Gene expression profile of malignant pleural mesothelioma (MPM) tumors show 3 molecular subgroups with significant deregulation of mitotic spindle assembly checkpoint (MSAC) pathway and microtubule network. MAD2L1, a key MSAC player, transcript and protein expression is upregulated in all tumors and epothilone B, a microtubule drug, shows excellent efficacy against MPM cancer cell lines.
Keywords: mesothelioma, microarray, network, therapeutics, MAD2L1, epothilone B
Abstract
Background
Malignant pleural mesothelioma (MPM) is a lethal neoplasm exhibiting resistance to most treatment regimens and requires effective therapeutic options. Though an effective strategy in many cancer, targeted therapy is relatively unexplored in MPM because the therapeutically important oncogenic pathways and networks in MPM are largely unknown.
Materials and methods
We carried out gene expression microarray profiling of 53 surgically resected MPMs tumors along with paired normal tissue. We also carried out whole transcriptomic sequence (RNA-seq) analysis on eight tumor specimens. Taqman-based quantitative Reverse-transcription polymerase chain reaction (qRT-PCR), western analysis and immunohistochemistry (IHC) analysis of mitotic arrest deficient-like 1 (MAD2L1) was carried out on tissue specimens. Cell viability assays of MPM cell lines were carried out to assess sensitivity to specific small molecule inhibitors.
Results
Bioinformatics analysis of the microarray data followed by pathway analysis revealed that the mitotic spindle assembly checkpoint (MSAC) pathway was most significantly altered in MPM tumors with upregulation of 18 component genes, including MAD2L1 gene. We validated the microarray data for MAD2L1 expression using quantitative qRT-PCR and western blot analysis on tissue lysates. Additionally, we analyzed expression of the MAD2L1 protein by IHC using an independent tissue microarray set of 80 MPM tissue samples. Robust clustering of gene expression data revealed three novel subgroups of tumors, with unique expression profiles, and showed differential expression of MSAC pathway genes. Network analysis of the microarray data showed the cytoskeleton/spindle microtubules network was the second-most significantly affected network. We also demonstrate that a nontaxane small molecule inhibitor, epothilone B, targeting the microtubules have great efficacy in decreasing viability of 14 MPM cell lines.
Conclusions
Overall, our findings show that MPM tumors have significant deregulation of the MSAC pathway and the microtubule network, it can be classified into three novel molecular subgroups of potential therapeutic importance and epothilone B is a promising therapeutic agent for MPM.
introduction
Malignant pleural mesothelioma (MPM) is an ill-understood aggressive malignant growth of the pleura surrounding the lungs with affected patients showing low median overall survival and poor response to most treatment regimens [1]. Histologically, there are three subtypes of MPM: epitheloid, sarcomatoid and biphasic or mixed, and MPM patients with sarcomatoid subtype show worse survival after surgery while those with epitheloid subtype show the best overall survival [2]. However, in all tumor stages, even with the best available chemotherapeutic treatment, the increase in median survival of MPM patient's is marginal [3]. Therefore, there is an urgent need to develop novel therapeutic strategy in this disease.
Gene expression studies in mesothelioma have identified oncogenes like CUL4A, c-jun, and MMP-14 [4–7], while there is evidence for the inactivation of tumor suppressor genes like NF2/merlin, p16/CDKN2A, p14/ARF, RASSF1A, LATS2 and more recently BAP1 [8]. Some of the important pathways involved in MPM pathogenesis include the NFκb, phosphatidylinositol-3-kinase (PI3K)-AKT, mitogen-activated protein kinase (MAPK) and Hippo along with frequent deregulation of EGFR and MET signaling [9, 10]. However, clinical trials targeting many of these pathways or targets have not yielded desirable results in terms of response to treatment [11].
The objectives of our study were to analyze robust gene expression profiles in MPM tumors to identify novel molecular pathways and networks that could also provide therapeutic targets. Here, we profiled 53 MPM tumors using high-density global transcriptomic microarrays along with paired normal specimens. Bioinformatic analysis of these tumor specimens identified the mitotic spindle assembly checkpoint (MSAC) pathway and microtubule network to be most significantly deregulated pathway and second-most significantly deregulated network in MPM, respectively. Our analysis also identified three subgroups of MPM tumors, exhibiting unique gene expression profiles with different therapeutic targets, which partially overlapped with tumor histotype. Moreover, we determined, using various target-specific small molecule inhibitors and cell viability studies, that the microtubule network is a viable candidate for targeted therapy of MPM.
materials and methods
tissue and cell-line specimens
Flash-frozen MPM tumor and adjacent nontumor tissue samples were obtained from surgical resections. Before RNA extraction, histology sections were reviewed to assess the percentage of tumor and that of malignant cells in the tumor specimens. For immunohistochemistry (IHC) analysis, a tissue microarray (TMA) of an independent set of 80 formalin-fixed and paraffin-embedded (FFPE) MPM tumor specimens was used. All tumor cases were staged and histologically subgrouped according to the World Health Organization classification [12]. The clinical and pathological characteristics of all tumor cases utilized for these analyses are presented in supplementary Table S1, available at Annals of Oncology online. The MPM cell lines were obtained from ATCC, Sigma-Aldrich Corp., DSMZ and collaborators.
