cluster 0.2 of the serine threonine kinases) from three smaller CDK-related clusters (clusters 6.0, 7.0, and 10.0). The reasons for this difference are not clear, however, the other small TribeMCL clusters (2, 3, 5, 8, 9, and 11) represent proteins with specialized roles (Table 2). The fragmentation of the CDKs may reflect an underlying difference in their biology, however, an artefact of the clustering cannot be ruled out. These clusters contain a large number of genomic predictions. In cluster 6.0, four of the seven genes are predicted. Similarly, in cluster 7.0, three of five genes are predicted. In both cases, these are predicted genes from the Ensembl family ENSMUSF00000000078. Whether these predictions are somehow skewing the clusters is unknown.
For the mammalian protein phosphatases, a similarly comprehensive classification scheme is not available. A recent whole-genome analysis on the protein phosphatase catalytic subunits of Arabidopsis identified 112 candidate phosphatase sequences (Kerk et al. 2002); these were split into 69 pp2c, 18 DUSP, 23 serine/threonine, 1 tyrosine, and 1 low molecular weight tyrosine phosphatase. The small number of tyrosine phosphatases in Arabidopsis does not reflect the situation in mouse in which the tyrosine phosphatases represent the largest class of phosphatases. To assess the quality of the clustering for the tyrosine phosphatase clusters, we compared them with those identified by Andersen et al. (2001) (http://science.novonordisk.com/PTP/database.asp). There was good overall agreement with clusters separating into receptor type and nonreceptor type phosphatases, however, both cluster 0.2 and 0.4 contained both receptor and nonreceptor-type members.
Novel Phosphoregulators Within the FANTOM2 Libraries
To assess the impact of the RIKEN FANTOM2 sequences, we only considered those for which the FANTOM sequence was the only source of a predicted peptide. Many of the other sequences presented here are also identified by RIKEN, but there is supporting evidence from another source. Some of these are also novel sequences, however, for the purpose of simplifying this assessment, these are considered as separate.
Starting with the phosphatase domain-containing sequences, we identified 8 homologs and 19 novel transcripts (Table 3). Novel transcripts for one MAPK phosphatase and two HSSH/slingshot-related dual-specificity phosphatases were identified in cluster 1. A transcript for a novel pp2c type phosphatase was identified in cluster 4. We also identified three smaller phosphatase-like clusters containing novel transcripts.
Cluster 6 contains PTEN (phosphatase and tensin)-like proteins. PTEN is an important PIP3 phosphatase. The cluster contained PTEN and an additional three proteins, one prediction for a cyclin G-associated kinase (human GAK has a tensin and phosphotyrosine motif—O14976), one known, annotated as tyrosine phosphatase and one novel transcript with tensin homology. As to whether these constitute members of the PTEN family is not clear.
Cluster 7 contains four cdc14 protein tyrosine phosphatase sequences, one of these is a predicted gene by Ensembl, one is a homolog of a human gene, and two are novel transcripts. Cluster 11 contains three sequences of unknown function, they all contain a protein phosphatase-2C motif, they cluster separately from the pp2c phosphatase cluster 4 and separately from the structurally similar pp2c-like, pyruvate dehydrogenase phosphatase cluster 17. The remaining novel transcripts include a protein that clusters with the ecto-5′ nucleotidase, CD73 (cluster 15), and two transcripts with similarity to dual-specificity phosphatase DUSP13.
Within the serine/threonine kinase cluster, there were 62 RIKEN-only sequences, 28 of these are completely novel transcripts. These novel transcripts include a doublecortin domain-containing kinase, SNF1-related kinases, calcium/calmodulin-dependent kinases, MAPK-activated protein kinases (cluster 0.1), nima-related kinases, and p21-activated kinases (cluster 0.4). Within the tyrosine kinase cluster, there were 11 RIKEN only sequences, 6 of which are novel transcripts, including an ephrin-like kinase (cluster 1.3).
Within the remaining sequences, there were a number of clusters containing homologs and novel transcripts. Clusters and novel transcripts of note include a casein-kinase 1 cluster (3), containing 12 CK1-related sequences; we identified three homologs not observed previously in mouse, a novel kinase related to EIF-2 α kinase (cluster 20), and an Il-1 receptor-associated kinase (IRAK) cluster (8) containing a hypothetical predicted by Ensembl, two novels, a homolog, and a known.
