Abstract
The transcriptional coactivator and histone acetyltransferase (HAT) p300 acetylates the four core histones and other transcription factors to regulate a plethora of fundamental biological processes including cell growth, development, oncogenesis and apoptosis. Recent structural and biochemical studies on the p300 HAT domain revealed a Theorell-Chance, or “hit-and-run”, catalytic mechanism. Nonetheless, the chemical mechanism of the entire reaction process including the proton transfer (PT) scheme and consequent acetylation reaction route remains unclear. In this study, a combined computational strategy consisting of molecular modeling, molecular dynamic (MD) simulation and quantum mechanics/molecular mechanics (QM/MM) simulation was applied to elucidate these important issues. An initial p300/H3/Ac-CoA complex structure was modeled and optimized using a 100 ns MD simulation. Residues that play important roles in substrate binding and the acetylation reaction were comprehensively investigated. For the first time, these studies reveal a plausible PT scheme consisting of Y1394, D1507 and a conserved crystallographic water molecule, with all components of the scheme being stable during the MD simulation and the energy barrier low for PT to occur. The two-dimensional potential energy surface for the nucleophilic attack process was also calculated. The comparison of potential energies for two possible elimination half-reaction mechanisms revealed that Y1467 reprotonates the coenzyme-A leaving group to form product. This study provides new insights into the detailed catalytic mechanism of p300 and has important implications for the discovery of novel small molecule regulators for p300.
Keywords: Epigenetics, p300, Proton Transfer, Histone Acetylation, QM/MM
Introduction
Histone modifications such as methylation, acetylation, ADP-ribosylation, ubiquitination, citrullination, and phosphorylation regulate specific and distinct functional outputs of genomes at the transcription level1, 2. Among all these modifications, histone acetylation mediated by histone acetyltransferases (HATs), which occurs both on the unstructured tails and the globular core domains of histones, controls many aspects of chromatin structure and function3–6. In addition, HATs also exert various influences on the genome by catalyzing the lysine acetylation of many other cellular proteins including themselves7–9. Different HAT members show varying degrees of substrate specificity, a property consistent with a role in maintaining a dynamic, acetylation-based epigenetic code3, 10. Moreover, recent evidence raises an interesting possibility that an acetylation-based code may operate through both mitosis and meiosis, providing a possible mechanism for germ-line transmission of epigenetic changes10–13.
P300, one of the major HAT proteins as it controls the expression of numerous genes in a cell-type-specific and signal-dependent manner. Accordingly, p300 has been shown to play an important role in cellular proliferation, apoptosis, and embryogenesis14–16. More recently, p300 has been shown to play a role in regulating embryonic stem (ES) cell pluripotency by modulating the expression of the pluripotency marker Nanog17, and to modulate the specification of liver progenitors over pancreas progenitors18. Additional data demonstrates that abnormal p300 function is associated with deregulated target gene expression, and is implicated in inflammation, cancer, cardiac hypertrophy, and genetic disorders such as the Rubinstein-Taybi syndrome, making p300 an attractive therapeutic target19–21. Because of these associations, small molecular p300 modulators may apply to determining the direction of stem cell differentiation18, be useful as probes to study p300 function22–25 for developing new chemical entities for therapeutic use26–28. Rational design of small molecules to interfere with p300 function on the basis of the catalytic mechanism and transition state structures may represent a feasible and efficient way to achieve these goals29.
Most HATs utilize two distinct mechanisms to catalyze the acetyl-transfer reaction. One is ternary complex mechanism, in which the cofactor acetyl coenzyme A (Ac-CoA) and the substrate bind to the enzyme simultaneously followed by the acetyl transfer reaction in the HAT/Ac-CoA/substrate ternary complex30. The other mechanism is a ping-pong mechanism, in which the acetyl group is covalently attached to the enzyme before being transferred to the substrate31. One major HAT family, GCN5, was shown to use the ternary complex mechanism32–34, whereas another major HAT family, MYST, was shown to employ the ping-pong mechanism35 or the ternary complex mechanism36. Intriguingly, it was reported that neither of these mechanisms applies to p300. Instead, p300 was shown to use a Theorell-Chance, or “hit-and-run”, catalytic mechanism37. As a special case of the ternary mechanism, ternary complex is also generated in Theorell-Chance mechanism. While distinct from the classic ternary mechanism, Theorell-Chance is characterized that the ternary complex never accumulates and the steady-state concentrations of the ternary complex is kinetically insignificant. Following the association of Ac-CoA, the protein substrate binds transiently to the p300 surface, allowing the lysine residue to snake through the enzyme active site to receive the acetyl group, followed by rapid protein dissociation. However, answers to several important questions regarding the detailed molecular mechanism of catalysis remain unanswered: 1)how does the lysine substrate bind to the enzyme; 2) how is the ε-amino group of the substrate lysine deprotonated prior to the reaction; 3) what is the chemical pathway for the entire catalytic process and which structural elements of p300 are critical for this process.
In this study, a p300/H3/Ac-CoA ternary structure model was constructed on the basis of the crystal structures of the p300/Lys-CoA and GCN5/H3/Ac-CoA complexes. Then a 100 ns MD simulation was performed on the complex model to obtain a suitable structural configuration for further investigations. The recognition network of substrate H3 was analyzed and compared with the experimental data. Meanwhile, two possible proton transfer (PT) schemes were proposed based on structural analysis, and the scheme consisting of Y1394, D1507 and a crystallographic water molecule important for the PT was validated by MD simulations and QM/MM calculations. Subsequently, the entire acetylation reaction pathway proceeding through a ternary complex was fully investigated by QM/MM calculations. Two residues in the active site that were previously shown to be important for catalysis, Y1467 and W1436, were also investigated. In summary, these computational studies provide a deeper understanding of Ac-CoA cofactor and lysine substrate binding, the PT pathway and the acetylation reaction mechanism of p300 and its related HAT proteins. The snapshots of the transition states of the acetyl-transfer reaction offers important insights for the design of small molecule modulators for regulating p300 function.
