Abstract
Together with the hemichordates, sea urchins represent basal groups of nonchordate invertebrate deuterostomes that occupy a key position in bilaterian evolution. Because sea urchin embryos are also amenable to functional studies, the sea urchin system has emerged as one of the leading models for the analysis of the function of genomic regulatory networks that control development. We have analyzed a total of 107,283 cDNA clones of libraries that span the development of the sea urchin Strongylocentrotus purpuratus. Normalization by oligonucleotide fingerprinting, EST sequencing and sequence clustering resulted in an EST catalog comprised of 20,000 unique genes or gene fragments. Around 7000 of the unique EST consensus sequences were associated with molecular and developmental functions. Phylogenetic comparison of the identified genes to the genome of the urochordate Ciona intestinalis indicate that at least one quarter of the genes thought to be chordate specific were already present at the base of deuterostome evolution. Comparison of the number of gene copies in sea urchins to those in chordates and vertebrates indicates that the sea urchin genome has not undergone extensive gene or complete genome duplications. The established unique gene set represents an essential tool for the annotation and assembly of the forthcoming sea urchin genome sequence. All cDNA clones and filters of all analyzed libraries are available from the resource center of the German genome project at http://www.rzpd.de.
The sea urchin system has a long and extremely successful history as a model organism and continues to be the animal model of choice for many embryologists because of the easy and unlimited availability of masses of eggs and sperm. For example, mechanisms of general biological importance such as egg activation, fertilization, calcium signaling, cell cycle, and exocytosis have been elucidated in sea urchins (Swann and Parrington 1999; Zimmerberg et al. 1999; Whitaker and Larman 2001). To fully exploit the potential of the urchin system, that is, to identify molecular mechanisms that underlie cell movements during gastrulation, axis-specification, cell and tissue differentiation, or signaling mechanisms (Angerer and Angerer 2003), a unique collection of the majority of the genes of this organism is necessary. In the past years, the sea urchin system has emerged as one of the leading models for the analysis of the function of genomic regulatory networks that control embryonic development (Arnone and Davidson 1997; Davidson et al. 2002a). Through a combination of array screens and quantitation of changes of expression of regulatory genes in perturbed embryos, an immensely interwoven system of cross-regulatory mechanisms has arisen. The capacity of such screens within a given regulatory network, like the endomesoderm network (Davidson et al. 2002b), will be significantly increased if they can be performed on an array that includes cDNA clones representing all sea urchin genes. Such a collection of cDNAs is also an indispensable tool to jump from identified genes to the relevant cis-regulatory elements on the genome. Although the upcoming genome sequence will be the prerequisite for such in-depth analysis, the availability of an EST-based unique gene set is an essential complement to enable gene identification, annotation, and assembly of the genome of the sea urchin Strongylocentrotus purpuratus. Because sea urchins occupy an important phylogenetic position as one of the most basal deuterostomes, the study of the gene content of their genome or the composition of their transcriptome is essential to analyze the origin of deuterostomes and the divergence of bilaterians.
We have therefore started the identification and analysis of the sea urchin embryo transcriptome. Oligonucleotide finger-printing (ONF) was used to normalize 107,283 cDNA clones, mostly from random primed libraries, prior to EST sequencing. Here we report the number of distinct transcript classes that we have identified for each developmental stage and across all stages and the sequences generated based on the above results. We assess the transcript diversity and completeness of the gene catalog that we have generated via homology searches, protein domain prediction, and their classification into gene families that include orthologs from protostomes (Caenorhabditis elegans and Drosophila melanogaster), deuterostomes (Ciona intestinalis), and vertebrates (man, mouse, Fugu).
The role of gene duplications as a major process of increasing the genetic material available to an organism and in this sense increasing diversity is becoming increasingly appreciated with the completion of each new genome sequence. As a significant step in evolution has been the divergence of protostomes and deuterostomes, we investigate the gene duplication activity that might have played a role at this point of bilaterian evolution. Thus, we compare the copy rates of genes included in the sea urchin catalog that we generated with the copy rate of genes in the sea squirt (C. intestinalis) and the vertebrate (man, Fugu) genomes. Using this comparison, we identify (1) genes duplicated in both Ciona and sea urchin that might represent duplications that happened in the common ancestor of both organisms, (2) genes appearing as a single copy in the sea urchin catalog while occurring in multiple copies in Ciona and vertebrates that potentially represent chordate- or vertebrate-specific duplications.
RESULTS
Choice of Developmental Stages and cDNA Libraries
To get an overview of the repertoire and the temporal expression of sea urchin genes, we selected several developmental stages that span sea urchin embryonic development. A preliminary analysis of the sequence complexity of the unfertilized egg has been previously described (Poustka et al. 1999). To complement the maternally expressed sequence content identified in that study, we analyzed several additional libraries: (1) An early-cleavage-stage library at the fourth to fifth cleavage (7 h of development), at which territorial information becomes organized into the distinct blastomeres using both maternal and very early zygotically derived genetic information. (2) A midblastula-stage library (20 h of development), at which major endomesodermal and oralaboral axis patterning mechanisms occur. (3) A midgastrula library (40 h of development). At this stage, most of the germ-layer specifications have occurred. The gastrula stage is especially important to detect transcripts involved in cell-cell signaling, cell movements, cell adhesion, and morphogenesis. (4) As one of the most interesting aspects of sea urchin (and echinoderm) development is the establishment of a pentaradially symmetric adult within the rudiment of the bilateral embryo, a library of 2-3-wk larva was constructed and analyzed. At this time of development, the left hydrocoel and the vestibule have already fused to form the rudiment, and a new genetic program patterning the future adult sea urchin is active. The extent of the overlap between the transcriptomes of this developmental stage and early embryonic development is unknown, but it has been shown that differences in patterning the embryo and the adult exist, for example, in the expression of Hox genes (Arenas-Mena et al. 2000).
As has been shown earlier (Davidson 1986; Poustka et al. 1999), long noncoding 3′-UTRs can hamper the identification of protein coding sequence in oligo(dT)-primed cDNA libraries from sea urchins. To obtain EST sequences from the underrepresented, often coding, central portions of mRNAs, the libraries analyzed in this study were constructed by random priming. Although through this strategy the normalization success of ONF is expected to be somewhat lower compared with conventional oligo(dT)-primed libraries (Poustka et al. 1999), it is nevertheless expected that sequences of multiple ESTs derived from random-primed clones can in many cases reconstitute a full-length cDNA. Furthermore, as the insert sizes in the cDNA libraries were kept at an average size of 1-1.5 kb, a complete insert sequence is expected to be obtained in many cases by the generation of 5′ and 3′ ESTs using universal primers. Random priming also has the advantage that most clones are expected to lack a poly(A) tail, and hence sequencing from the 3′-end is facilitated. To estimate the poly(A) content of random primed clones, an oligo(dT) primer was hybridized to macroarrays carrying these clones. About 5% of the clones gave strong signals indicating the presence of poly(A) tracts (data not shown).
Normalization, EST Sequencing and Clustering
A total of 107,283 clones were simultaneously analyzed by ONF as described earlier (Meier-Ewert et al. 1998; Herwig et al. 1999, 2002; Poustka et al. 1999; Clark et al. 2001). To prevent overclustering by ONF, the clustering stringency was trained on the real representation of the cDNA clones representing 30 different genes, whose representation in the libraries was determined by backhybridization to the macroarrays of the used libraries. As a result, 35,238 different clusters of cDNA clones were identified (Fig. 1).