Mouse Phosphoregulators
An advantage of examining both the protein kinases and protein phosphatases of mouse was to glimpse a global view of how these enzymes, which catalyze opposing reactions, could operate throughout the cell and how they are used in so many control systems. The most obvious observation was the high number of kinases in comparison to phosphatases. This is also the case in other eukaryotes, including yeast and man. A number of workers have commented on this previously, with the suggestion that protein kinases have higher specificities than protein phosphatases, however, this has been challenged (Zhang et al. 2002). There is clear evidence of substrate specificity by some phosphatases; consider the MKPs (map kinase phosphatases), which are specific for MAP kinases, and the cdc25 phosphatases specific to cyclin/CDK complexes.
The simplest explanation for this observation, however, is that a phosphatase is only required in systems in which the target protein needs to return to an unphosphorylated state. As mentioned in the introduction, there are alternative control mechanisms such as secondary modifications, which can modulate the activity of a phosphorylated protein, or in the case of ubiquitination, target it for destruction. Examples in which dephosphorylation is likely to be important are ones in which a protein has a long half life, and during that half life, it needs to cycle dynamically between phosphorylated and unphosphorylated forms; alternatively, cases in which there is a need for very fine grain control, opposing kinases, and phosphatases may compete to determine the activity of a given protein. An example of this is the competing activities of Wee1 kinase and cdc25B phosphatase on mitotic cyclin/cdk complexes (Russell and Nurse 1986).
Conclusion
The FANTOM2 project highlights the advantages of a transcriptome-based approach. FANTOM2 has been able to confirm transcript prediction from the genome, but additionally, it has identified transcripts missed by the Ensembl geneprediction pipeline. Some cases confirmed a Genscan prediction, whereas others identified completely undetected transcripts. Gene prediction algorithms applied to genomic sequence generally depend on a gene model, transcripts that break the model enhance our understanding of genes and how to better model them. This highlights the complementary nature of the transcriptome and the genome. Transcriptional evidence is necessary for defining coding regions within the genome and confirming genomic predictions. Conversely, genomic sequence is necessary for identifying potential transcripts and determining gene structure, in particular, exon-intron boundaries and promoter sites.
By mining sequence annotations for specific InterPro motifs, we have identified 561 candidate protein kinases and 162 protein phosphatases. Using TribeMCL protein sequence similarity clustering, we were able to separate the different classes of protein kinase and phosphatase within the sets. TribeMCL also provided a certain level of quality control to the classes assigned by InterPro motif detection, by allowing us to distinguish true kinase and phosphatase members from related gene family members. This separated the sets into 502 likely protein kinases and 96 likely protein phosphatases. Finally, the FANTOM libraries have provided us with the only transcript evidence for 13.5% of the protein phosphatases and 20% of the protein kinases described in the transcriptome. This represents a great resource, and the availability of full-length cDNAs from the FANTOM libraries will provide valuable clones necessary for functional confirmation of these genes in the future.
METHODS
Nonredundant Protein Sequences Used in This Analysis: RPS3, SWALL, and IPI
The RTPS group at RIKEN produced a nonredundant Representative Protein Set RPS3 that incorporates sequences from the public domain and the RIKEN FANTOM2 libraries. During the course of this study, the RPS set has undergone a number of updates; it currently stands at RPS 6 (http://fantom2.gsc.riken.go.jp). SWALL and the International Protein Index (IPI; http://www.ebi.ac.uk/IPI/) represent nonredundant protein sets available from EBI (http://srs.ebi.ac.uk). IPI is currently available for mouse and human, and merges entries from Ensembl, SWISS-PROT, TrEMBL, and REFSEQ. There is a large overlap between IPI and SWALL, however, SWALL has more partial sequences and does not incorporate Ensembl gene predictions.
Identification of Protein Kinases and Protein Phosphatases Within the FANTOM2 EST Sequences
Preceding the nonredundant kinase and phosphatase sets we detail in this work, a primary analysis was carried out on the raw FANTOM2 data that indicated a number of novel kinase and phosphatases were present. All sequences predicted by InterPro as containing a protein-kinase domain or a protein-phosphatase domain (Table 1) were inspected manually within the MATRICS annotation viewer and checked for similarity to known proteins. A small number of partial sequences identified in the curation phase were excluded from RPS3 (RPS had a requirement that the sequences be full length). These sequences were added back into the analysis prior to mapping.
