Abstract
The draft Fugu rubripes genome was released in 2002, at which time relatively few cDNAs were available to aid in the annotation of genes. The data presented here describe the sequencing and analysis of 24,398 expressed sequence tags (ESTs) generated from 15 different adult and juvenile Fugu tissues, 74% of which matched protein database entries. Analysis of the EST data compared with the Fugu genome data predicts that approximately 10,116 gene tags have been generated, covering almost one-third of Fugu predicted genes. This represents a remarkable economy of effort. Comparison with the Washington University zebrafish EST assemblies indicates strong conservation within fish species, but significant differences remain. This potentially represents divergence of sequence in the 5′ terminal exons and UTRs between these two fish species, although clearly, complete EST data sets are not available for either species. This project provides new Fugu resources, and the analysis adds significant weight to the argument that EST programs remain an essential resource for genome exploitation and annotation. This is particularly timely with the increasing availability of draft genome sequence from different organisms and the mounting emphasis on gene function and regulation.
The Japanese puffer fish (Fugu rubripes) was the second vertebrate genome to be completed to draft quality (Aparicio et al. 2002). Although this organism is intractable to experimental analysis, it is widely used as a tool in comparative genomic analyses (Barton et al. 2001; Rothenberg 2001; Brenner et al. 2002; Annilo et al. 2003; Goode et al. 2003; Nelson 2003; Yap et al. 2003). Indeed, partial sequence of a closely related fresh water puffer fish, Tetraodon nigroviridis, has been specifically promoted and used as a gene-finding tool (“Exofish”) for the human genome (Roest Crollius et al. 2000). Functional inferences based on interspecies sequence comparison have validated the use of comparative genomics (Makalowski and Boguski 1998), as evidenced by the increasing numbers of genomes in the sequencing pipelines, including Ciona (Dehal et al. 2002), mouse (Waterston et al. 2002), and zebrafish (http://www.ensembl.org/Danio_rerio) which are completed or well underway. Others are expected to follow and include Xenopus, sea urchin, and chicken.
The Fugu draft sequence indicates a total genome size of 365 Mb (Aparicio et al. 2002). This draft sequence represents 95% coverage of the genome in the form of unordered contigs, termed “scaffolds,” 80% of which contain two or more genes. Searches of the complete set of human predicted genes against the Fugu draft sequence produced strong matches for three-quarters of the data set, indicating large-scale conservation of gene content over 450 Myr of evolution (Kumar and Hedges 1998). The current Ensembl build of the Fugu genome sequence (v.12.2.1) comprises 35,180 Ensembl gene predictions and 38,510 predicted Ensembl gene transcripts (http://www.ensembl.org/Fugu_rubripes; referred to here as Fugu predicted genes [FPGs]). Annotation is based on ab initio gene predictions, homology, and gene prediction HMMs. Although many of these predicted genes show cross-phylum matches and are annotated as such via BLAST match results, there are still many predictions of novel genes. These may be Fugu- or fish-specific genes not shared by mammals or may reflect the fact that many vertebrate genes are highly derived. For example, many receptors or ligands identified in the mammalian immune and hematopoietic systems are either absent from Fugu or their orthologs are not easily identified by comparative BLAST analysis (Aparicio et al. 2002; Sambrook et al. 2002; K. Hultman and S. Johnson, unpubl.).
Although annotated gene prediction is increasingly accurate (Rogic et al. 2001,2002; Mathe et al. 2002), particularly if predictions from several programs are combined, conclusive identification and delimitation of coding regions is best provided by cDNA sequences. However, mass generation of full-length cDNA sequence is not an insubstantial challenge, and the most efficient method of generating cDNA resources is using single-pass sequencing of cDNA libraries, to generate expressed sequence tags (ESTs; Adams et al. 1995; Hillier et al. 1996; Gong 1999; Clark et al. 2001; Boardman et al. 2002). Given the current acknowledged value of the Fugu genome data, increased resources for transcriptional analysis and annotation pipelines will enhance the usefulness, especially for exploitation and data-mining of conserved noncoding regions. Therefore development of EST resources from this organism is a priority. This paper presents the results of the first major EST project conducted using this organism. The sequencing and analysis of 24,398 ESTs generated from 15 different cDNA libraries of adult ovary, fin, heart, intestine, skin, and muscle and juvenile whole body, spleen, gill, gut, gonad (undifferentiated), brain, eye, liver, and kidney is presented. The clones which were used to generate these ESTs are publicly available and represent a valuable resource for follow-up laboratory investigation by the wider community.
RESULTS
Overview of ESTs From All Libraries
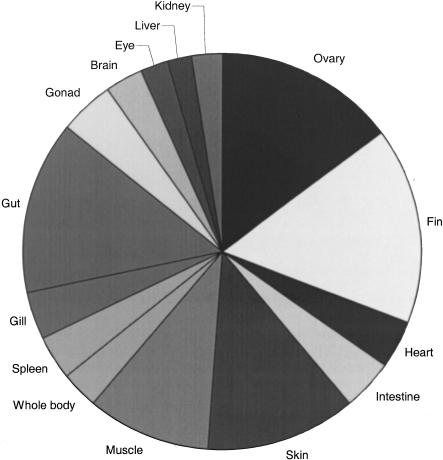
Fifteen cDNA libraries were produced from different tissues (ovary, fin, heart, intestine, skin, muscle [from an adult fish], whole body, spleen, gill, gut, gonad [undifferentiated], brain, eye, liver, and kidney [from a juvenile fish]). These were single-pass sequenced for ESTs, with each cDNA only sequenced once from the 5′ end of the clone, with average read lengths of 500 bp. Figure 1 provides a breakdown of the 24,398 EST sequences according to tissue origin.
