Abstract
The availability of the sequenced Drosophila melanogaster genome provides an opportunity to study sequence variation between copies within transposable element families. In this study,we analyzed the 624 copies of 22 transposable element (TE) families (14 LTR retrotransposons, five non-LTR retrotransposons, and three transposons). LTR and non-LTR retrotransposons possessed far fewer divergent elements than the transposons,suggesting that the difference depends on the transposition mechanism. However,there was not a continuous range of divergence of the copies in each class,which were either very similar to the canonical elements,or very divergent from them. This sequence homogeneity among TE family copies matches the theoretical models of the dynamics of these repeated sequences. The sequenced Drosophila genome thus appears to be composed of a mixture of TEs that are still active and of ancient relics that have degenerated and the distribution of which along the chromosomes results from natural selection. This clearly demonstrates that the TEs are highly active within the genome,suggesting that the genetic variability of the Drosophila genome is still being renewed by the action of TEs.
Transposable elements (TEs), which are repeated sequences able to move along the chromosomes, are major components of genomes (San Miguel et al. 1996; International Human Genome Sequencing Consortium 2001; Venter et al. 2001) and play a major part in the evolution of their host, notably by creating genetic variability (Shapiro 1999; Evgen'ev et al. 2000; Bowen and Jordan 2002). Their transposition mechanisms differ considerably, depending on the class to which they belong (LTR retrotransposons, non-LTR retrotransposons, transposons). The LTR and the non-LTR retrotransposons are first transcribed into an mRNA. The LTR retrotransposons are then retrotranscribed into a DNA molecule and finally inserted into the genome, whereas the non-LTR retrotransposons are retrotranscribed at the same time as they are inserted into the genome. The transposition process of both these retroelements thus leads to the creation of novel additional copies. The transposons, which are DNA-based elements, are excised from the genome before being reinserted at another site. As a result, a genome will include several copies of most of the TE families. Numerous studies of the number and localization of TEs in the genomes have been carried out in natural populations of Drosophila melanogaster and other sibling species (Biémont and Cizeron 1999). These studies have provided information about the dynamics of TEs and the forces that maintain them in genomes and populations (Biémont et al. 1997; Charlesworth et al. 1997). However, they have not provided any information about the polymorphism of the nucleic sequences of the elements, information that is necessary for any understanding of how TEs are transposed and regulated, and how they degenerate (by deletions, insertions, rearrangements, and divergence by substitutions) and are eventually eliminated from the genome. Some studies of the untranslated regulatory regions of retrotransposons show that there are differences in length probably corresponding to active and nonactive elements, and to regulatory elements (Csink and McDonald 1995; Jordan and McDonald 1998a; Costas et al. 2001). The nucleotide sequences of TEs can also reflect the relationships between different TE families because it is suspected that TEs may evolve by acquiring modules from different elements (Lerat et al. 1999). TEs present particular features, such as their AT-richness (Shields and Sharp 1989; Lerat et al. 2000, 2002a) and the specific dinucleotide pattern observed in some LTR retrotransposons (Lerat et al. 2002b), which are known to differ from those of the genes of their host genome. Because these characteristics of the TE sequences seem to be maintained by natural selection, we wondered whether they are still present in TEs that are no longer active or have degenerated.
Analyzing the full length LTR retrotransposons of the first release of the Drosophila genome in which the TE sequences were unfortunately not all of high quality, Bowen and McDonald (2001) reported close similarity between the TE copies. They suggested that these LTR retrotransposons had been transposed recently, in terms of millions of years. Release 3.0 of the sequenced genome of D. melanogaster now allows us to access all the copies of the TEs, which are now better in quality. We analyzed the sequences of 22 element families belonging to the three classes of TEs (14 LTR retrotransposons, 5 non-LTR retrotransposons, 3 transposons) on chromosome arms 2L, 2R, 3L, 3R, and X. These TEs had been chosen because of their significant copy numbers. In general, there is a high degree of homogeneity and a lack of divergent elements between the sequences of TEs within a given family, but when divergent elements do exist, they display a very low percentage of similarity to the full-length sequences. These findings suggest that TEs are highly active within the genome, and that the highly divergent copies reflect relics of ancient mobilizations.
RESULTS AND DISCUSSION
LTR-Retrotransposons
The bel-like Families
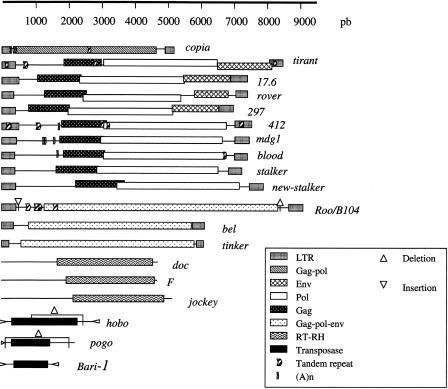
This group of families includes roo/B104, tinker, and bel (Frame et al. 2001). With 125 copies, the roo/B104 element has more copies than any other LTR retrotransposon. The level of similarity between these copies and the canonical element ranged from 95.31% to 100.0%. Thirty of the 36 copies that did not present any internal deletions seemed to be complete. Twenty-two sequences were solo LTR, 49 possessed internal deletions, 13 displayed both internal deletions and small insertions, and 2 displayed only insertions. Twelve of the copies including an insertion, displayed the same 12-bp insertion located at nucleotide 513 in the reference element, and 21 of those with deletions displayed the same 12–11-bp deletion at nucleotide 8422 in the reference element, suggesting that two groups of TEs transposed from each other (Fig. 1; Supplemental Materials available online at www.genome.org). Four copies of roo/B104 were interrupted by other elements: 2L_1643372 was interrupted by a doc element, 2R_9158204 by HMS-beagle, 2R_1432623 (the most divergent copy) by four elements (circe, BS, F, and DM88), and 2R_4540764 by hobo. Five copies presented internal rearrangements: 2L_6419126, 3L_13762171, and X_4731673 presented the same rearrangement in which the end of the sequence was inverted compared to the beginning, in 3R_23336300 the middle of the sequence was inverted, and in X_3356455 a portion of 683 bp was duplicated. The 2R_20192913 and 2R_20185276 copies had one LTR in common: the 5′ LTR of 2R_20192913 is the 3′ LTR of 2R_20185276. Four of the 22 solo LTRs found were shorter than the full-length LTR of the canonical element, and also displayed internal deletion and less similarity with the canonical LTR (95.97% in average). By using the Tandem Repeats Finder tool, we found diverse internal repeats in the roo/B104 canonical element sequence: an 18-bp repeat at nucleotide 1551 occurring 2.2 times, a 3-bp repeat at nucleotide 1068 occurring 28.7 times, a tandem repeat of 99 bp at nucleotide 725, and a region of 23-A repeats at nucleotide 981(Fig. 1; see Supplemental Materials). The presence and number of these repeats in the copies varied. The 18-bp and 99-bp repeats were either absent or present with the same rate of occurrence, depending on the copies. The 3-bp repeat and the (A)n region occurred different numbers of times.
