Abstract
For six mouse olfactory receptor genes from family 262 which are expressed in clustered populations of olfactory sensory neurons, the genomic as well as cDNA structures were deciphered. All genes contained several exons which in some cases were alternatively spliced. Immediately upstream of the transcription start sites, sequence motif blocks were identified that are highly conserved among olfactory receptor (OR) genes which are expressed in clustered neuronal populations. By means of electrophoretic mobility shift assays, it was demonstrated that segments of the motif block region interact with proteins extracted from nuclear fractions of the olfactory epithelium. Yeast one-hybrid screenings of an olfactory cDNA library led to the identification of a set of transcription factors that specifically bind to particular elements of the motif block region. The identified factors can be categorized into two types: One group is known to be involved in transcriptional initiation, and the second group represents factors involved in pattern formations. The identified components may contribute to govern the precise topographic expression pattern of olfactory receptor genes.
The mammalian olfactory system is able to recognize and discriminate a vast array of odorous compounds. This enormous capacity of molecular recognition is thought to be based on the multiplicity and diversity of a G-protein coupled receptor family. The receptor types are encoded by a large gene family (Buck and Axel 1991) which in mammals (such as mouse) comprises more than a thousand genes (Zhang and Firestein 2002), and in lower vertebrates (e.g., fish) about a few dozen genes (Ngai et al. 1993). Genes encoding the olfactory receptors (ORs) are found on most chromosomes; they reside in a variety of small and large clusters which can span up to 2 Mb and can contain as many as 100 individual genes (Sullivan et al. 1996; Buettner et al. 1998; Rouquier et al. 1998; Glusman et al. 2000a, 2001; Xie et al. 2000; Lane et al. 2001; Young et al. 2002; Zhang and Firestein 2002; for recent reviews, see Mombaerts 1999a,b, 2001; Young and Trask 2002). Despite the large size of the OR gene superfamily, individual olfactory sensory neurons (OSNs) are likely to express only a single or at most a few receptor genes from the repertoire (Malnic et al. 1999; Rawson et al. 2000); moreover, only one of the two alleles appears to be expressed in each neuron (Chess et al. 1994). In the nasal epithelium of mammals, OSNs expressing the same receptor type generally are not randomly distributed throughout the olfactory epithelium; they are either restricted to one of several broad expression zones (Ressler et al. 1993; Vassar et al. 1993; Strotmann et al. 1994b) or, like the cells expressing a member of the OR37 subfamily, are clustered within a small central region of the epithelium (Strotmann et al. 1992; 1994a).
Very little is known about the molecular parameters and processes underlying the complex transcriptional regulation of olfactory receptor expression. A detailed characterization of those DNA sequences which are involved in governing the proper spatial and temporal expression pattern of a distinct OR gene is an essential prerequisite for unraveling the mechanisms controlling expression of OR genes. Candidate regions for regulatory elements are the noncoding sequences of OR genes, most notably those conserved in OR genes which share the same expression pattern. Analyses of a few OR genes have not led to a coherent picture (Asai et al. 1996; Glusman et al. 2000b; Lane et al. 2001); this may be due to the diversity of the compared genes. To overcome this problem, the OR37 subfamily could be a model case, because it represents a small and well circumscribed group of genes with a special expression pattern (Kubick et al. 1997; Strotmann et al. 1994a; Strotmann et al. 2000). The recent completion of genome sequencing has enabled us to characterize the complete repertoire of the OR37 receptor subfamily of the mouse (Hoppe et al. 2003). It was found that a total of eight OR37 genes exist in the mouse genome (according to the new nomenclature of Zhang and Firestein [2002], the mOR37 genes belong to family 262). These highly related genes are organized in two clusters, a feature which appears to be rather unique for olfactory receptor subfamilies. Cluster I consists of five genes, which share a very high level of sequence identity (around 90%; Hoppe et al. 2000). Cluster II consists of three genes with less sequence similarity (about 60%). Using a comparative approach, we previously identified highly conserved sequence motifs immediately upstream of the transcription start site for the genes from cluster I; these sequence motifs represent potential binding sites for transcription factors (Hoppe et al. 2000). Two additional OR genes (mOR262-4 and -10) which are located in cluster I were found to be expressed in the same clustered manner; they also share the conserved 5′-sequence motifs (Hoppe et al. 2000).
In the present study we examined the 5′ genomic regions of the OR genes from the second `OR37' cluster, in an effort to identify characteristic, putative regulatory sequence motifs. A representative sequence was then employed to search for distinct transcription factors that may bind to these DNA motifs in the 5′ noncoding region of the clustered OR genes.
RESULTS
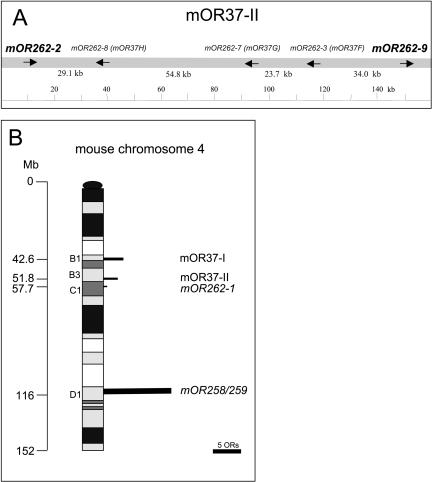
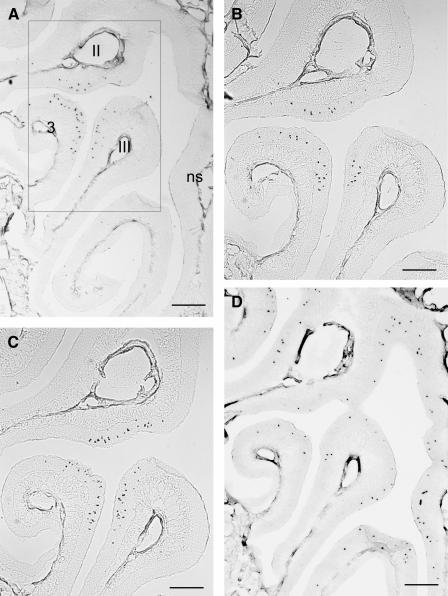
In order to analyze whether the `mOR37' genes in cluster II ('mOR37F', `mOR37G' and `mOR37H', named mOR262-3, mOR262-7, and mOR262-8, respectively, according to the nomenclature of Zhang and Firestein 2002) are colocalized with additional OR genes, the sequence 300 kb up- and downstream of the genomic locus was examined. The computer searches uncovered two OR genes (mOR262-2 and mOR262-9) flanking either side of the gene cluster (Fig. 1A). These two genes are also members of family 262, but both lack the `OR37'-characteristic insertion of six amino acids (Kubick et al. 1997) in the extracellular loop E3. The overall sequence identity of mOR262-2 and mOR262-9 with the `mOR37' genes is in the range between 53% and 59%, and thus they are considered members of different subfamilies. Assessing the sequence of an additional 300-kb region on either side uncovered no further OR genes, demonstrating that these five genes (mOR262-2, -3, -7, -8, and -9) form a distinct cluster. The question of whether the two genes mOR262-2 and -9 are expressed in clustered cell populations, like their relatives, was approached using in situ hybridization experiments. Coronal sections through the nasal cavity were incubated with specific antisense RNA probes. As shown in Figure 2A,B, cells expressing mOR262-2 or -9, respectively, are in fact located within the same small circumscribed region of the epithelium which was previously shown to be attributed to `mOR37' subtypes (Strotmann et al. 1992, 1994a). Cross-hybridizations can be excluded due to their low sequence identity and the hybridization conditions. Thus, all OR genes of chromosome 4 that have been analyzed to date are expressed in cell populations which are organized in a cluster.
Figure 1.
(A) Overview of the genomic organization of OR genes located at cluster mOR37-II (gene names are according to Zhang and Firestein [2002]; mOR37 notations are given in brackets). Genes marked in bold are receptors from family 262 (Zhang and Firestein 2002) which lack the insertion of amino acids in the E3 loop.Arrows indicate the transcriptional orientations.Intergenic distances are given below in kilobases; numbers are rounded off to the nearest decimal place.(B) Distribution of the genomic loci accommodating OR genes on mouse chromosome 4.Positions are given in megabases (Mb) and chromosomal bands.The length of bars represents the number of OR genes at each locus.
Figure 2.