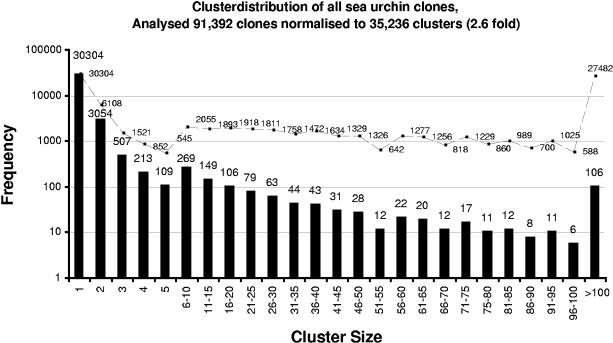
Figure 1.
Histogram of the size distribution of the oligonucleotide fingerprinting (ONF) clusters reflecting the abundance distribution of all clones across all libraries, that is, all analyzed developmental stages. The X-axis shows the cluster size (clusters containing more than five clones are grouped). The Y-axis represents the frequency of each cluster size group, which is also given at the top of the bars representing the group, overlaid with the actual number of clones in that category (i.e., 106 clusters of size >100 with 27,482 clones). In total, 91,392 clones were analyzed by ONF clustering (see Methods), and about a third of the clones (27,482) belong to the superprevalent class, summarized in only 106 clusters. A third of the clones (30,304) exist in only one copy in any of the libraries (presumably complex class transcripts). In total, 35,238 different clusters were identified, which indicates that a 2.6-fold normalization was achieved.
In total, 30,469 different clones (6126 from the egg library and 24,343 from the other random primed libraries) were sequenced from the 5′-end. Of these clones, 12,240 were sequenced from the 3′-end as well. Altogether, 42,709 high-quality reads were generated (Table 1).
Table 1.
Overview of the Number of cDNA Clones Analyzed, Normalized by Oligonucleotide Fingerprinting and Sequenced Across Various Libraries That Represent the Different Developmental Stages
| Stage | EGC | Cleavage | Blastula | Gastrula | Larva | Total |
|---|---|---|---|---|---|---|
| Clones analyzed by ONF | 6291 | 10,368 | 18,432 | 18,432 | 53,760 | 107,283 |
| Clones included in ONF clustering | 4689 | 9375 | 16,925 | 15,617 | 44,786 | 91,392 |
| Normalized in ONF clustering | 3098 1.5-fold | 4069 2.3-fold | 6557 2.6-fold | 6635 2.3-fold | 14,877 3.0-fold | 35,238 2.6-fold |
| Clones remaining as singletons (%) | 2673 (57%) | 3392 (36%) | 5421 (32%) | 5774 (37%) | 13,044 (29%) | 30,304 (33%) |
| Clones selected for sequencing | 6291 | 4091 | 6650 | 6659 | 15,746 | 39,437 |
| Total number of sequence reads | 6126 | 5546 | 9974 | 8095 | 12,968 | 42,709 |
| Number of 5′ reads | 6126 (100%) | 2894 (52.2%) | 5270 (52.8%) | 5604 (69.2%) | 10,575 (81.5%) | 30,469a (71.3%) |
| Number of 3′ reads | 0 | 2652 (47.8%) | 4704 (47.2%) | 2491 (30.8) | 2491 (18.5%) | 12,240 (28.7%) |
As an internal control, the nonredundant 6291 clones that were previously normalized from an unfertilized egg cDNA library (Poustka et al. 1999) were again subjected to ONF analysis. cDNA clones that were clustered together with those clones previously sequenced from the unfertilized egg cDNA library were rejected from sequencing because these transcripts were already identified. A total of 4689 of the unfertilized egg cDNA clones were included in the cluster analysis. If those were clustered again, they were assembled into 4098 clusters. This is a reduction in redundancy by only 12%, indicating the high normalization rate that can be achieved by ONF.
This number is equal to the total number of different clones sequenced
Overlapping ESTs were clustered following a two-step assembly strategy (see Methods), resulting in 27,993 independent sequence clusters, indicating that 92% of the preselected clones represent independent sequence. At this stage, some genes may be represented by more than one cluster corresponding to discrete nonoverlapping regions of a single transcript. Whenever a single clone had sequence reads from opposite ends, which were members of two different clusters, this information was used to merge the two clusters (or singletons). Following this clone-based merging of clusters/singletons, 20,172 nonredundant clusters remained. This number corresponds to the number of sea urchin genes for which we have created a sequence tag and is supported by considering the number of SWISS-PROT proteins having multiple hits among the sequence clusters. Before merging, 68% of the matched SWISS-PROT proteins had only one hit in the EST catalog; however, following merging 95% of the matched proteins were unique (BLAST e-value cutoff = e-40). About 7% of the ESTs contained simple repeats. To complete this sea urchin unigene catalog, other publicly available ESTs (primary mesenchyme cell ESTs [PMC project; Zhu et al. 2001]) were coclustered with our set. Out of 9399 PMC ESTs, 8938 ESTs fulfilled our quality criteria (see Methods). As a result of the coclustering, the total number of independent sequence clusters was nearly unchanged with 21,290. The following analyses are based on this set of clusters.
Assessment of Gene Coverage and Reconstitution of Full-Length Insert Sequences or Full ORFs
To estimate the coverage of the gene catalog, we compared the number of predicted genes from complete sequences of 42 S. purpuratus BACs (Davidson et al. 2002a) with the hit rate of ESTs. The 42 BACs that represent a total of 4.8 Mb of genomic sequence were analyzed using the gene-prediction program Genscan (Burge and Karlin 1997). A total of 264 genes were predicted on the BACs taking also single-exon predictions into account. Of these, using the stringent criterion of >90% identity over a minimum alignment length of 300 bp, 178 could be confirmed by an EST match, indicating that our catalog contains a tag for 67% of all genes in the sea urchin genome. There were an additional 37 EST matches outside of the predictions, indicating that by training the gene prediction programs to reduce false positives and negatives, the overlap between predictions and EST matches could be increased.
In all, 1545 EST clusters contained more than five reads. Although in this sense ONF (see Methods) is less efficient in normalizing random primed clones into distinct genes, the ESTs that constitute the large clusters partially overlap, thus allowing the reconstitution of the full-length sequence of genes without having to use expensive and labor-intensive primer walking sequencing. The mean sequence length in clusters with multiple ESTs was 939 bp. About 28% of the 5′ and 3′ reads of the same clones (3370 clones in 2620 clusters) directly overlapped and hence represent full-length insert sequences. The effect of combining ONF and using random primed clones is illustrated, for example, by cluster001767.a1.2, which represents a sea urchin ortholog of the Na+ and Cl- coupled neutral and basic amino acid transporter ATB0 (SP_RO:Q91Y60; Fig. 2D). In this, 46 EST reads derived from 40 different preselected clones comprised sufficient overlap to reconstitute a high-quality consensus sequence of 3907 bp covering the complete ORF of >600 amino acids, a short 5′, and an almost 2-kb 3′-UTR. Although this is also an example of a suboptimal normalization success for these genes, it is also an example of how random-primed clones can actually help to raise the chance of identifying a gene by reducing the number of matches due only to domains. A large number of such full-length reconstitutions, which, for example, are important for sequence-based functional predictions, were discovered and are visualized in the database described below.
