Abstract
Mitochondrial transcripts of two ascidian species were reconstructed through sequence assembly of publicly available ESTs resembling mitochondrial DNA sequences (mt-ESTs). This strategy allowed us to analyze processing and mapping of the mitochondrial transcripts and to investigate the gene organization of a previously uncharacterized mitochondrial genome (mtDNA). This new strategy would greatly facilitate the sequencing and annotation of mtDNAs. In Ciona intestinalis, the assembled mt-ESTs covered 22 mitochondrial genes (∼12,000 bp) and provided the partial sequence of the mtDNA and the prediction of its gene organization. Such sequences were confirmed by amplification and sequencing of the entire Ciona mtDNA. For Halocynthia roretzi, for which the mtDNA sequence was already available, the inferred mt transcripts allowed better definition of gene boundaries (16S rRNA, ND1, ATP6, and tRNA-Ser genes) and the identification of a new gene (an additional Phe-tRNA). In both species, polycistronic and immature transcripts, creation of stop codons by polyadenylation, tRNA signal processing, and rRNA transcript termination signals were identified, thus suggesting that the main features of mitochondrial transcripts are conserved in Chordata.
Expressed Sequence Tags (EST) are short single-pass sequence reads derived from cDNA clones selected randomly from cDNA libraries (Adams et al. 1991). As such, they represent a snapshot of the transcript inventory expressed in a given tissue, cell line, or developmental stage of an organism. For most model organisms, both EST and genome projects have been undertaken because the EST projects complement genome-sequencing efforts in the identification of new genes, the prediction and mapping of transcribed regions, and so on. In contrast to genomic sequences, ESTs are generally of low quality, poorly annotated, and highly redundant. Common EST features include ∼2% sequence error rate with high frequency of insertions and deletions, redundancy of sequences derived from highly expressed genes, and low representation of genes expressed at low levels. Moreover, ESTs may derive from unspliced immature mRNAs, alternative splicing and polyadenylation sites, cloning artifacts (chimerisms), and mitochondrial (mt) transcripts. Depending on tissue source and RNA extraction/fractioning methods, a remarkably high fraction of ESTs from a given cDNA library can be of mitochondrial origin (Hillier et al. 1996; Welle et al. 1999; Laveder et al. 2002). We can take advantage of mt-like ESTs to obtain information on mitochondrial transcription in organisms in which the mt genome (mtDNA) sequence is already known, or even to obtain data on mt gene organization and expression in organisms in which mtDNA sequence data are unavailable. Mitochondrial ESTs may help to correctly identify and/or to confirm mt gene boundaries, because at present, mt genes are mostly inferred by sequence similarity with previously published mtDNAs. Thus, initially incorrect gene annotations may be conserved and reproduced in newly sequenced mtDNA. Moreover, the boundaries of rRNA genes are commonly inferred as the entire region included between the two adjacent genes.
To date, the mitochondrial genomes of a wide range of organisms, from protozoa to fungi and animals, have been sequenced. However, transcription mechanisms have been investigated extensively in only a few species. It has been proposed that mitochondrial transcription is dependent on overall genome organization, rather than on phylogenetic lineage (Richard et al. 1998). Compact mt genomes, with no, or short intergenic spacers, are transcribed from few promoters to yield a small number of large polycistronic primary transcripts. These are further processed, mostly by tRNA removal (i.e., animal mtDNA, the green alga Prototheca wickerhamii, the fungi Aspergillus nidulans, and Neurospora crassa) (Ojala et al. 1981; Burger et al. 1985; Dyson et al. 1989; Wolff and Kuck 1996), but also by cleavage inside tRNAs (the red alga Chondrus crispus) (Richard et al. 1998, 1999) or at other specific primary or secondary structures (Burger et al. 1985; Dyson et al. 1989; Wolff and Kuck 1996). Large mt genomes with relaxed packaging of genetic information are transcribed into multiple small transcripts from individual promoters scattered over the genome (i. e., the yeast Saccharomyces cerevisiae and plants) (Costanzo and Fox 1990; Binder et al. 1996). The details of transcription have been investigated widely only in mammals (human and rodents), whereas partial data on mapping and processing of mt transcripts are available for a few other animals, such as sea urchin Paracentrotus lividus (Elliott and Jacobs 1989; Cantatore et al. 1990), and Drosophila (Berthier et al. 1986). In mammals, the promoters for transcription of the mtDNA heavy (H) and light (L) strands are located in the D-loop region and lead to the synthesis, for each strand, of a long primary transcript covering the whole or almost the whole genome. A third smaller transcript covers only the H-strand rRNA region. An early termination signal downstream of the 16S rRNA gene enhances the transcription of the rRNA region 15- to 100-fold compared with the rest of the mitochondrial genes (Christianson and Clayton 1986). The primary transcripts are processed to mature transcripts by excision of the tRNA sequences acting as punctuation points (Ojala et al. 1981). Mature mRNAs are monocistronic or bicistronic with two overlapping protein-coding genes, and carry poly(A) tails ∼50–60 nucleotides long (Hirsch and Penman 1973, 1974). The precursor containing the two rRNAs has been found in both the oligo(dT)-cellulose bound and unbound fraction (Montoya et al. 1983). The mature 12S and 16S rRNAs are both oligoadenylated; the A tail is of variable length, with the 3′-termini of 16S rRNA showing a greater heterogeneity and a longer A tail than the 12S rRNA (Dubin et al. 1981, 1982; Van Etten et al. 1983). An additional polyadenylated fraction of the 16S rRNA has also been found in human and hamster (Dubin et al. 1981; Montoya et al. 1983), but kinetic studies of human mt transcripts have excluded it from being a precursor of the mature rRNA (Gelfand and Attardi 1981). In general, the adenylation has been proposed to play a role in processing and termination of transcripts (Dubin et al. 1981; Montoya et al. 1983).
No data on mt transcription are available in Urochordata, which is the basal Chordata subphylum; moreover, the mtDNA has been completely sequenced only for the Ascidiacea Halocynthia roretzi (Yokobori et al. 1999). Such mtDNA is quite compact and contains 12 protein-coding genes, 2 rRNAs, and 23 tRNAs. Compared with the gene content of Chordata, this genome contains an additional tRNA-Gly gene, related to the use of a peculiar genetic code (Durrheim et al. 1993; Yokobori et al. 1993). Surprisingly, all of the genes are encoded on the G-rich strand, and the gene order is completely different from the almost-frozen gene order of other Chordata (Boore 1999; Saccone et al. 1999). Such features make the Urochordata a very interesting group to study the evolution of mtDNA, both at level of sequence and transcription/replication mechanisms. The availability of a significant number of EST sequences for two ascidian species, Halocynthia roretzi and Ciona intestinalis, allows the investigation of the power of mt-like ESTs in the study of transcription and processing mechanisms of mtDNA, the prediction of mitochondrial gene order and sequence of new mtDNAs, and in genome annotation.