Figure 1.
Tissue composition of the sequenced clones shown as a percentage of the total number of ESTs sequenced.
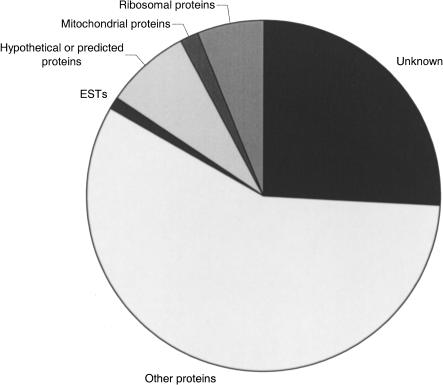
Seventy-four percent of the Fugu EST data set matched database entries in the SPTR database (BLAST bit score > 50; Fig. 2). The remaining 26% failed to match proteins in the SPTR database and therefore represented potentially novel sequences or UTRs of known genes. Of those matching SPTR entries, a small percentage (2%) were mitochondrial genes and 6% were ribosomal proteins. Additionally, 8% of the EST matches against the SPTR database were for hypothetical genes and other previously unsubstantiated ESTs, thus helping provide verification of these predictions.
Figure 2.
Fugu ESTs matching SPTR data using BLAST sequence similarity searching. Other proteins are those which matched SPTR entries and therefore comprise “known” genes.
Some of the libraries, specifically those denoted as KK/SS libraries in the Methods section, were generated from oligo-capping procedures to protect the 5′ ends of the transcripts against degradation prior to first-strand cDNA synthesis (Maruyama and Sugano 1994; Suzuki et al. 1997). As a result, a high percentage of ESTs from these libraries (21%) have BLAST matches to SPTR database entries that include the first SPTR amino acid. In most of these cases, the EST contains additional sequence 5′ of the alignment as well; that is, 5′ UTR (data not shown). However, identification of 5′ gene ends using such BLAST sequence similarity searches of coding sequence is a relatively crude method. Therefore this figure of 21% is probably an underestimate. In-depth analysis of selected ESTs (data not shown) indicates that the figure for full-length cDNAs may be as high as 33%.
Number of Identified Genes
Two methods were used to estimate the total number of Fugu genes identified by this EST program: (1) ICAtools (Parsons 1995), a sequence clustering tool, and (2) associating ESTs with their corresponding FPGs followed by clustering of residual ESTs based on their overlap. These efforts resulted in estimates of 9200 and 10,116 genes, respectively. Because Fugu is predicted to have 38,510 gene transcripts, this EST project identified transcript tags for between 25% and 28% of the Fugu predicted expressed gene set. The lower estimate for gene number identified by the EST project was obtained using ICAtools clustering, and this was regarded as an absolute minimum gene number, as ICAtools has a tendency to cluster paralogous sequences, as will be discussed later. The higher estimate, which is probably more accurate, was obtained first using BLAST to identify nearly identical matches of ESTs to the Fugu predicted gene (FPG) set, independent of the need for sequence overlap between ESTs. We found that 12,209 of the ESTs hit 4199 (10.8%) of the FPGs. However, this still left 12,472 Fugu ESTs without a match to FPGs. Phrap clustering of these remaining sequences then associated 6758 ESTs into 1433 clusters, with an average of 4.7 reads per cluster. Because each of these clusters contains more than one EST (and thus represents more than one cDNA), these clusters were taken as strong evidence for the presence of bona fide genes. Whether these identify new genes not in the FPG set, or instead identify 5′ UTRs of genes in the FPG data set is not clear. This still left 5714 singletons with no FPG match or cluster data. It should be remembered at this point that the Fugu genome is not complete and therefore not all ESTs would be expected to find a match against the genome data. Genomic contamination is often a minor problem with EST sequencing programs, and estimates from the WashU zebrafish EST project suggest that approximately 5% of EST clones result from genomic contamination or otherwise are unlikely to represent coding sequence (R. Waterman and S. Johnson, unpubl.). Although genomic contamination is likely to be highly library-dependent, assuming that those results hold for the libraries reported here, it is therefore expected that approximately 1230 (5% of the 24,398 EST clones) of the ESTs are also the result of genomic contamination. Thus the 5714 singletons may only represent somewhere in the order of 4484 (5714 – 1230) bona fide genes. With this adjustment, the number of Fugu genes identified in this project using this method totals 10,116 (4199 + 1433 + 4484).
Gene Diversity and Gene Discovery
An important issue in EST projects is the identification of libraries for more extensive analysis. None of the libraries reported here were generated with normalization methods. Accordingly, an understanding of library complexity (referred to in Table 1 as library diversity) and the probability that further sequencing from each will identify transcripts from genes without prior identified transcripts (referred to in Table 1 as library discovery) is of great utility. The two methods used previously to estimate gene number (ICAtools and FPG comparison followed by Phrap) were also used to analyze the gene discovery and diversity potential of each of the libraries (Table 1).
Table 1.
