Abstract
Autophagy is directed by numerous distinct autophagy-related (Atg) proteins. These transmit starvation-induced signals to lipids and regulatory proteins and assemble a double-membrane autophagosome sequestering bulk cytoplasm and/or selected cargos destined for degradation upon autophagosome fusion with a vacuole or lysosome. This Review discusses the structural mechanisms by which Atg proteins sense membrane curvature, mediate a PI(3)P-signaling cascade, and utilize autophagy-specific ubiquitin-like protein cascades to tether proteins to autophagosomal membranes. Recent elucidation of molecular interactions enabling vesicle nucleation, elongation, and cargo recruitment provides insights into how dynamic protein-protein and protein-membrane interactions may dictate size, shape, and contents of autophagosomes.
Main Text
Introduction
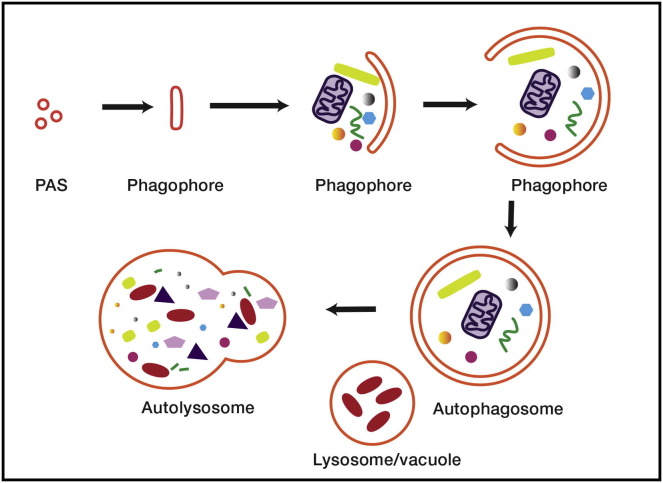
Macroautophagy (hereafter, “autophagy”) is a cellular self-consumptive pathway conserved throughout eukarya (Mizushima and Komatsu, 2011, Yang and Klionsky, 2010). Autophagy was first characterized as a way for single-celled organisms to survive starvation (Reggiori and Klionsky, 2013). From these humble evolutionary beginnings, the physiology of autophagy has expanded in the metazoa to encompass an ever-expanding repertoire of functions in health and disease (Boya et al., 2013, Mizushima et al., 2008). In autophagy, a double-membrane sheet, referred to as the phagophore, nucleates at the phagophore assembly site (PAS) (Rubinsztein et al., 2012, Weidberg et al., 2011). Yeast cells have a single PAS that is present constitutively. In mammalian cells, the counterpart of the PAS is less well defined, with autophagy likely initiated at multiple PAS structures that form and dissolve as needed. The phagophore grows by addition of membrane, most likely via vesicle fusion (Moreau et al., 2013). The growing phagophore takes on a cup-like shape, with the late phagophore resembling a fishbowl (Figure 1 ). Once the phagophore is sealed, it is referred to as the autophagosome (Figure 1). The autophagosome ultimately fuses with the vacuole or lysosome, resulting in degradation of the autophagosome and its contents. Amino acids and other metabolite degradation products are recycled by export through the lysosomal or vacuolar membrane.
Figure 1.
Autophagy
Autophagy is thought to commence with the clustering of membrane vesicles at the PAS. Membrane fusion leads to the formation of the open double-membrane sheet known as the phagophore. The phagophore expands and matures into the closed autophagosome. Finally, the autophagosome fuses with the lysosome, forming the autolysosome and leading to the degradation of the contents of the autophagosome.
In bulk autophagy, a portion of the cytosol is engulfed en masse by the phagophore. It is not known what sets the phagophore size and shape in this nonselective autophagy (Rubinsztein et al., 2012, Weidberg et al., 2011). Recently, an explosion of discoveries identified selective versions of autophagy devoted to numerous special purposes, such as the trafficking or clearance of damaged, unneeded, or toxic large cargos, including organelles such as mitochondria or peroxisomes (Shaid et al., 2013). Beyond these two major flavors of autophagy, subsets of the canonical autophagy machineries also regulate several autophagy-like pathways, such as biogenesis of the compartment for unconventional protein secretion (CUPS) in which an autophagosome-like compartment fuses with the plasma membrane rather than the lysosome (Bruns et al., 2011). Circadian rhythm-regulated degradation of photoreceptor outer segments, essential for retinal health, is orchestrated by a variant of autophagy that involves most of the Atg machinery but not the earliest components involved in phagophore nucleation (Kim et al., 2013b). Atg factors are involved in the formation of carriers derived from the endoplasmic reticulum, called EDEMosomes, a pathway hijacked by coronaviruses to form structures needed for viral replication (Reggiori et al., 2010). These and other examples of autophagy-like pathways have been reviewed recently (Bestebroer et al., 2013, Boya et al., 2013).
The cases noted above illustrate how autophagy and autophagy-like pathways are central to an astonishing array of diverse, fundamentally important physiological functions. This cries out for models clarifying physical and molecular mechanisms of autophagosome formation, which will be required for understanding regulation of these pathways and the prospects for their targeting by therapeutic interventions.
The membrane supply for autophagosome biogenesis apparently involves various sources at different stages in the process, with details undergoing vigorous debate. Membrane curvature also plays a role. The rim of the phagophore is highly curved, with a radius of curvature of 10–15 nm. This is energetically expensive to maintain (Hurley et al., 2010) and is on a size scale comparable to some Atg proteins themselves. The phagophore is more gently curved, with a radius of curvature of 150–450 nm in yeast. This is larger than the molecular size scale and energetically inexpensive on a local scale. The maintenance of this curvature over the entire phagophore is still an energetic challenge, however. This is especially the case in nonselective autophagy, in which adhesion to a specific substrate is not available to stabilize membrane curvature. The unique and complex structure of the phagophore suggests that equally unique and complicated molecular scaffolds might be required. Collectively, a recent acceleration of crystallographic, electron microscopy, and in vitro biochemical reconstitution studies of autophagy indicate that an integrated structural model for autophagy is achievable. Here, we review recent progress toward this goal and outline steps still requiring structural elucidation.
The Autophagic Parts List
The conserved core autophagy machinery, identified largely through yeast genetics (Mizushima et al., 2011, Reggiori and Klionsky, 2013), consists of nearly 40 autophagy-related (Atg) proteins mediating nonselective and/or selective autophagy. The components of this machinery, described in many excellent reviews (Mizushima et al., 2011, Reggiori and Klionsky, 2013, Rubinsztein et al., 2012), are summarized briefly here. Most Atg proteins function within dynamic networks of multiprotein complexes. Much of our current molecular understanding of autophagy is based on characterization of these Atg protein complexes and their roles in autophagy. Initiation of bulk autophagy in yeast requires Atg1, a protein kinase that participates in a complex with Atg13, Atg17, Atg29, and Atg31. The counterparts of these proteins are ULK1/2, ATG13, FIP200, and ATG101 in humans. The Atg1 complex, in turn, regulates a PI 3-kinase catalytic subunit Vps34, which functions in a complex with the protein kinase Vps15 and the regulatory protein Atg6 (Beclin 1 in humans). Vps34 forms both endosome and autophagy-specific complexes in yeast with Vps38 and Atg14, respectively (UVRAG and ATG14 in humans). Atg9 is the only conserved integral membrane protein in the pathway. Atg9 binds Atg17 and Atg2 and participates in early stages of autophagy through largely unknown biochemical mechanisms. The Atg2-Atg18 (ATG2A/B and WIPI-1-4 in humans) complex is targeted to autophagic membranes by the presence of Atg9 and PI 3-phosphate (PI(3)P). Although essential for autophagy, again, the biochemical functions of Atg2 and Atg18 are little understood. A distinctive set of Atg protein complexes revolve around a pair of ubiquitin-like protein (UBL) conjugation cascades that are specific for autophagy. The C terminus of one UBL, Atg8 in yeast and numerous orthologs in metazoans, becomes covalently linked through an E1-E2-E3 cascade to the lipid, phosphatidylethanolamine (PE), whereas the other autophagy-specific UBL, Atg12, upon its own conjugation forms part of the multiprotein E3 ligating Atg8 to PE. Atg8 ligation to PE serves multiple roles in autophagy, including in recruiting cargos and regulatory proteins to its marked membranes and in contributing to autophagosome biogenesis.
Much of the collective molecular mass of the Atg proteins is opaque in that it does not correspond to functionally annotated domains and motifs. To date, most biochemical investigation of Atg proteins has focused on the roles of recognizable catalytic and ubiquitin-like domains: the protein and lipid kinase regions of Atg1/ULK1/2 and Vps34 and the paralogs of the ubiquitin conjugation system. However, on a mass basis, much of these systems consists of helical solenoids, coiled-coils, β propellers, and intrinsically disordered regions. The proteins that scaffold 100 nm-scale structures such as coated vesicles and the nuclear pore consist mainly of helical solenoids and β propellers (Devos et al., 2004). These domains are present in the Vps15 subunit of the PI3KC3 complex of autophagy. Perhaps these domains could scaffold elements of the autophagosome. Unquestionably, a molecular mechanistic model for autophagy will depend on discovering what these noncatalytic domains are really doing and how the catalytic domains achieve autophagy-specific functions. Structural biology approaches are helping to answer these questions.