RNA extraction, gene profiling and transcriptome analyses
Total RNA was extracted from tissues and after assessment of its quality and quantity, it was labeled and hybridized on Affymetrix U133 plus 2.0 chips. RNA-sequencing (RNA-seq) was carried out on a SOLID 4.0 platform, and Bioscope 1.3.1 was used to align the reads and determine the RPKM values.
transcript and protein analysis of tissue and cell lysates
TaqMan RT-qPCR assays were carried out to determine MAD2L1 transcript levels with Glyceraldehyde-3-phosphatedehydrogenase (GAPDH) gene used as endogenous control. All samples were assayed in triplicate and ΔCT values were calculated between test gene and endogenous control for each sample. Protein from tissues and cell lines were extracted using published protocol. To assess relative levels of protein expression, β-actin expression was used for normalization and densitometric scans were analyzed using NIH's ImageJ application (http://rsb.info.nih.gov/ij/).
immunohistochemistry
FFPE tissue histology sections were immunostained and expression of MAD2L1 protein was quantified using a 4-value intensity score (0, 1+, 2+, 3+) and the percentage (0%–100%) of the extent of reactivity. The final score was obtained by multiplying the intensity and extent-of-reactivity values (range, 0–300).
cell viability assay
Cell viability was quantified using MTS assay on a 96-well plate reader to obtain IC50 data values for different drugs.
bioinformatics and statistical analyses
The details of the bioinformatics and subsequent statistical analysis of the gene expression data can be found in the supplementary Sweave Report, available at Annals of Oncology online. The microarray data [GSE51024] is available at the Gene Expression Omnibus (GEO) website (http://www.ncbi.nlm.nih.gov/geo/). Statistical analysis of qPCR, western blot data and IC50 data was carried out in Prism 6.0 (GraphPad Software, Inc., La Jolla, CA).
results
global gene expression profiling of MPM tumors
We carried out global transcriptomic microarray analysis using Affymetrix U133 plus 2.0 microarray chips on total RNA extracted from 53 cases of surgically resected MPM tumor tissues (comprising of 35 epitheloid, 12 biphasic and 6 sarcomatoid subtypes of tumors) with 38 paired nontumor tissue serving as controls. Bioinformatic analysis of the RNA microarray data involved using a linear regression model, which accounted for the branch effect seen in samples, to discover differentially expressed genes between normal and tumor pairs. About 2310 probesets representing 1746 genes or open reading frames (ORFs), at a highly significant false discovery rate (FDR) of equal to or smaller than 1E-09, were obtained (supplementary Table S2, available at Annals of Oncology online). Pathway analysis, using MetaCore software suite (GeneGo, Inc., Carlsbad, CA), of this set of genes revealed the metaphase checkpoint involving MSAC genes and kinetochore components to be most significantly deregulated pathway in MPM tumors (supplementary Table S3, available at Annals of Oncology online). There are 18 components of this pathway upregulated in MPM tumors including MAD2L1, budding uninhibited by benzimidazoles 1 homolog (BUB1), Baculoviral inhibitor of apoptosis repeat-containing 5 or Survivin (BIRC-5), Aurora Kinase A (AURKA) and others as detailed in supplementary Figure S1, available at Annals of Oncology online). This MSAC signature of 18 genes was also found in the top two pathways after performing gene set enrichment analysis for canonical pathways (http://www.broadinstitute.org/gsea/msigdb/genesets.jsp?collection=CP).