Figure 1.
Structures of the sequences of the canonical transposable elements. Structures of the reference elements zam, doc2, and doc3 are not represented because there were no copies of zam in the sequenced genome and because doc2 and doc3 were very similar to the canonical doc element. The deletions indicated for the hobo and pogo elements correspond to the internal deletions that gave rise to the regulatory elements of 1406 bp for hobo and 806 bp for pogo. The deletion and insertion indicated for the roo/B104 element correspond to the deletion and insertion of 12 bp found in several copies. The tandem repeats and the (A)n indicated on the elements correspond to repeats for which variants in the number of occurrences were found in some copies in comparison to the canonical element.
There were 10 copies of the tinker element, eight of which were complete and had on average 99.86% similarity to the reference, one copy had an internal deletion but had 99.77% similarity to the reference, and the last copy, which was 249-bp in length and had 85% similarity to the reference tinker element, seemed to be a solo-LTR. One of the complete copies contained several small insertions.
Four of the seven copies of bel were complete and shared 99.94% similarity with the reference bel element. Another copy shared 100% similarity but was only 74 bp in length, the first 1896 bases and the last 4156 bases being truncated. The two other copies were very divergent with mean similarities of about 87.10%.
The 412/mdg1-like Families
This group of families consists of five members: 412, mdg1, stalker, blood, and pilgrim (Costas et al. 2001). There were no copies of 412 and stalker elements on chromosome arm 2L. Twentyfive of the 30 copies of the 412 element were complete and displayed 99.81% similarity with the canonical 412. Of the other five copies, the 3L_9089318 copy had a very high level of similarity with the canonical element (99.84%), but the first 3955 bases had been deleted from its 5′ side. The 3L_20508657 and X_5264790 copies corresponded to solo-LTR with >99% similarity to the reference element. The X_5265729 possessed an internal deletion but also had a high level of similarity with 412. Finally, the 2R_73301 copy was a divergent element with <80.75% similarity to the reference. The main difference between the complete copies and the canonical element was the number of internal repeats within the LTR (32-bp repeat), the 5′ regulatory region (63-bp repeat), and the gag (15-bp repeat) and pol genes (21-bp repeat) (Fig. 1 and Supplemental Materials). Variants differed principally with regard to the number of repeats in their LTRs and regulatory region, and in a few cases in the gag gene. There were no differences in the number of repeats in the pol gene.
Among the 21 copies of the mdg1 element, 14 were complete and presented on average 99.21% similarity with the canonical mdg1, and the last copy, X_21195123, corresponded to a solo-LTR. The complete copies differed mainly in the number of occurrences of repeats: three regions of (A)n repeats in the 5′ regulatory region at positions 1226, 1286, 1556, and 1655 in the reference element, and a 14-bp repeat on the 3′ side of the sequence at position 6699. The other six sequences had no repeats; three of them were very divergent whereas the other three showed a very high level of similarity: the 2R_5697885 copy was truncated on its 5′ side, the middle of the sequence of the 3L_17679785 copy was inverted relative to its extremities, and the 2R_5683040 copy had an inversion (2688 bp) at the end of its sequence.
The blood element displayed 20 complete copies and one copy with a small 18-bp deletion at position 413. All sequences were very similar to the canonical element, with 99.92% similarity.
Eleven of the 18 copies of the stalker element displayed a high level of similarity (97.99%) to the canonical sequence. However, three of these 11 copies had regions with no similarity to the canonical stalker element. The corresponding region in the 3R_5130322 element showed 61.7% similarity with 412, suggesting that recombination had occurred between stalker and 412. The other seven copies, 3R-3555299, 3L_18659690, 3L_14774058, 2R_517895, X_1711698, X_21313234, and X_21320163 were more puzzling. On average, they showed 99% similarity to each other but only 80% similarity to stalker. However, most of them did not seem to be just divergent elements, because they were longer than the canonical stalker, which is 7256-bp long (versus 7881bp for 3R_3555299, 7895 bp for 3L_18659690, 7669 bp for 3L_14774058, 7671 bp for 2R_517896, and 7883 bp for X_1711698), and they had conserved 5′ and 3′ LTRs with no similarity to the known stalker LTRs, as well as possessing two open reading frames corresponding to the entire gag and pol genes (Fig. 1). We suggest that these seven sequences may be copies of a new TE that we have named new-stalker. This element must be very similar to stalker and potentially active. Four copies are clearly inactive: 2R_517896 contains an inserted nomad element, 3L_14774057 and X_21320163 possess an internal deletion, and X_21313234 seems to be a solo LTR. The estimated age of the copies of stalker and new-stalker (Table 1) suggests that stalker is probably no longer active, whereas new-stalker results from a recent event of mobilization.
Table 1.