Experimental section
Preparation of the simulation system
To date, the only crystal structure containing p300 is that of the p300/Lys-CoA complex. No p300/Ac-CoA/peptide complex structure is available. Therefore, we modeled a p300/H3/Ac-CoA complex based on the crystal structures of p300/Lys-CoA (PDB code: 3BIY)37 and GCN5/H3/Ac-CoA38 (PDB code: 1PU9) complexes. First, the lysine portion of the Lys-CoA was modified to an acetyl group to generate a p300/Ac-CoA structure. Second, the crystal structure of the GCN5 HAT bound to a 15-residue H3 peptide was aligned to p300/Ac-CoA by structural alignment with PyMOL plugin CEAlign39, then the H3 peptide was extracted to construct the p300/H3/Ac-CoA complex model. Since the H3 peptide is involved in the catalytic reaction of p300. and whether it locates on the top of the L1 loop or bound to the bottom of the binding site is still a question, therefore, we performed a series computational and structural analysis to select the H3 peptide conformation (see in the section of “The model selection of H3 conformation in the structure of the p300/H3/Ac-CoA model” in supporting information). Briefly, considering the structural and biophysical factors, the H3 substrate peptide binding to a shallow pocket on the top L1 loop will be modeled for further analysis.
The autoacetylation loop, which consists of L1520–L1580, was genetically deleted during crystallization and the resulting p300 HAT domain showed comparable acetyltransferase activity with the intact enzyme, thereby we didn’t include it in our model40, 41, 37. This model was then minimized using the AMBER force field42 with the following parameters: a distance-dependent dielectric function, a non-bonded cutoff of 8 Å, amber charges for the enzyme and H3 peptide, and Gastieger-Hückel charges for Ac-CoA and water molecules. The structures were minimized by the simplex method, followed by the Powell method to an energy gradient of < 0.05 kcal/(mol·Å). All procedures were performed using the Sybyl software package (Tripos, St. Louis, MO).
MD simulations
A 100 ns MD simulation was performed on the p300/H3/Ac-CoA system. Before simulations, this system was put into a suitably sized box with a 10 Å buffer distance between the solvent box wall and the nearest solute atoms. Then the box was solvated with the SPC (Simple Point Charge) water model. Each solvated system was submitted to energy minimization. Afterwards, counterions were added to the system to provide a neutral simulation system. The whole system was subsequently minimized again. The charges of the atoms of Ac-CoA were calculated by using the RESP method43 encoded in the AMBER suite program44 at the level of RHF/6-31G*. Covalent and non-bonded parameters for the ligand atoms were assigned, by analogy or though interpolation, from those already present in the AMBER force field.
MD simulations were carried out using the AMBER package (Version 10.0) with constant temperature, constant pressure (NPT), and periodic boundary conditions. The AMBER Parm99 force field42 was applied for the enzyme and H3 peptide. The Particle Mesh Ewald method45 was used to calculate the long-range electrostatics interactions. The non-bonded cutoff was set to 10.0 Å, and the non-bonded pairs were updated every 25 steps. The SHAKE method46 was applied to constrain all covalent bonds involving hydrogen atoms. Each simulation was coupled to a 300 K thermal bath at 1.0 atm of pressure (1 atm=101.3 kPa) by applying the algorithm of Berendsen et al47. The temperature and pressure coupling parameters were set as 1 ps. An integration step of 2 fs was set up for the MD simulations.
QM/MM calculations
The hybrid QM/MM mechanical approaches have been widely employed to enzymatic reaction studies48, 49. In this research, QM/MM calculations were performed by using a two-layered ONIOM method9 encoded in the Gaussian 03 program. In this approach, the molecular system under study is divided into an inner layer and an outer layer. The inner layer consists of the most critical elements of the system, and the rest of the system comprises the outer layer. the full system is called “real” and is treated with a low level of theory. The inner layer is termed “model” and is treated with both a low and a high level of theory. The total ONIOM energy EONIOM is given as following:
| (1) |
where E(high, model) is the energy of the inner layer (plus the link atoms) at the high level of the theory, E(low, real)is the energy of the entire system at the low level of theory, and E(low, model) is the energy of the model system at the low level of theory. Thus, the ONIOM method allows one to perform a high-level calculation on just a small, critical part of the molecular system and incorporate the effects of the surrounding elements at a lower level of the theory to yield a consistent energy expression on the full system.
The minimized p300/H3/Ac-CoA complex system optimized using the AMBER Parm99 force field was further optimized at the ONIOM (B3LYP/6-31G*:Amber) level to investigate the PT and acetylation mechanism. The quantum mechanics (QM) region for the PT consists of the methylene amonium CH2-CH2-NH3+ group of H3K14, the water molecule in the transfer wire (Wat1861 in the crystal structure), the side chain of Y1394 and D1507 (the proton shuttling residues), and the backbone of I1395 and W1436 (h-bonding with the proton shutting residues). The QM region for the acetylation reaction consists of the deprotonated side chain (-CH2-CH2-NH2) of H3K14, the -CH2-S-COCH3 group of Ac-CoA, the main chain of W1436, and side chain of Tyr1467. Each QM region was calculated by using the density functional theory with the B3LYP exchange-correlation functional and 6-31G* basis set. The remainder of the system (MM region) was treated with the AMBER Parm99 force field. A total of 5,512 atoms were included for the QM/MM calculations by using the ONIOM module as implemented in Gaussian 03. The electrostatic interactions between the QM and MM regions were calculated by using the electronic embedding method, which treats the polarization of the QM region by the MM region with scaled partial atomic charges of MM atoms, and the response of the QM region with the Merz-Singh-Kollman scheme for charge fitting so as to produce the changing partial charges of the QM atoms.
Results and discussions
The overall structure of the p300/H3/Ac-CoA model
Initially, the p300/H3/Ac-CoA model was constructed as described in materials and methods
The overall structure of p300 can be divided into two main parts according to the structural features of all HAT families, as shown in Figure 1: 1) the central core region, consisting of the α3 helix and β2–β5 strands that are structurally conserved in all HAT enzymes. This region participates in Ac-CoA binding and in templating the lysine side chain for acetyl-transfer 50, 51. 2) The flanking N- and C-terminal regions, which are structurally divergent between the different HAT subfamilies. These more variable regions have been shown to participate in histone substrate binding by forming a binding cleft that flanks the central core region50, 51. Structural comparison reveals that this cleft in p300 is highly acidic and shallower than the corresponding clefts in the Gcn5/PCAF and MYST HAT subfamilies. H-bond interactions and hydrophobic interactions between H3/p300 complex are also shown in Figure 1A and Figure 1B, respectively. p300 harbors an additional L1 loop in its N-terminal region, which is unique to this subfamily of HATs and participates in both Ac-CoA and lysine substrate binding. The autoacetylation loop, which consists of L1520–L1580, was genetically deleted during crystallization and the resulting p300 HAT domain showed comparable acetyltransferase activity with the intact enzyme, thereby we didn’t include it in our model37, 52, 53.