Mapping of Mouse Kinase and Phosphatase Sequences to a Mock Locus
As detailed in the Results section, sequences identified from the nonredundant protein databases, SWALL, IPI, and RPS3, were merged. Where possible, sequences were assigned Ensembl identifiers or MGI loci. These were extracted from cross-references found in the sequence entry, and if entries from different databases were derived from the same SWISS-PROT, TrEMBL, or REFSEQ entry, they also inherited the Ensembl or MGI locus. Those sequences for which there was no mapping information had their underlying EST sequence extracted. This EST sequence was then used in a BLASTN alignment with the Ensembl gene predictions. The best three hits for each EST were examined manually. In cases in which the alignment was convincing, the sequence entry was assigned the Ensembl gene to which it hit. Similarly, those sequences that failed to hit an Ensembl gene were used in a BLASTN alignment of the Ensembl Genscan predictions. Those sequences that failed to hit either, were assigned the original identifying EST accession number. The Ensembl sequences used are available for download from http://www.ensembl.org/Mus_musculus. The standalone version of BLAST used was BLASTALL available at http://www.ncbi.nih.gov/BLAST.
Selection of Representative Sequences for Downstream Analysis
After assigning all of the sequences to a mock locus, it was possible to sort the sequences and choose a representative. The longest peptide sequence that mapped to a given locus was extracted for cluster analysis. Annotations were examined from all sequences that mapped to the same locus. If the annotations generally agreed, the most informative annotation was taken.
Domain Architecture
InterPro domain predictions for all sequences used in this analysis were extracted from the database from which the sequence originated. Domain predictions on sequences from the SWALL and IPI sets were extracted using the sequence retrieval system SRS at EBI (Etzold and Argos 1993; http://srs.ebi.ac.uk).
TribeMCL Clustering
The representative sequences for each family were clustered using TribeMCL (Enright et al. 2002; http://www.ebi.ac.uk/research/cgg/tribe). The sequences were compared against themselves using an all-against-all BLASTP comparison. In this work, we used an expectation value cutoff of e-10 and used the BLOSUM62 matrix. The results of this blast comparison was then parsed and clustered by TribeMCL. We used an inflation value of five within TribeMCL to increase the granularity of the classifications. Mapping of cluster results to descriptions and domain combinations was carried out in Microsoft Excel.
CLUSTALV Clustering of Large TribeMCL Clusters
CLUSTALV (Higgins et al. 1992) was used to segment the serine/threonine kinase cluster, the tyrosine-kinase cluster, the tyrosine-phosphatase cluster, and the dual-specificity phosphatase cluster produced by TribeMCL. CLUSTALV was first used to produce multiple alignments for sequences within the larger clusters; it was then used to create phylogenetic trees using the Neighbor joining option, with 1000 bootstraps. The larger branches within these trees were then used to subclass the TribeMCL clusters.
Database Versions
Ensembl mouse release (v. 7.3b.3 12 July 2002). SWALL is updated weekly and IPI is updated monthly, the versions used to produce the nonredundant protein-kinase and protein-phosphatase sets were indexed the week of June 28 2002. RPS3—representative protein set 3 (http://fantom2.gsc.riken.go.jp).
InterPro Assignments
As detailed within the results, InterPro assignments for the initial identification of candidate sequences and for the later domain associations were extracted from the previously annotated data sets. This includes IPI, SWALL, FANTOM2, and RPS3. These assignments had been made using InterProScan, which are detailed at http://www.ebi.ac.uk/interpro.