Gene Diversity and Discovery Analyzed Using ICAtools (Displaying the Number of Singletons, Paired Sequences, Trios, and Those Present in Clusters of Four or More), and WU-BLAST
| Ovary | Fin | Heart | Intestine | Skin | Muscle | Whole body | Spleen | Gill | Gut | Gonad | Brain | Eye | Liver | Kidney | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Icatools | |||||||||||||||
| 1 | 53.6 | 28.7 | 47.7 | 48.6 | 46.6 | 20.4 | 26.7 | 48.7 | 47.7 | 38.6 | 34.9 | 66.5 | 57.8 | 28.9 | 46.9 |
| 2 | 8.3 | 6.0 | 5.8 | 8.9 | 9.7 | 4.8 | 6.9 | 5.4 | 8.9 | 8.3 | 6.8 | 6.9 | 10.3 | 8.3 | 9.4 |
| 3 | 2.8 | 2.5 | 2.0 | 3.0 | 2.9 | 1.3 | 3.4 | 3.2 | 4.3 | 2.9 | 3.0 | 2.1 | 2.0 | 3.4 | 3.4 |
| 4+ | 35.3 | 62.8 | 44.5 | 39.5 | 41.8 | 73.5 | 63.0 | 42.7 | 39.1 | 50.2 | 55.3 | 24.5 | 29.9 | 59.4 | 40.3 |
| Discovery | 0.33 | 0.14 | 0.31 | 0.26 | 0.25 | 0.10 | 0.13 | 0.28 | 0.27 | 0.21 | 0.18 | 0.35 | 0.26 | 0.17 | 0.23 |
| Diversity | 0.71 | 0.45 | 0.66 | 0.70 | 0.66 | 0.34 | 0.51 | 0.65 | 0.67 | 0.58 | 0.50 | 0.76 | 0.75 | 0.48 | 0.65 |
Diversity is defined as the number of different “genes” each library contributed, divided by the library size. Discovery is defined as the number of singletons in each library divided by library size.
Globally the redundancy was high, with 68.5% of ESTs present in 4+ copies, but there were clear differences among the libraries. The cluster patterns produced by ICAtools were largely reproduced when considering each library separately against the gene discovery and gene diversity ratios (Table 1). Both methods clearly showed that the muscle, whole body, and fin libraries were the most redundant, with highest diversity in the brain, eye, and ovary libraries. Another measure of redundancy is to define which genes comprise the largest clusters (data not shown), in this case, purely generated using ICAtools. This is also very useful information for future library production, as identification of highly represented clones provides sequences which can be used in a relatively simple and directed subtraction methodology. The most obvious candidates for this were cytokeratin in fin (comprising almost 18% of this particular library) and phosphoglycerate kinase and L-lactate dehydrogenase in muscle (comprising 9.06% and 7.51% of this library, respectively). Globally, the most common species were cytokeratin, beta globin, phosphoglycerate kinase, actin, and elongation factor 1α. This analysis was also a useful check for genomic contamination of repeat sequences. The five largest clusters from each tissue were BLAST sequence similarity-searched against the SPTR database, and those which did not produce significant matches were subsequently BLAST searched against the Fugu genome data. Of the latter, all mapped to unique locations in the genome, frequently 5′ to known genes, indicating their potential as 5′ UTRs.
Comparison of Fugu EST Clusters to Zebrafish EST Assemblies
With the sequencing of the zebrafish genome and the current availability of a large number of zebrafish ESTs (150,695) and clusters (25,184 assemblies from 156,067 ESTs generated from 105,565 clones), it was of interest to compare the Fugu and zebrafish data to determine how similar the two data sets were from two fish species, which diverged approximately 250 Myr ago. Matches to zebrafish ESTs would also verify Fugu EST clusters, particularly those with no match to FPGs. Overall, 14,931 Fugu ESTs or approximately 61% of the Fugu EST data set matched the current set of WashU zebrafish EST assemblies. Of the Fugu ESTs that matched FPGs (detailed in the section “Number of Identified Genes”), 82% also matched the WashU zebrafish EST assemblies (WZ assemblies, R. Waterman and S. Johnson, unpubl.). Of the Fugu ESTs that did not match FPGs, 40% of the non-FPG-hitting contigs match WZ assemblies, and 21.8% of non-FPG-hitting singletons match WZ assemblies. The low percentage of non-FPG singletons that match zebrafish EST clusters, compared to the number of non-FPG clusters that match zebrafish EST assemblies, may be due to genomic contamination, although current estimates of the latter suggest that this is not the whole picture, and the possibility cannot be ruled out that this class is enriched for poorly expressed Fugu-specific genes instead.
DISCUSSION
These EST data describe an important resource of cDNAs, which will allow more efficient exploitation of the Fugu genome data and added value for comparative genomics studies. Approximately one-third of the estimated number of Fugu predicted genes were tagged by the 24,398 ESTs generated within this project. This is remarkably efficient and is almost certainly due to the sampling of many libraries from different tissues.
The EST data were globally analyzed for the number of Fugu genes tagged by ESTs, content, and redundancy. In most of the analyses, two methods were used, ICAtools and an analysis based on comparison to the Fugu genome sequence and the number of predicted genes, followed by Phrap-based clustering of ESTs that failed to correspond to Fugu predicted genes. The results from these analyses give somewhat different estimates for gene number (9100 and 10,116, respectively), but similar estimates for gene diversity and discovery within each library (Table 1). The latter results are an important consideration in deciding how deep to sequence from each library, whether new libraries are needed, and how many ESTs are needed to adequately sequence the transcriptome. Muscle, whole body, and fin were the most redundant, whereas the brain, eye, and ovary libraries promised the highest gene discovery ratios and therefore present clear candidates for further sequencing. The data set for each library varied in terms of sample size (ranging from 444 for the eye library to 3916 for the fin library) due to clone availability, but the smaller data sets were still of sufficient size to estimate complexity. Analysis of the muscle library data set indicated that 200 clones produced a relatively accurate percentage for redundant clones. In general, the gene diversity and discovery of the libraries used through this stage of the project remain high. Additional libraries from other tissues may be necessary to expand the project and tag the majority of Fugu genes with ESTs.