Figure 1.
The overall structure of the p300/H3/Ac-CoA complex model. Cyan: N-terminal region; Magnet: central core region; Yellow: C-terminal region; Red: L1 loop. (A) H-bond interactions and (B) hydrophobic interactions between H3 and p300. Green sticks: p300; White sticks: H3 peptide.
To investigate the stability of the active site cavity, a 100-ns MD simulation was performed on the p300/H3/Ac-CoA complex. Firstly, the protonation states of key residues in active site and involved in PT were determined computationally by H++ program54. As shown in Table S1, the basic residues H3K14, Y1446 and Y1467 around the active site, and the PT shuttling residue Y1394 were largely protonated. While the depronation state was the dominate for D1444 and D1507, which are considered as the proton acceptor residues in PT process. Therefore, the calculation results were applied to the model used in MD simulation and the QM/MM study.
During the MD simulation, the root-mean-square deviation (RMSD) was monitored (from t= 0ps), using the backbone coordinates of the model structure as the reference (Figure 2A). In addition, to represent the stability of residues, the root-mean-square fluctuation (RMSF) of p300 in the complex model was also calculated. The B-factor (thermal factor) of p300 in the crystal structure of p300/Lys-CoA, was used as a measure for any type of displacement of an atom from its mean position, and was extracted and transformed to RMSF using the formula B=8π2*RMSF2/3 (Figure 2B). The RMSD tends to be flat after a 20 ns’ simulation, which indicates that structures from 20 to 100 ns are more reasonable. Notably, the RMSD of H3 is apparently higher than other components of the complex. Consistently, the average RMSD of apo-p300 is only about 1.8 Å while that of the complex is above 2.2 Å, likely due to the flexibility of the H3 peptide. This result is in good agreement with the Theorell-Chance mechanism, where the protein substrate is proposed to bind p300 for acetylation and leave rapidly before forming a stable ternary complex. The consistency between the theoretical and experimental RMSF shown in Figure 2B validates the quality of our model. We propose that the peaks of the RMSF curve represent the different loop regions of the protein. Interestingly, the, 23-resisue L1 loop (residues 1436–1459) is fairly stable during the simulation, displaying a lower RMSF value than other regions of p300 with defined secondary structure. This is consistent with the extensive interactions that the L1 loop makes with Ac-CoA in the p300/Lys-CoA crystal structure.
Figure 2.
(A) The time dependencies of the root-mean-square deviations (RMSDs) for the back bone atoms of the p300/H3/Ac-CoA complex, p300, H3 and Ac-CoA from their initial positions during the 100 ns simulation. (B) Residue fluctuations obtained by average residual fluctuations over 100 ns simulation and experimental result calculated from B factors of p300 (PDB code: 3BIY). The residue number of genetically deleted autoacetylation loop was represented by a gap. The color of the cartoon structure ranges from blue to red with increasing RMSF values.
H-bond and hydrophobic interaction analysis for the p300/H3/Ac-CoA model
To probe the molecular basis for H3 binding, both the H-bond and hydrophobic interactions between p300 and H3 were analyzed. First, the reaction residue H3K14 h-bonds strongly with the main chain oxygen atom of W1436. Its adjacent residue A315 h-bonds with P1439 and P1440 on the L1 loop, which contributes to further stabilize the H3K14 interaction. Second, two h-bond networks located at both ends of H3 together display important roles for anchoring the peptide chain. For one of these h-bonding networks, the electropositive side chains of terminal A7, R8 and K9 form an interaction network with the electronegative side chains of E1505, D1625 and D1628 from p300. This interaction is consistent with previous in vitro studies showing that the D1625R/D1628R mutant shows significantly decreased p300 HAT activity, further supporting the importance of this h-bond network for peptide substrate binding7. This interaction network provides a rationale for the observed preference for additional Lysine or Arginine residues two to three residues downstream or upstream of the primary acetylation site. For the other h-bonding network, R17, Q19 and A21 of H3 interact with E1442, K1456, I1457, K1459 and R1462 of p300, with most of these residues coming from the L1 loop. This same anchoring pattern (one h-bond network at each end of the histone) was also observed in the Gcn5 HAT subfamily38, 55. R8 and Q19 in the tGCN5/H3/Ac-CoA system and K166 and Q176 in the hGCN5/H3/Ac-CoA system h-bond with the enzyme at both ends of the H3 peptide, just like the corresponding K9 and Q19 residues in the p300/H3/Ac-CoA system. Third, Y1397 in the middle of the substrate binding cleft h-bonds with the main chain carbonyls of T11 and G12 next to the substrate lysine, which acts like a bridge to connect the substrate lysine and positively charged region mentioned above to help maintain the configuration of the H3 peptide.
Hydrophobic interactions between p300 and the H3 peptide are not only conducive to H3 binding, but also provide a favorable environment for nucleophilic attack of the deprotonated lysine side chain. It has been reported that in tGCN5 and hGCN5, there is a G-K-X-P recognition sequence in the middle of the histone peptide, which is responsible for most of the hydrophobic interactions between H3 and the enzyme38, 55. In our simulation, most hydrophobic contacts are located from G12 to P16 of the H3 peptide, which includes a G-K-A-P region. In the reaction center, H3K14 is surrounded by the hydrophobic side chains of Y1397, W1436, P1439, P1440 and Y1446. This highly hydrophobic environment could greatly facilitate the deprotonation of H3K14 and subsequent nucleophilic attack.
It is worth noting the structure alignment of different snapshots at a 20 ns interval also shows the fluctuations of H3 (Figure S6). Interestingly, the binding cleft is relatively shallow and polar comparing to GCN5 and Esa1 HATs. As required by the Theorell-Chance mechanism, the protein substrates associate weakly with the enzyme so that a rapid disassociation would occur and the ternary complex never accumulates during catalytic turnover. Taken together, the unstable and flexibility characteristic of the H3 peptide bound to p300 strongly supports the Theorell-Chance mechanism.