Acknowledgments
We thank the RIKEN Genome Exploration Research Group Phase I & II Team, Genomic Sciences Center, RIKEN, and the FANTOM consortium members. The Representative Protein Set RPS3 used in these analyses was generated by the RTPS group (RIKEN and FANTOM). The InterPro predictions were performed by Alexander A. Kanapin of the European Bioinformatics Institute.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.954803.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Ahmed, K., Gerber, DA., and Cochet, C. 2002. Joining the cell survival squad: An emerging role for protein kinase CK2. Trends Cell. Biol. 12: 226-230. [DOI] [PubMed] [Google Scholar]
- Airas, L., Niemela, J., Salmi, M., Puurunen, T., Smith, D.J., and Jalkanen, S. 1997. Differential regulation and function of CD73, a glycosyl-phosphatidylinositol-linked 70-kD adhesion molecule, on lymphocytes and endothelial cells. J. Cell. Biol. 136: 421-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, J.N., Mortensen, O.H., Peters, G.H., Drake, P.G., Iversen, L.F., Olsen, O.H., Jansen, P.G., Andersen, H.S., Tonks, N.K., and Moller, NP. 2001. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol. Cell. Biol. 21: 7117-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler, R., Attwood, T.K., Bairoch, A., Bateman, A., Birney, E., Biswas, M., Bucher, P., Cerutti, L., Corpet, F., Croning, M.D.R., et al. 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29: 37-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake, J.A., Richardson, J.E., Bult, C.J., Kadin, J.A., Eppig, J.T., and the Mouse Genome Database Group. 2002. The Mouse Genome Database (MGD): The model organism database for the laboratory mouse. Nucleic Acids Res. 30: 113-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changela, A., Ho, C.K., Martins, A., Shuman, S., and Mondragon, A. 2001. Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme. EMBO J. 20: 2575-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb, M.H. 1999. MAP kinase pathways. Prog. Biophys. Mol. Biol. 71: 479-500. [DOI] [PubMed] [Google Scholar]
- Dan, I., Watanabe, N.M., and Kusumi, A. 2001. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell. Biol. 11: 220-230. [DOI] [PubMed] [Google Scholar]
- Davezac, N., Baldin, V., Gabrielli, B., Forrest, A., Theis-Febvre, N., Yashida, M., and Ducommun, B. 2000. Regulation of CDC25B phosphatases subcellular localization. Oncogene 19: 2179-2185. [DOI] [PubMed] [Google Scholar]
- Ding, Y. and Dale, T. 2002. Wnt signal transduction: Kinase cogs in a nano-machine? Trends Biochem. Sci. 27: 327-329. [DOI] [PubMed] [Google Scholar]
- Drewes, G., Ebneth, A., Preuss, U., Mandelkow, E.M., and Mandelkow, E. 1997. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell 89: 297-308. [DOI] [PubMed] [Google Scholar]
- Enright, A.J. 2002. “Computational analysis of protein function in complete genomes.” Ph.D. thesis, pp. 60-76. University of Cambridge, Cambridge, UK.
- Enright, A.J., Van Dongen, S., and Ouzounis, C.A. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30: 1575-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzold, T. and Argos, P. 1993. SRS–an indexing and retrieval tool for flat file data libraries. Comput. Appl. Biosci. 9: 49-57. [DOI] [PubMed] [Google Scholar]
- The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I and II Team. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563-573. [DOI] [PubMed] [Google Scholar]
- Hanks, S. and Quinn, A.M. 1991. Protein kinase catalytic domain sequence database: Identification of conserved features of primary structure and classification of family members. Meth. Enzymol. 200: 38-62. [DOI] [PubMed] [Google Scholar]
- Higgins, D.G., Bleasby, A.J., and Fuchs, R. 1992. CLUSTAL V: Improved software for multiple sequence alignment. Cabios 8: 189-191. [DOI] [PubMed] [Google Scholar]
- Hofmann, T.G., Moller, A., Sirma, H., Zentgraf, H., Taya, Y., Droge, W., Will, H., and Schmitz, M.L. 2002. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 4: 1-10. [DOI] [PubMed] [Google Scholar]
- Hopfner, K.P., Karcher, A., Craig, L., Woo, T.T., Carney, J.P., and Tainer, J.A. 2001. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105: 473-485. [DOI] [PubMed] [Google Scholar]