In situ hybridizations of digoxigenin-labeled antisense riboprobes specific for representative OR genes from chromosome 4 on adjacent coronal sections through the mouse nasal cavity.Neurons expressing a gene from family 262 are detectable exclusively in three distinct areas of the sensory epithelium, located on endoturbinates II, III, and on ectoturbinate 3 (A,B,C).A representative gene from family 259 is distributed throughout the dorsal zone of the epithelium.(A) mOR262-2, (B) mOR262-9, (C) mOR262-1, (D) mOR259-9.Boxed area in (A) is shown in higher magnification in (B,C).ns, nasal septum.Scale bars = 500 μm in (A,D) and 200 μm in (B,C).
This raised the question of whether OR genes on this chromosome in general may be expressed in clustered cell populations. Chromosome 4 harbors two additional loci with OR genes: At position 57.7 Mb, there is a locus with only one gene (Fig. 1B); it turned out that this gene also belongs to family 262 (mOR262-1), but lacks the extended E3 domain. At a distance of 80 Mb, a fourth OR cluster is located comprising 16 genes (Zhang and Firestein 2002); they are all members of families 258 and 259 (Fig. 1B). In situ hybridization experiments demonstrated that mOR262-1 is indeed expressed in clustered neurons (Fig. 2C) which are located within the same central region of the nose as those from clusters I and II. In contrast, genes from the fourth locus were found to be expressed in zonal patterns, as shown for mOR259-9 in Figure 2D. Thus, three of the four loci on chromosome 4 accommodate OR genes with a clustered expression pattern.
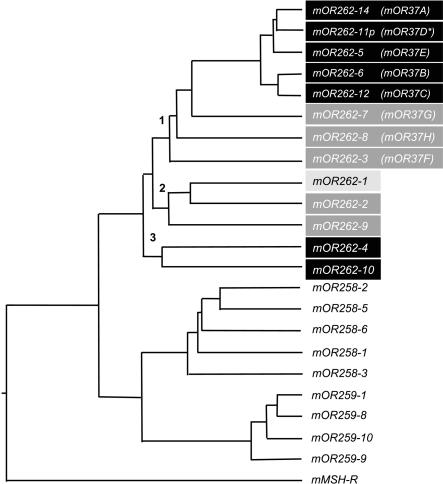
The relatedness of all genes on chromosome 4 was evaluated in more detail by means of phylogenetic analyses. As can be seen in Figure 3, all OR genes with a clustered expression pattern are closely related and not intermingled by OR genes with a zonal expression pattern. The genes with a clustered expression pattern can be categorized into three subgroups (1-3 in Fig. 3). All members of family 262 with an extended E3 loop ('mOR37' genes) are positioned on one branch (branch 1); their closest relatives are those family 262 members which are linked with cluster II as well as mOR262-1 (branch 2). The two OR genes associated with cluster I (branch 3) appear to be more distantly related.
Figure 3.
Phylogenetic tree based on the nucleotide sequences of representative OR genes located on chromosome 4.Family 262 is segregated on three different branches (1-3).Their cluster affiliation is indicated by shading: black, `mOR37' cluster I; medium gray, `mOR37' cluster II; light gray, single gene at position 57.7 Mb. The mouse melanocyte-stimulating hormone receptor (mMSH-R) was used as an outgroup.Pseudogenes are marked with an asterisk.Because the gene mOR37A was not given a new name in the study by Zhang and Firestein (2002), we named it mOR262-14.
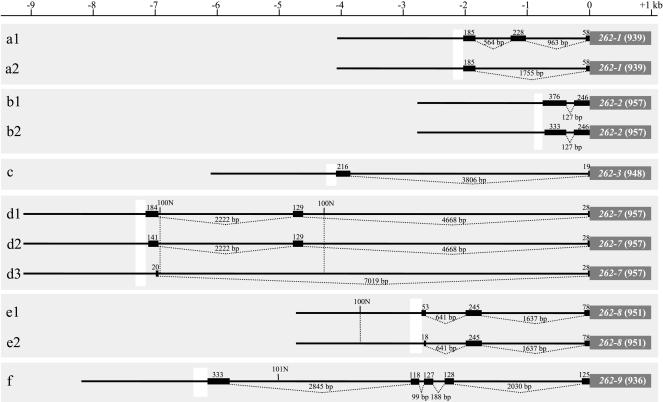
Our previous studies demonstrated that genes from cluster I (mOR262-4, -5, -6, 10, -11P, -12, and -14) share in their 5′- region a stretch of DNA with six well conserved motif blocks (Hoppe et al. 2000). These conserved sequences are located immediately upstream of their transcription start sites and are therefore considered putative regulatory elements. Consequently, the genes of cluster II were assessed for the 5′ noncoding sequences by RACE experiments and computer analyses. The emerging structures for the five genes in cluster II and mOR262-1 appeared to be rather variable (Fig. 4). The number of exons ranged from two to five; exons containing the coding sequence never contained any introns. The total length of introns was highly variable, ranging from 127 bp for mOR262-2 to 7019bp for mOR262-7. Except for mOR262-3 and mOR262-9, several splice variants were identified for each gene; do- nor and acceptor splice sites were found at the appropriate positions. The isoforms differed either in their usage of internal exons, for example, mOR262-1 (Fig. 4, a1, a2) or, in the majority of cases, in the length of their initial exon; the largest variety in length was found for the initial exon of mOR262-7 (Fig. 4, d1-d3). Pairwise comparisons of the noncoding exons revealed no significant homologies between the different genes (data not shown).
Figure 4.
Gene structures of mOR-262 genes from cluster II based on 5′ RACE experiments.All genes are aligned according to the initial methionine (position 0 kb).Boxes represent exons; lines below indicate intronic regions (lengths are given in kilobases).White blocks upstream of the initial exons mark the extension of conserved sequence stretches.Alternatively spliced isoforms are indicated by sequential numbers.Regions which are not yet sequenced are indicated with number of Ns.Numbers in brackets indicate the length of the coding sequence in bp.
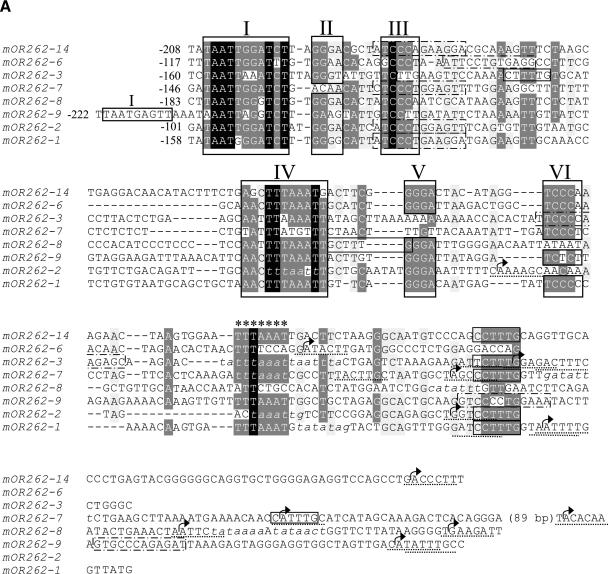
Determining the full extent of the mRNA allowed us to localize the transcription start site (TSS) for all cluster II genes and mOR262-1. Due to the highly variable gene structures, it was located between approximately 750 bp (mOR262-2) and 7200 bp (mOR262-7) upstream of the translation start. In order to identify putative expression control elements within this region, 1 kb upstream of the TSS was compared between the different genes using the programs T-COFFEE and Dialign2. Within this 1 kb, a stretch of about 170 bp was identified which shared a striking homology among the six genes (Fig. 5A). The sequence was located in close proximity (about 100-200 bp) upstream of the TSSs of the respective genes and was surrounded by DNA without any significant similarities (data not shown).
Figure 5.
(A) Nucleotide sequence alignment of the conserved sequences in the 5′ regions of OR genes expressed in clustered neuron populations. The alignment was generated using T-COFFEE and manually edited. Identical nucleotide positions in all sequences are shaded in black, highly conserved positions are shaded in gray. Numbered frames (I-VI) indicate characteristic sequence motif blocks; the repeated motif block I found in mOR262-9 is also boxed. Stars indicate an additional AT-rich stretch located close to the most upstream TSSs (arrows). Putative initiator elements are underlined; TATA boxes and TATA-box-like sequence stretches are indicated in italics. Olf-1-like sequence motifs are framed by dotted lines, E-boxes and E-box-like sequences by solid lines. The numbers indicate the distance in bp to the nearest TSS. (B) Array of the conserved sequence motifs identified in the 5′ region of clustered OR genes. Nucleotides spacing the sequence motifs are indicated by the double dotted lines. An X is included at positions where no motif block could be identified in one particular sequence, to uphold the conserved distance of the remaining blocks. The resulting consensus sequence of the conserved motif blocks I-VI is given below.
