Summary
Type VI secretion is critical for Vibrio cholerae to successfully combat phagocytic eukaryotes and to survive in the presence of competing bacterial species. V. cholerae type VI secretion system genes are encoded in one large and two small clusters. In V. cholerae, type VI secretion is controlled by quorum sensing, the cell-cell communication process that enables bacteria to orchestrate group behaviors. The quorum-sensing response regulator LuxO represses type VI secretion genes at low cell density and the quorum-sensing regulator HapR activates type VI secretion genes at high cell density. We demonstrate that the quorum-regulatory small RNAs (Qrr sRNAs) that function between LuxO and HapR in the quorum-sensing cascade are required for these regulatory effects. The Qrr sRNAs control type VI secretion via two mechanisms: they repress expression of the large type VI secretion system cluster through base pairing and they repress HapR, the activator of the two small type VI secretion clusters. This regulatory arrangement ensures that the large cluster encoding many components of the secretory machine is expressed prior to the two small clusters that encode the secreted effectors. Qrr sRNA-dependent regulation of the type VI secretion system is conserved in pandemic and non-pandemic V. cholerae strains.
Keywords: quorum sensing, small regulatory RNAs, type VI secretion system, Vibrio cholerae
Introduction
Quorum sensing is a cell-cell communication process that bacteria use to coordinate collective behaviors. Quorum sensing involves the production, release, detection, and response to extracellular signal molecules called autoinducers (Ng and Bassler, 2009). In Vibrio cholerae, which causes the life-threatening diarrheal disease cholera, two quorum-sensing pathways function in parallel (Miller et al., 2002; Zhu et al., 2002). At low cell density (LCD), when the concentrations of the two autoinducers are low, the quorum-sensing response regulator LuxO is phosphorylated (Figure 1). LuxO~P activates the transcription of genes encoding four quorum regulatory non-coding RNAs called Qrr1–4 (Lenz et al., 2004). At high cell density (HCD), when the concentrations of the two autoinducers are high, LuxO is not phosphorylated and, in this state, LuxO is unable to activate the transcription of qrr1–4 (Figure 1). The Qrr small RNAs (sRNAs) activate the translation of the LCD master regulator AphA and repress the translation of the HCD master regulator HapR. Thus, AphA is produced at LCD and HapR is produced at HCD (Rutherford et al., 2011; Shao and Bassler, 2012). At LCD, AphA activates expression of genes encoding major virulence factors such as the toxin-coregulated pilus (TCP) and cholerae toxin (CTX) (Skorupski and Taylor, 1999). At HCD, HapR represses TCP and CTX and HapR activates production of proteases that enable V. cholerae to disperse from the host back into the environment (Zhu et al., 2002; Hammer and Bassler, 2003).
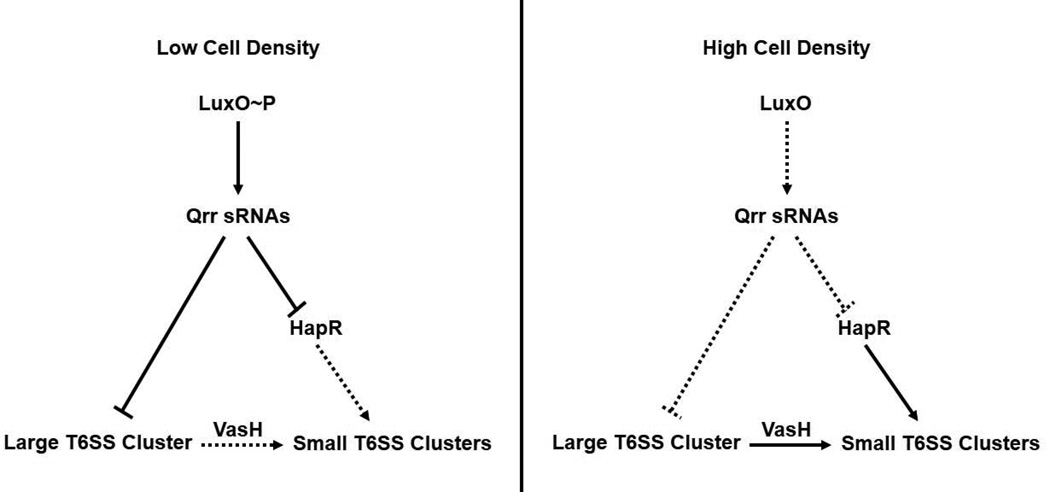
Figure 1. The Qrr sRNAs repress T6SS via two mechanisms.
