Abstract
Human Protein Reference Database (HPRD) is an object database that integrates a wealth of information relevant to the function of human proteins in health and disease. Data pertaining to thousands of protein-protein interactions, posttranslational modifications, enzyme/substrate relationships, disease associations, tissue expression, and subcellular localization were extracted from the literature for a nonredundant set of 2750 human proteins. Almost all the information was obtained manually by biologists who read and interpreted >300,000 published articles during the annotation process. This database, which has an intuitive query interface allowing easy access to all the features of proteins, was built by using open source technologies and will be freely available at http://www.hprd.org to the academic community. This unified bioinformatics platform will be useful in cataloging and mining the large number of proteomic interactions and alterations that will be discovered in the postgenomic era.
The past several years have witnessed an exponential increase in the amount of biological data, mainly due to development and application of high-throughput technologies including automated sequencing, gene expression microarrays, and mass spectrometry to characterize DNA, mRNA, and proteins, respectively (Brown and Botstein 1999; Pandey and Mann 2000; Hood and Galas 2003). With the sequence of the human genome sequence already available, a large majority of protein-coding genes has been identified that pave the way for additional genome-wide functional studies (Lander et al. 2001; Venter et al. 2001). Newer technologies such as protein microarrays, live cell microarrays, and RNAi hold the promise of systematically studying the entire human proteome to help understand the ultimate molecular machine —a human cell (Elbashir et al. 2001; Zhu et al. 2001; Ziauddin and Sabatini 2001). Nevertheless, the classical approach of performing individual biochemical and genetic experiments to assess the function of proteins and genes that has been followed for decades will continue to be just as valuable in the postgenomic era.
Systems biology is an emerging discipline that uses experimental approaches and computational biology to help understand biological phenomena on a global scale (Kitano 2002). For example, cellular networks such as signaling pathways provide us an opportunity to study the cell as a system. However, one of the key issues to tackle before the power of systems biology can be fully harnessed is to integrate genomic and proteomic data efficiently along with other sources of information such as the published literature (Birney et al. 2002). This integration will translate raw data into useful information, thereby expanding our knowledge about organisms. Recent advances in software technology permit the creation of suitable platforms for storage, analysis, and representation of complex biological information in such a manner that biologists can undertake a systems biology approach without being overwhelmed by the data (Navarro et al. 2003). Despite this, biological databases are still far from being such platforms because building databases that are comprehensive and scalable and that have intuitive and robust query systems requires a truly coordinated effort between biologists and computer professionals—something that is difficult to achieve. Although there exists a large number of protein databases, it is still difficult for biologists to sift through the available information and to synthesize it for analyzing and interpreting their own data. This is because of a number of factors, including redundancy in databases, erroneous entries, annotation errors, failure to account for alternative nomenclatures, lack of “interpreted information,” inability to query all fields of the data, and the paucity of experimental data incorporated into databases. In addition to this, blending of automated annotation with biological sequence databases could result in loss of clarity of the underlying biologically relevant information. For example, inclusion of novel gene transcripts predicted through gene prediction programs in databases that house experimentally determined transcripts could lead to confusion. Similarly, inclusion of predicted posttranslational modifications based on computer algorithms or sequence homology along with other experimentally derived data about proteins could lead to erroneous interpretation, or worse, unnecessary experimentation.
Therefore, we sought to create a protein-centric database that would serve as a comprehensive resource of information pertaining to human proteins, their characteristics, and functions. Access to such information would be invaluable to biologists in several ways. For instance, the type of domains found in proteins is generally predictive of their functional class or biological role. Posttranslational modifications such as phosphorylation and ubiquitination can drastically influence the activity of proteins and are commonly used as regulatory mechanisms in signal transduction pathways (Karin and Ben-Neriah 2000). Because proteins act in concert with other proteins, knowing the identity of interacting proteins along with the relevant binding sites can facilitate hypothesis-driven studies and elucidation of regulatory networks (Pawson and Nash 2003). The exact subcellular localization of proteins and their tissue distribution in the body is also pivotal to protein function (Nakai 2000). Finally, it is important to know whether any proteins are known to be associated with human diseases, as this might implicate them in certain pathways (Hanash 2003).