Any estimate of gene number from EST programs is largely dependent on the bioinformatics tools used to cluster the data. Estimates in this project varied from 9100 to 10,116. ICAtools, which gave the lower number, has a tendency to cluster paralogous sequences when using the default parameters. In-depth analysis of the whole-body library ICA-matches results compared to SPTR data revealed, for example, that the largest cluster with parent sequence similarity to α-hemoglobin was comprised of both hemoglobin α-chain and embryonic-type α-sequences. These two sequences share 72.7% amino acid sequence similarity and should tend to cluster independently; however, short stretches in excess of 50 bp with greater than 95% identity caused over-clustering. Similar over-clustering was also observed for other gene families. However, for rapid and simple cluster analysis ICAtools is useful, providing a measure of library redundancy and therefore an indicator of the efficiency of sequencing more clones. In contrast, the method based on comparison to predicted genes from the Fugu genomic sequence depends strongly on the efficiency of the gene predictor program to properly associate all of the exons from the same gene together in the same gene model. It also requires the availability of the genome data, an uncommon situation with most organisms.
The libraries described here are not normalized. However, they may be useful for generating gene expression profiles. Examples of some gene expression profiles across the Fugu libraries are given in Table 2. Some of these are highly specific, such as the ATP-dependent helicase ddx1, which was found tagged only in the ovary library, whereas others such as the 40s ribosomal protein S24 was tagged in 11 of the 15 libraries sampled. Of particular interest for gene annotation and discovery are the ESTs matching predicted genes with no ascribed function, such as the kiaa0922, flj22313, and cgi-51 proteins (Lai et al. 2000). Karsi et al. (2002) in their analysis of catfish skin cDNAs also noted many examples of ESTs with significant similarity to known sequences of unknown function in model systems such as human, mouse, cattle, Drosophila, and C. elegans. Although these sequences have no ascribed function, their conservation in mammals, fish, and invertebrates helps to provide evidence that these sequences have important functions conserved through many hundreds of millions of years of evolution. Identification of such conserved sequences between Fugu and human, or Fugu and other organisms, such as zebrafish, allows for more efficient annotation (in larger numbers) than that which can be currently obtained by experimental biology.
Table 2.
Examples of Expression Profiles Taken From the Fugu EST Libraries
|
Genes
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tissue | cgi-51 | apl | atpase6 | rps24 | kiaa0922 | ddx1 | lun-1 | flj22313 | epd-i | tmsb 12 |
| Ovary | +++++++ | ++ | ++ | ++ | ++ | |||||
| Fin | ++ | ++ | ||||||||
| Heart | ++ | ++ | + | |||||||
| Intestine | +++ | |||||||||
| Skin | + | + | ||||||||
| Muscle | ++++++ | ++ | ||||||||
| Whole body | ++ | + | ++ | |||||||
| Spleen | + | +++++++ | ||||||||
| Gill | +++ | ++ | ++ | |||||||
| Gut | + | ++ | + | + | ||||||
| Gonad | ++ | +++++ | ||||||||
| Brain | +++++ | + | ++ | +++ | ++ | |||||
| Eye | ++ | |||||||||
| Liver | ++ | + | ||||||||
| Kidney | ++ | + | +++ | |||||||
“+” denotes the presence of a single clone in a particular library. alp, actinin-associated LIM protein; atpase6, ATP synthase 6 (mitochondrial protein); rps244, 40s ribosomal protein S24; ddx1, ATP-dependent helicase; lun-1, Ring finger protein; epd-i, Ependymin I precursor; tmsb 12, Thymosin beta-12.
Almost 25% of the Fugu ESTs produced no BLAST matches against the SPTR database. This failure to match SPTR records could have been due to ESTs being derived from novel, Fugu, or fish-specific genes or that corresponding fish ESTs are simply not in the database. Although the zebrafish EST assembly database represents >100,000 clones, it is thought to identify only approximately 50% of zebrafish genes. Alternatively, this may reflect the fact that the Fugu EST sequences were all 5′ reads that may have been limited to 5′ UTR or noncoding or poorly conserved first exons. This could also be the reason why over 18% of ESTs that matched FPGs did not match any zebrafish WZ assemblies. Fugu and zebrafish diverged around 250 Myr (compared to human and mouse, which diverged around 80 Myr), and there are an increasing number of examples (M.S. Clark, unpubl.) where a full-length zebrafish cDNA sequence fails to identify the terminal 5′ exons of a gene in Fugu genomic sequence. Extrapolating the mammalian data, it is even less likely that there will be significant sequence similarity between the UTRs of Fugu and zebrafish, as a comparison of human and mouse UTRs produced only 67% and 69% nucleotide sequence identity for 5′ and 3′ UTRs, respectively (Makalowski et al. 1996), and an excess of sequence divergence in 5′ UTRs has been shown between human and chimpanzee (Hellmann et al. 2003).
Even with comparative genomics, finding genes in genomic sequence is a far from trivial problem. In general, approximately one-half of the genes can be found by homology, with the remaining relying on predictive methods for discovery (Mathe et al. 2002). Gene prediction programs are becoming increasingly accurate, with more than 90% of coding nucleotides correctly predicted (Burset and Guigo 1996; Claverie 1997; Guigo 1997; Burge and Karlin 1998; Haussler 1998; Rogic et al. 2001). Exact exon boundary definitions are only predicted with 70%–75% accuracy, whereas less than 50% of predicted genes correspond exactly to the actual transcripts (Dunham et al. 1999; Rogic et al. 2001). Most current gene prediction programs are trained on coding sequence and are poor at predicting alternative splice forms and noncoding regions such as noncoding RNAs, noncoding first exons, and UTRs. This situation is exacerbated when these UTRs have interspersed introns and also exhibit alternative splicing (Mathe et al. 2002). This presents as a large gap in the knowledge base, as recent estimates suggest that between 35% and 59% of human genes exhibit at least one alternative splice form (Modrek and Lee 2002) and 40% of human genes have completely noncoding first exons (Davuluri et al. 2001). A similar situation has been demonstrated in mouse, with further evidence presented to suggest that noncoding RNAs are a major component of the transcriptome (Okazaki et al. 2002). A similar situation would be expected in the other vertebrates.