- Kerk, D., Bulgrien, J., Smith, D.W., Barsam, B., Veretnik, S., and Gribskov, M. 2002. The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol. 129: 908-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.W., Kim, H.C., Kim, G.M., Yang, J.M., Boeke, J.D., and Nam, K. 2000. Human RNA lariat debranching enzyme cDNA complements the phenotypes of Saccharomyces cerevisiae dbr1 and Schizosaccharomyces pombe dbr1 mutants. NAR 28: 3666-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koretzky, G.A., Picus, J., Schultz, T., and Weiss, A. 1991.Tyrosine phosphatase CD45 is required for T-cell antigen receptor and CD2-mediated activation of a protein tyrosine kinase and interleukin 2 production. Proc. Natl. Acad. Sci. 8: 2037-2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostich, M., English, J., Madison, V., Gheyas, F., Wang, L., Qiu, P., Greene, J., and Laz, T.M. 2002. Human members of the eukaryotic protein kinase family. Genome Biol. 3: Research0043 [DOI] [PMC free article] [PubMed]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Larochelle, S. and Suter, B. 1995. The Drosophila melanogaster homolog of the mammalian MAPK-activated protein kinase-2 (MAPKAPK-2) lacks a proline-rich N-terminus. Gene 163: 209-214. [DOI] [PubMed] [Google Scholar]
- Lehman, J.A. and Gomez-Cambronero, J. 2002. Molecular crosstalk between p70S6k and MAPK cell signaling pathways. Biochem. Biophys. Res. Commun. 293: 463-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, A. and Blow, J.J. 2001. The origin of CDK regulation. Nat. Cell. Biol. 3: E182-E184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, K.A., Pitari, G.M., Kazerounian, S., Ruiz-Stewart, I., Park, J., Schulz, S., Chepenik, K.P., and Waldman, S.A. 2000. Guanylyl cyclases and signaling by cyclic GMP. Pharmacol. Rev. 52: 375-414. [PubMed] [Google Scholar]
- Maehama, T., Taylor, G.S., and Dixon, J.E. 2001. PTEN and myotubularin: Novel phosphoinositide phosphatases. Annu. Rev. Biochem. 70: 247-279. [DOI] [PubMed] [Google Scholar]
- Nigg, E.A. 2001. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell. Biol. 2: 21-32. [DOI] [PubMed] [Google Scholar]
- Pierantoni, G.M., Bulfone, A., Pentimalli, F., Fedele, M., Iuliano, R., Santoro, M., Chiariotti, L., Ballabio, A., and Fusco, A. 2002. The homeodomain-interacting protein kinase 2 gene is expressed late in embryogenesis and preferentially in retina, muscle, and neural tissues. Biochem. Biophys. Res. Commun. 290: 942-947. [DOI] [PubMed] [Google Scholar]
- Price, MA. and Kalderon, D. 2002. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by Glycogen Synthase Kinase 3 and Casein Kinase 1. Cell 108: 823-835. [DOI] [PubMed] [Google Scholar]
- The RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium. 2001. Functional annotation of a full-length mouse cDNA collection. Nature 409: 685-690. [DOI] [PubMed] [Google Scholar]
- Russell, P. and Nurse, P. 1986. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell 45: 145-153. [DOI] [PubMed] [Google Scholar]
- Taylor, G.S., Maehama, T., and Dixon, J.E. 2000. Inaugural article: Myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc. Natl. Acad. Sci. 97: 8910-8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testi, R. 1996. Sphingomyelin breakdown and cell fate. Trends Biochem. Sci. 21: 468-471. [DOI] [PubMed] [Google Scholar]
- Theodosiou, A. and Ashworth, A. 2002. MAP kinase phosphatases. Genome Biol. 3: Reviews3009.1-Reviews3009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima, F., Moriguchi, T., Wada, A., Fukuda, M., and Nishida, E. 1998. Nuclear export of cyclin B1 and its possible role in the DNA damage-induced G2 checkpoint. EMBO J. 17: 2728-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dongen, S. 2000. “Graph clustering by flow simulation.” PhD Thesis, University of Utrecht, The Netherlands.
- Yaffe, M.B. 2002. How do 14-3-3 proteins work?—Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 513: 53-57. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Zhou, B., and Xie, L. 2002. Modulation of protein kinase signalling by protein phosphatases and inhibitors. Pharmacol. Therap. 93: 307-317. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://fantom2.gsc.riken.go.jp; Functional annotation of mouse, RIKEN.
- http://pkr.sdsc.edu/html/pk_classification/pk_catalytic/pk_hanks_class.html; Hanks' kinase classification at The Protein Kinase Resource.
- http://science.novonordisk.com/PTP/database.asp; Structural and evolutionary relationships among protein tyrosine phosphatase domains. [DOI] [PMC free article] [PubMed]
- http://scop.mrc-lmb.cam.ac.uk/scop; Structural classification of proteins home page.
- http://srs.ebi.ac.uk; The Sequence retrieval system (SRS6).
- http://www.ebi.ac.uk/interpro; The InterPro home page.
- http://www.ebi.ac.uk/IPI/; International protein Index.
- http://www.ebi.ac.uk/research/cgg/tribe; Protein sequence clustering—TribeMCL.
- http://www.ensembl.org/Mus_musculus; Ensembl mouse genome home page.
- http://www.informatics.jax.org/; Mouse Genome Informatics (MGD).
- http://www.ncbi.nih.gov/BLAST; Basic local alignment search tool home page. [DOI] [PubMed]