The Reference Sequence Database (RefSeq) initiated by the National Center for Biotechnology Information (NCBI) serves as one of the central repositories for DNA and protein sequences and some of their characteristics (Pruitt et al. 2000). SWISS-PROT is another popular database that contains curated information about proteins (Boeckmann et al. 2003). However, several features of proteins that are crucial to understanding proteins and their functions are either not or inadequately addressed by existing databases. First, a large body of data on protein-protein interactions exists in humans, and yet, none of the databases covers this adequately. Second, although thousands of sites of posttranslational modifications have been determined experimentally, the protein sequences found in databases are not annotated with this information. Third, most databases do not provide enzyme/substrate relationships that are crucial for understanding molecular networks within cells. Fourth, although automatic prediction of domains from different programs is provided by several databases, it can be quite confusing to nonexperts to determine which predictions are correct and which ones are to be ignored. Lastly, information such as tissue distribution, subcellular localization, and disease association of proteins is available by searching literature databases but is not generally linked to database entries.
Our approach to solving this problem has been to create a database that integrates protein information from several sources and supports interoperability with other existing databases. Human Protein Reference Database (HPRD) is an object-oriented database developed with open source technologies. It has a user-friendly graphic interface that not only allows a biologist to query proteins but also permits browsing based on several parameters. Currently, it contains a completely nonredundant set of 2750 human proteins with ∼25,050 links to PubMed citations that allow a user to obtain additional information about the annotations. We anticipate that HPRD, which is freely available to the academic community, will become a unified bioinformatics platform that will not only allow easy storage and retrieval of protein data but also lead to systematic approaches to dissect the human and other proteomes.
METHODS
Sequence Selection
Almost all of the initial annotation was performed by scientists at the Institute of Bioinformatics (http://www.ibioinformatics.org), a nonprofit research organization, whose initial task was to create HPRD. The person who performed the initial curation and annotation is listed under credits as the initial annotator for each entry. A list of genes implicated in human diseases was obtained from the Online Mendelian Inheritance in Man (OMIM) database (Hamosh et al. 2002). In addition to this list, proteins that may have indirect involvement in human diseases were also considered for annotation. Other categories of genes were taken based on important classes of proteins, including signaling molecules, transcription factors, enzymes, ion channels, ligands, and receptors. Because of the redundancy of protein sequences, we chose only the longest isoform for every entry, and the shorter or other alternatively spliced variants were manually filtered out by using BLASTP and BLASTN programs available at the NCBI Web site (Altschul et al. 1997, 1990). We chose entries in the RefSeq database (with NP_ or NM_ prefixes for protein and DNA sequences, respectively) wherever possible. These sequences were used for further annotation. This was manually and individually repeated for every OMIM entry.
Annotation Strategy
The most important aspect of our annotation is that the literature extraction, analysis, and interpretation were performed by trained biologists. The starting point in each case was the OMIM database, and every entry was individually and manually annotated. Information about name, accession numbers, sequence features of mRNA, and protein sequence were obtained from RefSeq annotation; disease information was obtained from OMIM; and literature analysis from articles extracted from PubMed. Information about protein domain architecture was obtained by sequence analysis using SMART and Pfam programs as well as the literature (Schultz et al. 1998; Bateman et al. 2002). The information from SMART and Pfam was carefully interpreted by annotators who, in most cases, chose the default thresholds applied by these two programs. Protein domains obtained below the threshold were carefully inspected before rejecting them. The motifs were generally extracted directly from the literature, as many of them are not included in the domain prediction programs.
The information about protein-protein interactions was cataloged after a critical reading of the published literature. Exhaustive searches were done based on keywords and medical subject headings (MeSH) by using Entrez. The type of experiments that served as the basis for establishing protein-protein interactions was also annotated. Experiments such as coimmunoprecipitation were designated in vivo, GST fusion and similar “pull-down” type of experiments were designated in vitro, and those identified by yeast two-hybrid were annotated as yeast two-hybrid.
Posttranslational modifications were annotated based on the type of modification, site of modification, and the modified residue. In addition, the upstream enzymes that are responsible for modifications of these proteins were reported if described in the articles. The most commonly known and the alternative subcellular localization of the protein were based on the literature. The sites of expression of protein and/or mRNA were annotated based on published studies.
Software Development
HPRD was created by using BioBuilder, an annotation tool that we developed during the creation of this database (J.D. Navarro, S. Peri, C.K. Jonnalagadda, B.M. Vrushabendra, S. Vineeth, N. Talreja, A. Pandey, unpubl.). It is based on an extension of ZOPE (Z object publishing environment), an open source application server, which includes several advanced technologies such as ZOPE page templates and a robust object database. Instead of structured query language (SQL), the query engine for relational database management systems, we used ZCatalogs to query the ZOPE object database. ZCatalogs indexes the properties associated with every protein to provide rapid and efficient retrieval of objects or subcomponents of objects from the database. The schema of the database provides an overview of the objects used in the construction of HPRD and can be accessed at the HPRD Web site.