RESULTS
Ciona Mitochondrial Transcripts
A total of 996 EST sequences from Ciona intestinalis were assembled in 11 contigs, with 5 contigs forming two links (i.e., such non-overlapping contigs are linked by the presence of ESTs derived from the same cDNA clone; Table 1). The singletons represent only 2.2% of the assembled ESTs, and most of them (16 of 22) were selected by sharing of clone ID with mt-like ESTs, so are probably the results of cloning artifacts. Typical mitochondrial-encoded genes were unequivocally identified in most contigs using only the criterion of similarity in primary sequence and/or secondary structures (data not shown). The protein-gene boundaries were unambiguously identified in all contigs except for those containing ND5 and ND2. In the ND5 contig (Ci-10), the first 313 bp derive from a single EST, so the uncertain gene boundary identification might be explained by the intrinsic sequencing errors of ESTs, which were, in this case, not compensated by a consensus obtained from multiple ESTs. Similarly, the ND2 frame (Ci-5) showed a shift in a contig region formed by the assembly of only three ESTs. Two contigs, Ci-4 and Ci-9, resulted both similar to the mitochondrial 16S rRNA gene; however, Ci-4 shows only a 35-bp long region highly similar to animal 16S rRNAs, whereas Ci-9 is surprisingly almost identical over the whole length to the 16S rRNA of Halocynthia (99.35% identity). Considering that Ci-4 is part of a link that also includes ND1 and ND3 genes, we suggest that it actually represents the Ciona 16S rRNA gene, whereas Ci-9 may represent contamination sequences from Halocynthia.
Table 1.
Mapping of the Ciona intestinalis Mitochondrial-Like Contigs Inferred From EST Sequences on the Corresponding Complete mtDNA Sequence (AJ517314)
|
Contig
|
mtDNA
|
|||||
|---|---|---|---|---|---|---|
| No. EST | L (bp) | From.. .To | % Id | Gap | Genesa | |
| Link2/Ci-4 | 2 | 719 | 8177.. .8894 | 98.6 | 1 | 16S rRNA partial |
| Link2/Ci-11 | 30 | 945 | 6797.. .7740 | 99.6 | — | ND1 end + Nc |
| Link2/Ci-12 | 25 | 464 | 8915.. .9369 | 99.1 | — | I partial + ND3 + E |
| Ci-1 | 309 | 1646 | 11306.. .12909 | 99.2 | — | S2 partial + CO1 + Nc |
| Ci-36 | 525 | 3215 | 3459.. .6663 | 99.0 | 2 | A partial + CO2 + Cytb + P + ND4L + H + R + Q + CO3 + Nc |
| Ci-5 | 38 | 1689 | 1757.. .3434 | 99.6 | 1 | ATP6 partial + ND2 + Nc |
| Ci-8 | 35 | 1533 | 9456.. .10958 | 99.6 | — | ND4 + F + ATP8 |
| Ci-10 | 25 | 1697 | 13044.. .14724 | 99.3 | 6 | ND5 partial |
| Total | 989 | 11908 | 99.3 | 10 | ||
| Ci-9 | 3 | 620 | 12452.. .13074 | 99.3 | 2 | partial 16Sb |
| Link1/Ci-2 | 2 | 677 | 7799.. .8455 | 98.6 | 2 | CO2 endc |
| Link1/Ci-7 | 2 | 974 | 3337.. .4294 | 91.2 | — | ND2 endc |
For each link/contig are shown matching coordinates and percent identity (% Id) with the mtDNA of Ciona intestinalis.
tRNA genes are named with the one-letter code of the charged amino acid.
Comparison with Halocynthia roretzi mtDNA (AB024528).
Comparison with Paracentrotus lividus mtDNA (J04815).
Two other contigs were certainly not originated by Ciona sequences. Ci-2 and Ci-7, derived both from one clone of the same cDNA library (M.Branno, unpubl.), showed a surprisingly high nucleotide identity of 98.63% and 91.12% to sea urchin CO2 and ND2 genes, respectively. As reported in the EMBL entries, this cDNA library may be contaminated with unspecified eukaryotic sequences, therefore, our data strongly suggest the origin of these contigs from sea urchin, rather than from Ciona intestinalis.
The eight contigs considered as bona fide Ciona mitochondrial transcripts cover 11,908 bp, and their sequences were used to design primers specific for the amplification of the complete mtDNA of Ciona intestinalis (C. Gissi, F. Iannelli, and G. Pesole, in prep.). Figure 1 reports the Ciona mt contigs mapped on the completely sequenced Ciona intestinalis mtDNA. Such contigs match to 22 mitochondrial genes as follows:
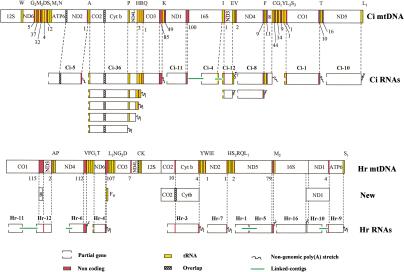
Figure 1.
Mapping of mt-like contigs/links on the complete mitochondrial genome of the corresponding organism, Ciona intestinalis (Ci) and Halocynthia roretzi (Hr). In both species, all genes are encoded on the same strand. Putative mitochondrial transcripts are indicated as RNAs and named in accordance with Tables 1 and 3. Links are connected by continuous green lines. Proposed modifications in the annotation of Halocynthia mtDNA are indicated as New. Partial genes are indicated as open boxes. Numbers below the mtDNA refer to the length (in base pairs) of the noncoding regions indicated in red. (ATP6) ATP synthase subunit 6; (8) ATP synthase subunit 8; (CO1–CO3) cytochrome c oxidase subunits; (Cyt b) cytochrome b; (ND1–ND6 and ND4L) NADH dehydrogenase subunits; (12S) small subunit ribosomal RNA; (16S) large subunit ribosomal RNA. tRNA genes, in yellow, are indicated by the amino acid transported. (L1) Leu (UUR); (L2) Leu (CUN); (S1) Ser (AGY); (S2) Ser (UCN); (G1) Gly (AGR); (G2) Gly (GGN); (M1) Met (AUG); (M2) Met (AUA).
an internal region of the 16S rRNA gene;
the complete coding region of eight protein genes (CO1, CO2, CO3, Cytb, ND2, ND3, ND4L, and ATP8);
the 3′-end region of four protein-coding genes (ND1, ND4, ND5, and ATP6);
six complete tRNA genes (tRNA-Glu, -Phe, -Pro, -His, -Arg, and -Gln);
three partial tRNA genes (tRNA-Ala, -SerUCN, and Ile).
The mtDNA sequencing confirmed the sequences and the gene order predicted from the putative transcripts inferred from ESTs and confirmed the contig Ci-4 to match the actual 16S rRNA gene. The assembled contigs resulted almost identically to the determined mtDNA sequence, showing >99% identity and only 10 indels (0.08%; Table 1).
Several contigs carried an ending poly(A) stretch confirmed by the assembly of a number of EST sequences (Table 2) not present in the genomic sequence (Fig. 1). It is noteworthy that such poly(A) mark exactly the 5′ end of a downstream tRNA (Fig. 1). Only the Ci-11 poly(A) downstream of ND1 was present in both the EST contig and the mtDNA sequence. Additional poly(A) stretches matching internal regions of contigs and absent in the contig consensus and genomic sequences were found in Ci-12, Ci-8, and Ci-36 (see Fig. 1 and Table 2). Even such internal poly(A) stretches started exactly at the 5′ end of a downstream tRNA gene (tRNA-Glu in Ci-12; tRNA-Phe in Ci-8; tRNA-Pro, -His, -Arg, and -Gln in Ci-36). Thus, the poly(A) stretches likely represent poly(A) tails added during the maturation process of a longer transcript, and are probably involved also in the maturation of tRNA transcripts.
Table 2.