The current gap in the ability of gene prediction programs to annotate complete gene structures reinforces the indispensable role of ESTs in genome annotation (Rogic et al. 2002). ESTs provide the means to identify transcription start sites and first exons of genes, especially when they are generated by the oligo-capping method (Suzuki et al. 1997,2002). Data from the human genome project verify the efficiency of EST-driven annotation. At high stringency, 70%–90% of all annotated genes were detected by near identity to EST sequences, and approximately half the alignments spanned multiple exons, thus aiding in the construction of gene predictions and elucidation of alternative splicing (Bailey Jr. et al. 1998). In acknowledgment of the important contribution ESTs can make to genome annotation, the Ensembl team is in the process of integrating EST data into Ensembl gene-building (Hubbard et al. 2002). The Fugu EST data have been made available to the Fugu Ensembl team for use in future annotation programs.
A complementary problem to identifying the transcribed portions of genes is that of identifying the cis-regulatory sequences involved in promoting gene expression (e.g., promoters and enhancers). The Fugu genome offers a particularly attractive model for identifying the promoter and enhancer elements, due to the relatively small intragenic regions compared to other vertebrate genomes. Because most cis-acting elements are found 5′ of the transcript, or in the first intron (Mignone et al. 2002), accurate prediction of the regions in which these elements are found depends on identifying the transcribed portion of the gene. As first introns are often longer than average (Maroni 1996), promoters and transcription start sites may be well upstream of the ATG start codon. As described above, methods for identifying noncoding transcribed elements, lacking the constraints that coding sequence provides to gene structure prediction, are particularly difficult to develop. EST projects provide direct, experimental evidence for the transcribed and exonic portions of genes, thus limiting the possible region in which purely regulatory sequence is to be found. One practical use delimiting the boundary between promoters and enhancers, on one hand, and the transcription start site, on the other, lies in the experimental analysis of zebrafish development. Driving green fluorescent protein (GFP) as a lineage or cell marker reflecting expression of a gene requires cloning the entire promoter and enhancer region of the desired gene in front of GFP coding sequence. Unlike Fugu, the intergenic regions in the zebrafish genome are quite large and difficult to predict. As a consequence, large regions of the zebrafish promoter and enhancer region, perhaps more than can be cloned in conventional plasmids, are often needed to give specific and meaningful expression. This problem is partially alleviated by recombining GFP into BAC clones (Liu et al. 2003), but this solution is less amenable to high-throughput methodologies. An attractive alternative for zebrafish experimental biology is to use compact Fugu promoters to provide specific control of GFP expression in transgenic zebrafish. Indeed, use of Fugu promoters has already been successfully used to drive GFP expression in appropriate patterns in mouse (Brenner et al. 2002; Griffin et al. 2002; Camacho-Hubner et al. 2002). Efficient utilization of this idea for a large number of genes, in a more high-throughput manner, requires some knowledge of the 5′ UTR of the transcript. This initial Fugu EST project, which identifies likely 5′ ends for more than 10,116 different genes, will greatly facilitate using Fugu genomic sequence to develop transgenic zebrafish or mice expressing GFP in gene-specific expression patterns.
With the large push to sequence more and more genomes (many of which will only be completed to draft standard), there is not a corresponding and relatively cheap effort to match the genome sequencing with EST projects to help with annotation. The present data provide experimental evidence for a large fraction of Fugu genes, with 5′ ATG sites identified in approximately 33% of the clones sequenced. These data will provide significant new resources for experimental and computational biologists exploiting the Fugu genome sequence.
METHODS
CDNA Library Construction
Two sets of libraries were constructed for this project. The first set was constructed by G. Elgar, S. Warner, and J. Hills at the RFCGR (formerly the HGMP-RC), Hinxton, Cambridge. The tissues used in these libraries were whole body, spleen, gill, gut, gonad, brain, eye, liver, and kidney. The RNA for the libraries was extracted using the QIAGEN Rneasy Midi Prep System. First-strand cDNA was prepared using the Stratagene cDNA Synthesis kit with the addition of XhoI/EcoRI linkers. The inserts were directionally cloned (5′–3′) into EcoRI/XhoI-cut pBluescript II KS+ (Stratagene) in XL2-Blue MRF E. coli cells (Stratagene). Each library has an estimated average insert size of 1 kb. Clones are available from http://www.hgmp.mrc.ac.uk/geneservice/reagents/index.shtml.
A second set of libraries, denoted KK/SS, was constructed by Sumio Sugano, Koichi Kawakami, Masahide Sasaki, Yutaka Suzuki, Kiyoshi Kikuchi, and Shugo Watabe (University of Tokyo, Institute of Medical Science and Laboratory of Aquatic Molecular Biology and Biotechnology). The fish were obtained from the Tokyo Metropolitan Central Wholesale Market, Japan. The tissues used in these libraries comprised ovary, fin, heart, intestine, skin, and muscle. Total RNA was extracted using Trizol (Life Technologies) with RNeasy (QIAGEN). Poly A+ RNA was isolated using Oligo-Tex (Nippon-Roche). The libraries were 5′ capped double-stranded cDNA prepared according to Suzuki et al. (1997) and Maruyama and Sugano (1994). The inserts were directionally cloned (T3 [5′] to T7 [3′]) into a modified pBluescript-FL vector (cloning sites: R site 1: PflM I [CCANNNNNTGG], R site 2: PflM I [CCANNNNNTGG]). The BamHI-SmaI sites were converted to BamHI-PflMI-SfiI-PflMI sites (SmaI is destroyed). Other parts of the vector were unmodified in host DH10B (T1 phage-resistant). Each library has an estimated average insert size of 2–3 kb+. Distribution information can be found through the I.M.A.G.E. Consortium/LLNL: info@image.llnl.gov.