BioBuilder has a content management system in which the information contained in HPRD can be easily modified through an HTML interface. The interface is designed such that the annotation process is greatly facilitated. BioBuilder was used for the administration of review of annotation that was carried in different places of the globe. The information associated with every protein object can be obtained in an extensible markup language (XML) format. The utility of the XML format is that it is a structured data format and lends itself well to importing from and exporting to different types of database systems. This format provides a unique architecture that can be efficiently used to parse the data and export to other file formats, and provides for easy data integration. In this respect, it is important to note that the evolving standards for microarray and proteomic data also use XML for their implementation.
RESULTS AND DISCUSSION
The diversity of protein-related data, in contrast to nucleotide sequence data, presents unique issues that must be taken into account before designing an optimum database system. During the development of HPRD, it was of utmost importance for us to ensure that every category of data was searchable and that the database was visual and user-friendly. We chose to develop a database that is based on a bioinformatics analysis by expert biologists as well as extensive curation of the published literature. With the participation of the biomedical community, this database will be extended to include all the known human proteins. The database is publicly available and can be accessed at http://www.hprd.org. Below, we will discuss some of the unique features of HPRD.
Human Disease Genes in HPRD
In the current release of HPRD, we have annotated a total of 2750 protein sequences. This number includes proteins encoded by 1484 genes that represent all genes with allelic variants that are annotated as linked to a human disease in OMIM database (Hamosh et al. 2002). Our database therefore serves as a nice protein-centric complement to the OMIM database that provides extensive annotations about genes and their variations associated with human diseases.
Asking Biological Questions
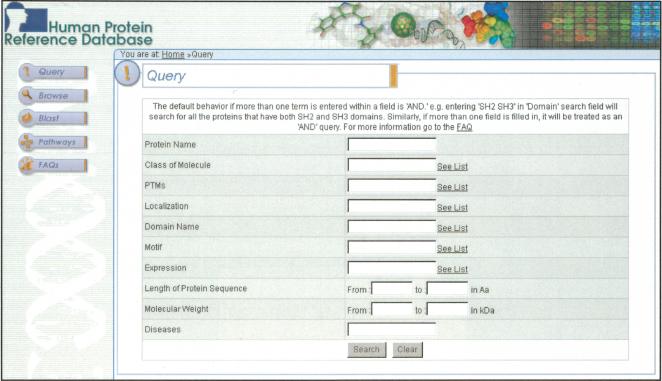
The Web page for the query has been designed to be as simple to use as possible without losing precision. Figure 1 shows a screenshot of the query page, indicating that the proteins can be retrieved based on name, type of protein, domain structure, posttranslational modification, size, localization, function, or involvement in disease. In addition to the query page, a user can find the proteins of interest in three other ways: by browsing through the categories of molecules, by using BLAST to search for homologous proteins, or by visualizing protein networks. The browse mode allows the user to navigate through categories of proteins that belong to a certain class (e.g., G protein-coupled receptors; tyrosine phosphatases), that contain certain domains and motifs (e.g., Sushi domains; RGD motif), that undergo posttranslational modifications (e.g., palmitoylation), or that are localized to certain subcellular compartments (e.g., lysosomes). Using the BLAST feature not only provides the user with an e-value but also displays the domain structure of the retrieved protein(s), which facilitates an intuitive visualization of the protein. The protein pathways provide a graphic view of protein-protein interaction data contained in HPRD and will be discussed in greater detail below.
Figure 1.
The query page of HPRD. A screenshotof HPRD query system shows differentfields by which a user can effectively search the database. The query system allows Boolean searches across the entire database. Pop-up lists that allow a user to choose the terms for querying are provided for most fields.