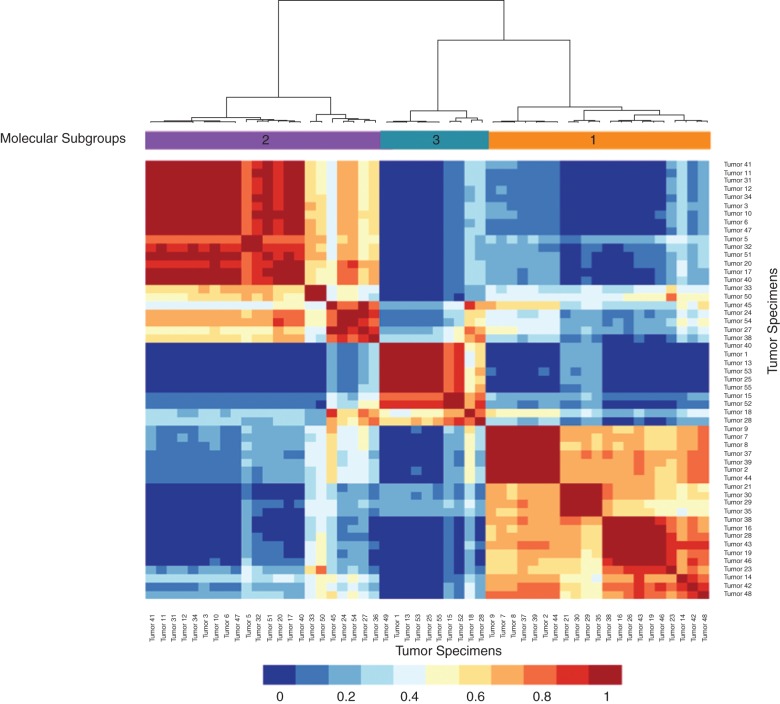
Interestingly, robust cluster analysis of the probesets (500–2000 probesets randomly chosen at an FDR of 0.01, i.e. from a total of 20 000 probesets) defined three molecular subgroups of tumors with differential transcript expression (Figure 1 and supplementary Sweave Report, available at Annals of Oncology online). These three gene expression-defined groups did not correlate with the histological subtypes and had variable number of histotypes. The epitheloid MPMs were present to differing extent between the three groups, but were mostly represented within subgroup 1, which contains 19 of the 35 epitheloid tumors. Subgroup 2 had most of the biphasic tumors (7 of 12), while subgroup 3 had most of the sarcomatoid cases (4 of 6). However, we did not find any significant association between the distribution of the MPM tumors in the three molecular subgroups with patient's clinico-pathological features like overall survival (supplementary Sweave Report, available at Annals of Oncology online).
Figure 1.
Robust cluster analysis identifies three molecular subgroups of mesothelioma tumors. In a process repeated 1000 times, a uniform number of probesets (500–2000) are randomly selected from a set selected at the FDR level of 0.01 (a total of 20 000 probesets). During each process, whenever any two samples are grouped together, it is recorded to generate the consensus matrix which denotes the average time any two samples are grouped together in each robust clusters. The consensus matrix is used as the similarity matrix to define the final clusters. The intensity bar below denotes the values of color coding used in the cluster map.
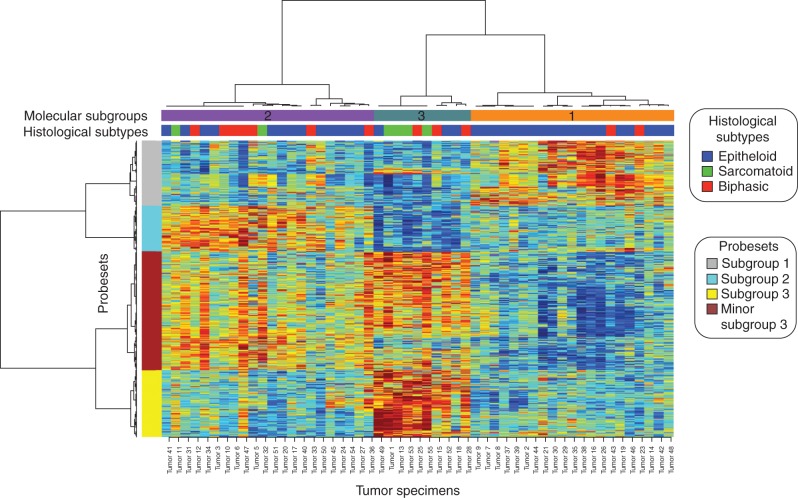
Comparing all probesets between these three subgroups of tumors cases, using ANOVA at FDR level of 1E-04, resulted in 1636 probesets, representing 1616 genes or ORFs, differentially expressed between them (Figure 2 and supplementary Sweave Report, available at Annals of Oncology online and supplementary Table S4, available at Annals of Oncology online). Altogether the analysis of these three subgroups of tumors revealed that many of the genes included in the MSAC pathway are present at their highest levels of expression within the molecular subgroup 3 (13 of 18). Also subgroup 3 tumor cases show two sets of probesets a major subgroup 3 and a minor subgroup 3.
Figure 2.
Heat map using ANOVA identifies probesets/genes different among the three molecular groups. There are actually two different expression groups within subgroup 3—one called subgroup 3 while the other called minor subgroup 3. These three subgroups correlate partially with histology but did not correlate with the run date and therefore not accounted by any batch effect. Mitotic spindle assembly checkpoint (MSAC) genes are differentially expressed with highest level of expression in subgroup 3 and minor subgroup 3 while other expression groups have upregulation of specific pathways.