Phagophore Initiation: The Atg1/ULK1/2 Complex
The earliest acting Atg complex is named for its protein kinase subunit, Atg1, which is essential for autophagy initiation in yeast. At least in yeast, however, the Atg1 kinase activity is not required for the very earliest steps (Cheong et al., 2008). Rather, the C-terminal early autophagy targeting/tethering (EAT) domain, conserved in human ULK1/2, seems to have a key role in membrane targeting (Chan et al., 2009), vesicle tethering (Ragusa et al., 2012), and coassembly with other subunits (Ragusa et al., 2012, Yeh et al., 2011). Upon starvation, yeast Atg1 coassembles with Atg13, Atg17, Atg29, and Atg31. Dephosphorylation of Atg13 following Tor protein kinase inactivation is thought to trigger Atg1 complex assembly and activation.
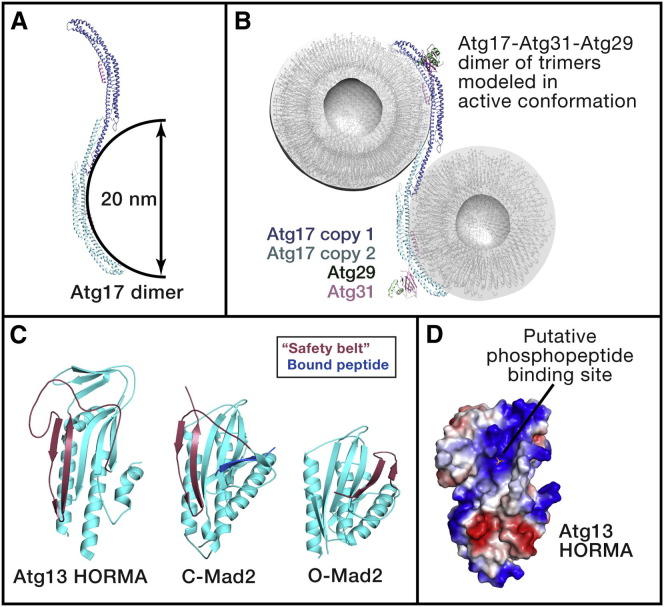
Though structures are not available for Atg1, all or part of its four partner subunits have been crystallized. None of these other subunits contain familiar catalytic or structural motifs, making them terra incognita in a bioinformatics sense. Atg17-Atg31-Atg29 form a constitutive complex. Atg31 is the structural bridge linking Atg17 to Atg29; hence, the nomenclature places Atg31 before Atg29. Atg17 is crescent shaped, with an arc whose radius of curvature is about 10 nm (Chew et al., 2013, Mao et al., 2013, Ragusa et al., 2012) (Figure 2 A). This is striking because the earliest vesicles to arrive and cluster at the PAS are also highly curved, with radii of 15–30 nm (Mari et al., 2010, Yamamoto et al., 2012). The only comparable class of crescent-shaped proteins are the members of the BAR domain superfamily, which bind to curved lipid vesicles and tubes (Frost et al., 2009). This leads to the compelling expectation that Atg17 will have a similar role in sensing or scaffolding highly curved membranes. Atg17 is a dimer, such that the two crescents together form the shape of the letter S. Atg17 dimerization is absolutely required for formation of the PAS for starvation-induced autophagy (Ragusa et al., 2012). Modeling suggests that the Atg17 double crescent could tether two vesicles for approach within about 2 nm of each other (Ragusa et al., 2012). The Atg17-Atg31-Atg29 complex, however, has no vesicle-tethering activity in vitro. How then could vesicle tethering be achieved? A plausible explanation stems from the finding that the N-terminal half of Atg29, together with Atg31, comprise a structurally novel fold that sterically obstructs the concave face of the crescent in the crystallized complex and thus prevents vesicle binding. Modeling suggests that the Atg29-Atg31 subcomplex could be displaced by appropriate activating signals (Ragusa et al., 2012), but this remains to be demonstrated. The Atg29 C-terminal domain is intrinsically disordered and is probably not involved in regulating access to the Atg17 crescent. Rather, the Atg29 C-terminal region is the locus of phosphoregulatory sites that control binding to Atg11 (Mao et al., 2013). The most important broad-brush lesson to emerge from structural studies of the Atg17-Atg31-Atg29 portion of the Atg1 complex is the importance of scaffolding elements within autophagy catalytic complexes. These elements likely control the physical positioning of the membrane vesicles that feed the growing phagophore. It is tempting to hypothesize that they guide the trajectory of the growth of the phagophore. It also seems likely that conformationally flexible portions of complexes mediate dynamic regulation.
Figure 2.
The Atg1 Autophagy Initiation Complex
(A) Atg17 is the main scaffold for the Atg1 complex and for the assembly of the PAS for bulk autophagy in yeast. This image shows the complementarity between the shape of the Atg17 dimer (with Atg29 and Atg31 removed for clarity) and a pair of 20 nm diameter vesicles (Ragusa et al., 2012).
(B) A model for the putative disinhibited conformation of the Atg17-Atg31-Atg29 complex bound to two 20 nm vesicles. Atg29 and Atg31 were moved from their crystallographic positions, which sterically collide with the docked vesicles, into a conformation where they do not interfere with binding.
(C) Structure of the HORMA domains of Atg13 (left) (Jao et al., 2013) and Mad2 (Sironi et al., 2002), with the latter in the closed and open conformations, respectively. The conformationally variable safety belt region is shown in red, and a peptide from Mad1 bound to Mad2 is shown in the central panel in blue.
(D) Surface model of the Atg13 HORMA domain (blue is electropositive and red electronegative) and crystallographic sulfate ion as a marker for a putative phosphopeptide-binding site (Jao et al., 2013).
Atg13 is a major locus of nutritional regulation of the Atg1 complex. Atg13 is a substrate for phosphorylation by Tor in yeast (Scott et al., 2000) and humans (Chang and Neufeld, 2009, Ganley et al., 2009, Hosokawa et al., 2009, Jung et al., 2009). Atg13 is the bridge between Atg1 and the other subunits in the complex and thus has a pivotal role in holding the complex together and regulating activity in a nutrient-dependent manner. The C-terminal two-thirds of Atg13 are predicted to be intrinsically disordered, and this region contains the loci of regulatory phosphorylation by Tor and other kinases. The N-terminal third of Atg13 is required, perhaps indirectly, for the recruitment of the Atg14-containing PI 3-kinase complex (Jao et al., 2013). The crystal structure of this region shows that Atg13 is a member of the HORMA (Hop1p, Rev1p and Mad2) domain family (Jao et al., 2013). The HORMA domain is best characterized in the context of the spindle checkpoint complex, where the HORMA domain protein Mad2 is a conformational switch (Mapelli and Musacchio, 2007). The Mad2 HORMA folds into two different structures, known as O-Mad2 and C-Mad2. In the conformational transition, β strands 7–8′ and the long β6–β8 loop rearrange from one side of the domain to the other, a dramatic refolding event (Figure 2B). The crystallized HORMA domain of Atg13 corresponds to the C-Mad2 structure, and it is unknown whether Atg13 is capable of transitioning into the “O” state. The Atg13 HORMA domain contains a potential C-state-specific phosphopeptide-binding site, predicted on structural grounds (Figure 2C). It has also been proposed that the Atg13 HORMA domain binds to PI(3)P (Karanasios et al., 2013); however, the proposed basic residues are scattered over the protein surface in a manner that is not consistent with formation of a specific binding site. This observation is intriguing because protein phosphorylation is central to autophagy induction, yet the Atg proteins contain none of the canonical phosphopeptide-binding domains.
Many of the conclusions reached from structural studies of the yeast Atg1 complex probably apply to the mammalian ULK1 complex too. The domain structure of ULK1 mirrors that of Atg1, and the membrane targeting role of the ULK1 EAT domain seems to be conserved (Chan et al., 2009). The architecture of yeast and human Atg13 is conserved, as is the overall structure of its N-terminal HORMA domain. However, individual HORMA basic residues implicated in function are not. The consensus view is that human FIP200, a large coiled-coil protein, has an early scaffolding role analogous to that of Atg17. This concept is plausible and attractive, though direct evidence at the structural and biochemical levels is still needed.
Beclin 1 and the PI3K Complex
Vps34 is the only phosphatidylinositol 3-kinase (PI3K) in yeast and is the only class III PI3K in human cells. Class III PI3Ks synthesize PI(3)P. Functional diversity is achieved through formation of alternative PI3KC3 complexes that incorporate the Vps34 catalytic subunit. The Atg14-containing form of the complex is uniquely involved in autophagy, whereas the Vps38/UVRAG-containing complex has a central role in endosome maturation. PI3KC3 complexes consist of the three common subunits Vps34, Vps15, and Atg6/Beclin 1 and one, but not both, of Vps38/UVRAG and Atg14/Barkor. Vps34 is the catalytic subunit responsible for phosphorylation of PI to PI(3)P. Vps15 is a large (>160 kDa) protein that contains a protein kinase domain, a helical repeat domain, and a β-propeller domain. Atg6/Beclin 1 is a core element of both complexes, notwithstanding its fame in autophagy. Atg6 contains an N-terminal unstructured region, a coiled coil, and a C-terminal α + β evolutionary conserved domain (ECD). Atg14 and Vps38 are coiled-coil proteins that partner with Atg6. With the exception of the helical repeat domain of Vps15, the structures of the other components of this complex are known either from crystal structures or can be inferred by homology to other known structures. The big unanswered structural question is how the different components are arranged relative to one another in space.