Domains
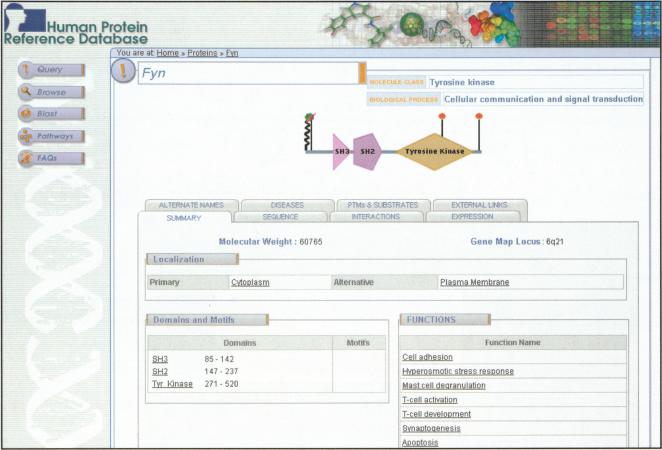
A knowledge of protein domain architecture is essential to assess the function and class of a protein. In the current release, we have annotated 417 protein domains predicted by SMART, Pfam, and the literature. Motifs are generally shorter elements of a protein that are important in protein-protein interactions and function (e.g., coiled-coil; nuclear localization signal). We have annotated 18 types of protein motifs. As more proteins are annotated, these numbers are expected to increase. A screenshot of the molecule page shows the domain architecture along with posttranslational modifications of a cytoplasmic tyrosine kinase, Fyn (Fig. 2).
Figure 2.
A screenshotshowing the molecule page of “Fyn” in HPRD. The molecule page shows a graphic representation of Fyn with its protein domains as polygons and sites of posttranslational modifications as vertical straightor wavy lines (colored symbols atthe end of lines representdifferent posttranslational modifications). Moving the cursor over the graph shows detailed domain names, range of amino acids, type of modification, and site of modification. The tabs guide the user to other annotated fields of the molecule. The summary tab shows a description of molecular weight, locus, subcellular localization, domain architecture details, and functions. For every molecule, the left panel shows links to query, browse, BLAST, and pathways pages. Most annotations are linked to corresponding PubMed articles that are accessible by clicking on the text.
Posttranslational Modifications
A large majority of proteins undergo modifications during or after translation. These co- or posttranslational modifications are crucial for protein stability, sorting, and function. Many modifications are directly related to diseases, and as such, information about these modifications is an important resource to investigate the function of proteins. However, there is a paucity of databases that provide exhaustive information about protein modifications and their representation in the context of protein domains. We have extracted 23 different types of protein modifications (Table 1) from the literature and annotated the modified residue and the enzymes responsible for the modifications. Phosphorylation is one of the most intensively studied protein modifications and occurs on serine, threonine, and tyrosine residues in vertebrates (Hunter 1998). It plays a vital role in cell growth, differentiation, and signal transduction and has important implications in the development of many diseases, including cancers. We have manually extracted a total of >1100 experimentally determined phosphorylation events. Another widespread posttranslational modification is glycosylation (Hart 2003). Glycoproteins have important functions in cell processes, such as protein sorting, immune recognition, receptor binding, and pathogenicity. We have annotated >190 glycosylation sites in the proteins annotated thus far. All posttranslational modifications are depicted visually on the graph of each molecule. In addition to these two modifications, several other types of modifications listed in Table 1 can be used to query or browse the database.
Table 1.
Posttranslational Modifications
| Type of modification | Number of modifications | Number of proteins |
|---|---|---|
| Acetylation | 37 | 16 |
| ADP ribosylation | 6 | 4 |
| Alkylation | 4 | 3 |
| Amidation | 8 | 2 |
| Carboxylation | 45 | 6 |
| Disulphide bridge | 136 | 41 |
| Glycation | 33 | 11 |
| Glycosylation | 191 | 54 |
| GPI anchor | 7 | 6 |
| Hydroxylation | 7 | 2 |
| Methylation | 47 | 23 |
| Myristoylation | 12 | 11 |
| Neddylation | 1 | 1 |
| Nitration | 6 | 2 |
| Palmitoylation | 33 | 19 |
| Phosphorylation | 1104 | 305 |
| Prenylation | 12 | 10 |
| Proteolytic cleavage | 146 | 71 |
| S-Nitrosylation | 4 | 4 |
| Sulfation | 23 | 8 |
| Sumoylation | 16 | 11 |
| Transglutamination | 5 | 2 |
| Ubiquitination | 17 | 8 |
The number of sites annotated by various posttranslation modifications is shown.