This 170-bp region was next examined for the characteristic sequence motifs, most notably such motifs which were previously identified in the putative promotor regions of OR genes in cluster I (Hoppe et al. 2000). An alignment of the conserved sequence region from all cluster II genes and mOR262-1 with that of representatives from cluster I (Fig. 5A) revealed a remarkable similarity which is particularly high within the motif block regions. The highest degree of conservation between all genes is found in motif blocks I, III, and IV. Based on the newly included genes, motif block I could even be extended by two nucleotides compared to the previous determination. Thus, all of the genes which are expressed in clustered neuron populations indeed share characteristic motif blocks in their 5′-upstream region.
In this comparative approach, an additional highly conserved segment emerged; it is located following the motif block region and consists of an AT-rich stretch (stars in Fig. 5A). Although this DNA segment is not a typical TATA box, its position relative to the transcription site, which is in the range of -20 to -40 for the genes mOR262-1, -2, -3, and -7, is the site where a TATA box is usually positioned. However, a “classical” TATA-box core sequence could only be identified in the putative promotor region of mOR262-1 and -8. All regions were examined for the presence of alternative motifs that may contribute to the initiation of transcription. This effort led to the identification of distinctive initiator elements for most analyzed genes including mOR262-6, -9, and -14 (Fig. 5A). Figure 5B shows the organization of the motif blocks for the various genes; it becomes obvious that not only the sequences of the motif blocks are conserved, but also their relative positions, especially within the first tandem of blocks (I-III). The distance between the blocks is slightly more variable in the second tandem (IV-VI).
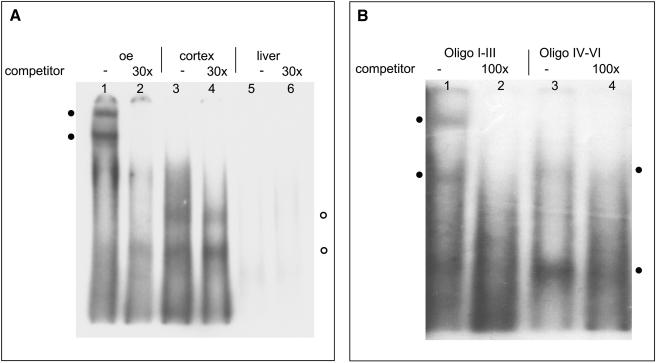
Database searches revealed that some of the conserved motifs represent consensus binding sequences for distinct transcription factors (Hoppe et al. 2000). However, it is not known whether they are relevant for governing the receptor expression, because knowledge of olfactory transcription factors is still very limited. In a first step to evaluate the relevance of the 5′-sequence motifs, we performed electrophoretic mobility shift assays (EMSAs) using the motif block region upstream of mOR262-6 as a representative sequence. A labeled double-stranded oligonucleotide containing the motif blocks of mOR262-6 was incubated with proteins extracted from nuclei isolated from the olfactory epithelium, as well as from cortex or liver for control. As shown in Figure 6A (lane 1), incubation of the oligonucleotide probe with protein extract from the olfactory epithelium resulted in two retarded bands of different size. In the presence of an excess of unlabeled oligonucleotide, which would compete for complex formation, no retarded bands were seen (Fig. 6A, lane 2). Proteins extracted from other tissues either bound, but could not be competed with excess of unlabeled oligos (Fig. 6A, lanes 3,4) or showed no binding (Fig. 6A, lanes 5,6). These results thus demonstrate that proteins from nuclei of cells from the olfactory epithelium interact with the 5′-DNA sequence of mOR262-6.
Figure 6.
(A) Characterization of protein interactions with sequence motif blocks I-VI from the putative promotor region of mOR262-6 using EMSAs. Each lane contains 10 fmol of 32P-labeled motif block DNA and was incubated with different nuclear extracts.(Lanes 1,2) 5 μg protein from olfactory epithelium; (lanes 3,4) 10 μg from cortex; (lanes 5,6) 20 μg from liver.Lanes (2,4,6) were pre-incubated with a 30-fold excess of unlabeled competitor DNA.(•) Shifted bands which are eliminated using the competitor DNA.(○) Bands which cannot be blocked by competitor.(B) Characterization of protein interactions with sequence motif blocks I-III (lanes 1,2) and IV-VI (lanes 3,4) from mOR262-6 using EMSAs.Each lane contains 10 fmol of 32P-labeled DNA and was incubated with 10 μg of nuclear extract from olfactory epithelium.Lanes (2,4) were pre-incubated with a 100-fold excess of unlabeled competitor DNA.(•) Shifted bands resulting from interactions of proteins with either motif blocks I-III (left) or motif blocks IV-VI (right).
Because the typical six motif blocks can be grouped into two pairs, which are organized in a tandem-like arrangement (see Fig. 5B; Hoppe et al. 2000), each subregion was next analyzed individually for its capability to interact with proteins from the olfactory epithelium. As shown in Figure 6B (lanes 1,2), incubation of nuclear proteins with motifs I-III again resulted in two bands; incubation with motifs IV-VI resulted in two bands as well; one of similar size as with motif I-III, but very weak and one significantly smaller (Fig. 6B, lanes 3,4). These results thus demonstrate that both repeats of the motif blocks bind proteins from the olfactory epithelium.
To determine the molecular identity of proteins that interact with these sites, we performed a yeast one-hybrid screen. The genetically modified yeast strain YM4271, which carries two copies of the motif block sequence of mOR262-6 upstream of the LacZ-reporter, was transformed with a cDNA library from the olfactory epithelium fused to the GAL4 activation domain. Two independent screening approaches led to 25 colonies, and six of them were confirmed as true positives by retransfection (Table 1).
Table 1.
Transcription Factors Identified in Yeast One-Hybrid Screen Interacting With the Putative mOR262-6 Promotor Region and Distinct Motif Block Regions
|
β-galactosidase activity
|
||||
|---|---|---|---|---|
| mOR262-6 complete | I + II | II + III | V + VI | |
| Ptx-1 | +++ | — | + | + |
| BEN | ++ | — | — | ++ |
| Olf-1/EBF-like-2 (OS) | ++ | — | — | — |
| Alx3 | + | +++ | — | — |
| LH-2 | + | + | — | — |
| AP-2β | + | — | + | — |
Sequencing and subsequent BLAST searches revealed that all six genes were coding for proteins which represent distinct transcription factors. The proteins encoded by the genes were classified as strong to weak activators of gene transcription on the basis of β-galactosidase reaction intensity. The strongest activation was observed for a clone encoding the `pituitary homeobox 1-factor' (Ptx-1; Drouin et al. 1998). Only slightly weaker staining was obtained with the `binding factor for early enhancer' (BEN; Bayarsaihan and Ruddle 2000) and the `olfactory neuron specific factor 1/early B-cell factor-like-2' (O/E-2(0S); Wang et al. 1997). The `aristaless-like homeobox 3' (Alx-3; ten Berge et al. 1998), the `limhomeobox-2 factor' (LH2/Lhx2; Xu et al. 1993), and the `activator protein 2β' (AP-2β; Moser et al. 1995) were also strong activators.
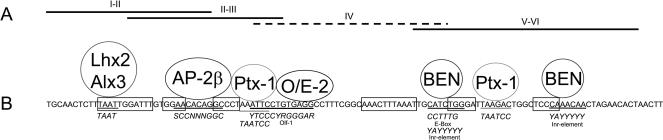
To characterize the protein-DNA interaction of these factors in more detail, the motif block region was divided into subregions (Fig. 7A), and corresponding yeast strains were generated. The strain containing motif block IV was unemployable due to strong self-activation. The six factors which interacted with the complete motif block region were retransfected into these strains. Most factors displayed a strong activation of the reporter gene when interacting with a particular subregion (Table 1), and interestingly, the binding of Alx-3 to the subregion containing motif blocks I+II led to a significantly stronger reaction compared with the complete motif block region. In contrast to all other factors, O/E-2(0S) only binds to the complete motif block region or to subregion IV.
Figure 7.
(A) Representation of oligonucleotides spanning the different segments of the motif blocks I-VI which were used in the yeast one-hybrid studies.The oligonucleotide containing motif block IV (dotted line) could not be utilized due to auto-activation.(B) Scheme of interactions for distinct transcription factors with the motif block region from mOR262-6.The positions of the respective motif blocks are indicated by boxes.The transcription factors are positioned at sites of their most likely interaction (underlined sequence).The consensus binding sequences from the literature are indicated below the motif block region in italics.