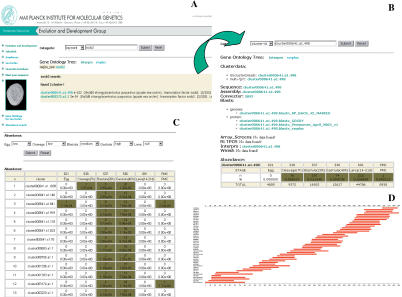
Figure 2.
Example of the interface of the sea urchin database. (A) Using the Soxb2 gene name as a keyword, a list of clusters matching this query is displayed. (B) Selecting one of the listed clusters, all the information relevant for this cluster is displayed. By selecting the cluster data field, information such as all clonenames, read directions, trace identifiers, and so on for all the clones in the cluster are given. The number and percentage of all clones in the cluster as well as the developmental stage from which they are derived is displayed in the abundance field at the bottom, which allows a quick overview of when the gene of interest is expressed. (C) Furthermore, selection of EST clusters according to the developmental stage from which the respective cDNA clones were isolated can also be performed. As an example, a list of clusters consisting of clones expressed at low levels in the egg and cleavage stage, medium levels in blastula stage, and high levels in gastrula but not expressed in the larva is shown. (D) A graphical representation of the overlap of the ESTs assigned to an EST cluster is provided for each of the clusters contained in the database. As an example, the 3907-bp alignment of all 46 reads derived from 40 different clones of the cluster001767.a1.2 that represents a sea urchin ortholog of the Na+ and Cl- coupled neutral and basic amino acid transporter ATB0 (SWISS-PROT entry number: SP_RO:Q91Y60) is shown. Bars represent the EST sequences. On the left the sequencer trace identifiers are given, which can be used to retrieve a specific EST sequence.
Expression Differences Between Developmental Stages Based on ONF Counts
Counting the number of cDNA clones within an ONF cluster representing a given gene results in information about its relative level of expression. As the origin of an EST is monitored, conclusions on the temporal pattern of expression can be gained, that is, when a gene is definitely expressed, and when it is likely to be expressed either not at all or at very low levels. Each EST cluster is associated with the number of clones in the ONF cluster from which a given clone was selected. Counting the sizes of ONF clusters allows the identification of genes putatively expressed in a stage specific manner. For each ONF cluster, we used a two-sided binomial test for the hypothesis that the cDNA abundance of a given library (i.e., developmental stage) in this cluster is comparable to the overall abundance of this library. A significant deviation from this hypothesis, indicated by a low P-value of the test, identifies clusters that show under- or overrepresentation of cDNAs from that library/developmental stage. Multiple ONF clusters were grouped together for counting transcript frequency if they were merged in the sequence-clustering step or if they were found to represent the same gene, as determined from BLAST search analysis. Between the embryonic stages, only few statistically relevant differences could be found. Genes that are overrepresented in the maternal libraries compared with other embryonic stages are, for example, cyclins a and b, cleavage stage histone h2a, and cdc14a2 phosphatase (Supplemental Table S1, available online at www.genome.org). However, several genes were identified that displayed significant differences of expression level between embryonic and larval stages of development. A selection of the most significantly overrepresented or underrepresented genes of the larva are given in Supplemental Table S2. Overrepresented genes could, for example, be genes that are necessary to develop the adult bauplan of the sea urchin. The analysis of such genes will be important to detect regulatory networks functioning in the development of the pentaradially symmetric urchin, in contrast to networks that function in the bilateral embryo. All these data were introduced into the database described below, so that a quick overview of when a gene is expressed is immediately visible.
Functional Annotation
At a BLAST e-value cutoff of ≤1e-06, 7146 consensus sequences displayed matches to proteins included in the SWISS-PROT database. Of these, 5114 could be annotated in at least one of the three broadly defined Gene Ontology (GO) functional classes (Biological Process, Molecular Function, or Cellular Component; Xie et al. 2002). Of these, 2426 were categorized in all three main categories (Fig. 3). A comparison of functional annotations between different stages of development did not show any large differences. In addition to the annotation through BLAST searches versus SWISS-PROT, all sequence clusters were also annotated to the GO classes via the protein domains they contain. Each cluster was scanned against the InterPro domain database (Apweiler et al. 2001). In total, 7087 of 19,821 translations contained at least one known domain. We found 1104 different protein or domain families in total. We annotated 440 additional clusters based on an InterPro match, totaling to 5554 clusters that could be annotated using GO. We have visualized this data in the database described below by integrating the GO annotation into a tree from which any functional class can be selected to display and retrieve the relevant sea urchin genes (Supplemental Fig. 1).
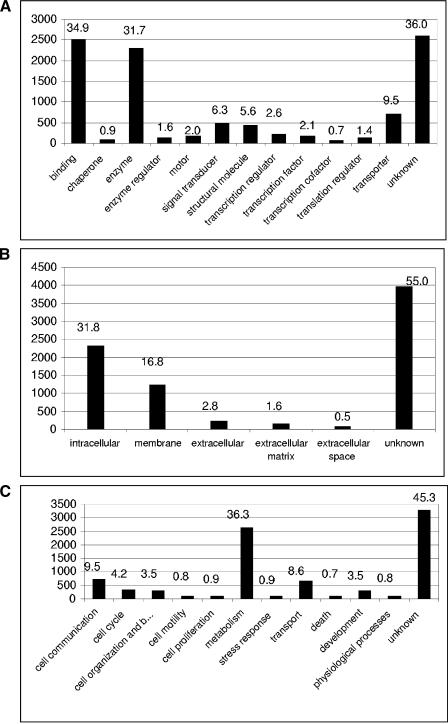
Figure 3.
Distribution of the EST consensus sequences into the main Gene Ontology (GO) functional classes. Of 7146 consensus sequences, 5114 could be annotated in at least one of the three Gene Ontology (GO) defined functional classes. The numbers above the bars represent the percentage of all sea urchin clusters with a significant match to the SWISS-PROT database that are classified in the given functional class. The height of the bars and the ordinate give the number of proteins per group. (A) 4570 proteins could be associated with a molecular function, (B) 3219 proteins with a potential cellular component, and (C) 3910 proteins were associated with a specific biological process. A total of 2426 proteins were annotated in all three main categories.
Phylogenetic Comparison of the Identified Sea Urchin Genes
BLAST searches were performed against a database that contained the predicted proteomes of six completed genomes of organisms that occupy key phylogenetic positions (see Methods). These were the proteomes of the protostomes of C. elegans and D. melanogaster, the basal chordate C. intestinalis, the teleost fish Fugu rubripes, and the mammals Homo sapiens and Mus musculus. We found that 7391 different clusters (35% of all 21,290) had at least one protein match (e-value ≤ 1e-7). A total of 3036 clusters have matches to all genomes analyzed (Table 2). Only 94 clusters show higher similarity to a protostome, either C. elegans or D. melanogaster, than to any vertebrate sequence in the set. Approximately 1993 clusters (27%) have a match to deuterostomes only, whereas 202 clusters have matches to Ciona only. The fact that the majority of sea urchin genes have higher sequence similarity to deuterostome rather than protostome genes and the high degree of sequence similarity of these matches show clearly the closer phylogenetic position of the sea urchin to deuterostomes.