Identification of rational PT scheme
As the positive charged state of reaction residue H3K14 was the dominating form under physiological environment (Table S1), the first step of the acetylation reaction should involves the deprotonation of the lysine side chain. For the Esa1 MYST HAT, the lysine proton is directly delivered to a conserved glutamate residue (E338)35. In contrast to Esa1, in the reaction center of p300 there is no electronegative residue within 5 Å of the ε-N atom of the target lysine, which leads to a hypothesis for the existence of a long-range mechanism in the PT process. In this case, the proton acceptor may be far from the lysine, the proton donor, and the long-range PT is usually mediated by waters or residues containing a hydroxyl group56, 57. We also note that in the GCN5 HAT, the proton is delivered to a conserved glutamate (E173 in yGCN533, E122 in tGCN538 and E80 in hGCN555). In particular, for tGCN5, a conserved water molecule is ideally located to shuttle a proton from the reactive lysine to E122, representing a long-range transfer scheme. A similar mechanism may be employed in p300. Analysis of the p300/Lys-CoA crystal structure (PDB code: 3BIY) indicates that the nearest electronegative residues that could be the proton acceptors around the reaction center are D1444 and D1507. Three crystal water molecules, Wat1777, Wat1861 and Wat1878, and two residues with a hydroxyl group, Y1394 and Y1446, may also serve as the intermediate proton carriers. Based on this comparison and analysis, we proposed two possible PT modes showed in scheme 1.
Scheme 1.
Two proposed PT schemes.
MD simulations were incompatible with scheme (b) since the positions of Wat1777 and Wat1878 were not stable enough to facilitate PT. In contrast, Wat1861 in scheme (a) was quite stable during the entire 100 ns simulation time. The distances between the heavy atoms involved in scheme (a) were all steady and close enough for the PT (Figure 3A). Detailed MD simulation analysis suggested that the high stability of scheme (a) is induced by a strong h-bond interaction between Wat1861 and the backbone carbonyl oxygen of I1395, which retains the location of the water molecule. In addition, the strong h-bond interaction between H3K14 and W1436 may help align the departing proton toward the water molecule, which is also apparently facilitated by the hydrophobic interactions between the same two residues (Figure 3B). Moreover, a hydrophobic local environment contributes to increase the pKa of the D1507 side chain, allowing it to function as a general base. An analogous favorable environment was not present in scheme (b), which instead showed remarkable fluctuation. The importance of residues Y1394 and D1507 in the PT mechanism of scheme a is supported by mutational studies. Specifically, Y1394F, D1507N and D1507A mutations significantly impair p300 acetylation activity37. In summary, due to the instability of the two water molecules in scheme (b), and the significantly more favorable MD simulation results with scheme (a) along with the supportive mutational studies, we propose that scheme (a) is used for PT lysine deprotonation for the first step of the p300 acetylation reaction.
Figure 3.

(A) the time dependencies of the distances between heavy atoms composing the PT wire of scheme (a) during the 100 ns simulation. (B) The interaction network that stable the configuration in scheme (a). The atoms directly involved in the transfer wire are shown as spheres.
QM/MM calculations were employed to validate PT scheme (a) and further investigate the subsequent chemical mechanism. The QM region included Wat1861, the side chain of H3K14, Y1394 and D1507, the backbone of I1395 and W1436, where I1395 h-bonds with Wat1861 to keep its position, W1436 h-bonds with H3K14 to orient the -NH3+ group, and W1436 contributes to the hydrophobic environment that facilitates lysine deprotonation as mentioned above. When systematically lengthening (at 0.05 Å intervals) the N-H bond of the target lysine toward Wat1861 with each step subjected to a geometry optimization, the protons in Wat1861 and the hydroxyl group of Y1394 migrated in a concerted manner. As a consequence, the hydroxyl hydrogen was bound to D1507, while the other two hydrogen atoms were abstracted by oxygen atoms, Oη and O, of Y1394 and water, respectively, generating a protonated D1507, a neutral Y1394, and a deprotonated H3K14. A potential energy profile captured the concerted nature of the PT reaction (Figure 4): the proton movement has a barrier of 0.65 kcal/mol, while the product (P) is 10.15 kcal/mol lower in energy than the reactant (R). This energy profile displays a notably low PT barrier and a dramatic energy decrease in the immediate products, which leads us to conclude that such a single-direction proton shuttle is energetically inescapable, or even intrinsic.
Figure 4.

The potential energy barrier for the PT process. The structures of reactant (R) and product (P) are displayed.
The PT schemes in three major HAT subfamilies (Gcn5/PCAF, MYST and p300) were compared by structure alignment. Interestingly, the PT processes in all three HAT subfamilies are common in transfer direction, but differ in molecular transfer scheme (Figure S7). The locations of E338 of Esa1, E122 of tGCN5 and Y1394 of p300 mapped very well. However, the conserved proton acceptor residues among these enzymes belong to different secondary structure elements. E338 is located on the end of the β10 strand of the C-terminal region of Esa1, and E122 in tGCN5 is located on the β4 strand of the core region. For p300, D1507 is found on the short loop before the α6 helix of the N-terminal region. It is also notable that the complexity of the three schemes is different. In Esa1, the proton is directly delivered to E338. In tGCN5, the proton is transferred to E122 by a water molecule. While in p300, the proton is shuttled to Y1394 on the β4 strand by a water molecule, and then the hydrogen atom from the hydroxyl group of Y1394 is delivered to D1507. In summary, the PT process in Esa1 is direct from substrate to enzyme, tGCN5 uses a water-mediated pathway, and p300 employs the most complex scheme, in which both a water molecule and a shuttle residue Y1394 are involved.
The catalytic mechanism of the acetylation reaction
The acetylation reaction can be divided into two steps: an addition half-reaction and an elimination half-reaction. In the addition half-reaction, nitrogen atom (N3) of H3K14 nucleophilically attacks the carbonyl carbon atom (C4) of Ac-CoA, promoting this carbon atom to change hybridization state from sp2 to sp3. Subsequently, the N3-C4 bond forms and the hydrogen (H2) of H3K14 is transferred to the oxygen atom (O5) of the acetyl portion of Ac-CoA, forming an intermediate. In the elimination half-reaction, normally it is the hydroxyl group of the intermediate that protonates CoAS- to generate the CoASH product, as found for N-acetyltransferases58, 59. However, the side chain hydroxyl group of Y1467 can also function as the general base to protonate the CoAS- since it forms a direct hydrogen bond to the sulfur atom of Ac-CoA (Scheme 2). The two steps and the two possibilities of the elimination half-reaction were investigated by QM/MM simulation. The reactant system for this simulation of the acetylation reaction was constructed based on the optimization of the product structure of the PT process.
Scheme 2.