Total number of TE copies detected in the chromosome arms 2L, 2R, 3L, 3R, and X
|
Number of copies on chromosome arm
|
CS
|
DS
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Name | Access number of canonical sequences | 2L | 2R | 3L | 3R | X | Identical copies/complete copiesa/Total copies | % | Average age (Myr) | % | Average age (Myr) |
| LTR retrotransposons | mdg1 | X59545b | 3 | 8 | 4 | 3 | 3 | 18/14/21 | 99.13 (0.07) | 0.25±0.043 | 85.39 (17) | 4.56±1.29 |
| 412 | X04132b | 0 | 6 | 11 | 5 | 8 | 29/25/30 | 99.77 (0.02) | 0.07±0.05 | 80.75 | 7.19 | |
| stalker | AF420242b | 0 | 1 | 4 | 3 | 3 | 11/1/11 | 97.99 (7.23) | 1.49±0.89 | - | - | |
| new-stalker | - | 0 | 1 | 2 | 1 | 3 | 7/3/7 | 99.80 (0.002) | 0.062±0.044 | - | - | |
| blood | X04671b | 11 | 2 | 2 | 5 | 1 | 21/20/21 | 99.92 (0.001) | 0.023±0.009 | - | - | |
| tirant | X93507b | 3 | 3 | 4 | 5 | 4 | 19/15/19 | 99.93 (0.008) | 0.021±0.03 | - | - | |
| zam | AJ000387b | 0 | 0 | 0 | 0 | 0 | - | - | - | - | - | |
| copia | M11240b | 13 | 5 | 5 | 4 | 4 | 30/26/31 | 99.88 (0.007) | 0.036±0.027 | 78.00 | 6.87 | |
| 297 | X03431b | 13 | 8 | 6 | 8 | 19 | 44/11/54 | 99.34 (0.014) | 0.21±0.04 | 86.23 (12.58) | 4.30±1.11 | |
| 17.6 | X01472b | 0 | 3 | 5 | 2 | 4 | 14/2/14 | 99.46 (0.14) | 0.17±0.12 | - | - | |
| rover | AF492764b | 0 | 1 | 0 | 2 | 2 | 4/1/5 | 99.21 (1.43) | 0.25±0.38 | 82.00 | 5.60 | |
| roo/B104 | AL031366b | 20 | 25 | 25 | 25 | 30 | 125/30/125 | 99.61 (0.82) | 0.12±0.30 | - | - | |
| bel | U23420b | 0 | 1 | 2 | 0 | 4 | 5/4/7 | 99.94 (0.002) | 0.019±0.015 | 87.10 | 4.58 | |
| tinker | EBIc | 1 | 3 | 2 | 3 | 1 | 9/8/10 | 99.85 (0.007) | 0.047±0.026 | 85.00 | 5.31 | |
| Non-LTR retrotransposons | jockey | M22874b | 9 | 18 | 14 | 14 | 17 | 66/0/72 | 99.63 (0.11) | 0.20±0.22 | 84.28 (3.85) | 4.91±0.61 |
| F | M17214b | 9 | 13 | 11 | 10 | 3 | 39/5/46 | 99.41 (2.80) | 0.10±0.16 | 83.55 (6.36) | 5.14±0.79 | |
| doc | X17551b | 15 | 5 | 17 | 10 | 5 | 50/19/52 | 99.81 (0.08) | 0.06±0.09 | 88.53 | 3.58 | |
| doc2 | BDGPd | 0 | 2 | 0 | 0 | 0 | 1/0/2 | 98.44 | 0.49 | 85.00 | 4.69 | |
| doc3 | BDGPd | 1 | 5 | 1 | 0 | 0 | 7/0/7 | 95.61 (0.81) | 2.05±0.83 | - | - | |
| Transposons | hobo | M69216b | 16 | 6 | 1 | 12 | 7 | 19/0/42 | 99.97 (0.002) | 0.01±0.015 | 87.84 (8.79) | 3.80±0.93 |
| pogo | X59837b | 12 | 5 | 7 | 11 | 8 | 43/5/43 | 99.88 (0.07) | 0.04±0.08 | - | - | |
| bari-1 | X67681b | 2 | 1 | 0 | 2 | 0 | 1/3/5 | 99.85 (0.007) | 0.04±0.026 | 81.02 | 5.93 | |
CS: % of similarity of conserved sequences
DS: % of similarity of diverged sequences
The variances of CS and DS are in parentheses
Copies with all functional features, and without internal deletions or truncations
Genbank accession number
EBI reference (European Bioinformatics Institute)
BDGP reference (Berkley Drosophila Genome Project)
The 297 and 17.6 Families
As can be seen in Table 1, there were more copies of the 297 element than of the 17.6 element in the genome. Eleven of the 54 copies of 297 were complete, with 99.36% similarity with the canonical element; 28 presented internal deletions but had a high level of similarity (99.33%); the 3R_4724013 copy contained an inserted 1731 element; the X_18485351 copy presented an internal rearrangement; 10 were very divergent with, on average, only 86% similarity to the reference element, and three were only conserved solo LTRs. One of the divergent copies appeared to be another solo-LTR.
There were only 14 copies of the 17.6 element, 12 of which were incomplete, and two complete, but all copies had 99.46% similarity to the canonical element. While searching for 17.6 copies, we detected five elements with <70% similarity to both the 17.6 and the 297 reference elements. These copies cannot be degenerate 17.6 or 297 elements because they have complete ORFs and complete identical LTRs. A BLASTN search on GenBank showed that these copies corresponded to the new recently reported rover element (Kaminker et al. 2002; accession number AF492764).
The tirant and zam Families
No full-length copy of the zam element, of which only one copy was found in the genome (Rizzon et al. 2002), was detected on the chromosome arms of the sequenced genome. Only a 287-bp fragment was detected on chromosome 3L. In contrast to zam, we found 19 copies of tirant, all having >99% similarity to the reference element. Three copies showed internal deletions. The tandem repeat of 19 bp in the LTRs (Viggiano et al. 1997) was found in all but one copy, 2L_21010933, which was the most divergent copy. The 102-bp repeat localized in the regulatory region of the reference element, which occurred six times (Viggiano et al. 1997), was also found in all the copies, apart from X_6075641, but variants were found that had two to six repeats (Fig. 1; Supplemental Materials).