Table 2.
BLAST Search Summary Against a Database Containing the Proteomes of Five Sequenced Genomes
| Species | Number of sea urchin clusters matching | Median e-value |
|---|---|---|
| Homo sapiens | 6479 | 1.0e-21 |
| Mus musculus | 5269 | 1.7e-16 |
| Fugu rubripes | 6082 | 1.6e-20 |
| Ciona intestinalis | 5583 | 1.8e-20 |
| Drosophila melanogaster | 4954 | 6.65e-14 |
| Caenorhabditis elegans | 4181 | 4.15e-10 |
Only the highest score of a sea urchin sequence cluster to the respective organism was taken into account.
Approximately 20% (145) of the 675 predicted Ciona genes that are reported to have no clear homolog in protostomes but match vertebrate proteins (see online Supplemental material of Dehal et al. 2002) display significant matches to a sea urchin cluster. Out of 2101 predicted Ciona-specific genes, 519 (25%) matched a sea urchin cluster. These results indicate that at least one-quarter of the genes thought to be chordate-specific as judged by the analysis of the Ciona genome sequence (Dehal et al. 2002) were already present at the base of deuterostome evolution.
Comparison of Gene Copy Rates Between Sea Urchins and Chordates
The estimation of the extent of duplication of the sea urchin genes included in our catalog was carried out in two steps that involved: (1) the classification of the sea urchin consensus sequences using a platform of 3453 orthologous groups that include genes found as single copy in the proteomes of C. elegans and D. melanogaster (CD families) only, as well as in C. elegans, D. melanogaster, and Saccharomyces cerevisiae (CDY families; Panopoulou et al. 2003) and (2) the counting of the number of genes per orthologous group for each of the participating organisms. Whereas the CDY gene families represent many genes functioning in ubiquitous ancestral cellular processes, the CD groups allow at least in part the discrimination between these genes and genes that might be specific for multicellular organisms. The condition of being single copy in all the compared invertebrate genomes allows the selection of genes that remained unduplicated at least up to the protostome-deuterostome split.
In total, 2782 sea urchin clusters were classified into 1532 CD/CDY groups on the basis of probable orthology. As a result, 1037 CD/CDY groups contained a single sea urchin gene, indicating that 67.6% of the gene families represented by the CD/CDY groups probably remained unduplicated in the sea urchin genome (Table 3). In comparison, following assignment of 5198 Ciona genes (32% of all predicted genes of the Ciona genome) to 2941 orthologous CD/CDY groups, ∼61% of the CD/CDY groups had a single Ciona ortholog (Table 3). Previously 10,209 human genes were assigned in a similar manner to 3044 CD/CDY groups, of which 1156 (38%) contain a single human ortholog as reported (Panopoulou et al. 2003). The above allocation of genes to orthologous groups can also be expressed as average gene copy rate. Thus, the average copy rate of sea urchin genes per CD/CDY cluster is 1.8, which is very similar to the copy rate for Ciona (1.76). These numbers for the invertebrate genomes of sea urchin and Ciona approximate the ratio of 1.6 found for amphioxus (Panopoulou et al. 2003), whereas the respective gene copy rate for the human genes is 3.4. If the calculation of gene copy rates is based only on the 1337 CD/CDY groups that contain orthologs from all three compared organisms (sea urchin, Ciona, man; Table 4), the values for the gene copy rates are 1.8 (sea urchin), 2.5 (Ciona), and 4 (man), indicating that deuterostome invertebrate genomes contain fewer gene duplications than vertebrate genomes.
Table 3.
Comparison of the Number of Sea Urchin and Ciona Orthologs Assigned in the CD/CDY Orthologous Groups
| CD/CDY group size | Number of CD/CDY groups for urchin/ciona | Percentage of CD/CDY groups for urchin/Ciona |
|---|---|---|
| 1 | 1037/1808 | 67.6/61.4 |
| 2 | 284/550 | 18.5/18.7 |
| 3 | 107/218 | 6.98/7.41 |
| 4 | 38/118 | 2.48/4.01 |
| 5 | 23/53 | 1.50/1.80 |
| 6 | 12/45 | 0.78/1.53 |
| 7 | 10/31 | 0.65/1.05 |
| 8 | 4/26 | 0.26/0.88 |
| 9 | 2/22 | 0.13/0.75 |
| 10 | 1/20 | 0.07/0.68 |
| >10 | 13/50 | 0.84/0.42 |
A total of 2782/5198 sea urchin/Ciona genes were assigned in 1532/2941 CD/CDY groups, respectively. CD/CDY groups containing more than 10 orthologs are grouped. The average copy rates per CD/CDY cluster are calculated from these numbers as 1.8 for sea urchin and 1.76 for Ciona.
Table 4.
Comparison of the Numbers of Sea Urchin Genes to the Numbers of their Ciona and Human Orthologs Assigned in the Same CD/CDY Group of Protostome and Yeast Single-Copy Genes
| Compared organisms | Number of CD/CDY groups | Number of genes | Copy rate |
|---|---|---|---|
| Sea urchin:human | 1447 groups shared | 2489:5777 | 1.7:4.0 |
| 1:1 | 390 (26.95%) | ||
| 1:>1 | 586 (40.49%) | ||
| >1:1 | 72 (4.97%) | ||
| Sea urchin:Ciona | 1394 groups shared | 2432:3498 | 1.7:2.5 |
| 1:1 | 610 (43.75%) | ||
| 1:>1 | 320 (22.9%) | ||
| >1:1 | 128 (9.18%) | ||
| Ciona:human | 2740 groups shared | 5805:9289 | 2.1:3.4 |
| 1:1 | 837 (30.54%) | ||
| 1:>1 | 842 (30.72%) | ||
| >1:1 | 185 (6.75%) | ||
| Sea urchin:Ciona:human | 1337 groups shared | 2348:3363:5411 | 1.8:2.5:4 |
| 1:1>1 | 307 (22.96%) | ||
| 1:>1:>1 | 239 (17.87%) | ||
| 1:>1:1 | 65 (4.86%) | ||
| >1:1:1 | 44 (3.29%) | ||
| 1:>1:>Ciona | 145 (10.84%) | ||
| su = ci and hum > su = ci | 357 (26.70%) | ||
| 1:1:1 | 279 (20.86%) |
As an example, ∼11% of the single-copy sea urchin genes are duplicated in both Ciona and human, but have significantly more copies in human than Ciona. The 26.70% of all sea urchin genes that were assigned to the CD/CDY system have the same copy number in Ciona but have more duplicates in human, and 20.86% of the shared single-copy CD/CDY genes are found as single copy in all three organisms. Pairwise comparison of groups that are in common between two of the organisms shows elevated duplication frequencies in vertebrates more clearly, as 43.75% of the single-copy sea urchin genes are found also as single copy in Ciona, whereas only 26.95% are present as single copy in the human genome. Accordingly, we found 40.49% and 30.72% of the sea urchin and Ciona single-copy genes, respectively, duplicated in the human genome. The copy rates are calculated from the number of genes per orthologous (CD/CDY) group.