Number of ESTs With poly(A) Stretches at the Ends of the Ciona Contigs and Stop Codon of the Protein-Coding Gene Located Immediately Before the poly(A) Stretch
| Contig | Genes | No. poly(A) ESTs | Stop codon |
|---|---|---|---|
| Ci-1 | CO1-Nc | 16 | |
| Ci-12 | ND3 | 1 | TAA |
| ND3-E | 9 | ||
| Ci-8 | ND4-Nc | 2 | |
| ND4-Nc-F-Nc-ATP8 | 6 | TA(A) | |
| Ci-36 | CO2-Cytb | 7 | TA(A) |
| CO2-Cytb-P-ND4L* | 6 | T(AA) | |
| CO2-Cytb-P-ND4L* | 12 | TAG | |
| CO2-Cytb-P-ND4L-H-Nc | 1 | ||
| CO2-Cytb-P-ND4L-H-Nc-R | 1 | ||
| CO2-Cytb-P-ND4L-H-Nc-R-Q-Nc-CO3-Nc | 17 | ||
| Ci-10 | ND5 | 5 | T(AA) |
| Ci-11 | ND1-Nc | 4 | |
| Ci-5 | ATP6-ND2-Nc | 2 |
tRNA genes are indicated by the amino acid transported. Noncoding regions are indicated as Nc. Parentheses indicate A bases belonging to the poly(A) tail. Asterisk indicates alternative polyadenylation sites at the stop codon of ND4L gene.
In some contigs, the poly(A) stretches were implicated in the creation of complete stop codons from incomplete T or TA codons (Table 2). In Ci-36, the Cytb frame stops with a TAG codon inside the following tRNA-Pro gene; however, an internal poly(A) tail starts 9 bp upstream of such a TAG codon, creating a TAA stop codon from an incomplete TA codon. Without the transcription data reported here, a 9-bp overlap between Cytb and tRNA-Pro would probably have been proposed. In the same contig, two alternative poly(A) start sites can be identified in the stop codon of the ND4L gene; one suggests the usage by ND4L of an incomplete T/TA stop codon (6 ESTs), and the other indicates the usage of a complete TAG stop codon (12 ESTs). In Ci-10 and Ci-8, the TAA stop codon of ND5 and ATP8 could also be created by polyadenylation.
Two cases of overlapped genes, expressed as transcripts with no poly(A) tails downstream of the first ORF, suggest the presence of mature bicistronic transcripts.
In Ci-5, the use of a start ATA codon in the ND2 frame implies an overlap of 31 bp with ATP6, but only a 1-bp overlap is observed if the start codon is the in-frame downstream ATG.
In Ci-36, CO2 and Cytb are also overlapped. The first 29 bp of the Cytb frame overlap the end of the CO2 frame. Seven possible start codons are present in the overlapping region, including a single ATG codon that would create an overlap of 25 bp.
The mtDNA sequence confirmed that overlaps were found in all cases of adjacent protein-coding genes with no tRNA genes in between. In Ci-36, the overlap of a single nucleotide between tRNA-Arg and tRNA-Gln could be resolved by polyadeanylation of the upstream transcript.
Transcribed noncoding regions (Nc) longer than 20 bp were identified at the 3′ end of Ci-36 and Ci-11 contigs, that is, downstream of CO3 and ND1, respectively (Fig. 1).
Halocynthia Mitochondrial Transcripts
A total of 453 Halocynthia ESTs were assembled in 4 contigs and 4 links highly similar to the Halocynthia mtDNA sequence (Table 3). Only the 5′ portion of Hr-link2 showed a statistically highly significant similarity to a nonmitochondrial gene (a mouse glycoprotein; Tblastx E-value: 8e-66), whereas other contigs and links were easily mapped on the mitochondrial genome of Halocynthia (Fig. 1). Together, they contain the partial or complete regions of 13 mitochondrial genes (tRNA-Thr, 16S rRNA, and 11 protein-coding genes, including the previously undetected ATP8 gene).
Table 3.
Mitochondrial-Like Contigs and Links Obtained From the Assemble of Halocynthia roretzi EST Sequences
| From...To
|
|||||||
|---|---|---|---|---|---|---|---|
| Name | EST No. | L (bp) | Contig | mDNA | % Id | Genes | No. poly(A) ESTs |
| Link1/Hr-1 | 3 | 698 | 1...698 | 10219...10921 | 96.4 | ND5 partial | — |
| Link1/Hr-5 | 3 | 700 | 1...683 | 11242...11925 | 96.1 | ND5 end + Nc | 2 |
| Link2/Hr-2 | 2 | 731 | — | — | — | Mouse glycoprotein (AJ012754) | — |
| Link2/Hr-6 | 2 | 612 | 1...579 | 3044...3624 | 97.1 | ND4 end + Nc | 2 |
| Hr-3 | 19 | 1788 | 1...1766 | 6809...8544 | 97.6 | CO2 end + Cytb | 8 |
| Hr-4 | 2 | 542 | 10...520 | 3890...4399 | 94.3 | T end + ND6 + Nc | 1 |
| Hr-7 | 4 | 956 | 1...930 | 8915...9839 | 97.4 | ND2 end | 2 |
| Link3/Hr-9 | 3 | 705 | 1...683 | 14027...14702 | 98.5 | ND1 end + ATP6 | 2 |
| Link3/Hr-10 | 1 | 562 | 557...1 | 13114...13671 | 96.2 | 16S rRNA end + ND1 start | — |
| Link4/Hr-11 | 1 | 571 | 1...566 | 181...748 | 98.6 | CO1 partial | — |
| Link4/Hr-12 | 1 | 641 | 1...641 | 1424...2065 | 99.2 | COI end + ATP8 + ND3 | — |
| Hr-16 | 412 | 1146 | 21...1121 | 11998...13100 | 99.0 | 16S rRNA partial | 151 |
| Total | 453 | 9652 | 97.5 | ||||
The boundaries of the similarity region between the contigs and the Halocynthia roretzi mtDNA sequence (AB024528) and the respective identity percentage are reported. tRNA genes are indicated by the amino acid transported. (Nc) noncoding region. Gene boundaries according to Yokobori et al. (1999).
The number of ESTs with a poly(A) stretch is also reported for each contig.
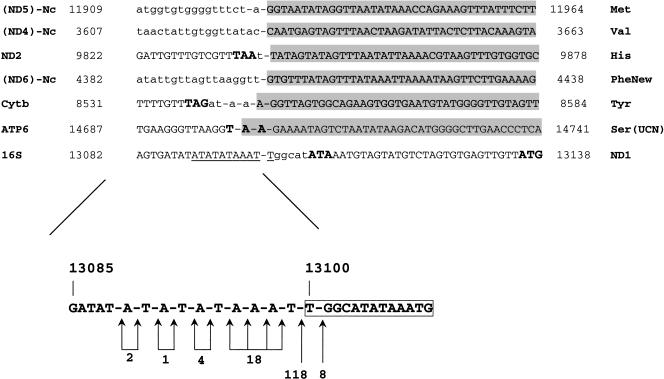
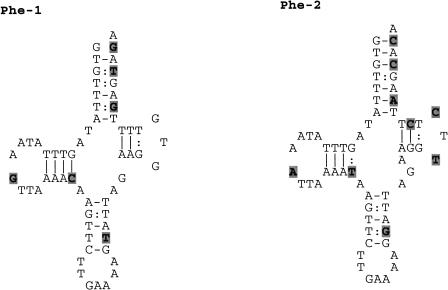
At the 3′ end of most contigs/links, a number of ESTs showed a poly(A) stretch absent in the mtDNA sequence (Table 3, Fig. 2). Such poly(A) stretch starts from 1–112 bp downstream of the stop codon of a protein-coding gene, implying the presence of a 3′ untranslated region in the transcript, but in the ATP6 contig in which the poly(A) stretch contributes to the formation of the TAA stop codon. The ATP6 gene is annotated in the mt genome with a 2-bp overlap with the next tRNA-Ser(UCN) gene (AB024528), a situation that implies the existence of two alternative processing/termination mechanisms to produce functional forms of both transcripts. The creation of the ATP6 stop codon by polyadenylation would exclude the overlap between the genes and allow the formation of functional transcripts of both genes with a single event. As found in Ciona, in all contigs, the poly(A) started exactly at the 5′ end of a tRNA gene, or, when the mtDNA template encoded A residues, was consistent with the 5′ end of a tRNA (Fig. 2). Only in the ND6 contig (Hr-4) have we found a poly(A) stretch starting inside a 107-bp long noncoding region (Nc). Careful analysis of this region showed that exactly downstream of this poly(A) start site, a typical tRNA cloverleaf structure with a 85.7% identity to the annotated tRNA-Phe can be found. Furthermore, the new tRNA-Phe gene shows a more canonical secondary structure than the previously identified gene (Fig. 3). The boundaries of the original tRNA-Phe and of the newly identified tRNA-Phe gene on the mtDNA sequence (AB024528) are 3696–3757 and 4400–4462, respectively.