Network analysis of differentially regulated set of genes, using MetaCore software suite (Gene Go, Inc., St Joseph, MI), showed the microtubule network to be the second-most significantly affected network in MPM tumors. There were 1990 network objects analyzed based on the list of significantly deregulated genes between tumor and paired normal samples, with tubulin showing the highest connectivity to network objects at a statistically significant FDR value of 0.05. The actual network results and networks generated in the software are illustrated in supplementary Figure S2, available at Annals of Oncology online.
mutational status of MSAC genes in MPM tumors
RNA-seq was carried out on eight tumors and one normal paired samples from the molecular subgroup 3 to assess the mutation status of MSAC genes. Mutations were not detected in any of the MSAC pathway genes. However, RNA-seq analysis helped us to confirm the microarray data on an independent platform. We compared the gene expression values obtained by Affymetrix U133 plus 2.0 array with the expression values obtained with RNA-seq platform. We found a significant correlation between the gene expression values for all probesets between the microarray and RNA-seq data (supplementary Figure S3, available at Annals of Oncology online).
RNA and protein expression of MAD2L1 in tumors and cell lines
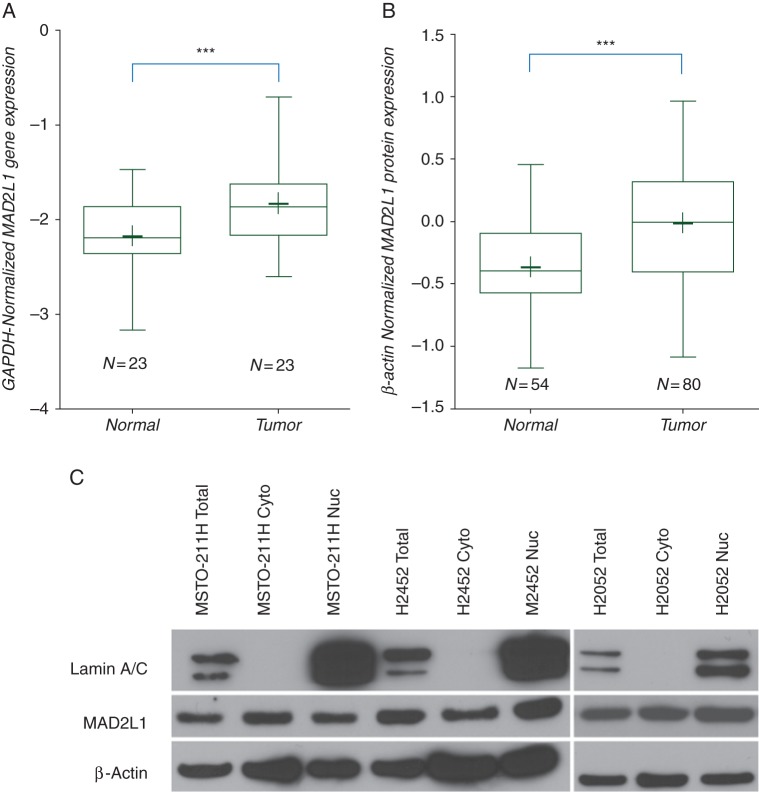
We decided to focus on MAD2L1 gene for our validation of microarray studies because it is a well-documented indicator of the deregulated status of the MSAC pathway. Its molecular role is to act as an alarm to signify that the MSAC is active and is the most stereotypical member of this pathway [13]. All tumors expressed high levels of MAD2L1 transcript compared with paired normal tissue (probeset 203362_s_at 3.09-fold high at P = 9.9E-10). The analysis of MAD2L1 transcript expression by RT-qPCR demonstrated that 23 MPM tumor samples had a significantly (P = 0.036) higher expression compared with their paired control samples (Figure 3A). The expression levels of Mad2L1 transcript by microarray correlates well to that obtained by using qPCR methodology (R2 = 0.62, P < 0.01, see supplementary Figure S4, available at Annals of Oncology online). In addition, protein expression analysis of lysates from 80 mesothelioma tumors and 54 nonmalignant samples showed a significant increase in mean value of MAD2L1 protein in tumor samples (Figure 3B). Similar results were obtained when we compared the 53 tumors and 38 normal samples included in the gene expression profiling experiment, see supplementary Figure S5, available at Annals of Oncology online). Expression of MAD2L1 protein in three mesothelioma cell lines showed high levels in both the cytoplasm and the nucleus without any preferential expression, although there might be more nuclear, than cytoplasmic, presence of MAD2L1 protein in H2452 cell line (Figure 3C).
Figure 3.