Analysis of the same common groups indicates that ∼23% of the genes duplicated in man might be man/vertebrate-specific duplication events as they are classified as single copy in sea urchin and Ciona. Approximately 18% (239 clusters) of the unduplicated sea urchin genes seem to be duplicated in Ciona and man, and hence these genes might have been duplicated in early deuterostome or even chordate evolution. To assess whether these clusters represent duplications common to chordates or deuterostomes, a phylogenetic analysis was carried out. Out of 239 groups, 60 contained enough sequence information for tree building (i.e., the sum of all overlapping regions of the alignment of all genes included in the group was longer than 80 amino acids after selection of conserved blocks using gblocks; see Methods). Of these, 24 trees could not be classified because of unresolved branches. Of the remaining 36 trees, 24 clearly showed that the genes represented by these groups were duplicated before the chordate/vertebrate split. Among those were, for example, a family of potassium channels and a family of nad(p)-dependent steroid dehydrogenases, which are essential components of cholesterol biosynthesis (Fig. 4; Nwokoro et al. 2001). In most cases, there are also significantly more vertebrate copies than chordate. This greater number of copies in vertebrates is in agreement with a higher tissue and organ complexity. However, because in most trees the sea urchin sequence is not at the root, it is likely that these genes were duplicated even before the separation of the echinoderm lineage. The other 12 trees (29%) represent genes that were duplicated independently in the two lineages after the vertebrate/chordate split. All phylogenetic trees made can be found on our Web page (http://www.molgen.mp-g.de/ag_seaurchin). Comparison of all CD/CDY copy rates common between either two or all three organisms are summarized in Table 4.
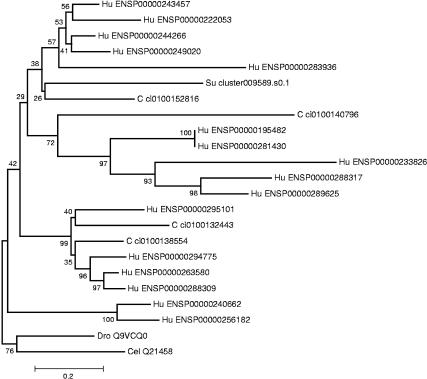
Figure 4.
Example of a neighbor-joining phylogenetic tree generated for a CD-type gene family that includes a single sea urchin and multiple Ciona and human orthologs. This CD group represents a family of potassium channels. Most of the branches contain a Ciona ortholog, and hence most of the genes of this family were duplicated after the protostome/deuterostome split but before the vertebrate/chordate divergence as the CD/CDY groups include orthologs of genes that are simultaneously single copy in C. elegans, D. melanogaster and yeast (S. cerevisiae). Because the sea urchin sequence does not root the tree, the expansions might have taken place before the separation of the echinoderm lineage. Additional vertebrate-specific expansions like in the example above were repeatedly observed in the selected CD/CDY groups. The C. elegans and D. melanogaster node was used as an outgroup for the tree. Numbers at branch points are confidence values derived from 1000 bootstrap resamplings of the alignment data. The sequence distance is indicated at the bottom as substitutions per site. Human genes are abbreviated by Hu followed by the Ensembl gene identifier (release 4.28; see Methods), Ciona genes are abbreviated by C followed by the Ciona gene model identifier number (JGI Release1; see Methods), and the sea urchin sequence is abbreviated by Su followed by the sequence cluster identifier that can be retrieved from our database. Other abbreviations are (Dro) D. melanogaster and (Cel) C. elegans.
A Sea Urchin Database
A major problem with large EST sets is that the relation of annotation and clone identity/availability is often lost when they are submitted to public databases. We have therefore annotated all ESTs that we generated with their putative gene ID, protein domain type, orthology relationship to protostome and deuterostome genes, sequence read direction, and alternative clone names (such as RZPD clone names for ordering additional clones) in a database (URCHIBASE: http://www.molgen.mpg.de/ag_seaurchin/) that includes largely independent sequence clusters, most of which represent individual genes (Fig. 2).
DISCUSSION
Sea urchins have been among the main model organisms used in embryological studies for more than a century. The attraction of the sea urchin system lies in its significant phylogenetic position as a basal deuterostome but also in its experimental advantages that enable functional studies such as those on calcium signaling, to studies intending to unravel major puzzles in developmental biology, like the mechanisms of axis specification or the characterization of cis-regulatory elements in the genome. However, the cloning of the relevant genes has been a time-consuming step for all above studies. We have generated a catalog of ∼20,000 nonredundant clustered EST consensus sequences, derived from 42,709 ESTs from five prenormalized cDNA libraries. During this study, ONF was applied for the first time on random primed cDNA clones. The objectives of this strategy were to (1) circumvent the generation of ESTs with low amounts of protein coding sequence, which is an inherent problem when oligo(dT)-primed sea urchin libraries are used; and (2) to generate a high percentage of overlapping sequences, which, in the ideal case, would cover the entire coding area of any given gene. As a result of using random primed libraries, higher coding potential for most clones was observed as compared with the oligo(dT)-primed egg library (see Methods for details). The availability of an extremely high number of 5′-ends of genes emerged as a large advantage of the ESTs derived from the random primed clones. This facilitates, for example, the rapid design of morpholino-substituted oligonucleotides, which have to be targeted to the translational start site of expressed genes to knock out translation (Heasman 2002). The cDNA libraries were constructed from embryonic stages that are spaced so that they cover major events during sea urchin embryogenesis. The investigation of different developmental stages allows the presentation of rough expression profiles for all identified genes. However, assuming that the EST catalog contains a tag for 50%-70% of all genes (see below), we expect that the representation of the rare class of transcripts is not as comprehensive as that of the more abundant ones. Because the prevalent transcripts are likely to be covered completely, we would expect 35% to 50% of the rare transcripts, for example, transcription factors, to be covered. More quantitative information on rare transcripts may be obtained by expression profiling experiments, a task that becomes achievable with the data set we have generated here.
The EST sequence set established here is the first for a basal nonchordate deuterostome on such a large scale. Together with hemichordates, echinoderms represent an important position in the evolution of deuterostomes as the most basal groups. Gene duplications are key processes in genome evolution (for reviews, see, e.g., Wolfe 2001; Prince and Pickett 2002; Wolfe and Li 2003). For example, it is believed that the sea urchin and amphioxus genomes represent archetypic genomes compared with vertebrate genomes, which are believed to have undergone two rounds of complete genome duplications—a hypothesis that is still controversial (Friedman and Hughes 2001; McLysaght et al. 2002; Panopoulou et al. 2003). By organizing the sea urchin clusters in gene families that are single-copy genes in CD/CDY, our results indicate that the majority (67%) of the sea urchin and 61.4% of Ciona genes can be found as single copy in their respective genomes. This result may indicate that an extensive, complete genome duplication has not occurred in sea urchin. Similar distributions of gene copy rates were found for amphioxus (Panopoulou et al. 2003). Nevertheless, 32% of the sea urchin genes might have been duplicated at least once. Among the 20 putatively most extensively duplicated genes in the sea urchin were found to be the BMP1/tolloid, multidrug resistance protein, and actin gene families (Supplemental Table S3). These genes appear to be also extensively duplicated in the Ciona, Fugu, and human genomes. The high frequency of duplicates in sea urchin shows that these most likely have been duplicated rapidly at the base of the deuterostome lineage after separation from the protostomes, as these genes exist as single copy in the protostomes C. elegans and D. melanogaster. These results may indicate that expansions of selective gene families but not complete genome duplications could have been important events at the divergence of protostomes and deuterostomes. Nevertheless, further sequencing of single genes or the whole genome sequence will be required to analyze this interesting aspect in detail.