Two possible acetylation schemes
The QM region included the side chain of the deprotonated H3K14, the -CH2-S-COCH3 group of Ac-CoA, the backbone of W1436, and the side chain of Y1467, considering their important roles during the reaction process. W1436 strongly h-bonds with the ε-nitrogen of the target lysine, which may contribute to orient the lysine amino group. Moreover, the aromatic side chain of W1436 together with the phenol group of Y1446 forms a hydrophobic environment that facilitating lysine deprotonation to trigger the PT process and acetylation reaction. Notably, this h-bond interaction may activate the reaction by making the reactive nitrogen atom partially negative and by stabilizing the intermediate. Indeed, this activation and stabilization mechanism is widespread in HAT proteins. For tGCN5, the backbone amide group of L126 forms a hydrogen bond with the carbonyl oxygen of the acetyl group of Ac-CoA so that the thioester carbonyl group can be polarized to achieve these activation and stabilization functions60. Y1467 coordinates with the sulfur atom of Ac-CoA, which may help position the carbonyl carbon. More importantly, Y1467 may also help activate the chemical bond between the sulfur atom and the carbonyl carbon. Finally, after the nucleophilic attack, Y1467 may also function as a general acid to protonate the CoASH leaving group as shown in scheme 2 (a).
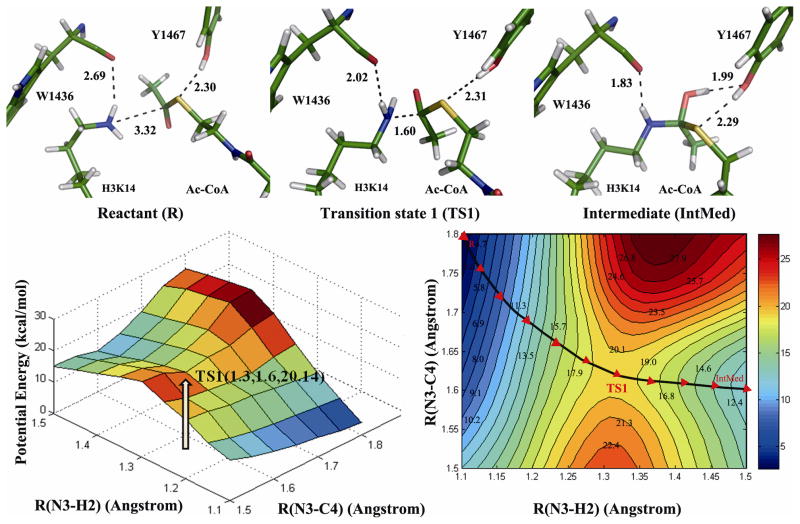
Along the nucleophilic attack reaction, the energies of the reactant (R), transition state (TS), and intermediate (IntMed) were determined by a two-dimensional QM/MM potential energy surface by defining the distances of R(N3-C4) and R(N3-H2) as the reaction coordinates (Figure 5). In the optimized reactant, R(N3-C4)=3.32 Å and R(N3-H2)=1.03 Å. In the optimized intermediate structure, the N-C bond is formed (R(N3-C4=1.43 Å)), the N3-H2 bond is broken, and C4=O5 of Ac-CoA changes to C4-O5H2. The calculated potential energy barrier of this step is 20.14 kcal/mol. The structure of the TS1 was determined by adiabatic mapping at the QM/MM level. In the TS, R(N3-C4)=1.60 Å and R(N3-H2)=1.30 Å. This structure illustrates that atom N3, H2, C4 and O5 were tuned to be in a plane to form a tetrahedron ring. The N3-H2 bond was partially broken, the N3-C4 bond was partially formed, and the C4-S6 bond was also partially broken simultaneously. Furthermore, the bond order of C4=O5 was transformed from double to single gradually during the whole process. Consistently, we also noticed that the two h-bond pairs (H3K14-W1436 and Ac-CoA-Y1467) mentioned above remained stable. This two h-bonds contributes to stabilizing the reactive components so that a energetically favored IntMed could properly formed. The distance between H3K14 and W1436 became even closer. This observation confirms the role of W1436 and Y1467 in stabilizing the IntMed.
Figure 5.
The potential energy surface (PES) and contour plot of the PES of the addition half-reaction pathway. Energy barriers of transition state 1 (TS1) is marked and structures of reactant (R), intermediate (IntMed) and product (P) are displayed.
As aforementioned, there are two possible mechanisms to protonate the leaving group after the intermediate is formed. To determine which scheme is more reasonable, we scanned the coordinates of S6-H2 and S6-H7 bond formation in both schemes (Figure 6). The energy barrier of scheme (a) is 12.08 kcal/mol, while for scheme (b) the energy barrier is 15.75 kcal/mol. Although the energy difference between these two schemes is not large, the lower energy barrier of scheme (a) indicates that the Y1467-meidated elimination half-reaction is more likely to occur. In addition, Y1467F showed a 430-fold reduction in V/K3737 also suggesting an important role of this residue in the reaction. Based on all these analyses, we consider scheme (a), in which Y1467 serves as the major general acid to protonate the leaving group, as the preferred reaction scheme. In TS2(a), the sulfur atom, the carbon atom of the acetyl group, the hydroxyl group of the IntMed, and the phenolic hydroxyl group of Y1467 forms a hexahedron ring together with two hydrogen bonds. In addition, the hydrogen bond between W1436 and the deprotonated lysine amino nitrogen is probably even stronger as the distance (1.77Å) between them is shorter than in the intermediate (1.83Å). Since a Y1467F mutant displays significantly reduced enzyme activity, it is mostly likely that, the acetylation reaction adopts scheme (a) to proceed. Altogether, the elimination half-reaction is mainly triggered by Y1467 and may be conducted directly by the IntMed when Y1467 is mutated.
Figure 6.
The potential energy barrier comparison of the two elimination half-reaction mechanisms. The structure of TS2(a) which is more favorable is displayed.
Conclusion
As the founding member of one of the major HAT subfamilies, p300 plays important roles in a plethora of biological processes, including cell growth, differentiation and transformation, oncogenesis and apoptosis. The catalytic mechanism of the p300 HAT has started to unfold through recent structural and biochemical studies. However, a complete delineation of the entire chemical pathway of the acetylation reaction remained unclear. Knowledge was lacking for some of the most critical components of the pathway, including the structure of the PT wire and the chemical mechanism of the acetyl transfer reaction. No study had been reported to date to address these important issues.