To define the putative interaction site for the identified transcription factors, the sequence of the mOR262-6 motif block region was assessed for stretches related to proposed consensus binding motifs for the identified factors. In fact, for each of the transcription factors, a consensus-related sequence motif could be identified in the particular subregion (Fig. 7B). For O/E-2(0S) it indeed turns out that subregion IV (Fig. 7B) contains such a consensus-related sequence motif for this factor. A comparison of the motif block regions of all clustered genes led to the identification of a consensus site for O/E-type transcription factors close to motif block III or VI, in most of the genes (Figs. 5A, 7B). For Alx-3 and Lhx-2, corresponding sites were identified in motif block I from each gene. Initiator elements or E-boxes which represent binding sites for BEN were present close to the TSS of each gene (Fig. 5A). In addition, consensus-related sites for AP-2β and Ptx-1 were found in each motif block region, although at slightly different positions (data not shown). Altogether, these data suggest that the appropriate factors can indeed interact with distinct positions of the motif block region.
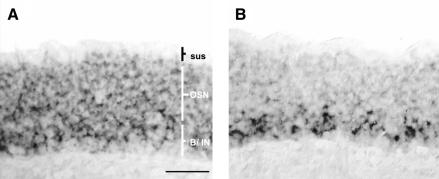
To localize and identify the cells in the olfactory epithelium which express the corresponding transcription factors, we performed in situ hybridization experiments, probing coronal sections through the nasal epithelium with gene-specific antisense RNA. For two of the factors, reactive cells could be visualized. The probe for O/E-2(0S) labeled cells throughout the layer of olfactory sensory neurons and basal cells, but was absent from the most apical layer, which contains the sustentacular cells (Fig. 8A). In contrast, LH-2-expressing cells were localized only in the deep layer of the epithelium (Fig. 8B), where basal cells and immature olfactory neurons are positioned. Cells reactive to both probes were not spatially restricted to a particular region of the nasal epithelium; rather they were found to be distributed throughout all turbinate structures.
Figure 8.
Expression patterns of O/E-2 (A) and LH-2 (B) in the olfactory epithelium revealed by in situ hybridization.Coronal sections were incubated with digoxigenin-labeled antisense riboprobes.The O/E-2 message is found in the layer of olfactory sensory neurons and basal cells. O/E-2 expression is absent from the sustentacular cell layer. LH-2-expressing cells are restricted to the deep layer of the olfactory epithelium, close to the basal membrane.sus, sustentacular cell layer; OSN, olfactory sensory neuron layer; B/IN, layer of basal cells and immature olfactory neurons.Scale bar, 50 μm.
DISCUSSION
In the present study we explored the genomic features of OR genes forming a small, highly related receptor family. Several conserved sequence motifs were identified upstream of the transcription start sites; these motifs are considered putative regulatory elements. We found that the region containing these elements in one representative gene in fact binds distinct transcription factors; moreover, we demonstrated that these factors are expressed in cells of the olfactory epithelium. These results suggest that at least some of the identified DNA motifs may represent important elements of the promotor region involved in the transcriptional control of these genes.
The identification of putative control elements immediately upstream of the transcription start is consistent with previous observations, suggesting that transcription of OR genes is initiated from a region up to 6.7 kb upstream of the initial exon (Asai et al. 1996; Qasba and Reed 1998). Recent studies using transgenic approaches even narrowed the region down to a few hundred basepairs and led to the concept that the control regions of OR genes may be compact (Vassalli et al. 2002).
The sequence motifs identified in this study share very high homology with those at the TSSs of previously documented clustered OR genes (Hoppe et al. 2000). The high degree of conservation for the motif blocks is in sharp contrast to the adjacent noncoding regions, which display virtually no sequence homology (Hoppe et al. 2000); these results are indicative of a strong negative selection pressure, acting on the conserved sequence motifs, and they suggest an important functional role of these elements.
The existence of highly conserved sequence motifs in the putative promotor region of the mOR262 genes seems to be in contrast to other OR genes; previous studies led to the identification of short and very diverse elements within the noncoding sequences of OR genes (Glusman et al. 2000b; Lane et al. 2001). Therefore, it was hypothesized that each OR gene may bear an individual transcriptional signature. The existence of highly conserved 5′ sequence motifs in all genes which are expressed in clustered neuron populations may indicate that these genes are controlled by other regulatory mechanisms than for those genes which are expressed in zonal patterns.
It has been hypothesized that distinct levels of control are required for a given neuron to choose one gene from the thousand available, and that this could be mediated by a combination of transcription factors (Chess 1998). The results of our gel-shift experiments and yeast one-hybrid studies demonstrate that the sequence upstream of the TSS of a clustered gene which contains the motif blocks indeed interacts with several transcription factors. Five of these identified factors had not previously been implicated in olfactory gene transcription. It was found that the proteins specifically bind to different subregions of the putative promotor region; thus it is conceivable that several of them may in a concerted action contribute to govern the temporal and topographical expression pattern of an OR gene.
On a first level of organization, the expression of OR genes in sensory neurons of the nasal neuroepithelium is believed to be governed by tissue-specific transcription factors (Reed 2000). Regarding the tissue-specific expression of the clustered OR genes, the interaction of O/E-2(0S) with the motif block region could be of particular relevance. O/E-2(0S) is a member of a small subfamily of “repeat helix-loop-helix” transcription factors which were originally identified by binding to two characteristic sequence motifs in the 5′ noncoding region of the OMP gene, a gene which is exclusively expressed in OSNs (Kudrycki et al. 1993). The socalled Olf-1 binding site was identified in the 5′ noncoding regions of several olfactory specific genes, such as Golf and adenylate cyclase type III (ACIII) (Wang and Reed 1993), leading to the hypothesis that the Olf-1 site plays a central role in governing the “olfactory phenotype” (Buiakova et al. 1994; Davis and Reed 1996) and to the identification of the sequence `YTCCCYRGG GAR' as consensus Olf-binding motif. Surprisingly, the motif block regions of clustered OR genes do not contain typical Olfconsensi, although an O/E-2(0S) factor interaction with that region was demonstrated for mOR262-6 in the present study. Previous studies indicated that the occurrence of strong Olf-consensi may indeed not be the case for many OR genes (Glusman et al. 1996, 2000b; Tsuboi et al. 1999). Our findings suggest that the factor O/E-2(0S) interacts with structurally different binding sites as well. It may be relevant in this context that important segments of the Olf-motif, such as a well conserved `CCC' halfside (Kudrycki et al. 1993) are present even in multiple copies within the putative promotor region of some of the clustered genes. In any case, the strong reporter gene activation of O/E-2(0S) suggests that it may be particularly important for controlling the expression of the clustered OR genes. This view is supported by the findings that in adult mice, O/E-2 is exclusively expressed in the olfactory epithelium, whereas other members of this transcription factor subfamily are found in a variety of tissues (Hagman et al. 1993; Wang et al. 1997).
Regarding the special topographic pattern of mOR262-expressing neurons, the identification of several homeobox-type transcription factors (Ptx-1, Alx-3, Lhx-2/LH-2) could be of particular relevance, because factors of this type are generally involved in pattern formation. In this context, the most likely interaction of LH-2 with motif block I is of particular interest; motif block I shows a striking homology to the so-called `rhodopsin-conserved-sequence-I' (RCSI; Papatsenko et al. 2001), which binds the homeobox factor Pax-6, a central element in the pattern formation during eye development in Drosophila and also in vertebrates (Sheng et al. 1997; Czerny et al. 1999; Hauck et al. 1999; Gehring 2002). Factors homologous to LH-2 are involved in pattern formations in Drosophila, for example, during wing formation (Rincon-Limas et al. 1999). Previous studies have shown that in mouse, LH-2 is exclusively expressed in neuronal cell populations (Xu et al. 1993). In Drosophila, an RCSI element together with a TATA box is responsible for the expression of rhodopsin genes in photoreceptor cells, whereas sequences further upstream determine the cell type in which a distinct rhodopsin isoform is expressed (Papatsenko et al. 1997). Based on the homology between motif block I and RCSI, it seems conceivable that a similar mechanism may be involved in the expression control of the clustered OR genes. Although a typical TATA box is missing in the putative promotor region of most of the OR genes, a so-called “initiator element” is located at or near the TSSs of all clustered genes. Such pyrimidine-rich sequences have been described as being used for generating the initiation complex in TATA-less promotors (Smale and Baltimore 1989). These sequences bind transcription factors of the TFII-I family; interestingly, BEN, a factor belonging to this family of transcription factors, was isolated in our yeast one-hybrid experiments and was shown to bind to the motif block region containing the corresponding consensus-like binding motif. It thus seems possible that this factor in concert with a homeobox factor may indeed be instrumental in governing the gene expression of OR genes in clustered neuron populations.