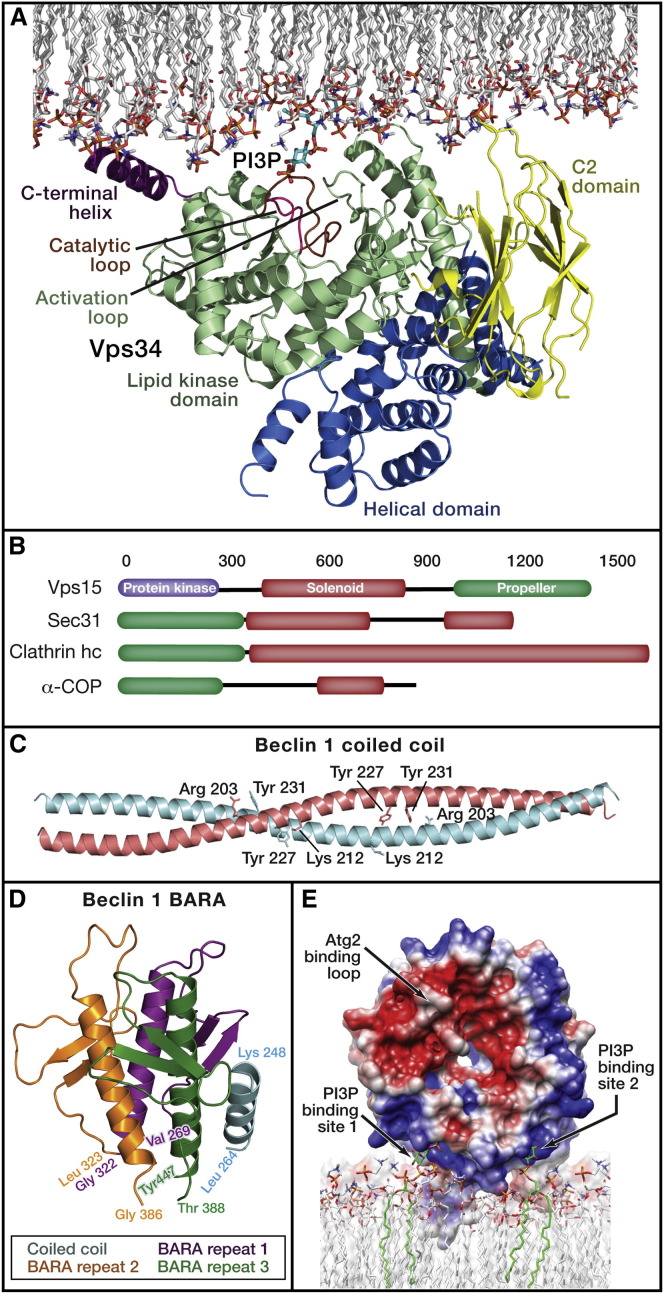
The Vps34 catalytic domain belongs to the eukaryotic kinase superfamily and has a typical kinase fold and is fused to a helical region (Miller et al., 2010) (Figure 3 A). The helical region appears to function as a spine for association with other domains and subunits. The helical region probably positions the phospholipid-binding C2 domain of Vps34 to bind to membranes such that the catalytic domain can access its substrate, the membrane-bound lipid PI. The structure of the Vps34 catalytic domain led to two significant insights. The first is that the C-terminal helix has a special role as both a membrane anchor and autoinhibitor. When Vps34 is not bound to the membrane, this helix blocks the ATP-binding site to prevent futile ATP hydrolysis. In the presence of the membrane, the helix moves to facilitate binding to the membranous substrate and, in so doing, unblocks the ATP site. The second is that the ATP-binding pocket of Vps34 is unusually constricted. This explains the specificity of the widely used Vps34 inhibitor 3-methyladenine (3MA). 3MA is smaller than most other PI3K inhibitors and fits snugly in the Vps34 pocket (Miller et al., 2010). Most inhibitors of the class I PI3Ks are too bulky to fit in this pocket.
Figure 3.
Complexes of PI(3)P Synthesis and Recognition
(A) Crystal structure of the catalytic core of Drosophila Vps34, consisting of the catalytic and helical domains (Miller et al., 2010). The C2 domain was not present in the crystal structure and was modeled as described on the basis of the C2 domain of a related PI 3-kinase (Miller et al., 2010). Vps34 is shown docked to the membrane in a conformation in which the C-terminal helix has been moved from its crystallographic position into its putative active conformation as bound to lipids.
(B) Most of the structure of the Vps15 subunit is unknown, with the exception of the propeller domain. The schematic shows that Vps15 resembles vesicle coat proteins in its overall domain architecture, with the addition of a protein kinase domain.
(C) The antiparallel coiled-coil dimer of beclin 1 (Li et al., 2012). Tyr residues that are phosphorylated by the EGFR are highlighted (Wei et al., 2013), as are basic residues that might potentially form stabilizing cross-dimer interactions with the phospho-Tyr.
(D) The pseudo 3-fold symmetric BARA domain of human beclin 1 (Huang et al., 2012). Each of the pseudo 3-fold repeats is colored differently. Four helices are present in this structure because it also includes the most C-terminal portion of the coiled-coil domain.
(E) Surface model of K. lactis Hsv2 (Baskaran et al., 2012, Krick et al., 2012), which serves as a structural model for Atg18 and human WIPI proteins. The two PI(3)P-binding sites are highlighted.
Vps15 comprises nearly half of the mass of PI3KC3 yet is the focus of less than 3% of the PI3KC3 literature. Vps15 consists of a catalytic domain with a predicted typical eukaryotic kinase superfamily fold, fused to a helical solenoid and a WD repeat β-propeller domain (Figure 3B). Only the latter domain has been crystallized (Heenan et al., 2009). The presence of helical solenoids and WD repeat domains in a single chain is characteristic of the structural proteins of nuclear pores and vesicle coats, including COPII (Devos et al., 2004, Lee and Goldberg, 2010). The significance of the analogy between the architecture of Vps15 and pore and coat constituents is currently unknown.
The ordered portions of Beclin 1, the coiled coil, and the ECD have been crystallized in a piecemeal basis. These domains are contiguous with one another, and the structures contain enough overlap to generate a composite model. The crystallized coiled coil of Beclin 1 consists of an antiparallel homodimer (Li et al., 2012) (Figure 3C). The structure of the homodimeric coiled coil of Beclin 1 shows a series of unfavorable contacts at the a and d positions that zipper the coils together (Li et al., 2012). Modeling suggests that the physiological pairing with Atg14 would lead to more favorable pairing. Two of the key pairing contacts in the Beclin 1 homodimer involve Tyr residues that are phosphorylated by the EGF receptor (EGFR) (Wei et al., 2013). Phosphorylation of these Tyr residues could potentially stabilize the Beclin 1 homodimer by introducing favorable electrostatic interactions across the dimer. By favoring the Beclin 1 homodimer over the Beclin 1-ATG14 heterodimer, this might explain autophagy suppression by EGF (Wei et al., 2013).
The C-terminal domain of human Beclin 1 and the corresponding β-α repeat autophagy-specific (BARA) domain of yeast Atg6 have been crystallized (Huang et al., 2012, Noda et al., 2012). The crystal structures show that this region has an elegant and novel fold in which three β-α repeats are arranged about a pseudo 3-fold axis (Figure 3D). The Beclin 1 C-terminal domain extends beyond the canonical ECD; thus, the ECD as traditionally defined is not a meaningful structural or functional unit. Therefore, we recommend the use of the term BARA for both Beclin 1 and Atg6. The BARA of Atg6 is essential for autophagy and for PAS targeting of PI3KC3, but it is not required for endosomal functions (Noda et al., 2012). Consistent with the requirement for PAS targeting in yeast, a C-terminal deletion within the BARA domain impairs membrane localization of the human ATG14-containing PI3KC3 (Fogel et al., 2013). It has been proposed that the Beclin 1 BARA directly binds to lipids (Huang et al., 2012), but it seems equally possible that this highly conserved domain binds to an upstream protein at the site of autophagy initiation. Though the functional binding partners of the BARA domain remain to be confirmed, the structures at least provide a framework for the key questions.