Model for Qrr sRNA control of T6SS in V. cholerae. At LCD, phosphorylated LuxO (LuxO~P) activates the production of the Qrr sRNAs. The Qrr sRNAs repress the HCD master regulator HapR, which is an activator of the two T6SS small clusters (VC1415–1421 and VCA0017–0021). The Qrr sRNAs also repress the T6SS large cluster (VCA0107–0124) directly through base pairing. The large cluster encodes a second activator, VasH (VCA0117), of the two small clusters. At HCD, LuxO is not phosphorylated. In this state, LuxO is unable to activate the production of the Qrr sRNAs, which leads to derepression of both HapR and derepression of the large T6SS cluster. VasH is produced from the large T6SS cluster. VasH and HapR activate expression of the small T6SS clusters.
Type VI secretion systems (T6SS) were recently discovered as virulence factor delivery systems in both pandemic and non-pandemic V. cholerae strains (Pukatzki et al., 2006). T6SS also exist in other bacterial species (Miyata et al., 2013). V. cholerae uses its T6SS to kill phagocytic eukaryotes via crosslinking of actin and to attack other bacteria by destroying cell wall structures (Pukatzki et al., 2006; Pukatzki et al., 2007; Ma et al., 2009; MacIntyre et al., 2010; Dong et al., 2013). T6SS+ cells counterattack other T6SS+ cells, and T6SS immunity protein-effector pairs prevent self-killing (Dong et al., 2013; Basler et al., 2013; Brooks et al., 2013).
In V. cholerae, T6SS proteins are encoded in one large cluster and two small clusters (Pukatzki et al., 2006; Zheng et al., 2011). The large cluster encodes structural components to build the secretion machinery and an essential regulator of T6SS, VasH, which functions together with RpoN to activate the two small clusters encoding the secreted effectors (Bernard et al., 2011; Kitaoka et al., 2011; Dong and Mekalanos, 2012). In pandemic V. cholerae strains, the quorum-sensing HCD master regulator HapR activates T6SS genes by binding to the promoter regions of the two small clusters (Ishikawa et al., 2009; Tsou et al., 2009). Disruption of the quorum-sensing response regulator LuxO, in the context of a null mutation in TsrA, which represses T6SS through an unknown mechanism, induces T6SS gene expression and effector secretion suggesting that LuxO is a repressor of the T6SS (Zheng et al., 2010). In V. cholerae O1 strain A1552, T6SS is also activated under high-osmolarity and low temperature conditions (Ishikawa et al., 2012). Here, we show that LuxO functions through the Qrr sRNAs to repress T6SS via two mechanisms: the Qrr sRNAs repress production of the T6SS activator HapR, which has the effect of decreasing expression of the two small T6SS clusters. Independently of HapR, the Qrr sRNAs repress T6SS by direct base pairing to the mRNA encoding the large T6SS cluster (Figure 1). Finally, we demonstrate that Qrr sRNA-dependent T6SS regulation is conserved in pandemic and non-pandemic V. cholerae strains.