Figure 2.
Location of the poly(A) stretches of Halocynthia contigs on the corresponding mtDNA sequence, with numbers referred to the positions on the mtDNA (AB024528). Dashes indicate the poly(A) start site. Presence of genes and/or noncoding regions (Nc) downstream and upstream of such sites is indicated. All of the alternative poly(A) start sites found in the 16S rRNA contig are reported in the lower box, with numbers below arrows referring to the ESTs supporting that start site. (Lowercase) Noncoding region; (grey background) tRNA sequences; (boxed sequence) rRNA transcription termination box; (bold) putative start codons and stop codons.
Figure 3.
Cloverleaf secondary structure of the annotated tRNA-Phe (Phe-1, location: 3694–3757) and the newly identified tRNA-Phe (Phe-2, location: 4400–4462) genes of Halocynthia roretzi mtDNA (AB024528). Nucleotide differences are reported in gray background.
In the 16S rRNA contig, there are at least six different sites in which poly(A) stretches start. All are concentrated in an AT-rich region, 11 bp long, located inside the predicted 16S rRNA gene, with the most frequent poly(A) start site found in 118 ESTs of a total of 151 covering this region (Fig. 2). The downstream sequence contains no tRNA structures, but there is a sequence showing a single mismatch to the conserved 13-mer TGGCAGAn5G rRNA transcription termination signal, which is responsible for the transcription termination downstream of the rRNAs in the human mtDNA (Christianson and Clayton 1986). In the whole Halocynthia mtDNA, this consensus termination box is found only once at position 13100. The multiple poly(A) start sites and the rRNA termination box clearly indicated a shorter 16S rRNA gene than has been proposed, with a 3′ end located about 50 bp upstream of the annotated end (Yokobori et al. 1999) (position 13100 against the annotated position 13165). Furthermore, this observation, together with the starting of ND1 contig at position 13109 inside the annotated 16S rRNA gene, would also suggest a longer ND1 gene (start position 13136 or 13106 instead of the annotated position 13166). Thus, the assembled ESTs also provided detailed information on gene boundaries.
The four noncoding regions longer than 20 bp, described by Yokobori et al. (1999), are all represented in the expressed sequences (Fig. 1), however, the 115-bp long noncoding region actually codes for the ATPase subunit 8 (C. Gissi, F. Iannelli, and G. Pesole, in prep.) and the 107-bp noncoding region downstream of ND6 is actually only 36 bp long, because of the presence in this region of the additional tRNA-Phe (Fig. 3).
In the contig Hr-3, the observation that there are no ESTs containing a poly(A) stretch after the 3′ end of CO2 gene suggests that CO2 and Cytb genes are on the same transcript. A final demonstration of this characteristic will come from the mapping of Halocynthia mitochondrial RNAs. Moreover, a careful analysis of this region shows that the Cytb frame has an ATG codon 21 bp upstream of the predicted start codon ATT (position 7431 instead of 7452), consistent with an 11-bp overlap between CO2 and Cytb. A longer Halocynthia Cytb gene is also supported by length and sequence similarity with the cow and chicken Cytb proteins (data not shown), whose amino acid sequences were validated by the resolution of the three-dimensional structure (Iwata et al. 1998; Zhang et al. 1998). Two other genes, ND1 and ATP6, were in the same link and reported as adjacent nonoverlapping genes in the mtDNA (Yokobori et al. 1999). Even in this case, it is possible to identify a potential overlap between the ND1 and the ATP6 frames. However, there is currently no further evidence in favor of a longer ATP6 gene.
Abundance of Mitochondrial Transcript
The analyzed ESTs derive from unmodified directional cDNA libraries constructed from a poly(A)+ RNA fraction (Makabe et al. 2001; Satou et al. 2002). Because the frequency of occurrence of a clone in an unaltered cDNA library depends on the abundance of the corresponding mRNA in the cell, we have an idea of the level of the mitochondrial polyadenylated transcripts in the different tissue sources of the cDNA libraries. Table 4 reports the number of mt-like clones for each library and for each contig/link of both ascidians. The Halocynthia ESTs (4160 sequences from 2289 clones) belong to a single cDNA library constructed from dechorionated fertilized eggs (Makabe et al. 2001). The Ciona ESTs (172,824 sequences from 97,748 clones) derive from 9 cDNA libraries obtained from fertilized eggs (Nishikata et al. 2001), cleavage stage embryos (Fujiwara et al. 2002), tailbud embryos (Satou et al. 2001), larvae (Kusakabe et al. 2002; M. Branno et al., unpubl.), adult (Ogasawara et al. 2002), testis (Inaba et al. 2002), and hemocytes (K. Shida et al. unpubl.).
Table 4.
Number of Mitochondrial-Like Clones Identified in the Halocynthia roretzi (Hr) and Ciona intestinalis (Ci) Published cDNA Libraries
| Ci
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene-Hr | Hr egg | Gene-Ci | Egg | CLS | TB | LV-1 | LV-2 | AD-1 | AD-2 | TS | Hem | Total |
| CO2 (sea urchin) | 1 | 1 | ||||||||||
| ND2 (sea urchin) | 1 | 1 | ||||||||||
| 16S rRNA (H. roretzi) | 3 | 3 | ||||||||||
| 16S rRNA | 249 | 16S rRNA | 1 | 1 | 2 | |||||||
| CO1/ND3 | 1 | CO1 | 37 | 25 | 54 | 6 | 19 | 44 | 185 | |||
| CO2/Cytb | 10 | CO2/Cytb/ND4L/CO3 | 57 | 58 | 99 | 2 | 4 | 11 | 81 | 312 | ||
| ND1/ATP6 | 3 | ND1 | 3 | 2 | 2 | 1 | 8 | 1 | 17 | |||
| ND2 | 3 | ATP6/ND2 | 1 | 6 | 3 | 11 | 1 | 22 | ||||
| ND3 | 7 | 7 | 2 | 16 | ||||||||
| ND4 | 2 | ND4 | 3 | 1 | 11 | 1 | 6 | 1 | 23 | |||
| ND5 | 3 | ND5 | 3 | 3 | 5 | 4 | 1 | 16 | ||||
| ND6 | 1 | |||||||||||
| Singletons | Singletons | 1 | 3 | 5 | 4 | 4 | 1 | 1 | ||||
| mt-like clones | 272 | mt-like clones | 104 | 96 | 184 | 3 | 11 | 33 | 0 | 156 | 4 | 591 |
| Sequenced clones | 2289 | Sequenced clones | 17,960 | 16,392 | 12,403 | 14,193 | 3637 | 25,286 | 66 | 5718 | 2057 | 97,748 |
| % mt-like clones | 11.88 | % mt-like clones | 0.58 | 0.59 | 1.48 | 0.02 | 0.30 | 0.13 | 0.00 | 2.73 | 0.19 | 0.60 |
(Egg) Fertilized eggs (Nishikata et al. 2001); (CLS) Cleavage stage embryo (Fujiwara et al. 2002); (TB) tailbud embryo (Satou et al. 2001); (LV-1) larval stage (Kusakabe et al. 2002). (LV-2) larval stage; (M. Branno et al. unpublished); (AD-1) adult (Ogasawara et al. 2002); (AD-2) adult (Simmen et al. 1998); (TS) testis (Inaba et al. 2002); (Hem) hemocytes (K. Shida unpubl.)