The Atg2-Atg18 Complex: The Receiving End of the PI(3)P Signal
The Atg2-18 complex is the most mysterious part of the core autophagy machinery. This complex binds to the edge of the phagophore (Graef et al., 2013, Suzuki et al., 2013b) and seems to be important for the expansion of the phagophore and for the recycling of the integral membrane protein Atg9. Atg2 is a very large protein whose sequence contains no informative motifs and whose structure is not known even at low resolution or in fragments. Fortunately, more is known about the smaller subunit of the complex, Atg18. Atg18 is a member of the phosphinositide binding seven-bladed β-propeller (PROPPIN) family. Most of these proteins, including Atg18, bind to both PI(3)P and PI(3,5)P2. It is currently thought that PI(3,5)P2 binding contributes to the nonautophagic function of Atg18 in vacuole homeostasis. In yeast, Atg18 is the only confirmed PI(3)P-specific lipid binding protein in autophagy. Crystal structures have been determined of another yeast PROPPIN, Hsv2 (Baskaran et al., 2012, Krick et al., 2012, Rogov et al., 2014, Watanabe et al., 2012), that is closely related to Atg18. These structures and their functional mapping onto Atg18 show that PI(3)P is bound at two sites, on propeller blades 5 and 6 (Figure 3E). A hydrophobic loop in blade 6 provides an additional membrane anchor to augment binding to PI(3)P-containing membranes. This loop is subject to phosphoregulation, which inhibits membrane binding (Tamura et al., 2013), probably through electrostatic repulsion. The propeller binds to membranes edge-on, such that the two flat faces of the propeller are both available to interact with other proteins. Atg2 binds via blade 2 (Rieter et al., 2013, Watanabe et al., 2012). Most of the key residues described above are well conserved in the human Atg18 orthologs WIPI-1–4, and it is expected that these structural interactions will be conserved as well. Having obtained a toehold in the mechanistic understanding of the Atg18 part of the complex, it is hoped that progress on the Atg2 part will pick up speed.
Atg12 and Atg8: Autophagy UBLs Linking Regulators and Cargos to Autophagosomal Membranes
The best structurally characterized components of autophagy revolve around the UBLs. Budding yeast have two distinctive autophagy UBLs, Atg12 and Atg8 (Ichimura et al., 2000, Mizushima et al., 1998). Higher eukaryotes maintain a single Atg12 but display a massively expanded family of Atg8 orthologs in two clades, LC3 (typically LC3A, B, and C, with multiple isoforms and/or splice variants in some organisms) and GABARAP (e.g., GABARAP, GABARAPL1, and GABARAPL2), whose functions generally share overall common features with the simpler yeast Atg8 pathway (reviewed in Rogov et al. [2014]). Accordingly, here we refer to “Atg8/LC3” to describe properties of the family as a whole. Atg12 and Atg8/LC3 adopt structures related to ubiquitin, with a globular β-grasp domain consisting of a four-stranded β sheet packing against two α helices and a flexible C-terminal tail culminating in a Gly that becomes covalently modified (Sugawara et al., 2004, Suzuki et al., 2005). Atg8/LC3 has two additional, distinctive N-terminal helices.
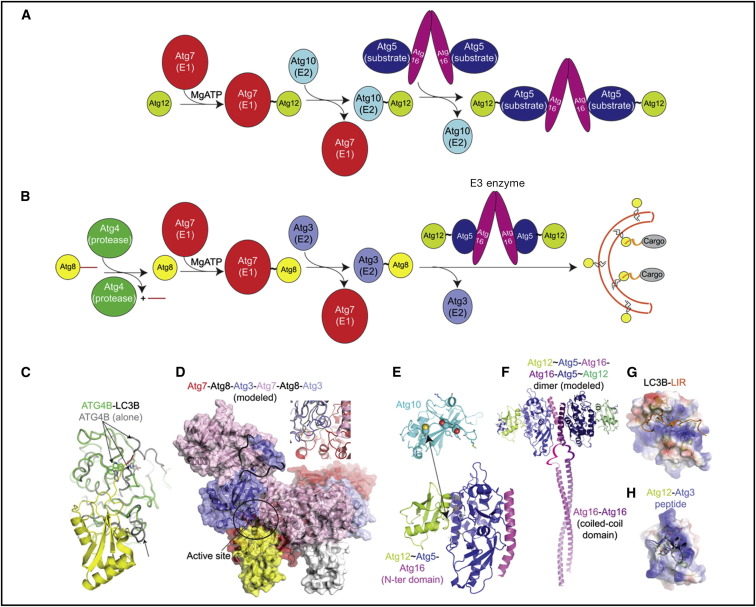
As with UBLs such as ubiquitin, Atg12 becomes C-terminally isopeptide bonded to a specific Lys on a target protein, Atg5, via an enzymatic cascade (Figure 4 A). This involves E1 (Atg7) and E2 (Atg10) enzymes, which catalyze formation of the Atg12∼Atg5 conjugate (here, “∼” refers to covalent complex) (Mizushima et al., 1998, Shintani et al., 1999). Mammalian ATG12 was also reported to regulate mitochondrial homeostasis and cell death via ligation to a different autophagy-specific E2, ATG3 (Radoshevich et al., 2010), though roles of ATG3∼ATG12 adducts remain poorly understood.
Figure 4.
Structures, Mechanisms, and Functions of Ubiquitin-like Protein Conjugation Cascades in Autophagy
(A) Atg12 ligation pathway progresses first via a thioester-linked Atg7∼Atg12 intermediate and then via a thioester-linked Atg10∼Atg12 intermediate, from which Atg12 is ligated to Atg5, which binds Atg16.
(B) Atg8/LC3 ligation pathway involves processing by Atg4, activation by Atg7, conjugation to Atg3, and ligation to phosphatidylethanolamine (PE) facilitated by the Atg12∼Atg5-Atg16 oligomer. Atg8/LC3 (represented in yellow circles) ligation to PE promotes expansion of the phagophore membrane and recruits cargos to autophagosomes.
(C) Structural superposition of free ATG4B (gray) and a complex between ATG4B (green) and the precursor form of LC3 (yellow, with C-terminal extension in red). The ATG4 Cys-His-Asp catalytic triad and Trp clamp are shown in sticks (Kumanomidou et al., 2006, Satoo et al., 2009, Sugawara et al., 2005). Arrows highlight regions of ATG4B conformational activation.
(D) Structural model for Atg8 (yellow and white) transfer between Atg7 (the two protomers in the homodimer colored red and pink) and Atg3 (two bound per Atg7 homodimer, colored slate and light blue) (Hong et al., 2011, Kaiser et al., 2013, Noda et al., 2011). One of two active sites is circled, from which Atg8 (yellow) is transferred between the active site of Atg7 (red) to Atg3 (slate) bound to the N-terminal domain of the opposite Atg7 in the dimer (pink). Missing Atg3 loops, ribbons. Inset, close-up of active site, superimposed with free Atg3 to highlight conformational activation. Modeling suggests similar transfer of Atg12 to Atg10.
(E) Structures of K. marxianus Atg10 (cyan) and human Atg12 (lime)∼Atg5 (blue)-Atg16 (magenta) (Hong et al., 2012, Noda et al., 2013, Otomo et al., 2013, Yamaguchi et al., 2012). Arrow highlights the Atg3 cysteine, from which Atg12 is transferred, to the Atg5 target. Spheres, Atg10’s Cys; positions that can be crosslinked to Atg5; sticks, residues implicated in Atg5 binding (from yeast Atg12, lime; from human and K. marxianus Atg5, gray and white).
(F) Model of an Atg12∼Atg5-Atg16 dimer, based on crystal contacts Atg12∼Atg5-Atg16 (N-terminal domain) structures (Metlagel et al., 2013, Otomo et al., 2013), connected by ribbons to Atg16 coiled coil (Fujioka et al., 2010).
(G) Surface of LC3B colored by electrostatic potential, bound to LIR motif from p62 (orange) (Ichimura et al., 2008, Pankiv et al., 2007).
(H) Surface of Atg12 colored by electrostatic potential, bound to Atg3 (slate) flexible region (Metlagel et al., 2013).
Unlike other UBLs, Atg8/LC3 family members are the only UBLs currently known to be ligated to a lipid-phosphatidylethanolamine (PE) and potentially other lipids (Ichimura et al., 2000). Atg8/LC3∼PE adducts are incorporated into the growing phagophore and autophagosomes, serving as hubs for transient and/or sustained recruitment of interacting proteins with the membrane during autophagosome biogenesis.
Atg8/LC3 conjugation has many distinctive features (Figure 4B). Atg8/LC3 family members are initially synthesized as C-terminally extended precursors, which are processed by a protease (Atg4) to expose a C-terminal Gly (Kirisako et al., 2000). The E1-E2-E3 ligation cascade for Atg8/LC3 is tied to the Atg12 pathway. Atg12 and Atg8/LC3 are both activated by Atg7 (Ichimura et al., 2000, Mizushima et al., 1998). Atg8/LC3 is transferred from Atg7’s catalytic Cys to that of the Atg3 (Ichimura et al., 2000). Atg8/LC3 is subsequently transferred to PE in a reaction facilitated by the Atg12∼5 conjugate acting as an E3 enzyme (Hanada et al., 2007). In vivo, Atg16 forms an oligomeric complex with Atg12∼Atg5, promoting Atg8/LC3∼PE formation (Kuma et al., 2002, Mizushima et al., 1999). Incorporation of Atg8/LC3∼PE adducts is critical for expansion of the phagophore and recruitment of specific regulatory proteins and cargos to autophagosomes (Kaufmann et al., 2014, Weidberg et al., 2010, Xie et al., 2008; also reviewed in Rogov et al., 2014). Mechanisms regulating Atg12∼Atg5 conjugation are emerging, and Atg8/LC3 lipidation is induced upon starvation. Ultimately, Atg4 cleaves Atg8/LC3 from PE on the exterior of autophagosomes (Kirisako et al., 2000). This may liberate Atg8/LC3-nucleated assemblies and free Atg8/LC3 to return into the conjugation cascade.