Transcript and protein analysis of MAD2L1 in tumors and cell lines. (A) qPCR analysis of MAD2L1 message in MPM tumor samples compared with normal paired samples. Twenty-three paired samples were analyzed for MAD2L1 expression using qPCR assays with GAPDH as normalizing endogenous controls and 2−ΔCt were plotted on a log10 scale to compare between the sets. The box plot shows expression in the tumors was significantly higher compared with paired normal control samples with a 3-fold difference in the means (***P = 0.0036). The ‘+’ marks the mean for the box plots. (B) MAD2L1 protein expression in tumor tissue. Western blots of tumor (N = 80 cases) and normal (N = 54 cases) were probed for MAD2L1- and β-actin-specific antibodies. After scanning, quantizing and normalizing the autoradiograms, the tumors show 2.7-fold increase in mean value of MAD2L1 protein, compared with β-actin (***P < 0.0001). The normalized intensity values were plotted on the Y-axis on a log10 scale. The ‘+’ marks the mean for the box plots. (C) Western blot analysis of three MPM cell lines. Protein lysates isolated from different cellular compartments were probed with MAD2L1- and control biomarkers-specific antibodies. The nuclear (Nuc), cytoplasmic (Cyto) and total (Total) levels of MAD2L1 protein are depicted in the blot. Nuclear lamin A and Nuclear lamin C serve as controls for presence of nuclear proteins while β-actin serves as loading control.
MAD2L1 protein expression analysis in tumor TMAs
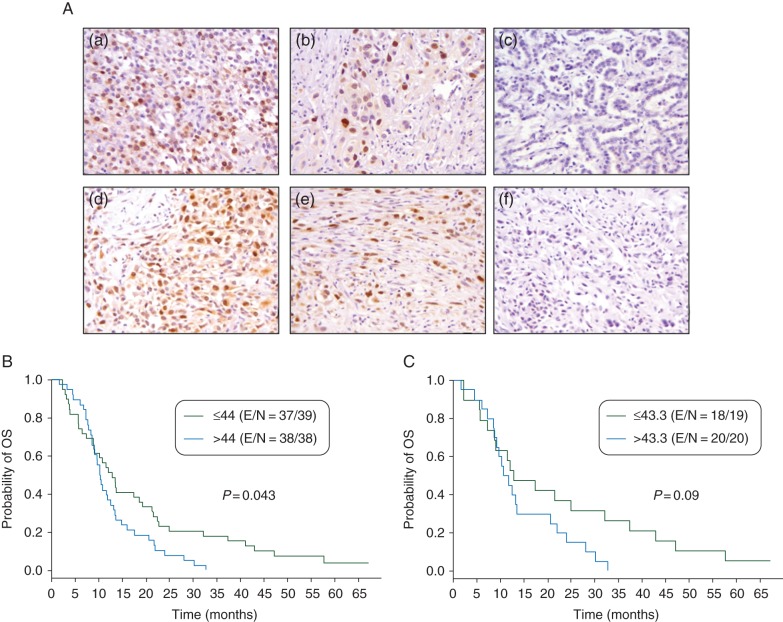
To better characterize the expression and localization of MAD2L1 in MPM tumors, we investigated the IHC expression of this protein in an independent set of 80 tumors placed in TMAs (supplementary Table S1 and Figure 4A). The expression of MAD2L1 in the cytoplasm of malignant cells was significantly higher in epitheloid tumors (P = 0.046) compared with the other two MPM histotypes. We did not find any correlation between MAD2L1 expression and the pathological stage of tumors; however, MPM with lymph node metastasis (mean score = 57.1) demonstrated a significantly (P = 0.011) higher cytoplasmic MAD2L1 expression than tumor without metastasis (mean score = 33.2). In univariate analysis, using the median score of expression as cutoff, higher nuclear MAD2L1 expression associated significantly (P = 0.043) with lower rates of overall survival in all MPM patients, and there was a trend (P = 0.09) to similar findings in patients with epitheloid histology tumors (Figure 4B and C). However, MAD2L1 IHC expression did not associate significantly with survival rates in the MPM patients in the multivariate analysis, adjusting by gender, pathological stage and lymph node status.
Figure 4.
MAD2L1 protein expression in mesothelioma tumors by IHC. (A) Representative histotypes of epithelioid type (a, ×200; b, ×400; c, ×200), and sarcomatoid type (d, ×200; e, ×400; f, ×200). Cases depicted in pictures a, b, d and e show positive nuclear moderate and strong positive nuclear signal of MAD2L1 protein in malignant tumor MPM cells. Cases depicted in pictures c and f show negative staining in malignant cells. (B) Kaplan–Meier showing overall survival of all MPMs by nuclear MAD2L1 expression using the median expression (44.0) as cutoff. (C) Kaplan–Meier showing overall survival of epitheloid MPMs by nuclear MAD2L1 expression using the median (43.3) expression as cutoff.