In this study, molecular modeling was applied to construct the p300/H3/Ac-CoA system. A 100ns MD simulation was carried out on this model to obtain the most suitable configurations for further study. The h-bond interactions between p300 and the H3 peptide were mainly formed by the electronegative residues E1505, D1625, D1628 of the substrate binding cleft and E1442, K1456, I1457, K1459 and R1462 of the L1 loop. In addition to residues G12 to P16 of the H3 peptide, which include a conserved G-K-A-P sequence in HAT substrates, hydrophobic interactions are mediated by non-polar part of Y1397, W1436, P1439, P1440 and Y1446 of the L1 loop. Structure analysis reveals two possible “proton-wires” for the deprotonation process of substrate H3K14. Consistent with previous mutation data, our MD simulation indicates that the proton wire consisting of H3K14, Wat1861, Y1394 and D1507 is highly likely. Detailed analysis suggests that a strong h-bond interaction between Wat1861 and the backbone carbonyl oxygen of I1395 might be the major stabilization factor for this wire by fixing the location of the water molecule. The extremely low potential energy in the QM/MM calculation provides additional support for this PT wire. Finally, QM/MM calculations were also performed to investigate the acetylation mechanism. The two-dimensional potential energy barrier of the addition half-reaction and structures of two TSs and IntMed were obtained. In particular, for the elimination half-reaction both potential energy barrier calculations and previous experimental data support the model whereby the protonation of the CoAS- group by Y1467 is more energetically favorable than protonation by the reaction IntMed.
In summary, our research elucidated several essential aspects of the acetylation reaction catalyzed by p300. The rational PT scheme was proposed and validated for the first time. Detailed knowledge of the configurations of the two TSs and IntMed do not only shed light on the mechanism research of other HAT families, but also opens up a novel avenue to discover a new family of small molecule effectors for p300. These small molecules can be designed specifically focused on specifically blocking the PT wire, interfering with the addition half-reaction or affecting the protonation of the leaving group on the basis of the TS structures.
Supplementary Material
Acknowledgments
This work was supported the Hi-Tech Research and Development Program of China (2012AA020302), the National Natural Science Foundation of China (81230076, 91229204, 91029704 and 21210003), the National Science and Technology Major Project “Key New Drug Creation and Manufacturing Program” ((2013ZX09507-004, 2013ZX09507001 and 2014ZX09507002-005-012). Computation resources were partially supported by Computer Network Information Center, Chinese Academy of Sciences and Shanghai Supercomputing Center.
Footnotes
Supporting Information Available: The model selection description of L1 loop in the structure of the p300/H3/Ac-CoA, Figure S1–9 and Table S1 including H3 peptide structural alignment during the MD simulation, structure mapping of the PT processes in three HAT subfamilies, RMSFs of MD simulation on different models, the interaction between L1 Loop and neighboring residues, and the predicted pKa for ionizable residues of the active site and PT scheme with different internal dielectric constant. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Fullgrabe J, Kavanagh E, Joseph B. Histone Onco-Modifications. Oncogene. 2011;30:3391–3403. doi: 10.1038/onc.2011.121. [DOI] [PubMed] [Google Scholar]
- 2.Turner BM. Defining an Epigenetic Code. Nat Cell Biol. 2007;9:2–6. doi: 10.1038/ncb0107-2. [DOI] [PubMed] [Google Scholar]
- 3.Shahbazian MD, Grunstein M. Functions of Site-Specific Histone Acetylation and Deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 4.Vempati RK, Jayani RS, Notani D, Sengupta A, Galande S, Haldar D. P300-Mediated Acetylation of Histone H3 Lysine 56 Functions in DNA Damage Response in Mammals. J Biol Chem. 2010;285:28553–28564. doi: 10.1074/jbc.M110.149393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tropberger P, Pott S, Keller C, Kamieniarz-Gdula K, Caron M, Richter F, Li G, Mittler G, Liu ET, Buhler M, et al. Regulation of Transcription through Acetylation of H3k122 on the Lateral Surface of the Histone Octamer. Cell. 2013;152:859–72. doi: 10.1016/j.cell.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 6.Sharma S, Liu J, Wei JQ, Yuan HJ, Zhang TF, Bishopric NH. Repression of Mir-142 by P300 and Mapk Is Required for Survival Signalling Via Gp130 During Adaptive Hypertrophy. EMBO Mol Med. 2012;4:617–632. doi: 10.1002/emmm.201200234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karukurichi KR, Cole PA. Probing the Reaction Coordinate of the P300/Cbp Histone Acetyltransferase with Bisubstrate Analogs. Bioorg Chem. 2011;39:42–47. doi: 10.1016/j.bioorg.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karukurichi KR, Wang L, Uzasci L, Manlandro CM, Wang Q, Cole PA. Analysis of P300/Cbp Histone Acetyltransferase Regulation Using Circular Permutation and Semisynthesis. J Am Chem Soc. 2010;132:1222. doi: 10.1021/ja909466d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen JH, Li Q. Life and Death of Transcriptional Co-Activator P300. Epigenetics-Us. 2011;6:957–961. doi: 10.4161/epi.6.8.16065. [DOI] [PubMed] [Google Scholar]