Atg4: An Atg8/LC3 Processing and Deconjugating Enzyme
In yeast, Atg8 processing and deconjugation are catalyzed by a single Atg4 (Kirisako et al., 2000). In higher eukaryotes, the Atg4 family is expanded with four members in humans (ATG4A–D), whose differences are poorly understood. Structures of ATG4A and ATG4B alone and of complexes between ATG4B and LC3B (Kumanomidou et al., 2006, Satoo et al., 2009, Sugawara et al., 2005) together reveal: (1) Atg8/LC3 recognition by ATG4B, (2) the basis for cleavage, and (3) Atg8/LC3-induced conformational changes that activate ATG4B. In brief, interactions between ATG4 and Atg8/LC3’s ubiquitin-like domain and C-terminal tail bury ∼1,680 and 940 Å2, respectively. Atg8/LC3’s tail extends across an ATG4 groove, with the neo C-terminal Gly positioned on one side by an ATG4 “regulatory loop” clamping the penultimate LC3 Phe and on the other side by a conserved ATG4 Trp securing LC3’s scissile bond into ATG4’s Cys-His-Asp catalytic triad (Figure 4C). Atg8/LC3 apparently activates its cleavage through promoting ATG4 conformational changes that: (1) dislodge ATG4’s C-terminal region from the LC3-binding site, (2) reposition the regulatory loop, and (3) displace ATG4’s N-terminal loop for postcleavage liberation of the severed LC3 region or PE.
Atg12 and Atg8 Activation by the E1 Atg7
With a C-terminal Gly, Atg8 and Atg12 are primed for three-step activation by Atg7 (Ichimura et al., 2000, Mizushima et al., 1998). First, Atg7 “activates” the otherwise inert UBL C terminus in a MgATP-dependent reaction, by catalyzing C-terminal adenylation of the UBL. Second, Atg7’s catalytic Cys attacks the activated UBL C terminus, AMP is released, and a covalent, presumably thioester-bonded Atg7∼UBL intermediate is produced, (Brownell et al., 2010, Ichimura et al., 2000, Mizushima et al., 1998). Third, each autophagy UBL is transferred from Atg7’s Cys to that of its cognate E2, resulting in thioester-linked Atg12∼Atg10 or Atg8/LC3∼Atg3 intermediates. More than a dozen structures of Atg7 or its domains—complexes with MgATP, Atg8, Atg10, and Atg3—provide insights into many aspects of this process (Hong et al., 2011, Kaiser et al., 2013, Noda et al., 2011, Taherbhoy et al., 2011, Yamaguchi et al., 2012).
Atg7 is a multidomain homodimer (Figure 4D) (Fan et al., 2011, Noda et al., 2011, Taherbhoy et al., 2011). The N-terminal domain engages surfaces of the E2 enzymes, Atg10 and Atg3, distal from their active sites (Hong et al., 2011, Kaiser et al., 2013, Noda et al., 2011, Taherbhoy et al., 2011, Yamaguchi et al., 2012). Atg7 binding apparently rearranges Atg3’s active site to expose the catalytic Cys. Atg7’s N-terminal domain remotely presents an E2 catalytic Cys to Atg7’s active site, which is located in a separate central domain.
Atg7’s central domain structurally resembles the adenylation domain from canonical E1 enzymes, with the exception of being a symmetric homodimer in which each protomer can activate a UBL molecule (Hong et al., 2011, Noda et al., 2011, Taherbhoy et al., 2011). Structures between Atg7’s central domain binding and Atg8 showed Atg8’s C terminus juxtaposed with the α phosphate from ATP (Hong et al., 2011, Noda et al., 2011). A conserved E1 Asp coordinates a magnesium ion, which in turn coordinates ATP’s phosphates to promote transfer of AMP from ATP to Atg8’s C terminus. Unlike other E1 enzymes, Atg7’s central domain also contains a “crossover/catalytic Cys” loop that crosses the domain, displays the Atg7 catalytic Cys, and adopts distinct structures for distinct functions. Thematically resembling Atg8/LC3-induced conformational changes in Atg4, Atg8 apparently promotes its own activation by (1) dislodging Atg7’s crossover/catalytic Cys from the Atg8-binding site and (2) prompting loop repositioning to promote catalysis. In an Atg7-Atg8-MgATP crystal structure, the “crossover/Cys loop” clamps Atg8’s C terminus in the adenylation active site, with the catalytic Cys facing away from the active site so as not to interfere with the adenylation reaction (Noda et al., 2011). The loop can also adopt a conformation in which the active site faces Atg8’s C terminus, presumably for attacking Atg8∼AMP to form the thioester-bonded Atg7∼Atg8 intermediate (Hong et al., 2011). Ultimately, Atg7’s “crossover/catalytic Cys” loop is again remodeled (Kaiser et al., 2013) to transfer Atg12 or Atg8/LC3 in “trans” from the active site of one Atg7 protomer to Atg10 or Atg3, respectively, bound to the opposite protomer of the Atg7 homodimer (Hong et al., 2011, Kaiser et al., 2013, Noda et al., 2011, Taherbhoy et al., 2011, Yamaguchi et al., 2012).
Ligation of the Autophagy UBLs
After forming Atg12∼Atg10 and Atg8∼Atg3 intermediates, mechanisms underlying ligation diverge. Atg12 is ligated without an E3 enzyme via Atg10 directly targeting Atg5 (Figure 4E). The detailed basis for Atg12 transfer from Atg10 to Atg5 remains unknown. Nonetheless, clues have been provided by nuclear magnetic resonance spectroscopy (NMR), mutagenesis, and crosslinking to identify Atg10 and Atg5 residues involved in their interaction (Hong et al., 2012, Matsushita et al., 2007, Yamaguchi et al., 2012). Mapping important residues suggests that Atg5 binds the concave surface surrounding Atg10’s active site (Yamaguchi et al., 2012). Enzyme assays also suggest that the Atg12 portion of the Atg10∼Atg12 intermediate also may recruit Atg5 (Yamaguchi et al., 2012). Interestingly, several Atg5 residues that are critical for ligation to Atg12 surround the Atg5-Atg12 interface in the Atg12∼Atg5-Atg16 (N-terminal domain) complex (Noda et al., 2013, Otomo et al., 2013, Yamaguchi et al., 2012). It is tempting to speculate that Atg12 and Atg5 form similar noncovalent contacts both before and after ligation, though future studies will be required to reveal details of an Atg10∼Atg12-Atg5 intermediate.
Atg8/LC3 transfer from Atg3’s catalytic cysteine to PE is accelerated by the Atg12∼Atg5 conjugate acting as an E3 enzyme (Hanada et al., 2007). Biochemical data indicate that Atg12∼Atg5 promotes structural rearrangement of Atg3’s active site residues, in keeping with the conformational activation observed by interactions of other enzymes in the autophagy UBL pathways (Sakoh-Nakatogawa et al., 2013). Although there is no structure of an Atg12∼Atg5 complex with full-length Atg3, significant insights into their interaction have been derived from subcomplexes. Mutations based on Atg12∼Atg5-Atg16N complex structures revealed numerous surfaces contributing to Atg8 ligation to PE (Figure 4) (Fujioka et al., 2010, Matsushita et al., 2007, Noda et al., 2013, Otomo et al., 2013). In the case of human proteins, a surface at the distal tip of Atg12 was found responsible for high-affinity binding to Atg3 (Otomo et al., 2013), as revealed in a crystal structure of part of a “flexible region (FR)” from human Atg3 bound to Atg12∼Atg5-Atg16N (Metlagel et al., 2013). Notably, a nearby Atg3 region binds Atg7, suggesting that Atg3 shuttles back and forth between E1 and E3 during Atg8/LC3 lipidation (Kaiser et al., 2013, Qiu et al., 2013, Taherbhoy et al., 2011). Different studies identified additional important Atg12∼Atg5-Atg16N surfaces, which may further recruit or activate the Atg3∼Atg8/LC3 intermediate, recruit PE, or play other roles (Fujita et al., 2008, Kaufmann et al., 2014, Romanov et al., 2012).