The comparison of the copy rates of sea urchin genes to those in Ciona and man, indicate similar gene duplication frequencies in sea urchins compared with Ciona. Taking all CD/CDY clusters into account, we found average copy rates of 1.8 for sea urchin and Ciona. For each CD/CDY group with an identified ortholog in all of the three (deuterostome) genomes, we found an average of 1.8 sea urchin, 2.5 Ciona, and 4 human orthologs. We therefore suggest that the sea urchin, similar to Ciona, might have approximately half the number of genes of human. Although the ortholog counts for urchin and Ciona are similar for this group of CDY clusters, it shows some discrepancy if compared with the gene counts of ∼27,000 for urchin (Cameron et al. 2000) and ∼16,000 for Ciona (Dehal et al. 2002). However, both gene counts are based on different methods and at present are possibly too far apart, with corrections to be made on both sides. Assuming that we are still missing half the protein-coding sequences of sea urchin genes, we expect that the average number of sea urchin orthologs in CD/CDY families that are shared among all the compared deuterostomes will move closer to that found for Ciona. If the gene counts for Ciona and sea urchin are assumed to be correct, then an alternative possibility to explain the discrepancy between copy rate and gene count would be that the genome of Ciona could have extra gene-specific duplications of genes in the selected CD/CDY set. Also possible, although rather unlikely, would be that the sea urchin has gained many completely new genes present neither in Ciona nor CDY.
Although the EST catalog is not complete, we believe that this data set is comprehensive enough to obtain a good estimate of the duplication frequencies as described above. For example, we estimate that our catalog contains a tag for up to 50%-70% of all sea urchin genes, because half of the CD/CDY gene families have at least one sea urchin ortholog (1532 of 3453), whereas the predicted genes from the Ciona genome contain orthologs for 2941 CD/CDY groups, and judging from the high number of predicted genes on 42 completely sequenced BACs that are already represented by an EST match (67%). Because there are only 37 cDNA sequence hits outside the gene predictions, only very few of these hits are likely due to random background. These 37 hits outside of gene predictions could be caused by 3′-UTRs being predicted badly, owing to genomic contamination in the cDNA library and the imperfections in the gene prediction. If the quality of the predictions could be improved by “teaching” on sea urchin DNA, some of the hits outside the present predictions would probably become verified, further increasing the hit rate. The estimate would therefore go up (and the small random background down), if the reliability of the gene prediction program could be improved further. For this reason, we are fairly confident that the data represent a tag for 50%-70% of all genes of the sea urchin.
Finally, the large EST resource described here will be an essential tool to assemble and annotate the forthcoming sea urchin genome sequence (Pennisi 2002). It is also a prerequisite to analyze the partly established networks of gene regulation in much greater detail and will allow the linkage of differentially expressed genes to their respective promoter regions on the genome. Nevertheless, in addition to the effort described here, further large EST sequencing projects are needed, especially full-length ORF sequencing projects, that is, from clones derived from libraries constructed using the SMART technology (Wiemann et al. 2001). Such libraries have been constructed and will be analyzed in the future (A.J. Poustka, unpubl.). Clones from the clusters described here will in future also be used for gene expression profiling experiments, ensuring that large amounts of data will be collected on the same physical set of resource clones.
METHODS
cDNA Library Construction and Normalization by Oligonucleotide Fingerprinting
All cDNA libraries were prepared from 1μg of poly(A)+ RNA using the GIBCO Superscript plasmid system for cDNA synthesis and plasmid cloning (GIBCO BRL Life Technologies, Cat. No. 18248-013). The unfertilized egg library was constructed using an oligo(dT) primer [5′-gactagttctagatcgcgagcggccgccc(t)15-3′]. The random primed libraries were constructed using the same kit and primer concentrations but with a random primer [aaaggaaggaa aaaagcggccgctaca(n)6T]. All other steps were performed as described in the manufacturer's manual. Clone arraying and library handling were performed as described (Clark et al. 1999). Filter production, oligonucleotide hybridizations, data processing, and quality control were carried out as described earlier (Poustka et al. 1999; Clark et al. 2001). Clustering was performed as described in Herwig et al. (1999). From the described random primed libraries, 100,992 cDNA clones were selected for ONF analysis. Together with the 21,925 clones from the unfertilized egg library, a total of 122,917 clones were ONF-analyzed. In total, 91,392 were used for the cluster analysis, as 31,608 clones were removed that never showed positive hybridization results, which is mostly because of unsuccessful PCR amplification, low transfer efficiency to the filter membranes, or short insert size of the respective clones. The overall normalization success of ONF on random primed cDNA libraries was found to be lower compared with similar experiments using oligo(dT)-primed libraries. The average normalization of the oligo(dT)-primed unfertilized egg library was 3.5-fold (Poustka et al. 1999), and the average normalization for the random primed libraries was 2.6-fold.
For clone selection for sequencing, a consensus fingerprint for each ONF cluster was calculated from the individual finger-prints of the clones within that cluster. Subsequently, the clone closest to the consensus fingerprint was selected for sequencing as the best representative of that cluster. The second criterion for selecting clones for EST sequencing was based on the complexity of the fingerprint, with the aim of finding the longest clone within the cluster. This selection is achieved by selecting the clones that have additional positive hybridization results compared with the consensus fingerprint. In cases in which the consensus clone and the longest clone were different, both clones were selected. This was the case for 2090 clusters. In 89% of the cases in which two clones were selected from an ONF cluster, sequence analysis verified that the sequences overlapped by some part and hence represented the same gene.
EST Sequencing and Clustering
Sequencing reactions were carried out on PCR products, using ABI 377 or ABI 3700 capillary automatic sequencers with fluorescent dye terminator technology.
The EST clustering procedure was carried out in three steps: (1) The raw ABI sequencer files were clipped with respect to quality, repeats, and vector content. Quality clipping was based on phred (Ewing et al. 1998) quality values, that is, a window of size 20 bp was slid through the quality files from both sides, and the clip positions (left/right) were determined by the first window position, where a threshold of phred-val. 15 was exceeded. Vector masking was performed by cross_match (http://www.phrap.org/phrap_documentation.html). The average length of all high-quality single reads after this procedure was 550 bp. (2) By an ALL-vs-ALL comparison (BLAST), the ESTs (from step 1) were preclustered: Sequences overlapping each other by >80 bp were recursively grouped into preclusters. (3) The preclusters from step 2 were then used as input sequence sets for the final clustering step based on true sequence assembly (cap3). In many cases, the preclusters cannot continuously be merged, but are broken into smaller clusters. The cluster-IDs given by the clustering procedure (Perl script) reflect the original preclustering by using the same main-ID for all clusters of the same precluster. The clusters were named using secondary IDs. The average length of all clusters, including singletons was 680 bp.