The factors identified in this study represent the first set of molecularly defined transcription factors which interact with a putative control region of OR genes; they may represent essential components for the cis-element-mediated regulation of OR gene expression.
METHODS
In situ Hybridization
Specific antisense riboprobes for OR genes and transcription factors were generated using the following primer combinations:
mOR262-1s: 5′-GAGGAAATGCCAGGAGAAAATGTCAC-3′ /
mOR262-1as: 5′-CATCCTTGGGAAAGTCATGGG-3′
mOR262-2s: 5′-GGGAAATTGGACCTCTGTCAC-3′ /
mOR262-2as: 5′-AGTTAAGATAGCACTGAAGGGG-3′
mOR262-3s: 5′-ATCGGACTTATTTGGAGG-3′ /
mOR262-3as: 5′-CCATCATTTGCCTAAGAGTC-3′
mOR262-7s: 5′-GGCGGGCATCCTCCTATGG-3′ /
mOR262-7as: 5′-CACTTACATCTTACTCAACG-3′
mOR262-8s: 5′-GATGGCTGACCGTGGATGAC-3′ /
mOR262-8as: 5′-CTGATGTCTCTAAGGAACG-3′
mOR262-9s: 5′-GGTGAGCTGAGTAGGTGAAG-3′ /
mOR262-9as: 5′-GCCTTTCAGATTTTCCGCAG-3′
mOR259-9s: 5′-TTCCCTTTCTGCCTCTATCG-3′ /
mOR259-9as: 5′-TGAATACTCCAGCTTCCCA-3′
Lhx-2s: 5′-ATCACCTCAACTGCTTCA-3′ /
Lhx-2as: 5′-AGCGTGGCATCCGAAGTC-3′
AP-2βs: 5′-GCGTCCCAAGCCATAGC-3′ /
AP-2βas: 5′-AATAGGTTCATGCCGCTG-3′
BENs: 5′-CCAGCAGAATATGACCCC-3′ /
BENas: 5′-CAGAGGTAGGTGAAGGATC-3′
O/E-2(OS)s: 5′-CGCTTTCGGCTGGGAGTC-3′ /
O/E-2(OS)as: 5′-TTGAATCTATGAGGCGGAC-3′
Ptx-1s: 5′-GCCCTTTGCGGTCCCCAC-3′ /
Ptx-1as: 5′-GTGCTGCTTGGACTTGAG-3′
The PCR reactions were performed with mouse genomic DNA as template in a Peltier PTC 200 Thermocycler (MJ Research) with the following time and temperature schedule: An initial 4-min denaturation at 96°C was followed by 35 cycles of denaturation at 96°C (1 min), annealing at 60°C (1 min), extension at 72°C (1 min), and a final extension at 72°C for 10 min; MasterTaq polymerase from Eppendorf was used for all amplifications. The PCR products were size-fractioned on agarose gels and cloned using the pGEM-T vector system according to the manufacturer's specifications (Promega). To remove the vector backbone prior to in vitro transcription, PCR reactions with T7out (5′-AATTGTAATACGACTCACTATAGG-3′) and SP6out (5′-CCAAGCTATTTAGGTGACACTATA-3′) primers were performed under the same conditions as above, except for primer annealing at 55°C. After separation of PCR products by agarose gel electrophoresis and purification using the E.Z.N.A. gel extraction kit (PEQLab), 250 ng of DNA was used for in vitro transcription.
Mice at 6-8-wks of age were killed by CO2 asphyxiation and decapitated. The lower jaw and top of the skull were removed. The tissue was embedded in tissue freezing medium and frozen on dry ice. Twelve-μm coronal sections through the nasal cavity were cut on a Jung CM 3000 cryostat (Leica), adhered to Starfrost microslides (Fisher), and fixed in 4% paraformaldehyde in 0.1 M NaHCO3 for 10 min. Hybridization conditions, posthybridization washes, and detection were as described (Strotmann et al. 1994b).
5′ RACE
To analyze the cDNA structure of the receptor genes, total RNA was extracted from mouse tissue using TRIZOL reagent (Life Technologies), chloroform-extracted, and isopropanol-precipitated. Total RNA was resuspended in water; to exclude contamination with genomic DNA, it was incubated with 3U of FPLC-pure DNAseI (Pharmacia) for 30 min at 37°C in a 20 μL volume of 40 mM Tris-HCl, pH 7.4, 6 mM MgCl2. Subsequently, DNAse was inactivated by incubation for 10 min at 70°C. For the full-length amplification of 5′-cDNA ends of the receptor genes, the GeneRacer kit (Invitrogen) was employed according to the manufacturer's specifications. The gene-specific primers were:
mOR262-1Race: 5′-ATGGGGGTTTGAAGGCGTGAATC-3′
mOR262-1Racenes: 5′-CAGAACAATCTCTAACCTCGGG TAC-3′
mOR262-2Race: 5′-GCTGAGGAAAAAGTACATGGGAG TG-3′
mOR262-2Racenes: 5′-CATCAGCAGACACAGCACAAA TACGG-3′
mOR262-3Race: 5′-GTGTGCAGGTGAGAGTCGAAAAT AC-3′
mOR262-3Racenes: 5′-GCCATTGCCCACTAGGATAAC TG-3′
mOR262-7Race: 5′-TGGATGACTACTGCGATAAGAACG-3′
mOR262-7Racenes: 5′-GAAAGCCCCACCAAGAGAAATTC TGTC-3′
mOR262-8Race: 5′-GTGAAGACGGGAATCAAAGATGCT TGC-3′
mOR262-8Racenes: 5′-GTATCCAGCAAGACCCAAAA GAAGG-3′
mOR262-9Race: 5′-GATAATGAGCAGACTGTTTCCCAGG-3′
mOR262-9Racenes: 5′-GAGGCAGAGCACAAATAG GAAAAGC-3′
For the amplification, 1 μL cDNA was employed as template in a first amplification round. PCR was performed on an MJResearch Peltier Thermocycler PTC 200 with the following temperature profile: 1 min 40 sec 94°C, 19 cycles of 30 sec 94°C, 40 sec 70°C-0.5°C/cycle, 1 min 30 sec 68°C, and an additional 25 cycles at the same conditions as above except for an annealing at 60°C. The reaction was completed by a final extension of 10 min at 68°C. For these PCR reactions 0.4 μL Titanium Taq DNA Polymerase (BD Biosciences Clontech) was used in a total 50μL reaction cocktail containing 32 ng/μL bovine serum albumin; 10 μM of each primer, 10 μM d-NTPs each, and 1× concentrated Titanium Taq buffer. Then, 1 μL of reaction products was used in a second-round PCR using the nested primers under the following PCR conditions: 2 min 94°C, 25 cycles of 30 sec at 94°C, 30 sec at 65°C, and 2 min at 68°C; the reaction was completed by a final extension of 10 min at 68°C. The reaction mixture was the same as described above except for not using BSA.
PCR products were subcloned into pGEM-T vector (Promega) and sequenced using TaqI polymerase and fluorescently labeled dideoxy-terminator chemistry on an automated sequencer (model 310, Applied Biosystems).
Electrophoretic Mobility Shift Assays (EMSAs)
Nuclear extracts from mouse tissues were prepared essentially as described with minor modifications (Dignam et al. 1983; Kudrycki et al. 1993). Briefly, the tissues were hand homogenized with 4 volumes of homogenization buffer [0.25 M sucrose, 15 mM Tris-HCl, pH 7.9, 60 mM KCl, 15 mM NaCl, 5 mM EDTA, 1 mM ethylene glycolbis (β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA), 0.15 mM spermine, 0.5 mM spermidine, 1 mM dithiothreitol (DTT), 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 2 μg of leupeptin per mL, 0.015 TIU of aprotinin per mL] in a Kontes all-glass homogenizer and centrifuged at 1000g. The pellets were resuspended in 4 tissue volumes of buffer A [10 mM N-2-hydroxyethylpiperazine-N′-2 ethenesulfonic acid (HEPES; pH 7.9), 1.5 mM MgCl2,10 mM KCl, 0.5 mM DTT, 0.05 mM PMSF, 1.25 μg of leupeptin per mL, 0.006 TIU of aprotinin per mL] and centrifuged at 2500g. The pellets were resuspended in 4 volumes of buffer A, homogenized after 10 min incubation on ice as before, and centrifuged at 25,000g. The pelleted nuclei were resuspended in 1 tissue volume of 1 M KCl mixed with 1 tissue volume of buffer C (20 mM HEPES, pH 7.9, 25% glycerol, 1.5 mM MgCl2, 1 mM EDTA, and containing DTT, PMSF, leupeptin, and aprotinin at the same concentration as in the homogenization buffer), mixed by pipetting up and down, and lysed by 30 min of rocking. The lysate was centrifuged at 25,000g, and the supernatant was dialyzed against dialysis buffer (10 mM Tris-HCl, pH 7.9, 1 mM EDTA, 5 mM MgCl2, 50 mM KCl, 10% glycerol, 3 mM DTT, 0.3 mM PMSF), and aliquots were stored at -80°C after a final step of centrifugation at 25,000g for 10 min.