Atg8/LC3-AIM/LIR Interactions: Dynamic Protein Recruitment to the Phagophore and Autophagsomal Membranes
Atg8/LC3∼PE localizes partner proteins to the autophagasomal membrane. Atg8/LC3-binding proteins often display an “Atg8-interacting motif (AIM)” or “LC3-interacting region (LIR)” sequence of Trp-x-x-Leu/Ile, often with adjacent acidic residues (Ichimura et al., 2008, Noda et al., 2008, Pankiv et al., 2007, Rozenknop et al., 2011). The AIM/LIR motif adopts a β-strand structure that incorporates into the Atg8/LC3 β sheet (Hain et al., 2012, Ichimura et al., 2008, Kondo-Okamoto et al., 2012, Noda et al., 2008, Pankiv et al., 2007, Rogov et al., 2013, Rozenknop et al., 2011, Suzuki et al., 2013a, Thielmann et al., 2009, von Muhlinen et al., 2012, Weiergräber et al., 2008, Yamaguchi et al., 2010). In the absence of a partner, Atg8/LC3 adopts a closed conformation, with an expansive basic surface. However, in complexes with AIM/LIR peptides, the Atg8/LC3 structure opens two hydrophobic pockets, with one embracing the Trp and the other binding the Leu/Ile from the AIM/LIR motif (Suzuki et al., 2013a). Atg8/LC3 family members use these pockets to recruit a plethora of variant AIM/LIR sequences, in some cases involving regulation and/or distinctions among Atg8/LC3 orthologs (Behrends et al., 2010, Rogov et al., 2014). For example, the “CLIR” Ile-Leu-Val-Val sequence from CALCOCO2/NDP52 binds selectively to LC3C, with the Ile docking in the “canonical Trp-binding pocket” and a Val in the “canonical Leu-binding pocket” (von Muhlinen et al., 2012). In contrast, the Ser-Phe-Val-Glu-Ile sequence from Optineurin requires Ser phosphorylation to contact a complementary basic surface and optimally engage Atg8/LC3 orthologs (Rogov et al., 2013). Of note, the AIM/LIR-binding region of Atg8/LC3 also recruits non-AIM/LIR sequences that adopt alternative structures to fill the two hydrophobic pockets (Noda et al., 2011). This plasticity enabled phage-display selection of a high-affinity peptide that adopts a distinctive structure upon binding to GABARAP (Weiergräber et al., 2008). Interestingly, the structural mode of partner protein recruitment appears to be conserved between Atg8/LC3 and Atg12: the recent structure of an Atg16-Atg5-Atg12-Atg3FR complex revealed architectural similarities between Atg12-Atg3 and Atg8/LC3-AIM/LIR complexes (Metlagel et al., 2013).
A major role of autophagy UBL pathways is recruiting autophagy receptors, which bridge cargos and Atg8/LC3∼PE at autophagosomal membranes (reviewed in (Rogov et al., 2014, Schreiber and Peter, 2014). Autophagy receptors typically display an Atg8/LC3-binding AIM/LIR and another domain for recruiting cargo for degradation. Structural studies have revealed numerous mechanisms by which autophagy receptors recognize cargos. Many receptors bind ubiquitin and promote autophagic degradation of ubiquitinated proteins, organelles, or pathogenic microbes (Shaid et al., 2013). A distinctive autophagy receptor is CALCOCO2/NDP52, which in addition to binding LC3C through its CLIR, displays a peptide motif recognizing LGALS8/galectin 8, which in turn bind sugars exposed on vesicles upon Salmonella-induced damage (Kim et al., 2013a, Li et al., 2013, Thurston et al., 2012).
The second major role for Atg8/LC3 ligation to PE is in expansion of the phagophore membrane, apparently through multiple mechanisms. Atg8/LC3 binds AIM/LIR or related sequences in other Atg proteins, most likely to orchestrate interactions regulating autophagosome assembly (Behrends et al., 2010). For example, Atg8/LC3 binding to Atg1 (or ULK1 and 2 in higher eukaryotes) and Atg13 enables tethering upstream signals to the growing phagophore (reviewed in Rogov et al., 2014). Atg8/LC3 also binds AIM/LIR-like sequences in UBL conjugation enzymes, including in Atg4, Atg7, Atg3, and Atg12∼Atg5-Atg16. As examples, Atg4B’s N terminus displays a variant LIR that binds to an LC3-Atg4B complex in the adjacent asymmetric unit in the crystal (Satoo et al., 2009). Likewise, Atg7’s intrinsically disordered extreme C-terminal sequence is not observed in crystals, but NMR studies revealed this binding Atg8 in a manner that is mutually exclusive with Atg8 engaging the Atg7 active site (Noda et al., 2011). Intriguingly, Atg12’s AIM is not a linear motif but is instead three-dimensional, comprising a Trp near the C terminus and a Val within Atg12’s globular domain. Atg12’s AIM apparently binds Atg8∼PE anchored in a membrane (Kaufmann et al., 2014). Atg12∼Atg5-Atg16 dimerization through Atg16’s coiled coil (Fujioka et al., 2010) would enable one arm of this E3 to bind membrane-embedded Atg8 and the other to recruit Atg3∼Atg8 to ligate Atg8 to PE localized within adjacent membrane (Kaufmann et al., 2014). Interestingly, autophagy receptors outcompete Atg12∼Atg5-Atg16 for the AIM-binding site on Atg8. This latter finding provides a rationale for how Atg8 ligation on the convex surface of autophagosomes can lead to membrane expansion while sparing Atg8∼PE on the concave side for cargo recruitment (Kaufmann et al., 2014). Although the roles of the autophagic membrane are only beginning to emerge, future structural and biophysical studies will undoubtedly identify exciting and unprecedented mechanisms by which autophagy UBL localization and functions are integrated by dynamic, synergistic, and competitive interactions of autophagy UBL proteins, their partners, and the lipids to which Atg8/LC3 are anchored.
Concluding Perspectives
Structural studies have elucidated high-resolution details of protein domains and subcomplexes regulating and directing many critical aspects of autophagy. Recent structures or models have provided unprecedented details into mechanisms for sensing membrane curvature, generating and sensing a PI(3)P signal, and underlying distinctive UBL conjugation that provides a membrane-linked platform for protein interactions during autophagosome biogenesis. Many of the structures revealed multisite interactions—either between proteins or between protein assemblies and membranes—that ensure that function is tied to correct Atg subcomplex architecture.
A major constraint in modeling autophagy stems from limited knowledge as to how information from Atg1/ULK1, the Beclin 1/PI(3)P pathway, WIPI proteins, and autophagy UBL cascades are physically integrated. Nonetheless, the existing structures provide clues to how components may be coordinated. Of note, Atg protein-interaction domains are embedded within conformationally fluctuating assemblies. As observed for components of the UBL conjugation machineries, even the well-folded domains of Atg proteins display loop rearrangement that dynamically controls Atg protein function upon complex assembly. The capacity for Atg proteins and subcomplexes to undergo structural remodeling is likely amplified by the elusive, extensive intrinsically disordered segments found in numerous Atg proteins. Disordered regions likely mediate dynamic, multisite interactions modulated by avidity, posttranslational modifications, or lipidic environments. Furthermore, although competing intermolecular interactions—such as Atg29-Atg31 sterically occluding Atg17’s membrane binding site or mutually exclusive Atg8/LC3 interactions with AIMs/LIRs from different proteins—are known, details controlling which complexes prevail at different stages of autophagy remain largely elusive. Another major gap in knowledge is understanding the roles of the numerous membranes associated with regulation and execution of autophagy. Of note, with the structure of the Atg17-Atg31-Atg29 portion of the Atg1 complex (Ragusa et al., 2012) and reconstitution of an Atg8-5-12-16 scaffold based on homotypic interactions via Atg16’s coiled coil (Kaufmann et al., 2014), the field is making progress toward understanding Atg protein organization on the tens of nanometer scale relevant to membrane interactions associated with autophagosome biogenesis. We anticipate that future structural studies will reveal reciprocal regulation of Atg proteins and their membrane partners, crosstalk between different portions of the pathway, and mechanisms prioritizing overlapping protein-protein interactions establishing the hierarchy of distinctive events orchestrating autophagy. The mechanism of autophagy is taking shape!
Acknowledgments
S. Baskaran and M. Ragusa are thanked for generating figures. B.A.S. acknowledges the Howard Hughes Medical Institute and NIH R01GM077053, P30CA021765, and ALSAC for support.
Contributor Information
James H. Hurley, Email: jimhurley@berkeley.edu.
Brenda A. Schulman, Email: brenda.schulman@stjude.org.