The copia Family
We detected 31 copies of copia. Twenty-six were complete and showed >99% similarity to the canonical sequence. Five copies were incomplete: 3L_12915389 and X_13761465 had an internal deletion; 2R_1510206 was short (1376 bp) and very divergent (78% similarity to the reference), and 2L_20133886 and 2L_20135490 were very short (325 bp) but were very similar to the reference element (99.70%). The 2L_20133886 and 2L_20135490 copies were located in the same region of the genome. They were immediately followed by a fragment of a pogo element on the reverse strand, and a fragment of a hoppel element on the direct strand. The fragment of hoppel had diverged from the canonical sequence, whereas the pogo fragment, which corresponded to the first 1420 bp, was very similar to the reference pogo (99.72%). The two copia-pogo-hoppel sequences were identical, suggesting that they could result from a duplication event. The canonical copia element is known to possess two tandem repeats (Csink and McDonald 1995): one 28-bp repeat at nucleotide 341, the other a 108-bp repeat at nucleotide 2589 (Fig. 1). All the copies in the sequenced genome possessed the second repeat, but 12 copies did not possess the first repeat. Moreover, there was variation in the numbers of a T-repeat in two regions, located at nucleotide 354 and 378 in the reference element (Fig. 1).
Non-LTR Retrotransposons
None of the 72 copies of the jockey element was complete, but 66 presented a high level of similarity to the canonical sequence (99.63%). All the copies were truncated on their 5′ side. The longest copies were >5000 bp in length (the reference element being 5154 bp in length). Six copies shared <85% similarity, and some had small internal deletions. The 2R_13422204 copy was interrupted by a roo element inserted at position 1340.
Thirty-nine of the 46 copies of the F element had 99.41% similarity to the canonical sequence, and the other seven were divergent, averaging <85% similarity to the reference. One of these divergent copies, 2R_513677, which showed 87.39% similarity to the reference F, contained an inserted copy of the newstalker element that was in turn interrupted by a nomad element (see above). Five F sequences appeared to be complete, the others generally displayed a truncation on their 5′ side, with four copies also displaying internal deletions and one copy being truncated on its 3′ side.
Nineteen of the 52 copies of the doc element appeared to be complete compared to the reference element, 31 copies were truncated on the 5′ side and had on average 99.90% similarity to the canonical element, and two copies were divergent, with a similarity level of about 88.53%. The 2L_14447792 and the 2L_19323155 copies had similarities to the reference doc of 99.54% and 100%, respectively, but both were interrupted by another element: hopper and blood, respectively. Two other types of doc element have been reported: doc2 and doc3 (Berkeley Drosophila Genome Project; http://www.fruitfly.org/p_disrupt/TE.html). The doc3 element displayed seven copies, all of which were truncated on the 5′ side and possessed numerous internal deletions. However, the percentage of similarity was high (95.61% on average). There were only two copies of the doc2 element, with 98.44% and 85% similarity to the reference element, respectively. No copy of these two elements was detected on the X chromosome.
Transposons
There were large numbers of copies of hobo and pogo (42 and 43 copies, respectively), whereas there were only five copies of bari-1.
The 42 copies of hobo were all incomplete, despite an almost complete copy on the X chromosome, which displayed only a small deletion. Eighteen 1406-bp copies had a single internal deletion, but a high level of similarity to the canonical element (99.99%). Such short copies, currently described as hobo regulators (Boussy and Daniels 1991), could still be trans mobilized by full-length elements because they still possess intact inverted terminal repeats (ITR) at each extremity (Boussy and Daniels 1991). The length of the other 23 copies ranged from 59 to 2067 bp. These copies were divergent, with a percentage similarity to the canonical sequence ranging from 80% to 91.86%. Two kinds of strain have been reported concerning the hobo element: E strains, which lack the canonical elements, and H strains, which contain both complete and defective hobo elements, with a majority of 1.5 kb defective elements (Streck et al. 1986). The sequenced Drosophila genome therefore corresponds to an H strain, even though no complete hobo element was found. Two possible explanations for the large number of similar defective elements (Blackman and Gelbart 1989) have been advanced: (1) the transposition rate could be higher than the internal deletion rate; (2) there could be a greater trend toward the amplification of defective rather than complete elements. The second hypothesis is based on the observation that defective elements still possess intact ITRs. The observation that these defective copies are very young (Table 1) is also in favor of this second hypothesis.
Five of the 43 pogo copies were complete with 99.90% similarity to the canonical element. Twenty seven copies were 186 bp in length, presented a single internal deletion, had a high level of similarity to the reference element (99%), and possessed complete ITRs. Nine copies of differing lengths (between 1068 and 1456 bp) had internal deletions, but once again, they had a high level of similarity to the reference (99.34%). Finally, the two last copies corresponded to the pogo fragments inserted near two copia fragments (see above). The presence of similar defective elements of the pogo element can be explained by the two hypotheses proposed above for hobo.
Five bari-1 elements were found, three being complete copies with 99.88% similarity to the reference element. One of the other two copies was a divergent copy, with 81.02% similarity to the reference, and the other possessed internal deletions and insertions, but displayed a high level of similarity to the reference. No copy of bari-1 was detected on the X chromosome.
Divergence and Evolution
Elements of the transposon class had the most degenerate copies, whereas retrotransposons tended to conserve full-length copies. When the retrotransposons did have an internal deletion, their sequence was often still very similar to the canonical element. For all retrotransposons, the few divergent copies observed were located near the centromeric region of each chromosome arm. For all TE classes, the TE copies that were interrupted by other elements were also found near the centromeric regions.
The LTR retrotransposons, 412, mdg1, tirant, roo/B104, and copia possess internal repeats. The copies have been found to display differences in the number of repeat occurrences or the absence of some repeats. Polymorphism in the occurrence of the repeats was observed even among sequences with high similarity to one another or to the canonical element. This variability in the occurrence of repeats is probably related to the activity of the copies, as has been reported for Tnt1 in tobacco (Casacuberta et al. 1995; Vernhettes et al. 1998).
There was no progressive divergence between TEs copies, and so we were not able to investigate the evolution of codon usage and dinucleotide pattern (Lerat et al. 2002a,b) according to the degree of divergence. The divergent copies were indeed either so similar that there were no differences in codon usage or dinucleotide pattern, or so distant that no coding part could be determined. Various hypotheses could account for this lack of a spectrum of divergence.