It is striking that the mt clones represent ∼12% of the Halocynthia egg library, with 10% of mt 16S rRNA transcripts, whereas the Ciona egg library contains only 0.58% of mitochondrial transcripts, with no 16S rRNA sequences. This difference is even more striking when considering that the clones screened in the Ciona egg cDNA library were about eight times more abundant than that screened in the Halocynthia egg library. This situation could be partially explained by the presence of the motif A5GA3GA7 inside of the Halocynthia 16S rRNA gene, which could be responsible for internal priming of the 16S trancripts by the oligo(dT) primers. In Halocynthia, 66 of 412 16S rRNA-like ESTs (16%) start/stop at such sites. On the contrary, in the 16S rRNA gene of Ciona, the motif is modified to GA8GACGA4, but none of the two 16S rRNA-like ESTs stop or start at this site. An additional explanation is related to the slight difference in the cDNA source. The egg cDNA libraries were obtained from dechorionated (Halocynthia) and not dechorionated (Ciona) eggs, therefore, the RNA of the follicle/test cells present in the chorion may distort the egg expression profile. Finally, a real difference between the two ascidian species in the polyadenylation/expression of 16S rRNA in fertilized eggs could also be hypothesized.
Between the Ciona cDNA libraries, the mitochondrial clones are more abundant in testis and tailbud embryos, where their percentages were 2.7% and 1.5%, respectively. Furthermore, in both cases, the mt ESTs were represented homogeneously between all of the mitochondrial contigs identified. The mtDNA may be highly expressed in these tissues, or a mtDNA dosage effect may be responsible for the relative abundance of mt transcripts.
The only mitochondrial gene absent from both the Halocynthia and Ciona EST collections was the 12S rRNA. Given the deep EST coverage of the Ciona cDNA libraries and the features of the 12S rRNA transcript in metazoa, the most likely explanation is that this transcript is not polyadenylated in ascidians, however, this hypothesis has to be demonstrated experimentally.
DISCUSSION
The mitochondrial origin of the assembled ESTs is supported by several observations. The analyzed ESTs derive from the random sequencing of unmodified cDNA libraries constructed from the total poly(A)+ RNA fraction, without reduction for the most abundant transcripts using normalization or subtraction approaches (Makabe et al. 2001). Metazoan mitochondrial transcripts are usually polyadenylated, and in some tissues constitute a large fraction of the total mRNAs, thus, it has been found that they constitute a large fraction in cDNA libraries obtained from tissue sources with a high density of mitochondria or mtDNA overexpression (Welle et al. 1999; Laveder et al. 2002). The nucleotide identity of assembled ESTs to the corresponding mtDNA is higher than 94% in Halocynthia and higher than 99% in Ciona. The small differences between mtDNA and EST contigs can be accounted for by the mean 2% EST sequencing errors (Hillier et al. 1996) and the frequent polymorphic sites found in the mtDNA. Furthermore, the availability of ∼40 times more EST clones for Ciona compared with Halocynthia explains the higher accurary of the predicted mt transcripts in Ciona. The similarity of some links to both nuclear and mitochondrial genes, or to mitochondrial sequences of other species can be easily explained by contaminations and cloning artifacts, given that the proportion of chimeric ESTs may be quite high in some cDNA libraries (Hillier et al. 1996). The possibility that the assembled ESTs derive from mitochondrial pseudogenes located in the nuclear genome (Numt) is very low. Numts were identified in a wide range of species (Bensasson et al. 2001), but up to now there is no evidence for their expression (Blanchard and Schmidt 1996). If the expressed sequences were actually Numts, we had to hypothesize their expression at a high level, because they were identified by superficial screening of cDNA libraries, and a recent gene transfer from mitochondria to nucleus, because of their high similarity to mtDNA. The identification of tRNAs with typical mitochondrial cloverleaf structures (Kumazawa and Nishida 1993; Helm et al. 2000) and of protein genes using the ascidian mt genetic code, so that they can be correctly translated only by the mitochondrial translation apparatus, further support the mitochondrial origin of such ESTs.
The mt-ESTs Provide Valuable Information on Mitochondrial Transcription and Processing
Precursor mitochondrial mRNAs appear rather frequently in various cDNA libraries (see Table 4). Several different precursors of mitochondrial transcripts have been also identified in rat RNA preparations (Tullo et al. 1994).
The ascidian mitochondrial transcripts appear polyadenylated, including the 16S rRNA. Most transcripts are polycistronic; both protein-coding and tRNA genes are often present in the same transcript. Two distinct pieces of evidence suggest the existence of mature bicistronic transcripts as follows: (1) the presence of overlapped ORFs in the genome; (2) the identification of transcripts containing almost the entire overlapped ORFs without traces of poly(A) stretches downstream of the first ORF (see Ciona contigs Ci-5 and Ci-36; Halocynthia Hr-3 contig). Although such hypothesis would need additional experimental validation, it is actually verified in mammals for ATP6/ATP8 and ND4/ND4L. Furthermore, the expression of overlapped ORFs by distinct monocistronic transcripts implies that the identified bicistronic transcripts are immature transcripts that are subsequently processed in one of the two possible mature functional transcripts.
The poly(A) tails are abutted on tRNA genes, suggesting that the tRNA cloverleaf structures are implicated in transcript maturation and polyadenylation. In some genes, the poly(A) tails also contribute to the creation of stop codons.
The mt-ESTs also provide interesting information on the transcription of rRNAs. In Halocynthia, the 16S rRNA transcript presents ragged 3′ ends with multiple poly(A) start site, and stops immediately upstream of a typical mitochondrial rRNA transcription termination signal (TRGCAKAN5G). This motif is well conserved near the 3′ end of 16S rRNA in a wide range of organisms, from protozoa to metazoa, independent of the identity of the adjacent gene (Valverde et al. 1994). In sea urchin, the rRNA termination box is located exactly where the 16S rRNA transcript terminates in vivo (Elliott and Jacobs 1989), at the same relative position inferred for Halocynthia from EST data. The rRNA transcription termination box has been found also in Ciona mtDNA within the tRNA-Ile sequence (C. Gissi, F. Iannelli, and G. Pesole, in prep.); however, no speculation on the polyadeylation sites of the 16S rRNA transcript can be presented in Ciona, because only two ESTs with no poly(A) stretches constitute the Ciona 16S rRNA contig (Table 1). It is striking to note that in Halocynthia, 16S rRNA transcripts represent >90% of total mt-like ESTs (Table 3), suggesting an overrepresentation of this molecule with respect to other mitochondrial mRNAs, as expected for the presence of a transcription termination signal. In both Ciona and Halocynthia cDNA libraries, no ESTs similar to the 12S rRNA were found, whereas ESTs similar to 16S rRNA were highly represented in Halocynthia, but not in Ciona cDNA libraries (Table 4). In species in which mt transcription was investigated, the two rRNAs were expressed at similar high levels, even when the genes were not adjacent, as occurs in sea urchin (Cantatore et al. 1990). The suggested coordinated expression of the rRNA genes is related to their role in ribosomal subunits. With regard to the polyadenylation of rRNA transcripts, the data are quite heterogeneous between species. In human and rodents, both rRNAs are oligoadenylated, but an additional, polyadenylated fraction of 16S rRNA was also found (Dubin et al. 1981, 1982; Montoya et al. 1983; Van Etten et al. 1983). In Xenopus laevis and rainbow trout, neither rRNAs are polyadenylated, but again in rainbow trout, a small percentage of 16S rRNA has been found in the poly(A)+ fraction (Rastl and Dawid 1979; Zardoya et al. 1995). On the contrary, in Drosophila, most of the 16S rRNA is polyadenylated and the 12S rRNA is predominantly not polyadenylated (Merten and Pardue 1981). The EST data clearly demonstrate that in both ascidian species, the 16S rRNA is polyadenylated. The lack of 12S rRNA-like ESTs suggest that this transcript is not polyadenylated or is expressed at very low levels. Furthermore, the additional role played by the Halocynthia 16S rRNA during the embryo development (Oka et al. 1998) could account for the abundance of 16S over 12S rRNA transcript, and its high proportion over the entire cDNA library of Halocynthia (Makabe et al. 2001). It would therefore be of interest to investigate the involvement of 16S rRNA in embryo development in Ciona.