small inhibitor studies targeting biomarkers based on microarray expression analysis
The presence of ‘druggable’ kinases expressed within different subgroups directed us to explore small molecule inhibitors targeting them in mesothelioma cell lines. Accordingly, we tested commercially available drugs specifically targeting five of these kinases: AURKA, NIMA-never in mitosis gene a-related kinase 2 (Nek2), CENP-A, Polo-like Kinase-1 (PLK1) and Kinesin family member 11 (KIF11) kinase. However, none of the cell lines tested was sensitive and had an IC50 value in the 10–100 µM range (data not shown). The microarray profiles also showed subgroup-specific upregulation of oncogenes like MET/hepatocyte growth factor receptor and fibroblast growth factor receptor 1 (FGFR1) in subgroup 1, while subgroup 3 showed increased expression of ribosomal protein S6 Kinase 1 (RPS6KA1), topoisomerase (DNA) II alpha (TOP2A) and AXL receptor tyrosine kinase. We tested small molecule inhibitors targeting these proteins in 14 mesothelioma cell lines (supplementary Figure S6, available at Annals of Oncology online). However, for all drugs (except the TOP2A inhibitor), the median IC50 was >5000 nM and individual IC50 was >1000 nM. Interestingly, though the TOP2A-specific inhibitor, etoposide, showed relatively ineffective cytotoxicity (median IC50 = 1928 nM) against most mesothelioma cell lines, it was effective against two of the sarcomatoid cell lines (DM3 IC50 = 1.6 nM and Mero-14 IC50 = 247 nM). This is significant in view of the fact that our microarray expression analysis showed that TOP2A expression was higher in the sarcomatoid-rich subgroup 3 tumors.
The network analysis of genes differentially expressed in the tumors suggested that targeting tubulin might be another viable option. Since taxane therapy has been mildly effective in clinical trials involving mesothelioma patients [14], we decided to explore novel nontaxane drugs. Accordingly, we exposed mesothelioma cell lines to epothilone B, a nontaxane microtubule inhibitor, which is a synthetic analog of natural metabolite produced by soil bacteria [15]. We found that epothilone B was by far most effective in inhibiting the proliferation of 14 MPM cancer cell lines with IC50 ranging from 0.01 to 27 nM and a median of 1.5 nM (supplementary Figure S6, available at Annals of Oncology online). Paclitaxel, though not as effective, did show potency in a certain subset of cell lines with IC50 ranging from 4 to 25285 nM and a median of 416 nM.
discussion
This is the first report illustrating that MPM tumors can be subdivided into three molecular subtypes based on differences in gene expression and pathways. In this study, we also show that MSAC pathway is the most significantly upregulated pathway in MPM tumors. Transcripts and levels of a key protein involved in the MSAC pathway, MAD2L1 was found upregulated in all MPM tumors, while higher nuclear levels of MAD2L1 protein correlated with lower overall survival in univariate analysis. We further demonstrate that the microtubule network is the second-most significantly affected network in these tumors and show that it can be therapeutically targeted by demonstrating the efficacy of a specific small molecule inhibitor targeting microtubules against viability of MPM cancer cell lines in culture.
Other profiling studies have also shown similar grouping of tumor specimens and, even in these studies, the tumor groups do not correlate or loosely correlate with histology [5, 6]. Also, unlike the unsupervised clustering method used by us, other groups have used an alternative method of using supervised groups, representing short and long overall survival of patients to reveal prognostic gene expression profiles distinct to each [6]. Importantly, although earlier profiling studies found some of the components of the MSAC pathway upregulated in MPM tumors this is the first study to show a robust involvement of many components of the pathway [7]. The physiological levels of MAD2L1 are critical in tumor development and its overexpression also causes increased recurrence of solid tumors by increasing chromosomal instability in a mouse lung cancer model [16, 17]. Moreover, a recent report suggests that MAD2L1 overexpression is causal for generating the phenotype of chromosomal instability especially when p53 and Rb pathways are inactive [18]. In support of the oncogenic function of MAD2L1 protein, our univariate analysis showed significant association of increased nuclear localization of MAD2L1 protein (where it exerts its known biological function) with decreased overall survival of MPM patients (Figure 4B and C).
This analysis agrees with observing higher expression of the protein in cytoplasm of epitheloid tumor cases, which show best overall survival among all histotypes. However, since metastatic cases also showed higher cytoplasmic expression of MAD2L1 protein further functional studies in cell lines are needed to explore if higher nuclear expression in cell lines would lead to more or less aggressive phenotype. Although we did not notice any dramatic differences in expression of MAD2L1 protein within the cytoplasmic and nuclear compartments of three mesothelioma cell lines (Figure 3C).
MAD2L1 protein undergoes phosphorylation, which affects its activity, but there are no antibodies commercially available to detect its phosphorylated form [19]. We explored the possibility of the presence of activating mutations in MAD2L1 by performing RNA-seq analysis on eight tumors of the subgroup 3 with highest levels of MSAC genes. However, we did not detect any mutations in this gene or any genes of the MSAC pathway. There have been only two reports (in breast cancer) in COSMIC database for mutations in MAD2L1 gene in all cancers tested [20]. At least one other profiling study has found increased levels of MAD2L1 transcript in MPM tumors. However, it did not report it since the level of expression of the transcript (fold change = 1.9) was close to but below the threshold of 2 [5]. Collectively, these observations suggest that it is the increased level of MAD2L1 wild-type protein, which is supporting tumorigenesis in MPM.