The Human Interactome
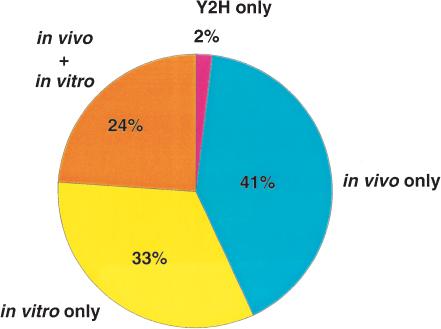
Proteins do not generally function as isolated entities but are often a part of larger protein complexes within a cell. A crucial aspect of any proteomic analysis lies in the elucidation of interacting proteins—the interactome—and in mapping the corresponding binding sites (Walhout and Vidal 2001). By understanding the proteins and their binding partners in the context of a network, a clearer insight into the functioning of a cell can be obtained. We have annotated >10,000 direct unique protein-protein interactions in HPRD that were derived from individual small-scale experiments published in the literature. Protein-protein interactions are sometimes difficult to evaluate because there is no established gold standard for determining protein-protein interactions, with each experimental method having its own limitations. False-positive results (biologically nonsignificant interactions) and false-negative results (missed biological interactions) can occur for a number of reasons. False positives can occur due to nonspecific binding, whereas false negatives can occur because of transient interactions, low abundance of protein expression, or inefficient extraction of certain types of proteins (e.g., hydrophobic proteins). Some of the important high-throughput methods such as yeast two-hybrid (Ito et al. 2000; Uetz et al. 2000) and coimmunoprecipitation and mass spectrometry (Gavin et al. 2002; Ho et al. 2002) have been used to systematically identify protein-protein interactions in yeast. Surprisingly, very little overlap was observed between the different methods implying that such interaction data from high-throughput studies must be interpreted with caution (Bader and Hogue 2002; Schachter 2002; von Mering et al. 2002). In this regard, it is notable that only 2% of the interactions contained in HPRD are derived solely from the yeast two-hybrid system, and more than two thirds of the interactions have been derived from experiments performed in vivo (Fig. 3). In light of the depth and the quality of experimental data, we expect that our database will make it easier to establish a benchmark database for human proteins.
Figure 3.
Distribution of types of experiments used to derive protein-protein interactions in HPRD based on reading of the literature.
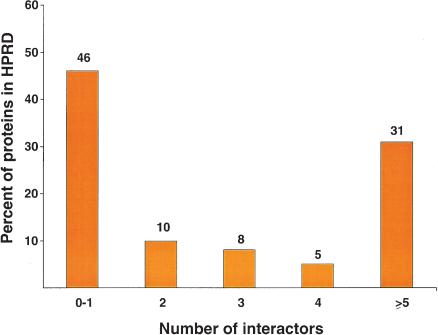
As an illustration, Grb2, is an adapter protein that is involved in diverse pathways ranging from cytoskeletal organization to proliferation to cell-cell communication. Our annotation revealed >200 interaction partners for this protein, which reflects a central role for this molecule as a linker in signal transduction pathways. One important aspect of our annotation of Grb2 was that although it is widely known to localize to the cytoplasm, a careful literature search revealed an alternate nuclear localization as well (Romero et al. 1998). Figure 4 shows the distribution of the number of interacting proteins per entry in HPRD, with the average number of interacting proteins being ∼3.7 per protein. Notably, 44% of proteins have three or more interacting proteins, and it is likely that with continued experiments to elucidate the function of these proteins, this number will increase significantly. Although it is difficult to estimate the total number of interactions in the human proteome, we believe that the minimum number of protein-protein interactions will be >200,000 if a gene count of ∼30,000 is assumed.
Figure 4.
A distribution of the number of interaction partners per protein in HPRD. The histogram shows the distribution of proteins with zero to one, two, three, four, or five or more interaction partners out of a total of 10,534 interactions.
Enzyme/Substrate Relationships
A large number of cellular processes in the cell are amplified by cascades of enzyme/substrate interactions. Typically, enzymes act on substrates to catalyze reactions ranging from phosphorylation or ubiquitination to proteolytic cleavage. In a given pathway, it is important to be aware of the enzymes and their substrates in addition to the other more stable protein-protein interactions that do not depend on any catalytic activity. However, no database currently offers a list of substrates and the enzymes that act upon them. We have annotated >1600 enzymes and substrates in our database. The site of modification on the substrate and the corresponding upstream enzyme is annotated for posttranslational modifications in most cases.
Protein Networks
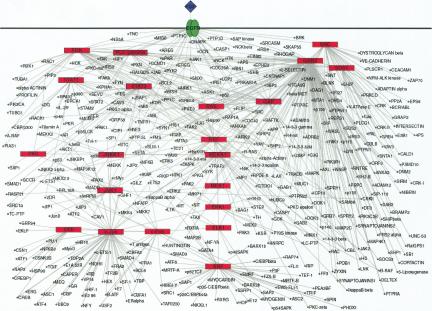
One major advantage of HPRD is that it allows us to construct protein interaction networks for different signaling pathways based on data contained in the database. Figure 5 shows the representation of the Epidermal Growth Factor (EGF) receptor pathway, in which the red boxes show the major proteins, and the circles in the network represent other interaction partners. The graph helps not only to visualize the protein interaction networks but also to potentially identify the function for a novel molecule by placing it in the context of a larger signaling network. Such networks reveal the complexity of patterns for a given class of molecules, pathway, or cellular process made possible by the availability of a large number of interacting proteins extracted from the literature. Thus far, we have generated nine signaling pathway networks and are in the process of expanding this list.