Long noncoding regions have been found between the expressed sequences in both ascidian mtDNAs. In Halocynthia, the two longest noncoding regions (112 and the 79 bp long) are both transcribed, apparently as 3′ untranslated regions of mRNAs (Fig. 1). Similarly, in Ciona, the 85-bp long noncoding region is expressed as 3′ untranslated region of the CO3 transcript, and the first part of the longest noncoding region (100 bp long) is also expressed (Fig. 1). Whether such noncoding regions act as regulatory sequences for ascidian mtDNA expression and/or replication is unknown. However, the lack of nontranscribed regulatory regions is not surprising in mtDNA. In mammals, both the noncoding regulatory regions, that is, the large D-loop containing region and the small L-strand replication origin, have been detected among the transcribed sequences as sense and antisense RNAs, and such transcripts are probably implicated in the regulation of mtDNA replication and expression (Sbisa et al. 1990, 1992).
Overall, the ascidian mt ESTs suggest that the mechanism of transcript maturation is conserved between ascidian and mammal mtDNAs. In both organisms, primary polycistronic transcripts are produced and then maturated through multiple endonucleolytic cuts, with the tRNA genes acting as punctuation signals in the processing (see Fig. 1).
The identification of mitochondrial ESTs is very useful for the definition of amplification/sequencing strategies for genomes not yet sequenced, such as the Ciona intestinalis mtDNA. About 80% of the Ciona mtDNA sequence and a partial map of gene organization were predicted from the mt EST data and allowed a fast amplification/sequencing of this genome. Both the sequences and gene order predicted from mt-ESTs were confirmed by the sequencing of the mtDNA and differ significantly from that of Halocynthia roretzi (C. Gissi, F. Iannelli, and G. Pesole, in prep.).
With or without the availability of complete mtDNA sequences, mt-ESTs facilitate mitochondrial gene annotation through the indication of overlapped protein-coding genes, poly(A) start sites, rRNA signal termination box, and 5′ tRNA ends (mostly corresponding to poly[A] start sites). With regard to the original annotation of Halocynthia mtDNA (Yokobori et al. 1999), the mt-ESTs allowed better definition of the boundaries of four genes (16S rRNA, ND1, ATP6, and tRNA-Ser) and the identification of an additional Phe-tRNA gene. In Ciona mtDNA, the mt-ESTs facilitated the annotation of tRNA genes as, in most cases, the 5′ tRNA end was marked by a poly(A) tail in the upstream transcript.
Our study demostrates that EST sequences are very useful for the analysis not only of the expression pattern of the nuclear genome, but also of the mitochondrial genome itself. Mitochondrial sequences can be identified in small and large EST collections, provided that the cDNA source is a tissue/developmental stage in which the mtDNA is abundant or highly expressed. The nature of these sequences as expressed tags and the high similarity to the mtDNA suggests that they are not nuclear pseudogenes. The mapping of the mitochondrial expressed sequences on the mt genome allows gene identification and a better definition of gene boundaries. In cases in which the mtDNA is unknown, mtEST assembly allows us to hypothesize, at least partially, the mt gene order and to accelerate mtDNA sequencing projects, facilitating the formulation of a cloning/amplification strategy. Independent of the availability of the mtDNA sequence, the EST data provide valuable information on mt transcription, allowing identification of polyadenylated processed and immature transcripts, mapping of polyadenylation sites, speculation of termination/processing signals, and raw quantification of mt transcript abundance.
The data reported here may require additional experimental validation, for example, to assess the polyadenylation status of the rRNA transcripts and the existence of mature bicistronic transcripts. However, the novel approach described here make valuable mt-EST data, generally an unused by-product of EST projects, provide significant clues to mitochondrial transcription and suitably address further experimental analyses.
METHODS
A total of 4160 Halocynthia roretzi ESTs derived from 2289 clones, and a total of 172,824 Ciona intestinalis ESTs from 97,748 clones were available in the EMBL database on April 2002. EST sequences highly similar to mitochondrial sequences were identified by in silico hybridization using the mtDNA of Halocynthia roretzi (AC no. AB024528) as a probe. BLAST searches (Altschul et al. 1990) were carried out against the Est_other Embl collection (April 2002). The searches were limited to either Ciona or Halocynthia sequences. Similarity to mitochondrial protein-coding genes was assessed by using TBLASTN. ESTs with statistically significant matches, as well as ESTs related to the former by clone ID, were collected, checked for vector contamination, and assembled using the Cap3 program (Huang and Madan 1999). Resulting alignments were checked carefully using the Consed (Gordon et al. 1998) and Seaview programs (Galtier et al. 1996). Nonoverlapping contigs sharing at least one clone ID were considered as derived from the same cDNA and were called links. EST sequences not included in contigs/links were indicated as singletons. To find all ESTs overlapping the original contigs, the ends of the mt-like contigs were used for an in silico chromosome walking, that is, such end sequences were used as probes for multiple cycles of analysis, each consisting of three steps as follows: (1) BLASTN against Ciona ESTs; (2) selection of ESTs with significant similarity and of EST sequences sharing clone IDs; (3) new EST assembly. This procedure allowed us to find all ESTs highly similar to and overlapping the ends of the original mt-like contigs, even if they did not resemble known mtDNA sequences. The final contigs were compared with and mapped onto the corresponding mitochondrial genomes. The ascidian protein-coding genes were defined according to the ascidian mitochondrial translation code (Durrheim et al. 1993; Yokobori et al. 1993), considering as potential start codons ATR (Met), ATT (Ile), TTG (Leu), and GTG (Val), as suggested by Yokobori et al. (1999). BLASTX against the nonredundant protein data base allowed the identification of protein-coding regions. The tRNA genes were identified using tRNAscan-SE (version 1.21) with the option of organellar tRNA structure search (Lowe and Eddy 1997) (http://www.genetics.wustl.edu/eddy/tRNAscan-SE/) and by manual inspection. Specific nucleotide patterns, such as the rRNA transcription termination box, were searched using the PatSearch program (Pesole et al. 2000; http://bighost.area.ba.cnr.it/BIG/Patsearch/). For each contig, lists of assembled ESTs, consensus sequences and alignments are available on request. The Ciona contigs/links were used to construct species-specific primers for the amplification of the Ciona mtDNA in five large overlapped fragments (C. Gissi, F. Iannelli, and G. Pesole, in prep.). The complete mtDNA sequence of Ciona intestinalis was submitted at the EMBL data base, AC number AJ517314.