References
- Baskaran S., Ragusa M.J., Boura E., Hurley J.H. Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy. Mol. Cell. 2012;47:339–348. doi: 10.1016/j.molcel.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C., Sowa M.E., Gygi S.P., Harper J.W. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestebroer J., V’kovski P., Mauthe M., Reggiori F. Hidden behind autophagy: The unconventional role of ATG proteins. Traffic. 2013;14:1029–1041. doi: 10.1111/tra.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P., Reggiori F., Codogno P. Emerging regulation and functions of autophagy. Nat. Cell Biol. 2013;15:713–720. doi: 10.1038/ncb2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell J.E., Sintchak M.D., Gavin J.M., Liao H., Bruzzese F.J., Bump N.J., Soucy T.A., Milhollen M.A., Yang X., Burkhardt A.L. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol. Cell. 2010;37:102–111. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- Bruns C., McCaffery J.M., Curwin A.J., Duran J.M., Malhotra V. Biogenesis of a novel compartment for autophagosome-mediated unconventional protein secretion. J. Cell Biol. 2011;195:979–992. doi: 10.1083/jcb.201106098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E.Y., Longatti A., McKnight N.C., Tooze S.A. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol. Cell. Biol. 2009;29:157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.Y., Neufeld T.P. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol. Biol. Cell. 2009;20:2004–2014. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Nair U., Geng J.F., Klionsky D.J. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 2008;19:668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew L.H., Setiaputra D., Klionsky D.J., Yip C.K. Structural characterization of the Saccharomyces cerevisae autophagy regulatory complex Atg17-Atg31-Atg29. Autophagy. 2013;9:1467–1474. doi: 10.4161/auto.25687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D., Dokudovskaya S., Alber F., Williams R., Chait B.T., Sali A., Rout M.P. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W., Nassiri A., Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) Proc. Natl. Acad. Sci. USA. 2011;108:7769–7774. doi: 10.1073/pnas.1016472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel A.I., Dlouhy B.J., Wang C.X., Ryu S.W., Neutzner A., Hasson S.A., Sideris D.P., Abeliovich H., Youle R.J. Role of membrane association and Atg14-dependent phosphorylation in beclin-1-mediated autophagy. Mol. Cell. Biol. 2013;33:3675–3688. doi: 10.1128/MCB.00079-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A., Unger V.M., De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y., Noda N.N., Nakatogawa H., Ohsumi Y., Inagaki F. Dimeric coiled-coil structure of Saccharomyces cerevisiae Atg16 and its functional significance in autophagy. J. Biol. Chem. 2010;285:1508–1515. doi: 10.1074/jbc.M109.053520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Itoh T., Omori H., Fukuda M., Noda T., Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley I.G., Lam H., Wang J., Ding X., Chen S., Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef M., Friedman J.R., Graham C., Babu M., Nunnari J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell. 2013;24:2918–2931. doi: 10.1091/mbc.E13-07-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hain A.U., Weltzer R.R., Hammond H., Jayabalasingham B., Dinglasan R.R., Graham D.R., Colquhoun D.R., Coppens I., Bosch J. Structural characterization and inhibition of the Plasmodium Atg8-Atg3 interaction. J. Struct. Biol. 2012;180:551–562. doi: 10.1016/j.jsb.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T., Noda N.N., Satomi Y., Ichimura Y., Fujioka Y., Takao T., Inagaki F., Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- Heenan E.J., Vanhooke J.L., Temple B.R., Betts L., Sondek J.E., Dohlman H.G. Structure and function of Vps15 in the endosomal G protein signaling pathway. Biochemistry. 2009;48:6390–6401. doi: 10.1021/bi900621w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.B., Kim B.-W., Lee K.-E., Kim S.W., Jeon H., Kim J., Song H.K. Insights into noncanonical E1 enzyme activation from the structure of autophagic E1 Atg7 with Atg8. Nat. Struct. Mol. Biol. 2011;18:1323–1330. doi: 10.1038/nsmb.2165. [DOI] [PubMed] [Google Scholar]
- Hong S.B., Kim B.W., Kim J.H., Song H.K. Structure of the autophagic E2 enzyme Atg10. Acta Crystallogr. D Biol. Crystallogr. 2012;68:1409–1417. doi: 10.1107/S0907444912034166. [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S.I., Natsume T., Takehana K., Yamada N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Choi W., Hu W., Mi N., Guo Q., Ma M., Liu M., Tian Y., Lu P., Wang F.-L. Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Res. 2012;22:473–489. doi: 10.1038/cr.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J.H., Boura E., Carlson L.A., Różycki B. Membrane budding. Cell. 2010;143:875–887. doi: 10.1016/j.cell.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Ichimura Y., Kumanomidou T., Sou Y.S., Mizushima T., Ezaki J., Ueno T., Kominami E., Yamane T., Tanaka K., Komatsu M. Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- Jao C.C., Ragusa M.J., Stanley R.E., Hurley J.H. A HORMA domain in Atg13 mediates PI 3-kinase recruitment in autophagy. Proc. Natl. Acad. Sci. USA. 2013;110:5486–5491. doi: 10.1073/pnas.1220306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C.H., Jun C.B., Ro S.H., Kim Y.M., Otto N.M., Cao J., Kundu M., Kim D.H. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser S.E., Qiu Y., Coats J.E., Mao K., Klionsky D.J., Schulman B.A. Structures of Atg7-Atg3 and Atg7-Atg10 reveal noncanonical mechanisms of E2 recruitment by the autophagy E1. Autophagy. 2013;9:778–780. doi: 10.4161/auto.23644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanasios E., Stapleton E., Manifava M., Kaizuka T., Mizushima N., Walker S.A., Ktistakis N.T. Dynamic association of the ULK1 complex with omegasomes during autophagy induction. J. Cell Sci. 2013;126:5224–5238. doi: 10.1242/jcs.132415. [DOI] [PubMed] [Google Scholar]
- Kaufmann A., Beier V., Franquelim H.G., Wollert T. Molecular mechanism of autophagic membrane-scaffold assembly and disassembly. Cell. 2014;156:469–481. doi: 10.1016/j.cell.2013.12.022. [DOI] [PubMed] [Google Scholar]
- Kim B.W., Hong S.B., Kim J.H., Kwon H., Song H.K. Structural basis for recognition of autophagic receptor NDP52 by the sugar receptor galectin-8. Nat. Commun. 2013;4:1613. doi: 10.1038/ncomms2606. [DOI] [PubMed] [Google Scholar]
- Kim J.Y., Zhao H., Martinez J., Doggett T.A., Kolesnikov A.V., Tang P.H., Ablonczy Z., Chan C.C., Zhou Z., Green D.R., Ferguson T.A. Noncanonical autophagy promotes the visual cycle. Cell. 2013;154:365–376. doi: 10.1016/j.cell.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Ichimura Y., Okada H., Kabeya Y., Mizushima N., Yoshimori T., Ohsumi M., Takao T., Noda T., Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo-Okamoto N., Noda N.N., Suzuki S.W., Nakatogawa H., Takahashi I., Matsunami M., Hashimoto A., Inagaki F., Ohsumi Y., Okamoto K. Autophagy-related protein 32 acts as autophagic degron and directly initiates mitophagy. J. Biol. Chem. 2012;287:10631–10638. doi: 10.1074/jbc.M111.299917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krick R., Busse R.A., Scacioc A., Stephan M., Janshoff A., Thumm M., Kühnel K. Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a β-propeller protein family. Proc. Natl. Acad. Sci. USA. 2012;109:E2042–E2049. doi: 10.1073/pnas.1205128109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A., Mizushima N., Ishihara N., Ohsumi Y. Formation of the approximately 350-kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J. Biol. Chem. 2002;277:18619–18625. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- Kumanomidou T., Mizushima T., Komatsu M., Suzuki A., Tanida I., Sou Y.S., Ueno T., Kominami E., Tanaka K., Yamane T. The crystal structure of human Atg4b, a processing and de-conjugating enzyme for autophagosome-forming modifiers. J. Mol. Biol. 2006;355:612–618. doi: 10.1016/j.jmb.2005.11.018. [DOI] [PubMed] [Google Scholar]
- Lee C., Goldberg J. Structure of coatomer cage proteins and the relationship among COPI, COPII, and clathrin vesicle coats. Cell. 2010;142:123–132. doi: 10.1016/j.cell.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.H., He L.Q., Che K.H., Funderburk S.F., Pan L.F., Pan N.N., Zhang M.J., Yue Z.Y., Zhao Y.X. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat. Commun. 2012;3:662. doi: 10.1038/ncomms1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Wandel M.P., Li F., Liu Z., He C., Wu J., Shi Y., Randow F. Sterical hindrance promotes selectivity of the autophagy cargo receptor NDP52 for the danger receptor galectin-8 in antibacterial autophagy. Sci. Signal. 2013;6:ra9. doi: 10.1126/scisignal.2003730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K., Chew L.H., Inoue-Aono Y., Cheong H., Nair U., Popelka H., Yip C.K., Klionsky D.J. Atg29 phosphorylation regulates coordination fo the Atg17-Atg31-Atg29 complex with the Atg11 scaffold during autophagy initiation. Proc. Natl. Acad. Sci. USA. 2013;110:E2875–E2884. doi: 10.1073/pnas.1300064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli M., Musacchio A. MAD contortions: conformational dimerization boosts spindle checkpoint signaling. Curr. Opin. Struct. Biol. 2007;17:716–725. doi: 10.1016/j.sbi.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Mari M., Griffith J., Rieter E., Krishnappa L., Klionsky D.J., Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J. Cell Biol. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita M., Suzuki N.N., Obara K., Fujioka Y., Ohsumi Y., Inagaki F. Structure of Atg5.Atg16, a complex essential for autophagy. J. Biol. Chem. 2007;282:6763–6772. doi: 10.1074/jbc.M609876200. [DOI] [PubMed] [Google Scholar]