Figure 5.
The EGF receptor signaling pathway generated by using the data contained in HPRD. The graph was drawn by using Pajek program and further manipulated manually (Batagelj and Mrvar 1998). The major signaling molecules involved in the EGF receptor pathway are labeled in red boxes. The interactors of the major proteins are shown as small circles. The graphs are generated in Scalable Vector Graphics (SVG), which allows the user to zoom in without any loss of resolution. Clicking any node links to the corresponding molecule page in HPRD.
BRCA1 as a Representative Entry in HPRD
We will provide BRCA1 as an example to illustrate the breadth and depth of annotation in HPRD and to highlight the importance of manual annotation of the entries. BRCA1 is a transcription factor that is mutated in certain forms of cancers and has been extensively studies over the past several years (Venkitaraman 2002). By a careful analysis of the literature, we were able to catalog 62 proteins that interact with BRCA1. The interacting domains/regions are indicated in most cases and can be easily visualized by clicking the “visualize interactions” button. For instance, the N terminus of BRCA1 is responsible for homodimerization with BRCA1 and for heterodimerization with BARD1, and its C-terminal region or BRCT domains mediate binding to BRCA2, RB, p53, CtIP, and RNA helicase A, among others. Eighty percent of the interactions were based on biochemical evidence that was derived from either in vivo or both in vitro and in vivo type of experiments. The remaining 20% of the interactions were based on in vitro interactions alone (including yeast two-hybrid), and none of that interaction data for BRCA1 was based on yeast two-hybrid data alone.
BRCA1 has been clearly demonstrated to be localized to the nucleus by several different groups. However, alternative localization to the cytoplasm and the centrosome can be easily found by browsing the BRCA1 entry in HPRD along with direct links to the primary literature. BRCA1 is annotated as phosphorylated on nine serine and one threonine residues by four different upstream kinases. Although it is widely known even to the nonspecialist that BRCA1 is implicated in breast and ovarian cancers, the entry in HPRD shows that it is also mutated in two other cancers, including prostate cancer. Similarly, although BRCA1 is well known to be expressed in breast and ovarian tissues, the HPRD entry indicates that it is also expressed in the thymus, testis, and lymphocytes in addition to several other tissues. In this regard, it is interesting to note that when BRCA1 was originally cloned as a candidate susceptibility gene for breast and ovarian cancers, it was clearly noted that it is more abundantly expressed in the thymus and testis than in breast and ovarian tissues (Miki et al. 1994). Finally, BRCA1 can be visualized as a direct interaction partner of a major signaling molecule in five out of nine receptor-mediated signaling pathways that are linked to HPRD, including IL-2, erythropoietin, and Fas receptor signaling pathways. This intimate involvement of BRCA1 in these hematopoietic pathways coupled to information about its expression in lymphocytes and thymus is very easily appreciated by using HPRD, which might otherwise be difficult to assimilate from a plethora of publications and numerous databases. It is hoped that inferences made from such connections would lead to generation of new hypotheses and experimentation to test them.
Conclusions
Our database complements other protein-protein interaction databases that are mostly focused on the yeast proteome, including BIND (Bader et al. 2001) and DIP (Xenarios et al. 2000), or on signaling proteins found in certain cell types such as that by the Alliance for Cell Signaling (Gilman et al. 2002). We have annotated 2750 proteins in total, including 10,534 unique protein-protein interactions. We have provided >25,000 PubMed links to various fields that direct a user to the relevant primary literature. Table 2 shows the statistics of the current release of HPRD: These numbers will grow with continued annotation efforts. It took ∼50,000 person hours over 8 months to develop the software and to read >300,000 research articles for the initial manual annotation of 2750 proteins currently residing in HPRD. Indeed, we anticipate completing annotation of ∼10,000 proteins by the end of 2003. However, a truly error-free and comprehensive database is impossible without the involvement of the biomedical community—no one can do a better job of annotations than those working on the proteins themselves. In this regard, we have provided a comment button for every molecule that will facilitate feedback from users. These user comments will allow us to correct any errors and to update published data concerning annotated proteins in addition to our own ongoing efforts to enrich and update the data. The information contained in HPRD is open to the scientific community—the source code of BioBuilder is freely available under the Lesser General Public License conditions.