Sequence Analysis
Homology searches were carried out using WU-BLAST (W. Gish, 1996-2003; http://blast.wustl.edu). The Proteome database used (Proteomes_April2003_v1) was built by combining the following protein data sets: Wormpep 97 (Sanger center, 20.2.2003 release), C. intestinalis Release1 (JGI, http://www.jgi.doe.gov), D. melanogaster (Ensembl release 11.3.1), H. sapiens (Ensembl release 4.28), M. musculus (Ensembl release Jan. 2002), and F. rubripes (Ensembl release 10.2.1). In total, this database contained 141,932 predicted proteins.
Assignments of sea urchin, Ciona, Fugu, and human genes to the CDY system were carried out as described in Panopoulou et al. (2003). The lists of 2101 putative Ciona-specific (http://genome.jgi-psf.org/ciona4/ascidian.txt) and 675 deuterostome/chordate (http://genome.jgi-psf.org/ciona4/chordate.txt) specific genes identifiers were downloaded from the science supplementary material Web server at http://www.sciencemag.org/cgi/content/full/298/5601/2157/DC1.
Protein alignments were generated with CLUSTAL W (Thompson et al. 1994). Conserved parts of the alignments were selected using Gblocks (Castresana 2000). The neighbor-joining method (Saitou and Nei 1987) was used to construct phylogenetic trees of only the alignments that were longer than 80 amino acids.
To estimate the proportion of coding sequences important for sequence-based functional predictions, the coding potential of each EST was investigated using the MZEF (Zhang 1997) and Genscan (Burge and Karlin 1997) programs. Because both these programs are designed to find genes in human genomic DNA, their performance in predicting coding sequences in sea urchin ESTs had first to be investigated. We found that 54.4% of all 5′ ESTs were predicted to be coding with either of the two programs (from 30,474 reads). To correct for low-quality reads, we extracted high-quality sequences by selecting reads with 500 or more bases (all sequences were quality clipped before any analysis). A total of 22,874 5′ reads fulfilled this criterion. Of these clones, 63% were found to be coding. The reliability of the above programs in predicting coding regions within sea urchin ESTs was tested by applying them to a set of ESTs with BLAST similarity match of e ≤ 10-40 to known proteins, therefore creating a set with equal or close to 100% coding sequence. Of 281 sequences that fulfilled this criterion, 82.2% were predicted by either of the programs as being coding. Therefore, we inferred that this computational approach to prediction of EST coding potential has a false-negative proportion of 17.8% and introduced this as a correction value to be added to the predictions. As a result of this correction, 80.9% of the 5′ and 72.5% of the 3′ reads were predicted to be coding.
Classification of ESTs into GO-defined functional classes was carried out via the keywords found in their SWISS-PROT matches and via InterPro domains detected. Detection of domains in the ESTs required the prior open reading frame prediction. To translate EST sequences, frameshift-sensitive programs were tested. The ESTScan (Iseli et al. 1999) and Framefinder (http://www.hgmp.mrc.ac.uk/~gslater/estateman/framefinder.html) programs were tested on a set of selected full-length gene sequences and the clustered EST set. The quality criteria tested were (1) the length of translated sequences that were plotted against each other and (2) the comparison of e-values as results from BLAST searches using translated sequences and BLASTX or the nucleotide sequences using T-BLASTX. The e-values and sequence length were investigated manually for ∼200 randomly chosen sequences. Framefinder was found to make a translation in any case even if it was unmeaningful (<50 amino acids), and a majority of translations were significantly shorter compared with ESTscan. ESTscan was finally used to translate all clusters.
The 42 completely sequenced S. purpuratus BAC sequences were analyzed using Genscan. These sequences are described in Davidson et al. (2002a) and were downloaded from the sea urchin genome Web site (http://jhegaala.caltech.edu/~t/transfer/bacs.tar.gz).
Computing and Database Management
Development and maintenance of the sea urchin database application was done on a DEC-Alpha computer with OSF-1 V4.0. The database was implemented with a sqlite database (http://www.sqlite.org) and a flat file system for accessing BLAST files. The database interface is created dynamically by CGI scripts (http://stein.cshl.org/WWW/software/CGI/) written in the Perl programming language (http://www.perl.org). The connection to the database is done via the DBI Perl module (http://dbi.perl.org/index.html) and the DBD::Sqlite module (http://search.cpan.org/perldoc?DBD::SQLite).
Resources
For each EST described in this project, trace control files allowing the visualization of the quality of each base, as well as cDNA clones and filter arrays, are available at the resource center of the German genome project (http://www.rzpd.de). The RZPD library number identifiers are 536 (40-h gastrula), 537 (20-h blastula), 538 (7 h cleavage), 621(unfertilized egg), and 691(larva). The RZPD link to order clones is also directly given in GenBank at NCBI.
Acknowledgments
We thank Matthew Clark, Pia Aanstad, Georg Otto, and Carola Burgtorf for discussions in the course of this project and Anahid Powell and Brian Cusack for help on the manuscript. We thank the RZPD for support with DNA filter production and Mario Drungowski for help with image analysis. This work was supported by the Max Planck Gesellschaft zur Förderung der Wissenschaften e.v. A.C. and larval library construction was supported by US NSF grant IBN-9982875.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1674103.
Footnotes
[Supplemental material is available online at www.genome.org. All data described are deposited in a searchable online database available at http://www.molgen.mpg.de/ag_seaurchin/. All sequences described in this study have been submitted to the GenBank EST section under accession nos. CD289359-CD297607, CD303636-CD324638, and CD330178-CD342180.]
References
- Angerer, L.M. and Angerer, R.C. 2003. Patterning the sea urchin embryo: Gene regulatory networks, signaling pathways, and cellular interactions. Curr. Top. Dev. Biol. 53: 159-198. [DOI] [PubMed] [Google Scholar]
- Apweiler, R., Attwood, T.K., Bairoch, A., Bateman, A., Birney, E., Biswas, M., Bucher, P., Cerutti, L., Corpet, F., Croning, M.D., et al. 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29: 37-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas-Mena, C., Cameron, A.R., and Davidson, E.H. 2000. Spatial expression of Hox cluster genes in the ontogeny of a sea urchin. Development 127: 4631-4643. [DOI] [PubMed] [Google Scholar]
- Arnone, M.I. and Davidson, E.H. 1997. The hardwiring of development: Organization and function of genomic regulatory systems. Development 124: 1851-1864. [DOI] [PubMed] [Google Scholar]
- Burge, C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78-94. [DOI] [PubMed] [Google Scholar]
- Cameron, R.A., Mahairas, G., Rast, J.P., Martinez, P., Biondi, T.R., Swartzell, S., Wallace, J.C., Poustka, A.J., Livingston, B.T., Wray, G.A., et al. 2000. A sea urchin genome project: Sequence scan, virtual map, and additional resources. Proc. Natl. Acad. Sci. 97: 9514-9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana, J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17: 540-552. [DOI] [PubMed] [Google Scholar]
- Clark, M.D., Panopoulou, G.D., Cahill, D.J., Bussow, K., and Lehrach, H. 1999. Construction and analysis of arrayed cDNA libraries. Methods Enzymol. 303: 205-233. [DOI] [PubMed] [Google Scholar]
- Clark, M.D., Hennig, S., Herwig, R., Clifton, S.W., Marra, M.A., Lehrach, H., Johnson, S.L., and the WU-GSC EST Group. 2001. An oligonucleotide fingerprint normalized and expressed sequence tag characterized zebrafish cDNA library. Genome Res. 11: 1594-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, E.H. 1986. Gene activity in early development, 3rd ed. Acadamic Press, Orlando, FL.