- 10.Turner BM. Histone Acetylation and an Epigenetic Code. Bioessays. 2000;22:836–45. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 11.Chen W, Bacanamwo M, Harrison DG. Activation of P300 Histone Acetyltransferase Activity Is an Early Endothelial Response to Laminar Shear Stress and Is Essential for Stimulation of Endothelial Nitric-Oxide Synthase Mrna Transcription. J Biol Chem. 2008;283:16293–16298. doi: 10.1074/jbc.M801803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Z, Chin YE, Zhang DD. Acetylation of Nrf2 by P300/Cbp Augments Promoter-Specific DNA Binding of Nrf2 During the Antioxidant Response. Mol Cell Biol. 2009;29:2658–2672. doi: 10.1128/MCB.01639-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hezroni H, Tzchori I, Davidi A, Mattout A, Biran A, Nissim-Rafinia M, Westphal H, Meshorer E. H3k9 Histone Acetylation Predicts Pluripotency and Reprogramming Capacity of Es Cells. Nucleus-Austin. 2011;2:300–309. doi: 10.4161/nucl.2.4.16767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das C, Lucia MS, Hansen KC, Tyler JK. Cbp/P300-Mediated Acetylation of Histone H3 on Lysine 56. Nature. 2009;459:113–U123. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodman RH, Smolik S. Cbp/P300 in Cell Growth, Transformation, and Development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 16.He H, Yu FX, Sun C, Luo Y. Cbp/P300 and Sirt1 Are Involved in Transcriptional Regulation of S-Phase Specific Histone Genes. PLoS One. 2011;6:e22088. doi: 10.1371/journal.pone.0022088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong XM, Jin Y. Critical Roles of Coactivator P300 in Mouse Embryonic Stem Cell Differentiation and Nanog Expression. J Biol Chem. 2009;284:9168–9175. doi: 10.1074/jbc.M805562200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu CR, Cole PA, Meyers DJ, Kormish J, Dent S, Zaret KS. Chromatin “Prepattern” and Histone Modifiers in a Fate Choice for Liver and Pancreas. Science. 2011;332:963–966. doi: 10.1126/science.1202845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gayther SA, Batley SJ, Linger L, Bannister A, Thorpe K, Chin SF, Daigo Y, Russell P, Wilson A, Sowter HM, et al. Mutations Truncating the Ep300 Acetylase in Human Cancers. Nat Genet. 2000;24:300–3. doi: 10.1038/73536. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh AK, Varga J. The Transcriptional Coactivator and Acetyltransferase P300 in Fibroblast Biology and Fibrosis. J Cell Physiol. 2007;213:663–671. doi: 10.1002/jcp.21162. [DOI] [PubMed] [Google Scholar]
- 21.Murata T, Kurokawa R, Krones A, Tatsumi K, Ishii M, Taki T, Masuno M, Ohashi H, Yanagisawa M, Rosenfeld MG, et al. Defect of Histone Acetyltransferase Activity of the Nuclear Transcriptional Coactivator Cbp in Rubinstein-Taybi Syndrome. Hum Mol Genet. 2001;10:1071–1076. doi: 10.1093/hmg/10.10.1071. [DOI] [PubMed] [Google Scholar]
- 22.Laurieri N, Crawford MH, Kawamura A, Westwood IM, Robinson J, Fletcher AM, Davies SG, Sim E, Russell AJ. Small Molecule Colorimetric Probes for Specific Detection of Human Arylamine N-Acetyltransferase 1, a Potential Breast Cancer Biomarker. J Am Chem Soc. 2010;132:3238–9. doi: 10.1021/ja909165u. [DOI] [PubMed] [Google Scholar]
- 23.Wisastra R, Ghizzoni M, Maarsingh H, Minnaard AJ, Haisma HJ, Dekker FJ. Isothiazolones; Thiol-Reactive Inhibitors of Cysteine Protease Cathepsin B and Histone Acetyltransferase Pcaf. Org Biomol Chem. 2011;9:1817–1822. doi: 10.1039/c0ob00464b. [DOI] [PubMed] [Google Scholar]
- 24.Dal Piaz F, Tosco A, Eletto D, Piccinelli AL, Moltedo O, Franceschelli S, Sbardella G, Remondelli P, Rastrelli L, Vesci L, et al. The Identification of a Novel Natural Activator of P300 Histone Acetyltranferase Provides New Insights into the Modulation Mechanism of This Enzyme. ChemBioChem. 2010;11:818–827. doi: 10.1002/cbic.200900721. [DOI] [PubMed] [Google Scholar]
- 25.Euser AS, Arends LR, Evans BE, Greaves-Lord K, Huizink AC, Franken IHA. The P300 Event-Related Brain Potential as a Neurobiological Endophenotype for Substance Use Disorders: A Meta-Analytic Investigation. Neurosci Biobehav Rev. 2012;36:572–603. doi: 10.1016/j.neubiorev.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 26.Khan SN, Khan AU. Role of Histone Acetylation in Cell Physiology and Diseases: An Update. Clin Chim Acta. 2010;411:1401–1411. doi: 10.1016/j.cca.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 27.Selvi BR, Mohankrishna DV, Ostwal YB, Kundu TK. Small Molecule Modulators of Histone Acetylation and Methylation: A Disease Perspective. Bba-Gene Regul Mech. 2010;1799:810–828. doi: 10.1016/j.bbagrm.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Selvi BR, Senapati P, Kundu TK. Small Molecule Modulators of Epigenetic Modifications: Implications in Therapeutics. Curr Sci. 2012;102:29–36. [Google Scholar]
- 29.Delvecchio M, Gaucher J, Aguilar-Gurrieri C, Ortega E, Panne D. Structure of the P300 Catalytic Core and Implications for Chromatin Targeting and Hat Regulation. Nat Struct Mol Biol. 2013;20:1040–6. doi: 10.1038/nsmb.2642. [DOI] [PubMed] [Google Scholar]
- 30.Lewendon A, Murray IA, Shaw WV, Gibbs MR, Leslie AG. Replacement of Catalytic Histidine-195 of Chloramphenicol Acetyltransferase: Evidence for a General Base Role for Glutamate. Biochemistry. 1994;33:1944–50. doi: 10.1021/bi00173a043. [DOI] [PubMed] [Google Scholar]
- 31.Thompson S, Mayerl F, Peoples OP, Masamune S, Sinskey AJ, Walsh CT. Mechanistic Studies on Beta-Ketoacyl Thiolase from Zoogloea-Ramigera - Identification of the Active-Site Nucleophile as Cys89, Its Mutation to Ser89, and Kinetic and Thermodynamic Characterization of Wild-Type and Mutant Enzymes. Biochemistry. 1989;28:5735–5742. doi: 10.1021/bi00440a006. [DOI] [PubMed] [Google Scholar]
- 32.Sternglanz R, Schindelin H. Structure and Mechanism of Action of the Histone Acetyltransferase Gcn5 and Similarity to Other N-Acetyltransferases. Proc Natl Acad Sci U S A. 1999;96:8807–8808. doi: 10.1073/pnas.96.16.8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanner KG, Trievel RC, Kuo MH, Howard RM, Berger SL, Allis CD, Marmorstein R, Denu JM. Catalytic Mechanism and Function of Invariant Glutamic Acid 173 from the Histone Acetyltransferase Gcn5 Transcriptional Coactivator. J Biol Chem. 1999;274:18157–18160. doi: 10.1074/jbc.274.26.18157. [DOI] [PubMed] [Google Scholar]