Table 2.
HPRD Statistics
| Feature | Number of proteins |
|---|---|
| Total number of annotated proteins | 2750 |
| Number of protein-protein interactions | 10,534 |
| Number of posttranslational modification events | 1900 |
| Number of types of domains and motifs | 435 |
| Number of substrates | 1610 |
The total number of entries in the various fields in HPRD are shown.
We plan to integrate publicly available microarray data into HPRD in the near future. This will facilitate a gene-centric view to determine whether the mRNA expression pattern of a given gene is reported to be altered by any published study. Microarray users can already take advantage of our detailed annotation to classify proteins in several ways to generate novel hypotheses or to narrow down the likely candidates involved in a biological process. This database will be a valuable resource for the proteomic community as well because a majority of posttranslational modifications have a fixed molecular mass that will allow more precise searches of the protein database. The information about site of expression, subcellular localization, and association with diseases will be invaluable in experiments involving subproteomes. The wealth of information of HPRD will play a crucial role in adopting integrative approaches to interpret high-throughput experimental data such as those derived from microarrays and proteomic experiments that are critically dependent upon existence of good databases. We believe that this database will strengthen efforts at careful manual analysis and interpretation of the role of genes and proteins in complex systems and will become a knowledge base for the human proteome in the near future.
Acknowledgments
Dr. Pandey serves as Chief Scientific Advisor to the Institute of Bioinformatics. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1680803.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader, G.D. and Hogue, C.W. 2002. Analyzing yeast protein-protein interaction data obtained from different sources. Nat. Biotechnol. 20: 991-997. [DOI] [PubMed] [Google Scholar]
- Bader, G.D., Donaldson, I., Wolting, C., Ouellette, B.F., Pawson, T., and Hogue, C.W. 2001. BIND: The Biomolecular Interaction Network Database. Nucleic Acids Res. 29: 242-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batagelj, V. and Mrvar, A. 1998. Pajek: Program for large network analysis. Connections 21: 47-57. [Google Scholar]
- Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S.R., Griffiths-Jones, S., Howe, K.L., Marshall, M., and Sonnhammer, E.L. 2002. The Pfam protein families database. Nucleic Acids Res. 30: 276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney, E., Clamp, M., and Hubbard, T. 2002. Databases and tools for browsing genomes. Annu. Rev. Genomics Hum. Genet. 3: 293-310. [DOI] [PubMed] [Google Scholar]
- Boeckmann, B., Bairoch, A., Apweiler, R., Blatter, M.C., Estreicher, A., Gasteiger, E., Martin, M.J., Michoud, K., O'Donovan, C., and Phan, I. 2003. The SWISS-PROT protein knowledge base and its supplement TrEMBL in 2003. Nucleic Acids Res. 31: 365-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, P.O. and Botstein, D. 1999. Exploring the new world of the genome with DNA microarrays. Nat. Genet. 21: 33-37. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494-498. [DOI] [PubMed] [Google Scholar]
- Gavin, A.C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J.M., Michon, A.M., and Cruciat, C.M. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141-147. [DOI] [PubMed] [Google Scholar]
- Gilman, A.G., Simon, M.I., Bourne, H.R., Harris, B.A., Long, R., Ross, E.M., Stull, J.T., Taussig, R., Arkin, A.P., and Cobb, M.H. 2002. Overview of the Alliance for Cellular Signaling. Nature 420: 703-706. [DOI] [PubMed] [Google Scholar]
- Hamosh, A., Scott, A.F., Amberger, J., Bocchini, C., Valle, D., and McKusick, V.A. 2002. Online Mendelian Inheritance in Man (OMIM): A knowledge base of human genes and genetic disorders. Nucleic Acids Res. 30: 52-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanash, S. 2003. Disease proteomics. Nature 422: 226-232. [DOI] [PubMed] [Google Scholar]
- Hart, G.W. 2003. Structural and functional diversity of glycoconjugates: A formidable challenge to the glycoanalyst. Methods Mol. Biol. 213: 3-24. [DOI] [PubMed] [Google Scholar]