- Metlagel Z., Otomo C., Takaesu G., Otomo T. Structural basis of ATG3 recognition by the autophagic ubiquitin-like protein ATG12. Proc. Natl. Acad. Sci. USA. 2013;110:18844–18849. doi: 10.1073/pnas.1314755110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S., Tavshanjian B., Oleksy A., Perisic O., Houseman B.T., Shokat K.M., Williams R.L. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science. 2010;327:1638–1642. doi: 10.1126/science.1184429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M.D., Klionsky D.J., Ohsumi M., Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Noda T., Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Moreau K., Renna M., Rubinsztein D.C. Connections between SNAREs and autophagy. Trends Biochem. Sci. 2013;38:57–63. doi: 10.1016/j.tibs.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Noda N.N., Kumeta H., Nakatogawa H., Satoo K., Adachi W., Ishii J., Fujioka Y., Ohsumi Y., Inagaki F. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- Noda N.N., Satoo K., Fujioka Y., Kumeta H., Ogura K., Nakatogawa H., Ohsumi Y., Inagaki F. Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol. Cell. 2011;44:462–475. doi: 10.1016/j.molcel.2011.08.035. [DOI] [PubMed] [Google Scholar]
- Noda N.N., Kobayashi T., Adachi W., Fujioka Y., Ohsumi Y., Inagaki F. Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy. J. Biol. Chem. 2012;287:16256–16266. doi: 10.1074/jbc.M112.348250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda N.N., Fujioka Y., Hanada T., Ohsumi Y., Inagaki F. Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation. EMBO Rep. 2013;14:206–211. doi: 10.1038/embor.2012.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo C., Metlagel Z., Takaesu G., Otomo T. Structure of the human ATG12∼ATG5 conjugate required for LC3 lipidation in autophagy. Nat. Struct. Mol. Biol. 2013;20:59–66. doi: 10.1038/nsmb.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Qiu Y., Hofmann K., Coats J.E., Schulman B.A., Kaiser S.E. Binding to E1 and E3 is mutually exclusive for the human autophagy E2 Atg3. Protein Sci. 2013;22:1691–1697. doi: 10.1002/pro.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoshevich L., Murrow L., Chen N., Fernandez E., Roy S., Fung C., Debnath J. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell. 2010;142:590–600. doi: 10.1016/j.cell.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragusa M.J., Stanley R.E., Hurley J.H. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151:1501–1512. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Klionsky D.J. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics. 2013;194:341–361. doi: 10.1534/genetics.112.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Monastyrska I., Verheije M.H., Calì T., Ulasli M., Bianchi S., Bernasconi R., de Haan C.A.M., Molinari M. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieter E., Vinke F., Bakula D., Cebollero E., Ungermann C., Proikas-Cezanne T., Reggiori F. Atg18 function in autophagy is regulated by specific sites within its β-propeller. J. Cell Sci. 2013;126:593–604. doi: 10.1242/jcs.115725. [DOI] [PubMed] [Google Scholar]
- Rogov V.V., Suzuki H., Fiskin E., Wild P., Kniss A., Rozenknop A., Kato R., Kawasaki M., McEwan D.G., Löhr F. Structural basis for phosphorylation-triggered autophagic clearance of Salmonella. Biochem. J. 2013;454:459–466. doi: 10.1042/BJ20121907. [DOI] [PubMed] [Google Scholar]
- Rogov V., Dötsch V., Johansen T., Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Romanov J., Walczak M., Ibiricu I., Schüchner S., Ogris E., Kraft C., Martens S. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012;31:4304–4317. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenknop A., Rogov V.V., Rogova N.Y., Löhr F., Güntert P., Dikic I., Dötsch V. Characterization of the interaction of GABARAPL-1 with the LIR motif of NBR1. J. Mol. Biol. 2011;410:477–487. doi: 10.1016/j.jmb.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Rubinsztein D.C., Shpilka T., Elazar Z. Mechanisms of autophagosome biogenesis. Curr. Biol. 2012;22:R29–R34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- Sakoh-Nakatogawa M., Matoba K., Asai E., Kirisako H., Ishii J., Noda N.N., Inagaki F., Nakatogawa H., Ohsumi Y. Atg12-Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site. Nat. Struct. Mol. Biol. 2013;20:433–439. doi: 10.1038/nsmb.2527. [DOI] [PubMed] [Google Scholar]
- Satoo K., Noda N.N., Kumeta H., Fujioka Y., Mizushima N., Ohsumi Y., Inagaki F. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009;28:1341–1350. doi: 10.1038/emboj.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber A., Peter M. Substrate recognition in selective autophagy and the ubiquitin-proteasome system. Biochim. Biophys. Acta. 2014;1843:163–181. doi: 10.1016/j.bbamcr.2013.03.019. [DOI] [PubMed] [Google Scholar]
- Scott S.V., Nice D.C., 3rd, Nau J.J., Weisman L.S., Kamada Y., Keizer-Gunnink I., Funakoshi T., Veenhuis M., Ohsumi Y., Klionsky D.J. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J. Biol. Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- Shaid S., Brandts C.H., Serve H., Dikic I. Ubiquitination and selective autophagy. Cell Death Differ. 2013;20:21–30. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Mizushima N., Ogawa Y., Matsuura A., Noda T., Ohsumi Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999;18:5234–5241. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi L., Mapelli M., Knapp S., De Antoni A., Jeang K.T., Musacchio A. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 2002;21:2496–2506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara K., Suzuki N.N., Fujioka Y., Mizushima N., Ohsumi Y., Inagaki F. The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells. 2004;9:611–618. doi: 10.1111/j.1356-9597.2004.00750.x. [DOI] [PubMed] [Google Scholar]
- Sugawara K., Suzuki N.N., Fujioka Y., Mizushima N., Ohsumi Y., Inagaki F. Structural basis for the specificity and catalysis of human Atg4B responsible for mammalian autophagy. J. Biol. Chem. 2005;280:40058–40065. doi: 10.1074/jbc.M509158200. [DOI] [PubMed] [Google Scholar]
- Suzuki N.N., Yoshimoto K., Fujioka Y., Ohsumi Y., Inagaki F. The crystal structure of plant ATG12 and its biological implication in autophagy. Autophagy. 2005;1:119–126. doi: 10.4161/auto.1.2.1859. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Tabata K., Morita E., Kawasaki M., Kato R., Dobson R.C., Yoshimori T., Wakatsuki S. Structural basis of the autophagy-related LC3/Atg13 LIR complex: Recognition and interaction mechanism. Structure. 2013;22:47–58. doi: 10.1016/j.str.2013.09.023. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Akioka M., Kondo-Kakuta C., Yamamoto H., Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J. Cell Sci. 2013;126:2534–2544. doi: 10.1242/jcs.122960. [DOI] [PubMed] [Google Scholar]
- Taherbhoy A.M., Tait S.W., Kaiser S.E., Williams A.H., Deng A., Nourse A., Hammel M., Kurinov I., Rock C.O., Green D.R., Schulman B.A. Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway. Mol. Cell. 2011;44:451–461. doi: 10.1016/j.molcel.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura N., Oku M., Ito M., Noda N.N., Inagaki F., Sakai Y. Atg18 phosphoregulation controls organellar dynamics by modulating its phosphoinositide-binding activity. J. Cell Biol. 2013;202:685–698. doi: 10.1083/jcb.201302067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thielmann Y., Weiergräber O.H., Mohrlüder J., Willbold D. Structural framework of the GABARAP-calreticulin interface—implications for substrate binding to endoplasmic reticulum chaperones. FEBS J. 2009;276:1140–1152. doi: 10.1111/j.1742-4658.2008.06857.x. [DOI] [PubMed] [Google Scholar]
- Thurston T.L.M., Wandel M.P., von Muhlinen N., Foeglein A., Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Muhlinen N., Akutsu M., Ravenhill B.J., Foeglein A., Bloor S., Rutherford T.J., Freund S.M., Komander D., Randow F. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol. Cell. 2012;48:329–342. doi: 10.1016/j.molcel.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Kobayashi T., Yamamoto H., Hoshida H., Akada R., Inagaki F., Ohsumi Y., Noda N.N. Structure-based analyses reveal distinct binding sites for Atg2 and phosphoinositides in Atg18. J. Biol. Chem. 2012;287:31681–31690. doi: 10.1074/jbc.M112.397570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y.J., Zou Z.J., Becker N., Anderson M., Sumpter R., Xiao G.H., Kinch L., Koduru P., Christudass C.S., Veltri R.W. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013;154:1269–1284. doi: 10.1016/j.cell.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H., Shvets E., Shpilka T., Shimron F., Shinder V., Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H., Shvets E., Elazar Z. Biogenesis and Cargo Selectivity of Autophagosomes. In: Kornberg R.D., Raetz C.R.H., Rothman J.E., Thorner J.W., editors. Volume 80. Annual Reviews; Palo Alto, CA: 2011. pp. 125–156. (Annual Review of Biochemistry). [DOI] [PubMed] [Google Scholar]
- Weiergräber O.H., Stangler T., Thielmann Y., Mohrlüder J., Wiesehan K., Willbold D. Ligand binding mode of GABAA receptor-associated protein. J. Mol. Biol. 2008;381:1320–1331. doi: 10.1016/j.jmb.2008.06.086. [DOI] [PubMed] [Google Scholar]
- Xie Z., Nair U., Klionsky D.J. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Noda N.N., Nakatogawa H., Kumeta H., Ohsumi Y., Inagaki F. Autophagy-related protein 8 (Atg8) family interacting motif in Atg3 mediates the Atg3-Atg8 interaction and is crucial for the cytoplasm-to-vacuole targeting pathway. J. Biol. Chem. 2010;285:29599–29607. doi: 10.1074/jbc.M110.113670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Noda N.N., Yamamoto H., Shima T., Kumeta H., Kobashigawa Y., Akada R., Ohsumi Y., Inagaki F. Structural insights into Atg10-mediated formation of the autophagy-essential Atg12-Atg5 conjugate. Structure. 2012;20:1244–1254. doi: 10.1016/j.str.2012.04.018. [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Kakuta S., Watanabe T.M., Kitamura A., Sekito T., Kondo-Kakuta C., Ichikawa R., Kinjo M., Ohsumi Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J. Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Klionsky D.J. Eaten alive: a history of macroautophagy. Nat. Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh Y.Y., Shah K.H., Herman P.K. An Atg13 protein-mediated self-association of the Atg1 protein kinase is important for the induction of autophagy. J. Biol. Chem. 2011;286:28931–28939. doi: 10.1074/jbc.M111.250324. [DOI] [PMC free article] [PubMed] [Google Scholar]