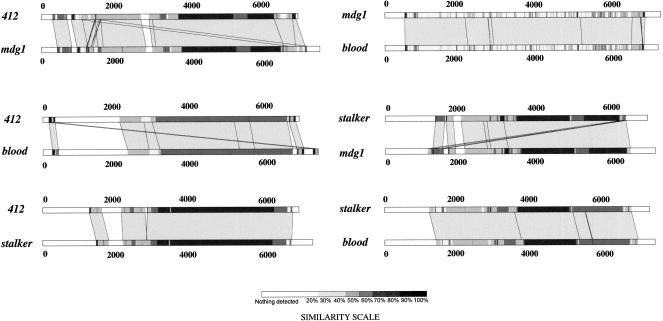
Relationships Between Different Sequences
The TEs of a given family could be derived from different ancestral elements. For example, the stalker element could be a mosaic element, because different regions of its genome have discordant phylogenetic relationships with the corresponding regions of other members of the 412/mdg1-like families (Costas et al. 2001) (Fig. 2). Similarly, the new-stalker element, reported above, could also be the product of recombination events. The new-stalker element is longer than stalker (Makarova 1997), and both have two identical LTRs at their extremities, although the two elements have different LTRs. This clearly suggests that new-stalker and stalker are two distinct elements, both of which are likely to be active. Another example of recombination is the evidence that the roo/B104 element shares similarity of sequence of its env gene to the zam element, indicating that roo/B104 may have captured its putative envelope coding region from a zam-like element (Frame et al. 2001). Such findings confirm that TEs could have acquired parts of their sequence from other TEs (Lerat et al. 1999). All these data suggest that a highly divergent element can give rise to another new element. This could explain the absence of divergent elements in the retrotransposon class. However, the question still remains how a divergent element, inevitably nonfunctional because of the presence of stop codons inside its ORFs and mutations in its LTRs, can give rise to autonomous copies.
Figure 2.
Lalnview representations of the LFasta results between the members of the 412/mdg1 family.
The 297 element is similar to the gag region of Tv1 of D. virilis but not to 17.6 of D. melanogaster. In contrast, it shares more features with the pol region of 17.6 than with that of Tv1. This suggests that the Tv1 element of D. virilis has been transferred to D. melanogaster and after recombining with 17.6 has given rise to D. melanogaster 297. Interestingly, rover also displays similarity between its gag gene and the gag gene of Tv1. Rover could therefore be another product of recombination between Tv1 and 17.6.
Maintenance and Turn Over
The absence of divergence among copies of the LTR and non-LTR retrotransposons could result from a rapid turnover that eliminates TE copies as soon as they become inactive. This is illustrated by the genome of Saccharomyces cerevisiae that contains only LTR retrotransposons characterized by a high degree of homogeneity between full-length sequences. This homogeneity suggests that active elements have been transposed, whereas inactive elements have been eliminated by LTR-LTR recombination (Jordan and McDonald 1998b; Kim et al. 1998), which would explain the high number of solo-LTRs in this genome (Jordan and McDonald 1999). However, the sequenced Drosophila genome possesses very few solo-LTRs, the highest number of such solo-LTRs being found in roo/B104 (22 solo-LTRs out of 125 sequences). One divergent solo-LTR was detected for tinker out of nine sequences, one for stalker out of 11 copies, one for 412 out of 30 copies, one for mdg1 out of 21 copies, and four for 297 out of 54 copies. This means that there are far fewer solo-LTRs in Drosophila than in yeast, in which solo-LTRs account for 85% of all the copies (Kim et al. 1998). A turn-over hypothesis in Drosophila would imply that the mechanism of TE elimination is different from that of LTR-LTR recombination. This would be possible if the rate of transposition is lower than that of the selection process that prevents TEs insertions (Biémont et al. 1997; Charlesworth et al. 1997).
Incomplete non-LTR retrotransposons, generally truncated on their 5′ side as a consequence of the transposition mechanism of this class of TEs (Hutchison III et al. 1989) are usually known as “Dead-On-Arrival” (DOA) elements. Such DOA elements have been found for jockey, doc, doc2, doc3, and F, although full-length copies have also been found for F. Petrov and Hartl (1997) have analyzed different copies of the non-LTR retrotransposon Helena in several Drosophila species. They found that copies of this TE have low nucleotide polymorphism, but a large number of internal deletions, which contribute to the high rate of DNA loss in the Drosophila genome. For F, doc, and jockey of the sequenced genome, however, we found internal deletions in only a few copies, the 5′ truncations being often the only difference from the reference element, although some very short conserved copies were found. In contrast, all the copies of doc2 and doc3, of which there were very few copies, had internal deletions. This suggests that the loss of DNA as a result of internal deletions within the Helena element (Petrov and Hartl 1997) is not a general feature of all the non-LTR retrotransposons. It is possible that doc2, doc3, and Helena are very ancient components of the genome, which lost their capacity to move long ago and are therefore vulnerable to deletions. This agrees with the age of doc3, which has been estimated to be 2 Myr (Table 1). In contrast, F, jockey, and doc, which are characterized by a relatively young age (Table 1), may have been active until recently, and have therefore avoided erosion. Non-LTR retrotransposons do not, therefore, generally provide a good estimation of the deletion pattern of the Drosophila genome.
Recent Transpositions of the LTR and Non-LTR Retrotransposons
The LTR and non-LTR retrotransposons of the sequenced genome may have been transposed recently, and therefore have not had enough time to diverge. This suggests that the very divergent members of these families are probably very ancient insertions, as seen in Table 1, maintained in the genome because they are neutral or because they are located in regions of the chromosomes (in this case, in the pericentromeric regions) where the recombination rate is low and selection therefore less efficient (Hill and Robertson 1966; Charlesworth et al. 1994). Such a hypothesis would imply that most of the LTR retrotransposons of the strain used for genome sequencing have recently moved at a significant rate, which is not an unrealistic assumption, as inbred lines may be subjected to sudden bursts of TE mobilization (Biémont et al. 1987, 1990). The presence of numerous degenerate copies of the transposons suggests either that this class of TEs was mobilized earlier than the retrotransposons or that degeneracy has resulted from their specific regulation mechanism (Brookfield 1991). This latter hypothesis is supported by the observation that the copies of the transposons that have internal deletions but present high similarity of their conserved sequence with the reference copy are very young (Table 1).