Partial Sequencing of 5′ Ends of cDNA Inserts
The RFCGR libraries sequenced at the RFCGR were sequenced using limiting dilutions of dNTPs and primers, as described at http://fugu.hgmp.mrc.ac.uk/Protocols/Biology/. Inserts were amplified using short T7 and T3 primers, and the PCR products were directly sequenced using cDNA1 primer 5′ggcgaattggagctc caccg3′ and Big Dye Terminator sequencing mix. The KK/SS libraries sequenced at the RFCGR were sequenced using the limiting dilution method described above, but with the addition of four times the designated amount of dNTPs. The inserts were amplified using T7 and T3 primers with a longer extension time of 3 min, and directly sequenced using a cDNA primer 5′cgctc tagaactagtggatcca3′ and Big Dye Terminator sequencing mix. The Department of Genetics at Washington University Medical School sequenced all clones directly from plasmid preparations, as described in Hillier et al. (1996) and Marra et al. (1999). On average, the length of sequence reads from each clone was 500 bp using both sequencing methodologies.
Data Availability
All EST data have been submitted to either EMBL or GenBank and are available in the public databases (EMBL, GenBank, and DDBJ). Fugu genomic data, including the ESTs, are available on the following sites: http://fugu.hgmp.mrc.ac.uk/, www.fugu-sg.org/, http://genome.jgi-psf.org/fugu6/fugu6.home.html. Genomic data are available at http://www.ensembl.org/Fugu_rubripes, and the ESTs have also been made available to the Fugu Ensembl team for annotation purposes. The Fugu EST sequences plus associated BLAST annotations can be accessed from the HGMP Web site, detailed above.
Bioinformatics
All ESTs were BLAST sequence similarity-searched using NCBI BLAST (Altschul et al. 1997). The BLASTX v. 2.2.3 program was used to search against the SPTR database v. 23 (Bairoch and Apweiler 2000). An e–-value of 1.0 or more was used to filter weak similarities in the BLAST searches. Additionally, the MSPCrunch program (Sonnhammer and Durbin 1994) was used to filter out matches with an e–-value cutoff of 1.0 or more. Homology of the Fugu data to sequences from other organisms in the databases was inferred if the BLASTX matches produced a bit score in excess of 50 and a raw value over 1e–05. To determine the number of ESTs matching amino acid 1 of SPTR matches, the top annotation lines of all of the SPTR entries showing a match over 50 and subject values of exactly 1 and also between 1 and 10 were extracted.
The ESTs were compared to two main data sets: the Ensembl build of the Fugu data (v.11.2.1) comprising 35,180 Ensembl gene predictions with an estimated 38,510 predicted Ensembl gene transcripts (www.ensembl.org/Fugu_rubripes). WU-BLAST was used to search the cDNAs against the Fugu genome data and matches were given, provided there was at least 96% identity over 125 bp. The Washington University zebrafish EST assemblies were also used. These comprised 25,185 assemblies from 156,067 ESTs from 105,565 clones. The assemblies are associations based on stringent sequence overlap and matepairs (R. Waterman and S. Johnson, unpubl.). The Fugu ESTs were BLAST searched against the zebrafish assemblies using an 1e–10 cutoff point. Clustering was performed using Phred–Phrap (Ewing and Green 1998; Ewing et al. 1998) and cluster.pl (a program developed “in house” at WashU by Richard E. Waterman), BLAST using 93% identity over 100 bp, WU-BLAST (W. Gish [1996–2003] http://blast.wustl.edu), or ICAtools (Parsons et al. 1992; Parsons 1995). For the gene discovery/gene diversity analysis, a “gene” was defined as the set of all ESTs that hit a Fugu transcript (FPG) at 96% identity over 125 bp, or a set of all non-FPG-hitting ESTs that clustered with each other by BLASTN at 93% identity over 100 bp, or an EST left as a singleton after not hitting any FPG and failing to cluster with any other ESTs.
Acknowledgments
S.S., K.K., M.S., K.K., S.W., and Y.S. thank Dr. Yuji Nagashima for obtaining the fish. This work was supported by an MRC grant (M.S.C., Y.J.K.E., A.T., G.E.); NIH DK55379 (S.L.J.); and grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (K.K., S.S., and S14104008 to S.W.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1691503. Article published online before print in November 2003.