Aside from exploring the biological basis for many features of MPM pathogenesis, we investigated if these subgroups also aid in identifying newer therapeutic options. The MSAC pathway is a highly druggable pathway with small molecules inhibitors available for targeting a number of protein kinases and motor kinesins [21]. However, inhibitors targeting five of these kinases did not show any efficacy in decreasing the viability of mesothelioma cell lines (data not shown). Also, the microarray data suggested other kinases, with subgroup-specific expression, as possible targets for, e.g. (i) subgroup 1 shows increased expression of MET and FGFR1 receptors, (ii) subgroup 3 also shows increased expression of RPS6KA1 kinases, TOP2A and AXL receptor. However, only one of these targets, TOP2A, showed efficacy in cell viability studies (supplementary Figure S6, available at Annals of Oncology online). Etoposide, the TOP2A inhibitor, showed increased cytotoxicity against two of the sarcomatoid cell lines, relative to others, suggesting that subgroup-specific gene signatures could guide therapeutic intervention.
Contrary to the results obtained against the MSAC pathway, we had great success with targeting the microtubule network—the second-most significantly affected network in all as well as subgroup 3 tumors. Increased expression of Tubulin genes like Tubulin alpha 4b (TUBA4B) and Tubulin beta 2b (TUBB2B) as well as proteins, which modulate its function like polymerization promoting protein (TPPP) is seen in all tumors. Microtubules could be targeted using taxane or nontaxane-based inhibitors, which affects its stability or polymerization capacity [22]. Analog of the nontaxane inhibitor, epothilone B, is used for treating patients with refractory metastatic breast cancer [23]. It binds the tubulin heterodimers and stabilizes the microtubules by decreasing its rate of dissociation causing cell-cycle arrest at G2-M phase leading to cell cytotoxicity and eventual apoptosis of cancer cells [24]. Though epothilone B binds at the same sites as paclitaxel, it has been shown to have better efficacy due to its simpler chemical structure and better water solubility [25]. We also found paclitaxel to have significant activity against these cell lines but overall the median IC50 dose was higher than that for epothilone B. Also, some cell lines were very resistant to paclitaxel but sensitive to epothilone B (supplementary Figure S6, available at Annals of Oncology online). This raises the interesting possibility that nontaxane microtubule-targeting therapy might prove to be of benefit in treatment of mesothelioma patients. The two recently published datasets of predictive biomarkers in cancer cell lines, Cancer Cell Line Encyclopedia (CCLE) [26] dataset with paclitaxel drug sensitivity data in 504 cancer cell lines and Genomics of Drug Sensitivity in Cancer (GDSC) dataset [27] with paclitaxel and epothilone B drug sensitivity data in 360 and 662 cancer cell lines respectively, will enable further exploration of predictive biomarkers in mesothelioma for these microtubule-targeting drugs.
funding
This work was supported by grants from the Department of Defense (W81XWH-07-1-0306 to IIW and AT), the National Cancer Institute (Cancer Center Support Grant CA-16672), Aileen Dillon Endowment for Mesothelioma Research, George Fleming Endowment for Mesothelioma Research, ASCO Career Development award K12 (CA088084 to AT) and IASLC Young Investigator Award (2011–2013 to MS).
disclosure
The authors have declared no conflicts of interest.
Supplementary Material
acknowledgements
The authors thank Humam Kadara for correlation analysis of gene expression and RNA-seq data, Zuoming Chu for help with RNA extractions and Laxmi Kakarala with TMA setup.