Preparation of double-stranded radiolabeled DNA was done either by PCR or annealing of sticky-end-generating oligonucleotides. The mOR262-6 motif block region was amplified from an appropriate BAC clone using the primer combination 5′-CGGAATTCAGACTCACAGAGAAGCGTC-3′ and 5′-CG AAGCTTGTTAGTGTTCTAGTTGTTTGGG-3′ and prepared by HindIII and EcoRI restriction endonuclease digestion of the cloned fragment out of pGEMT-vector. The following oligonucleotides were used for annealing reactions: 5 ′ -TCTGCAACTCTTTAATTGGATTTGTGGAACACAGGC CCTAAATTCC-3′ and 5′-TGTGGAATTTAGGGCCTGTGTTCCA CAAATCCAATTAAAGAGTTGC-3′ containing motif blocks I-III and 5′-TTTCGGCAAACTTTAAATTGCATCTGGGATTAA GACTGGCTCCCAAACAACTAG-3′ and 5′-TTCTAGTTGTTTGG GAGCCAGTCTTAATCCCAGATGCAATTTAAAGTTTGCCG-3′ containing motif blocks IV-VI. Annealing was done in a 20-μL reaction containing 1 μg of each oligonucleotide for 3 min at 65°C, 5 min at 37°C, and 10 min at room temperature. 5′ overhangs of all produced double-stranded DNA fragments were used for filling up 200-400 ng DNA by Klenow polymerase in a total volume of 20 μL containing 5 mM dTTP, dCTP, dGTP, 20 μCi [α-32P] dATP, and 1 U of Klenow enzyme for 30 min at 30°C. Reactions were stopped by adding 0.2 M EDTA, pH 7.5; excess nucleotides were removed with Micro Bio-Spin P-30 columns (Bio-Rad) according to the manufacturer's specifications.
Reaction mixtures (20-45 μL total) containing 10 mM Tris-HCl, pH 7.9, 30 mM NaCl, 7% glycerol, 4.3 mM MgCl2, 1 mM EDTA, pH 8.0, 0.18 mM PMSF, 1.8 mM DTT, 1.8 mg of aprotinin per mL, 1.2 mg of leupeptin per mL, 5 μg of poly(dI-dC), and 5-20 μg of protein extract were incubated on ice in the presence of 10 fmol double-stranded radiolabeled DNA. In the competition experiments, the appropriate DNA fragments were preincubated on ice with the binding reaction mixtures 10 min prior to addition of the 32P-labeled probe. After a 20-min incubation, the reaction mixtures were mixed with the loading buffer and directly subjected to electrophoresis on 5% polyacrylamide gels (29:1, acrylamide-bisacrylamide). Before loading the gel, a 1-h 15-min prerun at 120 V and 20 mA was carried out; the dried gel was analyzed by autoradiography.
Yeast One-Hybrid Screening
For the yeast one-hybrid screening, the Matchmaker One-Hybrid system (Clontech) was used according to the manufacturer's specifications. The bait plasmid `pLacZi-motif blocks' was constructed by ligation of three tandem copies of double-stranded oligonucleotides generated by PCR, each containing the entire motif block region from mOR262-6. For amplification of the motif block region, the following primer-combinations were used:
mOR262-6 EcoRI s: 5′-CGGAATTCAGACTCACAGA GAAGCGTC-3′ /
mOR262-6 SpeI as: 5′-CGACTAGTGTTAGTGTTCTAGTT GTTTGGG-3′
mOR262-6 SpeI s: 5′-CGACTAGTAGACTCACAGAGAAGC GTC-3′ /
mOR262-6 HindIII as: 5′-CGAAGCTTGTTAGTGTTCTAGT TGTTTGGG-3′
mOR262-6 HindIII s: 5′-CGAAGCTTAGACTCACAGAAGC GTC-3′ /
mOR262-6 XhoI as: 5′-CGCTCGAGGTTAGTGTTCTAGTT GTTTGGG-3′
Cloned PCR products were isolated from pGEM-T by using the appropriate restriction digestion enzyme combination, and purified DNA was then cloned into an EcoRI/XhoI-digested pLacZi Vector (Clontech). For generating the `bait'-plasmids `pLacZi-motif block I+II, II+III, IV, and V+VI' containing two tandem copies of distinct motif blocks, the following synthesized oligonucleotides were cloned into pLacZi EcoRV/XhoI vectors after annealing to each other as described above:
5′-AATTC[TGCAACTCTTTAATTGGATTTGTGGAAC ACAG]2C-3′ /
5′-TCGAG[CTGTGTTCCACAAATCCAATTAAAGAGT TGCA]2G-3′
5′-AATTC[GATTTGTGGAACACAGGCCCTAAAATT CCTG]2C-3′ /
5′-TCGAG[CAGGAATTTTAGGGCCTGTGTTCCACAAA TC]2G-3′
5′-AATTC[TTCCTGTGAGGCCTTTCGGCAAACTTTAAAT TGCATCT]2C-3′ /
5′-TCGAG[AGATGCAATTTAAAGTTTGCCGAAAGGCC TCACAGGAA]2G-3′
5′-AATTC[TGCATCTGGGATTAAGACTGGCTCCCAAAC AACTA]2C-3′ /
5′-TCGAG[TAGTTGTTTGGGAGCCAGTCTTAATCCCA GATGCA]2G-3′
One μg of the different bait plasmids was linearized with NcoI at the URA3 locus and integrated into the yeast genome of the strain YM4271 (Liu et al. 1993; Wilson et al. 1991) by homologous recombination according to the manufacturer's (Clontech) specifications.
The solid-state amplification of rat olfactory epithelium MATCHMAKER cDNA library (Clontech) was done with 1.5 × 106 cfu put in 1 L 2× LB-medium containing 0.3% SeaPrep Ultralow-Gelling Soft-Agarose (FMC BioProducts) and ampicillin (100 μg/mL) at a temperature of 20°-25°C. One hundred-μL aliquots of well mixed medium were plated on LBamp-plates for DNA concentration calculation. Twenty-mL aliquots of medium were pipetted in precooled tubes and incubated for 60 min on ice until gelling of the medium. After a 30-h incubation at 37°C, the medium was centrifuged for 5 min at 8000 rpm at room temperature twice. For plasmid isolation, the `plasmid-Giga-Kit' (QIAGEN) was used. The bacterial pellet was resuspended in 125 mL buffer I (10 mM EDTA, 100 μg/mL RNAse A, 50 mM Tris-HCl, pH 8.0), mixed with 125 mL buffer II (200 mM NaOH, 1% SDS), and incubated at room temperature for 5 min and mixed well with 125 mL of buffer III (2.55 M Kac, pH 4.8). Immediately after a 30-min incubation on ice and centrifugation at 18,000 rpm at 4°C, the supernatant was centrifuged again under the same conditions and then put on a Qiagen-Tip 10000 that was equilibrated with 754 mL of QBT-buffer (750 mM NaCl, 50 mM MOPS, pH 7.0, 1,15% Triton X-100, 15% ethanol) before use. Subsequently the column was washed twice with QC-buffer (1 M NaCl, 50 mM MOPS, pH 7.0, 15% ethanol), and the DNA was then eluated with 75 mL QF-buffer (1.25 M NaCl, 50 mM Tris-HCl, pH 8.5, 15% ethanol). Precipitated plasmid DNA (adding 0.7 vol isopropanol) was pelleted for 30 min at 4°C at 14,000 rpm. After subsequent washing (10 mL 70% ethanol), 5 mL TE-buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA) was added to the dried DNA.
The large-scale transformation with the rat olfactory epithelium MATCHMAKER cDNA library (Clontech) was carried out according to the manufacturer's specifications using 2 mg denatured salmon sperm DNA and 20 μg amplified library DNA.