Acquisition by Horizontal Transfer
An alternative explanation for the lack of divergent copies in LTR and non-LTR retrotransposons resulting from a recent transposition burst event of some TEs as proposed for the yeast and Drosophila genomes (Kim et al. 1998; Jordan et al. 1999; Terzian et al. 2000; Bowen and McDonald 2001), would be the acquisition of a new TE by horizontal transfer. However, although this is likely to be true for elements like 297, which possesses part of Tv1 of D. virilis, and rover, it is difficult to envisage the horizontal transfer of all the elements in the Drosophila genome, especially as we have evidence that some TEs are very ancient components of the genome (Biémont and Cizeron 1999). The estimated ages of the divergent copies indeed indicate that most TEs are very ancient components of the D. melanogaster genome, even present before the divergence of D. melanogaster from D. simulans, which is estimated ∼2.3 millions years ago (Li et al. 1999).
Conclusion
The high sequence homogeneity that we have observed for transposable element copies within most families is compatible with the theoretical predictions of various simple models of the dynamics of repeated sequences (Ohta 1985; Slatkin 1985; Brookfield 1986; Hudson and Kaplan 1986) in which a recent increase in transposition rate or a high rate of transposition, and hence a high turnover of TE sequences, has the effect of reducing divergence among TE copies. In addition to transposition rate, these models also take into account biased and unbiased gene conversion, TE family size, selection against TEs, and effective size of the host population. Differences between these parameters in various TEs account for the different patterns of homogeneity observed between copies within the different TE families analyzed in our study. The sequenced Drosophila genome thus appears to be composed of a mixture of very active TEs and of ancient relics that have degenerated and rearranged. Many rearrangements, such as those observed among the bel-like elements, could result from recombination between copies, and then duplication of the rearranged copies, for example, for the three copies of roo that showed the same kind of rearrangement. The observation that the highly divergent copies are mainly located in the pericentromeric regions of the chromosomes suggests that TE insertions, which are known to accumulate in these regions and to be less exposed to natural selection than insertions on the chromosome arms, are old components that have had time to degenerate (Biémont et al. 1997; Charlesworth et al. 1997). Recent waves of mobilization of some TEs, the acquisition of new TEs by horizontal transfer, and changes in the characteristics of the TEs or the host population, are therefore major factors that can modify the equilibrium state predicted by the models. However, whatever the mechanisms involved, a high level of homogeneity between TE sequences within a family clearly indicates a high level of activity of this family within the genome, suggesting that the genetic variability of the Drosophila genome is constantly being renewed by the action of TEs.
METHODS
TE Copy Extraction
The sequences of the D. melanogaster chromosome arms 2L, 2R, 3L, 3R, and X from the 3.0 release of the genome were retrieved from the Web site of the Berkley Drosophila Genome project (http://www.fruitfly.org/). This version is the first to present true nucleotide TE sequences. However, we only have information about the TEs localized on the euchromatin regions of the genome, because the heterochromatin, which accounts for 30%–40% of the genome, has not been sequenced because of technical difficulties (Myers et al. 2000). This is a drawback of the analysis because many TEs are embedded within the heterochromatin and may contribute to the global TE regulation copy numbers (Kidwell and Lisch 2000).
A reference bank of the complete sequences of canonical TEs was constituted using sequences from the Flybase database (http://flybase.bio.indiana.edu/) and from the European Bioinformatics Institute (http://www.ebi.ac.uk). Sequences of TEs in the chromosome arms were detected using the RepeatMasker program (A.F.A. Smit and P. Green, unpubl; http://repeatmasker.genome.washington.edu/cgi-bin/RM2_req.pl) with our reference bank of elements. Manual analyses were then performed to determine position of the analyzed TE copies.
Most families of the TEs were chosen because they had a large number of copies in the sequenced genome, but we also analyzed elements with low copy numbers such as bari-1, doc2, doc3, stalker, and rover. We therefore analyzed the bel, 412/mdg1, and 297/17.6 families of LTR retrotransposons, the copia element and the tirant element, which display variants in natural populations (Csink and McDonald 1995; Marsano et al. 2000), the non-LTR retrotransposons, jockey, F, and doc, and the transposons hobo, pogo, and bari-1. The elements and their copy number on the chromosome arms are listed in Table 1. The canonical pilgrim, belonging to the 412/mdg1 family (Costas et al. 2001), was not clearly identified. We did not take it into account in our analysis, even though several copies were detected. For the localization and features of the different transposable elements reported here, see Supplemental Materials.
Comparison of TE Sequences
Copies from a given TE family were compared to the canonical sequence (see Table 1 for accession numbers) using the LFasta program (Pearson and Lipman 1988), which performs local alignments between two sequences. A graphical representation of the LFasta data was produced using Lanlview software (Duret et al. 1996), which gives the percentage of similarity of the aligned regions. Searches for internal repeated regions in TE sequences were performed using the Tandem Repeats Finder tool (Benson 1999). For copies displaying similarity to more than one canonical element, a search of coding parts was performed using the ExPASy translate tool (http://www.expasy.org/) before carrying out a BLASTP analysis (Altschul et al. 1997) to identify the putative ORFs. To establish the internal deletion pattern of the sequences, multiple alignments were performed for the copies of each family using the SEAVIEW sequence editor (Galtier et al. 1996) and the BLASTN program (Altschul et al. 1997). The percentage of similarity of each copy with the reference element was computed using the GCG Wisconsin package (Womble 2000). For each transposable element family, we calculated the average percentage of similarity to the canonical element, according to the synonymous and nonsynonymous sites and the noncoding regions (see Supplemental Materials).