References
- Adams, M.D., Kerlavage, A.R., Fleischmann, R.D., Fuldner, R.A., Bult, C.J., Lee, N.H., Kirkness, E.F., Weinstock, K.G., Gocayne, J.D., White, O., et al. 1995. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature 377: 3-174. [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annilo, T., Chen, Z.Q., Shulenin, S., and Dean, S. 2003. Evolutionary analysis of a cluster of ATP-binding cassette (ABC) genes. Mamm. Genome 14: 7-20. [DOI] [PubMed] [Google Scholar]
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301-1310. [DOI] [PubMed] [Google Scholar]
- Bailey Jr., L.C., Searls, D.B., and Overton, G.C. 1998. Analysis of EST-driven gene annotation in human genomic sequence. Genome Res. 8: 362-376. [DOI] [PubMed] [Google Scholar]
- Bairoch, A. and Apweiler, R. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28: 45-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, L.M., Gottgens, B., Gering, M., Gilbert, J.G., Grafham, D., Rogers, J., Bentley, D., Patient, R., and Green, A.R. 2001. Regulation of the stem cell leukemia (SCL) gene: A tale of two fishes. Proc. Natl. Acad. Sci. 98: 6747-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boardman, P.E., Sanz-Ezquerro, J., Overton, I.M., Burt, D.W., Bosch, E., Fong, W.T., Tickle, C., Brown, W.R., Wilson, S.A., and Hubbard, S.J. 2002. A comprehensive collection of chicken cDNAs. Curr. Biol. 12: 1965-1969. [DOI] [PubMed] [Google Scholar]
- Brenner, S., Venkatesh, B., Yap, W.H., Chou, C.F., Tay, A., Ponniah, S., Wang, Y., and Tan, Y.H. 2002. Conserved regulation of the lymphocyte-specific expression of lck in the Fugu and mammals. Proc. Natl. Acad. Sci. 99: 2936-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C.B. and Karlin, S. 1998. Finding the genes in genomic DNA. Curr. Opin. Struct. Biol. 8: 346-354. [DOI] [PubMed] [Google Scholar]
- Burset, M. and Guigo, R. 1996. Evaluation of gene structure prediction programs. Genomics 34: 353-367. [DOI] [PubMed] [Google Scholar]
- Camacho-Hubner, A., Richard, C., and Beermann, F. 2002. Genomic structure and evolutionary conservation of the tyrosinase gene family from Fugu. Gene 285: 59-68. [DOI] [PubMed] [Google Scholar]
- Clark, M.D., Hennig, S., Herwig, R., Clifton, S.W., Marra, M.A., Lehrach, H., Johnson, S.L., and the WU-GSC EST Group. 2001. An oligonucleotide fingerprint normalized and expressed sequence tag characterized zebrafish cDNA library. Genome Res. 11: 1594-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie, J.M. 1997. Computational methods for the identification of genes in vertebrate genomic sequences. Hum. Mol. Genet. 6: 1735-1744. [DOI] [PubMed] [Google Scholar]
- Davuluri, R.V., Grosse, I., and Zhang, M.Q. 2001. Computational identification of promoters and first exons in the human genome. Nat. Genet. 29: 412-417. [DOI] [PubMed] [Google Scholar]
- Dehal, P., Satou, Y., Campbell, R.K., Chapman, J., Degnan, B., De Tomaso, A., Davidson, B., Di Gregorio, A., Gelpke, M., Goodstein, D.M., et al. 2002. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science 298: 2157-2167. [DOI] [PubMed] [Google Scholar]
- Dunham, I., Shimizu, N., Roe, B.A., Chissoe, S., Hunt, A.R., Collins, J.E., Bruskiewich, R., Beare, D.M., Clamp, M., Smink, L.J., et al. 1999. The DNA sequence of human chromosome 22. Nature 402: 489-495. [DOI] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186-194. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- Gong, Z. 1999. Zebrafish expressed sequence tags and their applications. Methods Cell Biol. 60: 213-233. [DOI] [PubMed] [Google Scholar]
- Goode, D.K., Snell, P.K., and Elgar, G.E. 2003. Comparative analysis of vertebrate Shh genes identifies novel conserved noncoding sequence. Mamm. Genome 14: 192-201. [DOI] [PubMed] [Google Scholar]
- Griffin, C., Kleinjan, D.A., Doe, B., and van Heyningen, V. 2002. New 3′ elements control Pax6 expression in the developing pretectum, neural retina and olfactory region. Mech. Dev. 112: 89-100. [DOI] [PubMed] [Google Scholar]
- Guigo, R. 1997. Computational gene identification: An open problem. Comput. Chem. 21: 215-222. [DOI] [PubMed] [Google Scholar]
- Haussler, D. 1998. Computational gene identification. Trends Biochem. Sci. Suppl. S: 12-15. [Google Scholar]
- Hellman, I., Zollner, S., Enard, W., Ebersberger, I., Nickel, B., and Paabo, S. 2003. Selection on human genes as revealed by comparisons to chimpanzee cDNA. Genome Res. 13: 831-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier, L.D., Lennon, G., Becker, M., Bonaldo, M.F., Chiapelli, B., Chissoe, S., Dietrich, N., DuBuque, T., Favello, A., Gish, W., et al. 1996. Generation and analysis of 280,000 human expressed sequence tags. Genome Res. 6: 807-828. [DOI] [PubMed] [Google Scholar]
- Hubbard, T., Barker, D., Birney, E., Cameron, G., Chen, Y., Clark, L., Cox, T., Cuff, J., Curwen, V., Down, T., et al.,2002. The Ensembl genome database project. Nucleic Acids Res. 30: 38-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsi, A., Cao, D., Li, P., Patterson, A., Kocabas, A., Feng, J., Ju, Z., Mickett, K.D., and Liu, Z. 2002. Transcriptome analysis of channel catfish (Ictalurus punctatus): Initial analysis of gene expression and microsatellite-containing cDNAs in the skin. Gene 285: 157-168. [DOI] [PubMed] [Google Scholar]
- Kumar, S. and Hedges, S.B. 1998. A molecular timescale for vertebrate evolution. Nature 392: 917-920. [DOI] [PubMed] [Google Scholar]