Colony-lift filter assays were carried out on nitrocellulose membranes (Schleicher and Schüll, 0.025 μm) according to the `Yeast Protocols Handbook' (#PT3024-1 version #PR91200) from Clontech Laboratories.
Plasmids were isolated from yeast clones by a method modified by Kaiser and Auer (1993). Two to three colonies were incubated in 5 mL of appropriate medium at 30°C for 48 to 64 h at 200 rpm. Yeast cells were resuspended in 1 mL of water subsequent to pelleting for 7 min at 3000 rpm and then mixed with 100 μL of glass beads (Sigma); after a 30-sec centrifugation and removal of the supernatant, 50 μL TE/SDS (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1% SDS) and 100 μL phenol/chloroform/isoamylalcohol (25:24:1) were added. Cracking of the yeast cells was performed by 10× alternate vortexing and cooling of the samples on ice for 30 sec. A 5-min centrifugation at 4°C at 14,000 rpm was followed by a purifying dialysis of the supernatant against double-distilled water.
Sequence Analyses
The chromosomal positions of the mouse OR genes were mapped using the Ensembl BLAST server (http://www.ensembl.org/; version March 3rd, 2003). More detailed analyses of the `mOR37-II' cluster organization and gene structures were subsequently performed using Genamics Expression (program version 1.080). Phylogenetic analyses were performed using the ClustalV method (Higgins and Sharp 1989) for sequence alignment; calculations of phylogenetic trees were completed with the neighbor-joining algorithm in MEGALIGN (DNAStar). The comparative analyses of promotor regions of OR genes analyzed in this study are based on the programs T-COFFEE (http://www.ch.embnet.org/software/Tcoffee.html) and Dialign2 (http://bioweb.pasteur.fr/seqanal/interfaces/dialign2.html).
Nomenclature of OR Genes
According to the mOR gene nomenclature of Zhang and Firestein (2002), the `mOR37' genes analyzed here were given the corresponding names: mOR37F (mOR262-3), mOR37G (mOR262-7), mOR37H (mOR262-8). In consideration of the similar special structural features of these genes compared to formerly described genes (Strotmann et al. 1999), we kept the `old' nomenclature also for these OR genes. Formerly described `OR37' genes and `OR37' related genes of cluster mOR37-I on mouse chromosome 4 now are named: mOR262-6 (mOR37B), mOR262-12 (mOR37C), mOR262-11-p (mOR37D), mOR262-5 (mOR37E), mOR262-4 (mOR17), mOR262-10 (mOR6). Because the gene mOR37A was not given a new name in the study by Zhang and Firestein (2002), we named it mOR262-14.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft Leibniz Programm and Sonderforschungsbereich (SFB) 495. We thank Tom Lambert for critically reading the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1372203.
References
- Asai, H., Kasai, H., Matsuda, Y., Yamazaki, N., Nagawa, F., Sakano, H., and Tsuboi, A. 1996. Genomic structure and transcription of a murine odorant receptor gene: Differential initiation of transcription in the olfactory and testicular cells. Biochem. Biophys. Res. Commun. 221: 240-247. [DOI] [PubMed] [Google Scholar]
- Bayarsaihan, D. and Ruddle, F.H. 2000. Isolation and characterization of BEN, a member of the TFII-I family of DNA-binding proteins containing distinct helix-loop-helix domains. Proc. Natl. Acad. Sci. 97: 7342-7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck, L. and Axel, R. 1991. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell 65: 175-187. [DOI] [PubMed] [Google Scholar]
- Buettner, J.A., Glusman, G., Ben-Arie, N., Ramos, P., Lancet, D., and Evans, G.A. 1998. Organization and evolution of olfactory receptor genes on human chromosome 11. Genomics 53: 56-68. [DOI] [PubMed] [Google Scholar]
- Buiakova, O.I., Krishna, N.S., Getchell, T.V., and Margolis, F.L. 1994. Human and rodent OMP genes: Conservation of structural and regulatory motifs and cellular localization. Genomics 20: 452-462. [DOI] [PubMed] [Google Scholar]
- Chess, A. 1998. Olfactory receptor gene regulation. Adv. Immunol. 69: 437-447. [DOI] [PubMed] [Google Scholar]
- Chess, A., Simon, I., Cedar, H., and Axel, R. 1994. Allelic inactivation regulates olfactory receptor gene expression. Cell 78: 823-834. [DOI] [PubMed] [Google Scholar]
- Czerny, T., Halder, G., Kloter, U., Souabni, A., Gehring, W.J., and Busslinger, M. 1999. Twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol. Cell 3: 297-307. [DOI] [PubMed] [Google Scholar]
- Davis, J.A. and Reed, R.R. 1996. Role of Olf-1 and Pax-6 transcription factors in neurodevelopment. J. Neurosci. 16: 5082-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam, J.D., Lebovitz, R.M., and Roeder, R.G. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11: 1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin, J., Lamolet, B., Lamonerie, T., Lanctot, C., and Tremblay, J.J. 1998. The PTX family of homeodomain transcription factors during pituitary developments. Mol. Cell Endocrinol. 140: 31-36. [DOI] [PubMed] [Google Scholar]
- Gehring, W.J. 2002. The genetic control of eye development and its implications for the evolution of the various eye-types. Int. J. Dev. Biol. 46: 65-73. [PubMed] [Google Scholar]
- Glusman, G., Clifton, S., Roe, B., and Lancet, D. 1996. Sequence analysis in the olfactory receptor gene cluster on human chromosome 17: Recombinatorial events affecting receptor diversity. Genomics 37: 147-160. [DOI] [PubMed] [Google Scholar]
- Glusman, G., Bahar, A., Sharon, D., Pilpel, Y., White, J., and Lancet, D. 2000a. The olfactory receptor gene superfamily: Data mining, classification, and nomenclature. Mamm. Genome 11: 1016-1023. [DOI] [PubMed] [Google Scholar]
- Glusman, G., Sosinsky, A., Ben Asher, E., Avidan, N., Sonkin, D., Bahar, A., Rosenthal, A., Clifton, S., Roe, B., Ferraz, C., et al. 2000b. Sequence, structure, and evolution of a complete human olfactory receptor gene cluster. Genomics 63: 227-245. [DOI] [PubMed] [Google Scholar]
- Glusman, G., Yanai, I., Rubin, I., and Lancet, D. 2001. The complete human olfactory subgenome. Genome Res. 11: 685-702. [DOI] [PubMed] [Google Scholar]
- Hagman, J., Belanger, C., Travis, A., Turck, C.W., and Grosschedl, R. 1993. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes & Dev. 7: 760-773. [DOI] [PubMed] [Google Scholar]
- Hauck, B., Gehring, W.J., and Walldorf, U. 1999. Functional analysis of an eye specific enhancer of the eyeless gene in Drosophila. Proc. Natl. Acad. Sci. 96: 564-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, D.G. and Sharp, P.M. 1989. Fast and sensitive multiple sequence alignments on a microcomputer. Comput. Appl. Biosci. 5: 151-153. [DOI] [PubMed] [Google Scholar]
- Hoppe, R., Weimer, M., Beck, A., Breer, H., and Strotmann, J. 2000. Sequence analyses of the olfactory receptor gene cluster mOR37 on mouse chromosome 4. Genomics 66: 284-295. [DOI] [PubMed] [Google Scholar]
- Hoppe, R., Breer, H., and Strotman, J. 2003. Organization and evolutionary relatedness of OR37 olfactory receptor genes in mouse and human. Genomics 82: 355-364. [DOI] [PubMed] [Google Scholar]
- Kaiser, P. and Auer, B. 1993. Rapid shuttle plasmid preparation from yeast cells by transfer to E. coli. Biotechniques 14: 552. [PubMed] [Google Scholar]
- Kubick, S., Strotmann, J., Andreini, I., and Breer, H. 1997. Subfamily of olfactory receptors characterized by unique structural features and expression patterns. J. Neurochem. 69: 465-475. [DOI] [PubMed] [Google Scholar]