- Davidson, E.H., Rast, J.P., Oliveri, P., Ransick, A., Calestani, C., Yuh, C.H., Minokawa, T., Amore, G., Hinman, V., Arenas-Mena, C., et al. 2002a. A genomic regulatory network for development. Science 295: 1669-1678. [DOI] [PubMed] [Google Scholar]
- Davidson, E.H., Rast, J.P., Oliveri, P., Ransick, A., Calestani, C., Yuh, C.H., Minokawa, T., Amore, G., Hinman, V., Arenas-Mena, C., et al. 2002b. A provisional regulatory gene network for specification of endomesoderm in the sea urchin embryo. Dev. Biol. 246: 162-190. [DOI] [PubMed] [Google Scholar]
- Dehal, P., Satou, Y., Campbell, R.K., Chapman, J., Degnan, B., De Tomaso, A., Davidson, B., Di Gregorio, A., Gelpke, M., Goodstein, D.M., et al. 2002. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science 298: 2157-2167. [DOI] [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- Friedman, R. and Hughes, A.L. 2001. Pattern and timing of gene duplication in animal genomes. Genome Res. 11: 1842-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman, J. 2002. Morpholino oligos: Making sense of antisense? Dev. Biol. 243: 209-214. [DOI] [PubMed] [Google Scholar]
- Herwig, R., Poustka, A.J., Muller, C., Bull, C., Lehrach, H., and O'Brien, J. 1999. Large-scale clustering of cDNA-fingerprinting data. Genome Res. 9: 1093-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herwig, R., Schulz, B., Weisshaar, B., Hennig, S., Steinfath, M., Drungowski, M., Stahl, D., Wruck, W., Menze, A., O'Brien, J., et al. 2002. Construction of a `unigene' cDNA clone set by oligonucleotide fingerprinting allows access to 25,000 potential sugar beet genes. Plant J. 32: 845-857. [DOI] [PubMed] [Google Scholar]
- Iseli, C., Jongeneel, C.V., and Bucher, P. 1999. ESTScan: A program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 138-148. [PubMed]
- McLysaght, A., Hokamp, K., and Wolfe, K.H. 2002. Extensive genomic duplication during early chordate evolution. Nat. Genet. 31: 200-204. [DOI] [PubMed] [Google Scholar]
- Meier-Ewert, S., Lange, J., Gerst, H., Herwig, R., Schmitt, A., Freund, J., Elge, T., Mott, R., Herrmann, B., and Lehrach, H. 1998. Comparative gene expression profiling by oligonucleotide fingerprinting. Nucleic Acids Res. 26: 2216-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwokoro, N.A., Wassif, C.A., and Porter, F.D. 2001. Genetic disorders of cholesterol biosynthesis in mice and humans. Mol. Genet. Metab. 74: 105-119. [DOI] [PubMed] [Google Scholar]
- Panopoulou, G., Hennig, S., Groth, D., Krause, A., Poustka, A.J., Herwig, R., Vingron, M., and Lehrach, H. 2003. New evidence for genome-wide duplications at the origin of vertebrates using an amphioxus gene set and completed animal genomes. Genome Res. 13: 1056-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi, E. 2002. SEQUENCING: Chimps and fungi make genome. Science 296: 1589b-1591. [DOI] [PubMed] [Google Scholar]
- Poustka, A.J., Herwig, R., Krause, A., Hennig, S., Meier-Ewert, S., and Lehrach, H. 1999. Toward the gene catalog of sea urchin development: The construction and analysis of an unfertilized egg cDNA library highly normalized by oligonucleotide fingerprinting. Genomics 59: 122-133. [DOI] [PubMed] [Google Scholar]
- Prince, V.E. and Pickett, F.B. 2002. Splitting pairs: The diverging fates of duplicated genes. Nat. Rev. Genet. 3: 827-837. [DOI] [PubMed] [Google Scholar]
- Saitou, N. and Nei, M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406-425. [DOI] [PubMed] [Google Scholar]
- Swann, K. and Parrington, J. 1999. Mechanism of Ca2+ release at fertilization in mammals. J. Exp. Zool. 285: 267-275. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker, M. and Larman, M.G. 2001. Calcium and mitosis. Semin. Cell Dev. Biol. 12: 53-58. [DOI] [PubMed] [Google Scholar]
- Wiemann, S., Weil, B., Wellenreuther, R., Gassenhuber, J., Glassl, S., Ansorge, W., Bocher, M., Blocker, H., Bauersachs, S., Blum, H., et al. 2001. Toward a catalog of human genes and proteins: Sequencing and analysis of 500 novel complete protein coding human cDNAs. Genome Res. 11: 422-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K.H. 2001. Yesterday's polyploids and the mystery of diploidization. Nat. Rev. Genet. 2: 333-341. [DOI] [PubMed] [Google Scholar]
- Wolfe, K.H. and Li, W.H. 2003. Molecular evolution meets the genomics revolution. Nat. Genet. 33 Suppl: 255-265. [DOI] [PubMed] [Google Scholar]
- Xie, H., Wasserman, A., Levine, Z., Novik, A., Grebinskiy, V., Shoshan, A., and Mintz, L. 2002. Large-scale protein annotation through gene ontology. Genome Res. 12: 785-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M.Q. 1997. Identification of protein coding regions in the human genome by quadratic discriminant analysis. Proc. Natl. Acad. Sci. 94: 565-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X., Mahairas, G., Illies, M., Cameron, R.A., Davidson, E.H., and Ettensohn, C.A. 2001. A large-scale analysis of mRNAs expressed by primary mesenchyme cells of the sea urchin embryo. Development 128: 2615-2627. [DOI] [PubMed] [Google Scholar]
- Zimmerberg, J., Coorssen, J.R., Vogel, S.S., and Blank, P.S. 1999. Sea urchin egg preparations as systems for the study of calcium-triggered exocytosis. J. Physiol. 520 Pt 1: 15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://blast.wustl.edu; WU-BLAST.
- http://dbi.perl.org/index.html; perl DBI-module homepage.
- http://genome.jgi-psf.org/ciona4/ascidian.txt; Ciona-specific gene identifiers.
- http://genome.jgi-psf.org/ciona4/chordate.txt; deuterostome/chordate-specific gene identifiers.
- http://jhegaala.caltech.edu/~t/transfer/bacs.tar.gz; sea urchin genome.
- http://search.cpan.org/perldoc?DBD::SQLite; perl DBD::SQLite-module.
- http://stein.cshl.org/WWW/software/CGI; perl CGI-module.
- http://www.expasy.org; Expert Protein Analysis System Molecular Biology Server.
- http://www.hgmp.mrc.ac.uk/~gslater/estateman/framefinder.html; Framefinder program.
- http://www.jgi.doe.gov; Joint Genome Institute.
- http://www.molgen.mpg.de/ag_seaurchin/; sea urchin database.
- http://www.perl.org; perl programming language.
- http://www.phrap.org/phrap_documentation.html; documentation on phrap.
- http://www.rzpd.de; Resource Center/Primary database of the German genome project.
- http://www.sciencemag.org/cgi/content/full/298/5601/2157/DC1; Science supplemental material Web server.
- http://www.sqlite.org; sqlite.