- Lai, C.H., Chou, C.Y., Ch'ang, L.Y., Liu, C.S., and Lin, W. 2000. Identification of novel human genes evolutionarily conserved in Caenorhabditis elegans by comparative proteomics. Genome Res. 10: 703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P., Jenkins, N.A., and Copeland, N.G. 2003. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13: 476-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makalowski, W. and Boguski, M.S. 1998. Evolutionary parameters of the transcribed mammalian genome: An analysis of 2,820 orthologous rodent and human sequences. Proc. Natl. Acad. Sci. 95: 9407-9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makalowski, W., Zhang, J., and Boguski, M.S. 1996. Comparative analysis of 1196 orthologous mouse and human full-length mRNA and protein sequences. Genome Res. 6: 846-857. [DOI] [PubMed] [Google Scholar]
- Maroni, G. 1996. The organisation of eukaryotic genes. Evol. Biol. 29: 1-19. [Google Scholar]
- Marra, M., Hillier, L., Kucaba, T., Allen, M., Barstead, R., Beck, C., Blistain, A., Bonaldo, M., Bowers, Y., Bowles, L., et al. 1999. An encyclopedia of mouse genes. Nat. Genet. 21: 191-194. [DOI] [PubMed] [Google Scholar]
- Maruyama, K. and Sugano, S. 1994. Oligo-capping: A simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene 138: 171-174. [DOI] [PubMed] [Google Scholar]
- Mathe, C., Sagot, M.F., Schiex, T., and Rouze, P. 2002. Current methods of gene prediction, their strengths and weaknesses. Nucleic Acids Res. 30: 4103-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignone, F., Gissi, C., Liuni, S., and Pesole, G. 2002. Untranslated regions of mRNAs. Genome Biol. 3: reviews 0004.1-0004.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek, B. and Lee, C. 2002. A genomic view of alternative splicing. Nat. Genet. 30: 13-19. [DOI] [PubMed] [Google Scholar]
- Nelson, D.R. 2003. Comparison of P450s from human and fugu: 420 million years of vertebrate P450 evolution. Arch. Biochem. Biophys. 409: 18-24. [DOI] [PubMed] [Google Scholar]
- Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563-573. [DOI] [PubMed] [Google Scholar]
- Parsons, J.D. 1995. Improved tools for DNA comparison and clustering. Comput. Appl. Biosci. 11: 603-613. [DOI] [PubMed] [Google Scholar]
- Parsons, J.D., Brenner, S., and Bishop, M.J. 1992. Clustering cDNA sequences. Comput. Appl. Biosci. 8: 461-466. [DOI] [PubMed] [Google Scholar]
- Roest Crollius, H., Jaillon, O., Bernot, A., Dasilva, C., Bouneau, L., Fischer, C., Fizames, C., Wincker, P., Brottier, P., Quetier, F., et al. 2000. Estimate of human gene number provided by genome-wide analysis using Tetraodon nigroviridis DNA sequence. Nat. Genet. 25: 235-238. [DOI] [PubMed] [Google Scholar]
- Rogic, S., Mackworth, A.K., and Ouellette, F.B. 2001. Evaluation of gene-finding programs on mammalian sequences. Genome Res. 11: 817-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogic, S., Ouellette, B.F., and Mackworth, A.K. 2002. Improving gene recognition accuracy by combining predictions from two gene-finding programs. Bioinformatics 18: 1034-1045. [DOI] [PubMed] [Google Scholar]
- Rothenberg, E.V. 2001. Mapping of complex regulatory elements by pufferfish/zebrafish transgenesis. Proc. Natl. Acad. Sci. 98: 6540-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J.G., Russell, R., Umrania, Y., Edwards, Y.J., Campbell, R.D., Elgar, G., and Clark, M.S. 2002. Fugu orthologues of human major histocompatibility complex genes: A genome survey. Immunogenetics 54: 367-380. [DOI] [PubMed] [Google Scholar]
- Sonnhammer, E.L. and Durbin, R. 1994. A workbench for large-scale sequence homology analysis. Comput. Appl. Biosci. 10: 301-307. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y., Yoshitomo-Nakagawa, K., Maruyama, K., Suyama, A., and Sugano, S. 1997. Construction and characterization of a full length-enriched and a 5′-end-enriched cDNA library. Gene 200: 149-156. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y., Yamashita, R., Nakai, K., and Sugano, S. 2002. DBTSS: Database of human transcriptional start sites and full-length cDNAs. Nucleic Acids Res. 30: 328-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Yap, W.H., Tay, A., Brenner, S., and Venkatesh, B. 2003. Molecular cloning of the pufferfish (Takifugu rubripes) Mx gene and functional characterization of its promoter. Immunogenetics 54: 705-713. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.ensembl.org/Danio_rerio; Zebrafish genome data.
- http://www.ensembl.org/Fugu_rubripes; Centralized site of Consortium Fugu genome data.
- http://www.hgmp.mrc.ac.uk/geneservice/reagents/index.shtml; European source of Fugu clones and reagents.
- http://fugu.hgmp.mrc.ac.uk/Protocols/Biology/; Cambridge Fugu genome group laboratory protocols.
- http://fugu.hgmp.mrc.ac.uk/; Home page of the Cambridge Fugu genome project.
- www.fugu-sg.org/; Home page of the Singapore Fugu genome project.
- http://genome.jgi-psf.org/fugu6/fugu6.home.html; Home page of the JGI Fugu genome project.
- http://www.ensembl.org/Fugu_rubripes; Centralized site of Consortium Fugu genome data.
- www.ensembl.org/Fugu_rubripes; Centralized site of Consortium Fugu genome data.
- http://blast.wustl.edu; Washington University BLAST archives.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All EST data have been submitted to either EMBL or GenBank and are available in the public databases (EMBL, GenBank, and DDBJ). Fugu genomic data, including the ESTs, are available on the following sites: http://fugu.hgmp.mrc.ac.uk/, www.fugu-sg.org/, http://genome.jgi-psf.org/fugu6/fugu6.home.html. Genomic data are available at http://www.ensembl.org/Fugu_rubripes, and the ESTs have also been made available to the Fugu Ensembl team for annotation purposes. The Fugu EST sequences plus associated BLAST annotations can be accessed from the HGMP Web site, detailed above.