- Ho, Y., Gruhler, A., Heilbut, A., Bader, G.D., Moore, L., Adams, S.L., Millar, A., Taylor, P., Bennett, K., and Boutilier, K. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180-183. [DOI] [PubMed] [Google Scholar]
- Hood, L. and Galas, D. 2003. The digital code of DNA. Nature 421: 444-448. [DOI] [PubMed] [Google Scholar]
- Hunter, T. 1998. The Croonin lecture 1997: The phosphorylation of proteins on tyrosin: Its role in cell growth and disease. Philos. Trans. R Soc. Lond. B Biol. Sci. 353: 583-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., Tashiro, K., Muta, S., Ozawa, R., Chiba, T., Nishizawa, M., Yamamoto, K., Kuhara, S., and Sakaki, Y. 2000. Toward a protein-protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc. Natl. Acad. Sci. 97: 1143-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin, M. and Ben-Neriah, Y. 2000. Phosphorylation meets ubiquitination: The control of NF-κB activity. Annu. Rev. Immunol. 18: 621-663. [DOI] [PubMed] [Google Scholar]
- Kitano, H. 2002. Computational systems biology. Nature 420: 206-210. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Miki, Y., Swensen, J., Shattuck-Eidens, D., Futrealm P.A., Harshman, K., Tavtigian, S., Liu, Q., Cochran, C., Bennett, L.M., Ding, W., et al. 1994. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266: 66-71. [DOI] [PubMed] [Google Scholar]
- Nakai, K. 2000. Protein sorting signals and prediction of subcellular localization. Adv. Protein Chem. 54: 277-344. [DOI] [PubMed] [Google Scholar]
- Navarro, J.D., Niranjan, V., Peri, S., Jonnalagadda, C.K., and Pandey, A. 2003. From biological databases to platforms for biomedical discovery. Trends Biotechnol. 21: 263-268. [DOI] [PubMed] [Google Scholar]
- Pandey, A. and Mann, M. 2000. Proteomics to study genes and genomes. Nature 405: 837-846. [DOI] [PubMed] [Google Scholar]
- Pawson, T. and Nash, P. 2003. Assembly of cell regulatory systems through protein interaction domains. Science 300: 445-452. [DOI] [PubMed] [Google Scholar]
- Pruitt, K.D., Katz, K.S., Sicotte, H., and Maglott, D.R. 2000. Introducing RefSeq and LocusLink: Curated human genome resources at the NCBI. Trends Genet. 16: 44-47. [DOI] [PubMed] [Google Scholar]
- Romero, F., Ramos-Morales, F., Dominguez, A., Rios, R.M., Schweighoffer, F., Tocque, B., Pintor-Toro, J.A., Fischer, S., and Tortolero, M. 1998. Grb2 and its apoptotic isoform Grb3-3 associate with heterogeneous nuclear ribonucleoprotein C, and these interactions are modulated by poly(U) RNA. J. Biol. Chem. 273: 7776-7781. [DOI] [PubMed] [Google Scholar]
- Schachter, V. 2002. Bioinformatics of large-scale protein interaction networks. Biotechniques (Suppl): 16-27. [PubMed]
- Schultz, J., Milpetz, F., Bork, P., and Ponting, C.P. 1998. SMART: A simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. 95: 5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz, P., Giot, L., Cagney, G., Mansfield, T.A., Judson, R.S., Knight, J.R., Lockshon, D., Narayan, V., Srinivasan, M., and Pochart, P. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623-627. [DOI] [PubMed] [Google Scholar]
- Venkitaraman, A.R. 2002. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108: 171-182. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- von Mering, C., Krause, R., Snel, B., Cornell, M., Oliver, S.G., Fields, S., and Bork, P. 2002. Comparative assessment of large-scale data sets of protein-protein interactions. Nature 417: 399-403. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J. and Vidal, M. 2001. Protein interaction maps for model organisms. Nat. Rev. Mol. Cell Biol. 2: 55-62. [DOI] [PubMed] [Google Scholar]
- Xenarios, I., Rice, D.W., Salwinski, L., Baron, M.K., Marcotte, E.M., and Eisenberg, D. 2000. DIP: The database of interacting proteins. Nucleic Acids Res. 28: 289-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H., Bilgin, M., Bangham, R., Hall, D., Casamayor, A., Bertone, P., Lan, N., Jansen, R., Bidlingmaier, S., and Houfek, T. 2001. Global analysis of protein activities using proteome chips. Science 293: 2101-2105. [DOI] [PubMed] [Google Scholar]
- Ziauddin, J. and Sabatini, D.M. 2001. Microarrays of cells expressing defined cDNAs. Nature 411: 107-110. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.hprd.org; Human Protein Reference Database.
- http://www.ibioinformatics.org; Institute of Bioinformatics home page.