- 34.Trievel RC, Rojas JR, Sterner DE, Venkataramani RN, Wang L, Zhou J, Allis CD, Berger SL, Marmorstein R. Crystal Structure and Mechanism of Histone Acetylation of the Yeast Gcn5 Transcriptional Coactivator. Proc Natl Acad Sci U S A. 1999;96:8931–6. doi: 10.1073/pnas.96.16.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan Y, Harper S, Speicher DW, Marmorstein R. The Catalytic Mechanism of the Esa1 Histone Acetyltransferase Involves a Self-Acetylated Intermediate. Nat Struct Biol. 2002;9:862–869. doi: 10.1038/nsb849. [DOI] [PubMed] [Google Scholar]
- 36.Berndsen CE, Albaugh BN, Tan S, Denu JM. Catalytic Mechanism of a Myst Family Histone Acetyltransferase. Biochemistry. 2007;46:623–629. doi: 10.1021/bi602513x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X, Wang L, Zhao KH, Thompson PR, Hwang Y, Marmorstein R, Cole PA. The Structural Basis of Protein Acetylation by the P300/Cbp Transcriptional Coactivator. Nature. 2008;451:846–850. doi: 10.1038/nature06546. [DOI] [PubMed] [Google Scholar]
- 38.Clements A, Poux AN, Lo WS, Pillus L, Berger SL, Marmorstein R. Structural Basis for Histone and Phosphohistone Binding by the Gcn5 Histone Acetyltransferase. Mol Cell. 2003;12:461–473. doi: 10.1016/s1097-2765(03)00288-0. [DOI] [PubMed] [Google Scholar]
- 39.Shindyalov IN, Bourne PE. Protein Structure Alignment by Incremental Combinatorial Extension (Ce) of the Optimal Path. Protein Eng. 1998;11:739–47. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 40.Karanam B, Jiang LH, Wang L, Kelleher NL, Cole PA. Kinetic and Mass Spectrometric Analysis of P300 Histone Acetyltransferase Domain Autoacetylation. J Biol Chem. 2006;281:40292–40301. doi: 10.1074/jbc.M608813200. [DOI] [PubMed] [Google Scholar]
- 41.Karanam B, Wang L, Wang D, Liu X, Marmorstein R, Cotter R, Cole PA. Multiple Roles for Acetylation in the Interaction of P300 Hat with Atf-2. Biochemistry-Us. 2007;46:8207–16. doi: 10.1021/bi7000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules (Vol 117: Pg 5179, 1995) J Am Chem Soc. 1996;118:2309–2309. [Google Scholar]
- 43.Bayly CI, Cieplak P, Cornell WD, Kollman PA. A Well-Behaved Electrostatic Potential Based Method Using Charge Restraints for Deriving Atomic Charges - the Resp Model. J Phys Chem. 1993;97:10269–10280. [Google Scholar]
- 44.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber Biomolecular Simulation Programs. J Comput Chem. 2005;26:1668–88. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Darden T, York D, Pedersen L. Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 46.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-Integration of Cartesian Equations of Motion of a System with Constraints - Molecular-Dynamics of N-Alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 47.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. Molecular-Dynamics with Coupling to an External Bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 48.Rooklin DW, Lu M, Zhang YK. Revelation of a Catalytic Calcium-Binding Site Elucidates Unusual Metal Dependence of a Human Apyrase. J Am Chem Soc. 2012;134:15595–15603. doi: 10.1021/ja307267y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hou GH, Cui Q. Qm/Mm Analysis Suggests That Alkaline Phosphatase (Ap) and Nucleotide Pyrophosphatase/Phosphodiesterase Slightly Tighten the Transition State for Phosphate Diester Hydrolysis Relative to Solution: Implication for Catalytic Promiscuity in the Ap Superfamily. J Am Chem Soc. 2012;134:229–246. doi: 10.1021/ja205226d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marmorstein R. Structure of Histone Acetyltransferases. J Mol Biol. 2001;311:433–444. doi: 10.1006/jmbi.2001.4859. [DOI] [PubMed] [Google Scholar]
- 51.Wang L, Tang Y, Cole PA, Marmorstein R. Structure and Chemistry of the P300/Cbp and Rtt109 Histone Acetyltransferases: Implications for Histone Acetyltransferase Evolution and Function. Curr Opin Struct Biol. 2008;18:741–747. doi: 10.1016/j.sbi.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karanam B, Jiang L, Wang L, Kelleher NL, Cole PA. Kinetic and Mass Spectrometric Analysis of P300 Histone Acetyltransferase Domain Autoacetylation. J Biol Chem. 2006;281:40292–301. doi: 10.1074/jbc.M608813200. [DOI] [PubMed] [Google Scholar]
- 53.Karanam B, Wang L, Wang DX, Liu X, Marmorstein R, Cotter R, Cole PA. Multiple Roles for Acetylation in the Interaction of P300 Hat with Atf-2. Biochemistry. 2007;46:8207–8216. doi: 10.1021/bi7000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A. H++: A Server for Estimating Pk(a)S and Adding Missing Hydrogens to Macromolecules. Nucleic Acids Res. 2005;33:W368–W371. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang J, Lu J, Lu D, Liang Z, Li L, Ouyang S, Kong X, Jiang H, Shen B, Luo C. Investigation of the Acetylation Mechanism by Gcn5 Histone Acetyltransferase. PLoS One. 2012;7:e36660. doi: 10.1371/journal.pone.0036660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hermann JC, Ridder L, Mulholland AJ, Holtje HD. Identification of Glu166 as the General Base in the Acylation Reaction of Class a Beta-Lactamases through Qm/Mm Modeling. J Am Chem Soc. 2003;125:9590–9591. doi: 10.1021/ja034434g. [DOI] [PubMed] [Google Scholar]
- 57.Hermann JC, Hensen C, Ridder L, Mulholland AJ, Holtje HD. Mechanisms of Antibiotic Resistance: Qm/Mm Modeling of the Acylation Reaction of a Class a Beta-Lactamase with Benzylpenicillin. J Am Chem Soc. 2005;127:4454–4465. doi: 10.1021/ja044210d. [DOI] [PubMed] [Google Scholar]
- 58.Qiao QA, Yang C, Qu R, Jin Y, Wang M, Zhang Z, Xu Q, Yu Z. A Density Functional Theory Study on the Role of His-107 in Arylamine N-Acetyltransferase 2 Acetylation. Biophys Chem. 2006;122:215–20. doi: 10.1016/j.bpc.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 59.Wang H, Liu L, Hanna PE, Wagner CR. Catalytic Mechanism of Hamster Arylamine N-Acetyltransferase 2. Biochemistry. 2005;44:11295–306. doi: 10.1021/bi047564q. [DOI] [PubMed] [Google Scholar]
- 60.Lin YX, Fletcher CM, Zhou JX, Allis CD, Wagner G. Solution Structure of the Catalytic Domain of Gcn5 Histone Acetyltransferase Bound to Coenzyme A. Nature. 1999;400:86–89. doi: 10.1038/21922. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.