- Kudrycki, K., Stein-Izsak, C., Behn, C., Grillo, M., Akeson, R., and Margolis, F.L. 1993. Olf-1-binding site: Characterization of an olfactory neuron-specific promoter motif. Mol. Cell. Biol. 13: 3002-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, R.P., Cutforth, T., Young, J., Athanasiou, M., Friedman, C., Rowen, L., Evans, G., Axel, R., Hood, L., and Trask, B.J. 2001. Genomic analysis of orthologous mouse and human olfactory receptor loci. Proc. Natl. Acad. Sci. 98: 7390-7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Wilson, T.E., Milbrandt, J., and Johnston, M. 1993. Identifying DNA-binding sites and analyzing DNA-binding domains using a yeast selection system. Methods in Enzymology. 5: 125-137. [Google Scholar]
- Malnic, B., Hirono, J., Sato, T., and Buck, L.B. 1999. Combinatorial receptor codes for odors. Cell 96: 713-723. [DOI] [PubMed] [Google Scholar]
- Mombaerts, P. 1999a. Odorant receptor genes in humans. Curr. Opin. Genet. Dev. 9: 315-320. [DOI] [PubMed] [Google Scholar]
- Mombaerts, P. 1999b. Seven-transmembrane proteins as odorant and chemosensory receptors. Science 286: 707-711. [DOI] [PubMed] [Google Scholar]
- Mombaerts, P. 2001. The human repertoire of odorant receptor genes and pseudogenes. Annu. Rev. Genomics Hum. Genet. 2: 493-510. [DOI] [PubMed] [Google Scholar]
- Moser, M., Imhof, A., Pscherer, A., Bauer, R., Amselgruber, W., Sinowatz, F., Hofstadter, F., Schule, R., and Buettner, R. 1995. Cloning and characterization of a second AP-2 transcription factor: AP-2 β. Development 121: 2779-2788. [DOI] [PubMed] [Google Scholar]
- Ngai, J., Chess, A., Dowling, M.M., Necles, N., Macagno, E.R., and Axel, R. 1993. Coding of olfactory information: Topography of odorant receptor expression in the catfish olfactory epithelium. Cell 72: 1-20. [DOI] [PubMed] [Google Scholar]
- Papatsenko, D., Sheng, G., and Desplan, C. 1997. A new rhodopsin in R8 photoreceptors of Drosophila: Evidence for coordinate expression with Rh3 in R7 cells. Development 124: 1665-1673. [DOI] [PubMed] [Google Scholar]
- Papatsenko, D., Nazina, A., and Desplan, C. 2001. A conserved regulatory element present in all Drosophila rhodopsin genes mediates Pax6 functions and participates in the fine-tuning of cell-specific expression. Mech. Dev 101: 143-153. [DOI] [PubMed] [Google Scholar]
- Qasba, P. and Reed, R.R. 1998. Tissue and zonal-specific expression of an olfactory receptor transgene. J. Neurosci. 18: 227-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson, N.E., Eberwine, J., Dotson, R., Jackson, J., Ulrich, P., and Restrepo, D. 2000. Expression of mRNAs encoding for two different olfactory receptors in a subset of olfactory receptor neurons. J. Neurochem. 75: 185-195. [DOI] [PubMed] [Google Scholar]
- Reed, R.R. 2000. Regulating olfactory receptor expression: Controlling globally, acting locally. Nat. Neurosci. 3: 638-639. [DOI] [PubMed] [Google Scholar]
- Ressler, K.J., Sullivan, S.L., and Buck, L.B. 1993. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell 73: 597-609. [DOI] [PubMed] [Google Scholar]
- Rincon-Limas, D.E., Lu, C.H., Canal, I., Calleja, M., Rodriguez-Esteban, C., Izpisua-Belmonte, J.C., and Botas, J. 1999. Conservation of the expression and function of apterous orthologs in Drosophila and mammals. Proc. Natl. Acad. Sci. 96: 2165-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquier, S., Taviaux, S., Trask, B.J., Brand-Arpon, V., van den, E.G., Demaille, J., and Giorgi, D. 1998. Distribution of olfactory receptor genes in the human genome. Nat. Genet. 18: 243-250. [DOI] [PubMed] [Google Scholar]
- Sheng, G., Thouvenot, E., Schmucker, D., Wilson, D.S., and Desplan, C. 1997. Direct regulation of rhodopsin 1 by Pax-6/eyeless in Drosophila: Evidence for a conserved function in photoreceptors. Genes & Dev. 11: 1122-1131. [DOI] [PubMed] [Google Scholar]
- Smale, S.T. and Baltimore, D. 1989. The “initiator” as a transcription control element. Cell 57: 103-113. [DOI] [PubMed] [Google Scholar]
- Strotmann, J., Wanner, I., Krieger, J., Raming, K., and Breer, H. 1992. Expression of odorant receptors in spatially restricted subsets of chemosensory neurones. Neuroreport 3: 1053-1056. [DOI] [PubMed] [Google Scholar]
- Strotmann, J., Wanner, I., Helfrich, T., Beck, A., and Breer, H. 1994a. Rostro-caudal patterning of receptor-expressing olfactory neurones in the rat nasal cavity. Cell Tissue Res. 278: 11-20. [DOI] [PubMed] [Google Scholar]
- Strotmann, J., Wanner, I., Helfrich, T., Beck, A., Meinken, C., Kubick, S., and Breer, H. 1994b. Olfactory neurones expressing distinct odorant receptor subtypes are spatially segregated in the nasal neuroepithelium. Cell Tissue Res. 276: 429-438. [DOI] [PubMed] [Google Scholar]
- Strotmann, J., Hoppe, R., Conzelmann, S., Feinstein, P., Mombaerts, P., and Breer, H. 1999. Small subfamily of olfactory receptor genes: Structural features, expression pattern and genomic organization. Gene 236: 281-291. [DOI] [PubMed] [Google Scholar]
- Strotmann, J., Conzelmann, S., Beck, A., Feinstein, P., Breer, H., and Mombaerts, P. 2000. Local permutations in the glomerular array of the mouse olfactory bulb. J. Neurosci. 20: 6927-6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, S.L., Adamson, C., Ressler, K.J., Kozak, C.A., and Buck, L.B. 1996. The chromosomal distribution of mouse odorant receptor genes. Proc. Natl. Acad. Sci. 93: 884-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge, D., Brouwer, A., el Bahi, S., Guenet, J.L., Robert, B., and Meijlink, F. 1998. Mouse Alx3: An aristaless-like homeobox gene expressed during embryogenesis in ectomesenchyme and lateral plate mesoderm. Dev. Biol. 199: 11-25. [DOI] [PubMed] [Google Scholar]
- Tsuboi, A., Yoshihara, S., Yamazaki, N., Kasai, H., Asai-Tsuboi, H., Komatsu, M., Serizawa, S., Ishii, T., Matsuda, Y., Nagawa, F., et al. 1999. Olfactory neurons expressing closely linked and homologous odorant receptor genes tend to project their axons to neighboring glomeruli on the olfactory bulb. J. Neurosci. 19: 8409-8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassalli, A., Rothman, A., Feinstein, P., Zapotocky, M., and Mombaerts, P. 2002. Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb. Neuron 35: 681. [DOI] [PubMed] [Google Scholar]
- Vassar, R., Ngai, J., and Axel, R. 1993. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell 74: 309-318. [DOI] [PubMed] [Google Scholar]
- Wang, M.M. and Reed, R.R. 1993. Molecular cloning of the olfactory neuronal transcription factor olf-1 by genetic selection in yeast. Nature 364: 121-126. [DOI] [PubMed] [Google Scholar]
- Wang, S.S., Tsai, R.Y., and Reed, R.R. 1997. The characterization of the Olf-1/EBF-like HLH transcription factor family: Implications in olfactory gene regulation and neuronal development. J. Neurosci. 17: 4149-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, T.E., Fahrner, T.J., Johnston, M., and Milbrandt, J. 1991. Identification of the DNA binding site for NGFI-B by genetic selection in yeast. Science 252: 1296-1300. [DOI] [PubMed] [Google Scholar]
- Xie, S.Y., Feinstein, P., and Mombaerts, P. 2000. Characterization of a cluster comprising approximately 100 odorant receptor genes in mouse. Mamm. Genome 11: 1070-1078. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Baldassare, M., Fisher, P., Rathbun, G., Oltz, E.M., Yancopoulos, G.D., Jessell, T.M., and Alt, F.W. 1993. LH-2: A LIM/homeodomain gene expressed in developing lymphocytes and neural cells. Proc. Natl. Acad. Sci. 90: 227-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, J.M. and Trask, B.J. 2002. The sense of smell: Genomics of vertebrate odorant receptors. Hum. Mol. Genet. 11: 1153-1160. [DOI] [PubMed] [Google Scholar]
- Young, J.M., Friedman, C., Williams, E.M., Ross, J.A., Tonnes-Priddy, L., and Trask, B.J. 2002. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum. Mol. Genet. 11: 535-546. [DOI] [PubMed] [Google Scholar]
- Zhang, X. and Firestein, S. 2002. The olfactory receptor gene superfamily of the mouse. Nat. Neurosci. 5: 124-133. [DOI] [PubMed] [Google Scholar]