references
- 1.Campbell NP, Kindler HL. Update on malignant pleural mesothelioma. Semin Respir Crit Care Med. 2011;32:102–110. doi: 10.1055/s-0031-1272874. [DOI] [PubMed] [Google Scholar]
- 2.Steele JP. Prognostic factors for mesothelioma. Hematol Oncol Clin North Am. 2005;19:1041–1052. doi: 10.1016/j.hoc.2005.09.009. vi. [DOI] [PubMed] [Google Scholar]
- 3.Kindler HL. Systemic treatments for mesothelioma: standard and novel. Curr Treat Options Oncol. 2008;9:171–179. doi: 10.1007/s11864-008-0071-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singhal S, Wiewrodt R, Malden LD, et al. Gene expression profiling of malignant mesothelioma. Clin Cancer Res. 2003;9:3080–3097. [PubMed] [Google Scholar]
- 5.Gordon GJ, Rockwell GN, Jensen RV, et al. Identification of novel candidate oncogenes and tumor suppressors in malignant pleural mesothelioma using large-scale transcriptional profiling. Am J Pathol. 2005;166:1827–1840. doi: 10.1016/S0002-9440(10)62492-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopez-Rios F, Chuai S, Flores R, et al. Global gene expression profiling of pleural mesotheliomas: overexpression of aurora kinases and P16/CDKN2A deletion as prognostic factors and critical evaluation of microarray-based prognostic prediction. Cancer Res. 2006;66:2970–2979. doi: 10.1158/0008-5472.CAN-05-3907. [DOI] [PubMed] [Google Scholar]
- 7.Crispi S, Calogero RA, Santini M, et al. Global gene expression profiling of human pleural mesotheliomas: identification of matrix metalloproteinase 14 (MMP-14) as potential tumour target. PLoS One. 2009;4:e7016. doi: 10.1371/journal.pone.0007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sekido Y. Molecular pathogenesis of malignant mesothelioma. Carcinogenesis. 2013;34:1413–1419. doi: 10.1093/carcin/bgt166. [DOI] [PubMed] [Google Scholar]
- 9.Sekido Y. Genomic abnormalities and signal transduction dysregulation in malignant mesothelioma cells. Cancer Sci. 2010;101:1–6. doi: 10.1111/j.1349-7006.2009.01336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zucali PA, Ceresoli GL, De Vincenzo F, et al. Advances in the biology of malignant pleural mesothelioma. Cancer Treat Rev. 2011;37:543–558. doi: 10.1016/j.ctrv.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Jakobsen JN, Sorensen JB. Review on clinical trials of targeted treatments in malignant mesothelioma. Cancer Chemother Pharmacol. 2011;68:1–15. doi: 10.1007/s00280-011-1655-3. [DOI] [PubMed] [Google Scholar]
- 12.Travis WD, M-HHK BE, Harris CC. Pathology and Genetics of Tumours of the Lung, Pleura. Thymus Heart. Lyon, France: International Agency for Research on Cancer (IARC): IARC World Health Organization Classification of Tumours; 2004. [Google Scholar]
- 13.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 14.Vorobiof DA, Rapoport BL, Chasen MR, et al. Malignant pleural mesothelioma: a phase II trial with docetaxel. Ann Oncol. 2002;13:412–415. doi: 10.1093/annonc/mdf046. [DOI] [PubMed] [Google Scholar]
- 15.Goodin S. Novel cytotoxic agents: epothilones. Am J Health Syst Pharm. 2008;65:S10–S15. doi: 10.2146/ajhp080089. [DOI] [PubMed] [Google Scholar]
- 16.Sotillo R, Hernando E, Diaz-Rodriguez E, et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sotillo R, Schvartzman JM, Socci ND, et al. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464:436–440. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schvartzman JM, Duijf PH, Sotillo R, et al. Mad2 is a critical mediator of the chromosome instability observed upon Rb and p53 pathway inhibition. Cancer Cell. 2011;19:701–714. doi: 10.1016/j.ccr.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S, Sun H, Ball HL, et al. Phosphorylation of the spindle checkpoint protein Mad2 regulates its conformational transition. Proc Natl Acad Sci USA. 2010;107:19772–19777. doi: 10.1073/pnas.1009000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Percy MJ, Myrie KA, Neeley CK, et al. Expression and mutational analyses of the human MAD2L1 gene in breast cancer cells. Genes Chromosomes Cancer. 2000;29:356–362. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1044>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 21.Bolanos-Garcia VM. Assessment of the mitotic spindle assembly checkpoint (SAC) as the target of anticancer therapies. Curr Cancer Drug Targets. 2009;9:131–141. doi: 10.2174/156800909787580980. [DOI] [PubMed] [Google Scholar]
- 22.Stanton RA, Gernert KM, Nettles JH, et al. Drugs that target dynamic microtubules: a new molecular perspective. Med Res Rev. 2011;31:443–481. doi: 10.1002/med.20242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toppmeyer DL, Goodin S. Ixabepilone, a new treatment option for metastatic breast cancer. Am J Clin Oncol. 2010;33:516–521. doi: 10.1097/COC.0b013e3181b9cd52. [DOI] [PubMed] [Google Scholar]
- 24.Rogalska A, Marczak A, Gajek A, et al. Induction of apoptosis in human ovarian cancer cells by new anticancer compounds, epothilone A and B. Toxicol In Vitro. 2013;27:239–249. doi: 10.1016/j.tiv.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 25.Downing KH. Structural basis for the interaction of tubulin with proteins and drugs that affect microtubule dynamics. Annu Rev Cell Dev Biol. 2000;16:89–111. doi: 10.1146/annurev.cellbio.16.1.89. [DOI] [PubMed] [Google Scholar]
- 26.Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang W, Soares J, Greninger P, et al. Genomics of drug sensitivity in cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.