Acknowledgments
This work was supported by the Progetto Giovani Ricercatori of the University of Milano, FIRB project (Ministero dell'Istruzione e Ricerca Scientifica, Italy), and by Telethon. We thank Cecilia Saccone, Aurelio Reyes, David Horner, and Flavio Mignone for valuable comments on the manuscript. The constructive comments of an anonymous referee were greatly appreciated.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1227803. Article published online before print in August 2003.
References
- Adams, M.D., Kelley, J.M., Gocayne, J.D., Dubnick, M., Polymeropoulos, M.H., Xiao, H., Merril, C.R., Wu, A., Olde, B., Moreno, R.F., et al. 1991. Complementary DNA sequencing: Expressed sequence tags and human genome project. Science 252: 1651–1656. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Bensasson, D., Zhang, D., Hartl, D.L., and Hewitt, G.M. 2001. Mitochondrial pseudogenes: Evolution's misplaced witnesses. Trends Ecol. Evol. 16: 314–321. [DOI] [PubMed] [Google Scholar]
- Berthier, F., Renaud, M., Alziari, S., and Durand, R. 1986. RNA mapping on Drosophila mitochondrial DNA: Precursors and template strands. Nucleic Acids Res. 14: 4519–4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder, S., Marchfelder, A., and Brennicke, A. 1996. Regulation of gene expression in plant mitochondria. Plant Mol. Biol. 32: 303–314. [DOI] [PubMed] [Google Scholar]
- Blanchard, J.L. and Schmidt, G.W. 1996. Mitochondrial DNA migration events in yeast and humans: Integration by a common end-joining mechanism and alternative perspectives on nucleotide substitution patterns. Mol. Biol. Evol. 13: 537–548. [DOI] [PubMed] [Google Scholar]
- Boore, J.L. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27: 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger, G., Helmer Citterich, M., Nelson, M.A., Werner, S., and Macino, G. 1985. RNA processing in Neurospora crassa mitochondria: Transfer RNAs punctuate a large precursor transcript. EMBO J. 4: 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantatore, P., Roberti, M., Loguercio Polosa, P., Mustich, A., and Gadaleta, M.N. 1990. Mapping and characterization of Paracentrotus lividus mitochondrial transcripts: Multiple and overlapping transcription units. Curr. Genet. 17: 235–245. [DOI] [PubMed] [Google Scholar]
- Christianson, T.W. and Clayton, D.A. 1986. In vitro transcription of human mitochondrial DNA: Accurate termination. Proc. Natl. Acad. Sci. 83: 6277–6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo, M.C. and Fox, T.D. 1990. Control of mitochondrial gene expression in Saccharomyces cerevisiae. Annu. Rev. Genet. 24: 91–113. [DOI] [PubMed] [Google Scholar]
- Dubin, D.T., Timko, K.D. and Baer, R.J. 1981. The 3′ terminus of the large ribosomal subunit “17S” RNA from hamster mitochondria is ragged and oligoadenylated. Cell 23: 271–278. [DOI] [PubMed] [Google Scholar]
- Dubin, D.T., Montoya, J., Timko, K.D., and Attardi, G. 1982. Sequence analysis and precise mapping of the 3′ ends of HeLa cell mitochondrial ribosomal RNAs. J. Mol. Biol. 157: 1–19. [DOI] [PubMed] [Google Scholar]
- Durrheim, G.A., Corfield, V.A., Harley, E.H., and Ricketts, M.H. 1993. Nucleotide sequence of cytochrome oxidase (subunit III) from the mitochondrion of the tunicate Pyura stolonifera: Evidence that AGR encodes glycine. Nucleic Acids Res. 21: 3587–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson, N.J., Brown, T.A., Ray, J.A., Waring, R.B., Scazzocchio, C., and Davies, R.W. 1989. Processing of mitochondrial RNA in Aspergillus nidulans. J. Mol. Biol. 208: 587–599. [DOI] [PubMed] [Google Scholar]
- Elliott, D.J. and Jacobs, H.T. 1989. Mutually exclusive synthetic pathways for sea urchin mitochondrial rRNA and mRNA. Mol. Cell. Biol. 9: 1069–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara, S., Maeda, Y., Shin, I.T., Kohara, Y., Takatori, N., Satou, Y., and Satoh, N. 2002. Gene expression profiles in Ciona intestinalis cleavage-stage embryos. Mech. Dev. 112: 115–127. [DOI] [PubMed] [Google Scholar]
- Galtier, N., Gouy, M., and Gautier, C. 1996. SEAVIEW and PHYLO_WIN: Two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12: 543–548. [DOI] [PubMed] [Google Scholar]
- Gelfand, R. and Attardi, G. 1981. Synthesis and turnover of mitochondrial ribonucleic acid in HeLa cells: The mature ribosomal and messenger ribonucleic acid species are metabolically unstable. Mol. Cell. Biol. 1: 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D., Abajian, C., and Green, P. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8: 195–202. [DOI] [PubMed] [Google Scholar]
- Helm, M., Brule, H., Friede, D., Giege, R., Putz, D., and Florentz, C. 2000. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA 6: 1356–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier, L.D., Lennon, G., Becker, M., Bonaldo, M.F., Chiapelli, B., Chissoe, S., Dietrich, N., DuBuque, T., Favello, A., Gish, W., et al. 1996. Generation and analysis of 280,000 human expressed sequence tags. Genome Res. 6: 807–828. [DOI] [PubMed] [Google Scholar]
- Hirsch, M. and Penman, S. 1973. Mitochondrial polyadenylic acid-containing RNA: localization and characterization. J. Mol. Biol. 80: 379–391. [DOI] [PubMed] [Google Scholar]
- ____. 1974. The messenger-like properties of the poly(A)plus RNA in mammalian mitochondria. Cell 3: 335–339. [DOI] [PubMed] [Google Scholar]
- Huang, X. and Madan, A. 1999. CAP3: A DNA sequence assembly program. Genome Res. 9: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba, K., Padma, P., Satouh, Y., Shin-I, T., Kohara, Y., Satoh, N., and Satou, Y. 2002. EST analysis of gene expression in testis of the ascidian Ciona intestinalis. Mol. Reprod. Dev. 62: 431–445. [DOI] [PubMed] [Google Scholar]
- Iwata, S., Lee, J.W., Okada, K., Lee, J.K., Iwata, M., Rasmussen, B., Link, T.A., Ramaswamy, S., and Jap, B.K. 1998. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science 281: 64–71. [DOI] [PubMed] [Google Scholar]
- Kumazawa, Y. and Nishida, M. 1993. Sequence evolution of mitochondrial tRNA genes and deep-branch animal phylogenetics. J. Mol. Evol. 37: 380–398. [DOI] [PubMed] [Google Scholar]
- Kusakabe, T., Yoshida, R., Kawakami, I., Kusakabe, R., Mochizuki, Y., Yamada, L., Shin-i, T., Kohara, Y., Satoh, N., Tsuda, M. et al. 2002. Gene expression profiles in tadpole larvae of Ciona intestinalis. Dev. Biol. 242: 188–203. [DOI] [PubMed] [Google Scholar]