The age of the copies was estimated using the method described in Bowen and McDonald (2001). Pairwise comparisons were made between each copy and the canonical element considered as an active element. The divergences were computed with the GCG Wisconsin package (Womble 2000) using the Kimura-2 parameter method. Ages were computed according to the formula T = K/(2r) where T is the time of divergence, K is the divergence, and r is the substitution rate (Li 1997). We used 0.016 for the value of synonymous substitutions per site per million years, as estimated for Drosophila (Li 1997). This rate value is a good estimator of neutral evolution because it concerns the less constrained regions of the genes. However, the ages estimated for the TE copies were an overestimation of the real ages because the real rate of substitution in TEs was underestimated.
The copies were identified according to the following convention: chromosome arm_nucleotide position of the beginning of the sequence.
Acknowledgments
This work was funded by the Centre National de la Recherche Scientifique (UMR 5558, GDR 2157) and the Association pour la Recherche sur le Cancer (contract 5428).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.827603.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print in July 2003.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, G. 1999. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 27: 573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biémont, C. and Cizeron, G. 1999. Distribution of transposable elements in Drosophila species. Genetica 105: 43-62. [DOI] [PubMed] [Google Scholar]
- Biémont, C., Aouar, A., and Arnault, C. 1987. Genome reshuffling of the copia element in an inbred line of Drosophila melanogaster. Nature 329: 742-744. [DOI] [PubMed] [Google Scholar]
- Biémont, C., Arnault, C., and Heizmann, A. 1990. Massive changes in genomic locations of P elements in an inbred line of Drosophila melanogaster. Naturwissenschaften 77: 485-488. [DOI] [PubMed] [Google Scholar]
- Biémont, C., Tsitrone, A., Vieira, C., and Hoogland, C. 1997. Transposable element distribution in Drosophila. Genetics 147: 1997-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman, R.K. and Gelbart, W.M. 1989. The transposable element hobo of Drosophila melanogaster. In Mobile DNA. (ed. D.E. Berg and M.M. Howe), pp. 523-529. American Society for Microbiology, Washington D.C.
- Boussy, I.A. and Daniels, S.B. 1991. Hobo transposable elements in Drosophila melanogaster and D. simulans. Genet. Res. 58: 27-34. [DOI] [PubMed] [Google Scholar]
- Bowen, N.J. and Jordan, I.K. 2002. Transposable elements and the evolution of eukaryotic complexity. Curr. Issues Mol. Biol. 4: 65-76. [PubMed] [Google Scholar]
- Bowen, N.J. and McDonald, J.F. 2001. Drosophila euchromatic LTR retrotransposons are much younger than the host species in which they reside. Genome Res. 11: 1527-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookfield, J.F.Y. 1986. A model of DNA sequence evolution within transposable element families. Genetics 112: 393-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookfield, J.F.Y. 1991. Models of repression of transposition in P-M hybrid dysgenesis by P cytotype and by zygotically encoded repressor proteins. Genetics 128: 471-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta, J.M., Vernhettes, S., and Grandbastien, M.A. 1995. Sequence variability within the tobacco retrotransposon Tnt1 population. EMBO J. 14: 2670-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., Sniegowski, P., and Stephan, W. 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371: 215-220. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., Langley, C.H., and Sniegowski, P.D. 1997. Transposable element distributions in Drosophila. Genetics 147: 1993-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costas, J., Valade, E., and Naveira, H. 2001. Amplification and phylogenetic relationships of a subfamily of blood, a retrotransposable element of Drosophila. J. Mol. Evol. 52: 342-350. [DOI] [PubMed] [Google Scholar]
- Csink, A.K. and McDonald, J.F. 1995. Analysis of copia sequence variation within and between Drosophila species. Mol. Biol. Evol. 12: 83-93. [DOI] [PubMed] [Google Scholar]
- Duret, L., Gasteiger, E., and Perrière, G. 1996. LalnView: A graphical viewer for pairwise sequence alignments. Comp. Applic. Biosci. 12: 507-510. [DOI] [PubMed] [Google Scholar]
- Evgen'ev, M.B., Zelentsova, H., Poluectova, H., Lyozin, G.T., Veleikodvorskaja, V., Pyatkov, K.I., Zhivotovsky, L.A., and Kidwell, M.G. 2000. Mobile elements and chromosomal evolution in the virilis group of Drosophila. Proc. Natl. Acad. Sci. 97: 11337-11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame, I.G., Cutfield, J.F., and Poulter, R.T.M. 2001. New BEL-like LTR retrotransposons in Fugu rubripes, Caenorhabditis elegans, and Drosophila melanogaster. Gene 263: 219-230. [DOI] [PubMed] [Google Scholar]
- Galtier, N., Gouy, M., and Gautier, C. 1996. SEAVIEW and PHYLO_WIN: Two graphic tools for sequence alignment and molecular phylogeny. Comp. Appl. Biosci. 12: 543-548. [DOI] [PubMed] [Google Scholar]
- Hill, W.G. and Robertson, A. 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8: 269-294. [PubMed] [Google Scholar]
- Hudson, R.R. and Kaplan, N.L. 1986. On the divergence of members of a transposable element family. J. Math. Biol. 24: 207-215. [DOI] [PubMed] [Google Scholar]
- Hutchison III, C.A, Hardies, S.C., Loeb, D.D., Shehee, W.R., and Edgell, M.H. 1989. LINEs and related retroposons: Long interspersed repeated sequences in the eucaryotic genome. In Mobile DNA. (eds. D.E. Berg and M.M. Howe), pp. 593-617. American Society for Microbiology, Washington D.C.
- International Human Genome Sequencing Consortium. 2001. Initial Sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Jordan, I.K. and McDonald, J.F. 1998a. Interelement selection in the regulatory region of the copia retrotransposon. J. Mol. Evol. 47: 670-676. [DOI] [PubMed] [Google Scholar]
- Jordan, I.K. and McDonald, J.F. 1998b. Evidence for the role of recombination in the regulatory evolution of Saccharomyces cerevisiae Ty element. J. Mol. Evol. 47: 14-20. [DOI] [PubMed] [Google Scholar]
- Jordan, I.K. and McDonald, J.F. 1999. The role of interelement selection in Saccharomyces cerevisiae Ty element evolution. J. Mol. Evol. 49: 352-357. [DOI] [PubMed] [Google Scholar]
- Jordan, I.K., Matyunina, L.V., and McDonald, J.F. 1999. Evidence for the recent horizontal transfer of long terminal repeat retrotransposons. Proc. Natl. Acad. Sci. 96: 12621-12625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker, J.S., Bergman, C.M., Kronmiller, B., Carlson, J., Svirkas, R., Patel, S., Frise, E., Wheeler, D.A., Lewis, S.E., Rubin, G.M., et al. 2002. The transposable elements of the Drosophila melanogaster euchromatin: A genomics perspective. Genome Biol. 3: RESEARCH0084.1–0084.20. [DOI] [PMC free article] [PubMed]
- Kidwell, M.G. and Lisch, D.R. 2000. Transposable elements and host genome evolution. Trends Ecol. Evol. 15: 95-99. [DOI] [PubMed] [Google Scholar]
- Kim, J.M., Vanguri, S., Boeke, J.D., Gabriel, A., and Voytas, D.F. 1998. Transposable elements and genome organization: A comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 8: 464-478. [DOI] [PubMed] [Google Scholar]
- Lerat, E., Brunet, F., Bazin, C., and Capy, P. 1999. Is the evolution of transposable elements modular? Genetica 107: 15-25. [PubMed] [Google Scholar]
- Lerat, E., Biémont, C., and Capy, P. 2000. Codon usage and the origin of P elements. Mol. Biol. Evol. 17: 467-468. [DOI] [PubMed] [Google Scholar]
- Lerat, E., Capy, P., and Biémont, C. 2002a. Codon usage by transposable elements and their host genes in five species. J. Mol. Evol. 54: 625-637. [DOI] [PubMed] [Google Scholar]
- Lerat, E., Capy, P., and Biémont, C. 2002b. The relative abundance of dinucleotides in transposable elements in five species. Mol. Biol. Evol. 19: 964-967. [DOI] [PubMed] [Google Scholar]
- Li, W. 1997. Mol. Evol. Sinauer, Sunderland, MA.
- Li, Y.J., Satta, Y., and Takahata, N. 1999. Paleo-demography of the Drosophila melanogaster subgroup: Application of the maximum likelihood method. Genes Genet. Syst. 74: 117-127. [DOI] [PubMed] [Google Scholar]
- Makarova, K.S. 1997. A small open reading frame of the stalker retrotransposon reveals a high similarity to the second small frame of the mdg1 retrotransposon. Genetika 33: 1016-1019. [PubMed] [Google Scholar]
- Marsano, R.M., Moschetti, R., Caggese, C., Lanave, C., Barsanti, P., and Caizzi, R. 2000. The complete Tirant transposable element in Drosophila melanogaster shows a structural relationship with retrovirus-like retrotransposons. Gene 247: 87-95. [DOI] [PubMed] [Google Scholar]
- Myers, E.W., Sutton, G.G., Delcher, A.L., Dew, I.M., Fasulo, D.P., Flanigan, M.J., Kravitz, S.A., Mobarry, C.M., Reinert, K.H., Remington, K.A., et al. 2000. A whole-genome assembly of Drosophila. Science 287: 2196-2204. [DOI] [PubMed] [Google Scholar]
- Ohta, T. 1985. A model of duplicative transposition and gene conversion for repetitive DNA families. Genetics 10: 513-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, W.R. and Lipman, D.J. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. 85: 2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov, D.A and Hartl, D.L. 1997. Trash DNA is what gets thrown away: High rate of DNA loss in Drosophila. Gene 205: 279-289. [DOI] [PubMed] [Google Scholar]
- Rizzon, C., Marais, G., Gouy, M., and Biémont, C. 2002. Recombination rate and the distribution of transposable elements in the Drosophila melanogaster genome. Genome Res. 12: 400-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel, P., Tikhonov, A., Jin, Y.K., Motchoulskaia, N., Zakharov, D., Melake-Berhan, A., Springer, P.S., Edwards, K.J., Lee, M., Avramova, Z., et al. 1996. Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765-768. [DOI] [PubMed] [Google Scholar]
- Shapiro, J.A. 1999. Transposable elements as the key to a 21st century view of evolution. Genetica 107: 171-179. [PubMed] [Google Scholar]
- Shields, D.C. and Sharp, P.M. 1989. Evidence that mutations patterns vary among Drosophila transposable elements. J. Mol. Biol. 207: 843-846. [DOI] [PubMed] [Google Scholar]
- Slatkin, M. 1985. Genetic differentiation of transposable elements under mutation and unbiased gene conversion. Genetics 110: 145-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streck, R.D., MacGaffey, J.E., and Beckendork, S.K. 1986. The structure of hobo transposable elements and their insertion sites. EMBO J. 5: 3615-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzian, C., Ferraz, C., Demaille, J., and Bucheton, A. 2000. Evolution of the gypsy endogenous retrovirus in the Drosophila melanogaster subgroup. Mol. Biol. Evol. 17: 908-914. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Meyers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Vernhettes, S., Grandbastien, M.A., and Casacuberta, J.M. 1998. The evolutionary analysis of the Tnt1 retrotransposon in Nicotiana species reveals the high variability of its regulatory sequences. Mol. Biol. Evol. 15: 827-836. [DOI] [PubMed] [Google Scholar]
- Viggiano, L., Caggese, C., Barsanti, P., and Caizzi, R. 1997. Cloning and characterization of a copy of Tirant transposable element in Drosophila melanogaster. Gene 197: 29-35. [DOI] [PubMed] [Google Scholar]
- Womble, D.D. 2000. GCG: The Wisconsin package of sequence analysis programs. Meth. Mol. Biol. 132: 3-22. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://repeatmasker.genome.washington.edu/cgi-bin/RM2_req.pl; RepeatMasker.
- http://www.fruitfly.org/index.html; the Berkeley Drosophila Genome Project 2000.
- http://www.ebi.uk/index.html; the European Bioinformatics Institute 2001.
- http://flybase.bio.indiana.edu/; A database of the Drosophila Genome.
- http://www.expasy.org/; ExPASy molecular biology server.