- Laveder, P., De Pitta, C., Toppo, S., Valle, G., and Lanfranchi, G. 2002. A two-step strategy for constructing specifically self-subtracted cDNA libraries. Nucleic Acids Res. 30: e-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, T.M.and Eddy, S.R. 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makabe, K.W., Kawashima, T., Kawashima, S., Minokawa, T., Adachi, A., Kawamura, H., Ishikawa, H., Yasuda, R., Yamamoto, H., Kondoh, K., et al. 2001. Large-scale cDNA analysis of the maternal genetic information in the egg of Halocynthia roretzi for a gene expression catalog of ascidian development. Development 128: 2555–2567. [DOI] [PubMed] [Google Scholar]
- Merten, S.H. and Pardue, M.L. 1981. Mitochondrial DNA in Drosophila. An analysis of genome organization and transcription in Drosophila melanogaster and Drosophila virilis. J. Mol. Biol. 153: 1–21. [DOI] [PubMed] [Google Scholar]
- Montoya, J., Gaines, G.L., and Attardi, G. 1983. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell 34: 151–159. [DOI] [PubMed] [Google Scholar]
- Nishikata, T., Yamada, L., Mochizuki, Y., Satou, Y., Shin-i, T., Kohara, Y., and Satoh, N. 2001. Profiles of maternally expressed genes in fertilized eggs of Ciona intestinalis. Dev. Biol. 238: 315–331. [DOI] [PubMed] [Google Scholar]
- Ogasawara, M., Sasaki, A., Metoki, H., Shin, I.T., Kohara, Y., Satoh, N., and Satou, Y. 2002. Gene expression profiles in young adult Ciona intestinalis. Dev. Genes Evol. 212: 173–185. [DOI] [PubMed] [Google Scholar]
- Ojala, D., Montoya, J., and Attardi, G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature 290: 470–474. [DOI] [PubMed] [Google Scholar]
- Oka, T., Amikura, R., Kobayashi, S., Yamamoto, H., and Nishida, H. 1998. Localization of mitochondrial large ribososmal RNA in myoplasm of the early ascidian embryo. Dev. Growth Differ. 41: 1–8. [DOI] [PubMed] [Google Scholar]
- Pesole, G., Liuni, S., and D'Souza, M. 2000. PatSearch: A pattern matcher software that finds functional elements in nucleotide and protein sequences and assesses their statistical significance. Bioinformatics 16: 439–450. [DOI] [PubMed] [Google Scholar]
- Rastl, E.and Dawid, I.B. 1979. Expression of the mitochondrial genome in Xenopus laevis: A map of transcripts. Cell 18: 501–510. [DOI] [PubMed] [Google Scholar]
- Richard, O., Bonnard, G., Grienenberger, J.M., Kloareg, B., and Boyen, C. 1998. Transcription initiation and RNA processing in the mitochondria of the red alga Chondrus crispus: Convergence in the evolution of transcription mechanisms in mitochondria. J. Mol. Biol. 283: 549–557. [DOI] [PubMed] [Google Scholar]
- Richard, O., Kloareg, B., and Boyen, C. 1999. mRNA expression in mitochondria of red alga Chondrus crispus requires a unique RNA-processing mechanism, internal cleavage of upstream tRNAs at pyrimidine 48. J.Mol. Biol. 288: 579–584. [DOI] [PubMed] [Google Scholar]
- Saccone, C., De Giorgi, C., Gissi, C., Pesole, G., and Reyes, A. 1999. Evolutionary genomics in Metazoa: The mitochondrial DNA as a model system. Gene 238: 195–209. [DOI] [PubMed] [Google Scholar]
- Satou, Y., Takatori, N., Yamada, L., Mochizuki, Y., Hamaguchi, M., Ishikawa, H., Chiba, S., Imai, K., Kano, S., Murakami, S.D., et al. 2001. Gene expression profiles in Ciona intestinalis tailbud embryos. Development 128: 2893–2904. [DOI] [PubMed] [Google Scholar]
- Satou, Y., Takatori, N., Fujiwara, S., Nishikata, T., Saiga, H., Kusakabe, T., Shin-i, T., Kohara, Y., and Satoh, N. 2002. Ciona intestinalis cDNA projects: Expressed sequence tag analyses and gene expression profiles during embryogenesis. Gene 287: 83–96. [DOI] [PubMed] [Google Scholar]
- Sbisa, E., Nardelli, M., Tanzariello, F., Tullo, A., and Saccone, C. 1990. The complete and symmetric transcription of the main non coding region of rat mitochondrial genome: In vivo mapping of heavy and light transcripts. Curr. Genet. 17: 247–253. [DOI] [PubMed] [Google Scholar]
- Sbisa, E., Tullo, A., Nardelli, M., Tanzariello, F., and Saccone, C. 1992. Transcription mapping of the Ori L region reveals novel precursors of mature RNA species and antisense RNAs in rat mitochondrial genome. FEBS Lett. 296: 311–316. [DOI] [PubMed] [Google Scholar]
- Simmen, M.W., Leitgeb, S., Clark, V.H., Jones, S.J., and Bird, A. 1998. Gene number in an invertebrate chordate, Ciona intestinalis. Proc. Natl. Acad. Sci. 95: 4437–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullo, A., Tanzariello, F., D'Erchia, A.M., Nardelli, M., Papeo, P.A., Sbisa, E., and Saccone, C. 1994. Transcription of rat mitochondrial NADH-dehydrogenase subunits. Presence of antisense and precursor RNA species. FEBS Lett. 354: 30–36. [DOI] [PubMed] [Google Scholar]
- Valverde, J.R., Marco, R., and Garesse, R. 1994. A conserved heptamer motif for ribosomal RNA transcription termination in animal mitochondria. Proc.Natl. Acad. Sci. 91: 5368–5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten, R.A., Bird, J.W., and Clayton, D.A. 1983. Identification of the 3′-ends of the two mouse mitochondrial ribosomal RNAs. The 3′-end of 16 S ribosomal RNA contains nucleotides encoded by the gene for transfer RNALeuUUR. J. Biol. Chem. 258: 10104–10110. [PubMed] [Google Scholar]
- Welle, S., Bhat, K., and Thornton, C.A. 1999. Inventory of high-abundance mRNAs in skeletal muscle of normal men. Genome Res. 9: 506–513. [PMC free article] [PubMed] [Google Scholar]
- Wolff, G.and Kuck, U. 1996. Transcript mapping and processing of mitochondrial RNA in the chlorophyte alga Prototheca wickerhamii. Plant Mol. Biol. 30: 577–595. [DOI] [PubMed] [Google Scholar]
- Yokobori, S., Ueda, T., and Watanabe, K. 1993. Codons AGA and AGG are read as glycine in ascidian mitochondria. J. Mol. Evol. 36: 1–8. [DOI] [PubMed] [Google Scholar]
- Yokobori, S., Ueda, T., Feldmaier-Fuchs, G., Paabo, S., Ueshima, R., Kondow, A., Nishikawa, K., and Watanabe, K. 1999. Complete DNA sequence of the mitochondrial genome of the ascidian Halocynthia roretzi (Chordata, Urochordata). Genetics 153: 1851–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zardoya, R., Perez-Martos, A., Bautista, J.M., and Montoya, J. 1995. Analysis of the transcription products of the rainbow trout (Oncorynchus mykiss) liver mitochondrial genome: Detection of novel mitochondrial transcripts. Curr. Genet. 28: 67–70. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Huang, L., Shulmeister, V.M., Chi, Y.I., Kim, K.K., Hung, L.W., Crofts, A.R., Berry, E.A., and Kim, S.H. 1998. Electron transfer by domain movement in cytochrome bc1. Nature 392: 677–684. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://bighost.area.ba.cnr.it/BIG/Patsearch/; PatSearch, a program for the detection of patterns and structure motifs in nucleotide sequences. [DOI] [PMC free article] [PubMed]
- http://www.genetics.wustl.edu/eddy/tRNAscan-SE/; tRNAscan-SE, a program for the prediction of tRNA genes.