Results
LuxO functions through the Qrr sRNAs to repress T6SS
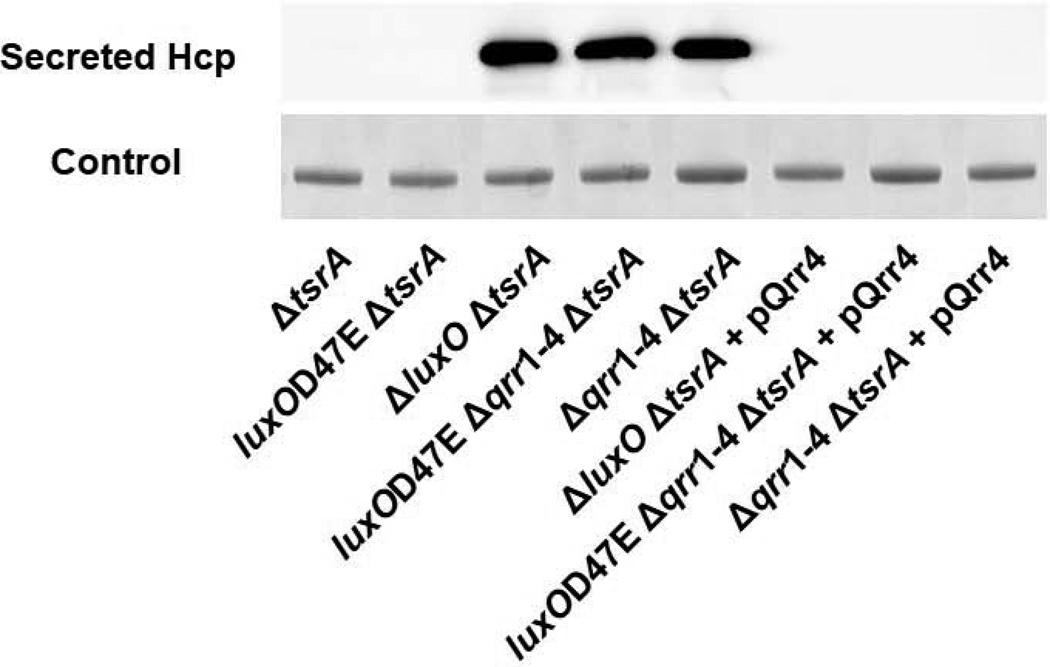
In the pandemic V. cholerae strain O1 El Tor C6706, the T6SS is repressed by the quorum-sensing response regulator LuxO and activated by the quorum-sensing HCD master regulator HapR (Ishikawa et al., 2009; Tsou et al., 2009; Zheng et al., 2010). Because the Qrr sRNAs function between LuxO and HapR in the quorum-sensing cascade, we wondered whether the Qrr sRNAs are involved in T6SS repression. To explore this possibility, we monitored secretion of the Hcp protein in WT and quorum-sensing mutant strains. Hcp secretion is commonly used as a hallmark of V. cholerae T6SS function. No Hcp secretion could be detected in V. cholerae strains carrying an intact tsrA gene, confirming previous findings that Hcp secretion is repressed by TsrA (Zheng et al., 2010). To circumvent this problem, we deleted tsrA in all of the strains examined. We also exploited LuxOD47E, an allele that locks LuxO into a form mimicking phosphorylated LuxO (Freeman and Bassler, 1999). LuxOD47E constitutively activates qrr expression. Hcp secretion could not be detected in the ΔtsrA strain and the luxOD47E, ΔtsrA strain, while Hcp secretion occurred in the ΔluxO, ΔtsrA strain (Figure 2). This result is consistent with previous findings that LuxO is a repressor of T6SS (Zheng et al., 2010). Both the luxOD47E, Δqrr1–4, ΔtsrA strain and the Δqrr1–4, ΔtsrA strain showed high level secretion of Hcp suggesting that the Qrr sRNAs are required for T6SS repression (Figure 2). Indeed, introduction of a plasmid expressing Qrr4 into the Hcp-secreting strains fully repressed Hcp secretion (Figure 2). These results suggest that, in the absence of the Qrr sRNAs, LuxO~P cannot control the T6SS. We conclude that LuxO functions through the Qrr sRNAs to repress T6SS.
Figure 2. LuxO functions through the Qrr sRNAs to repress T6SS.
Western blot showing secreted Hcp protein in V. cholerae O1 El Tor C6706 ΔtsrA (YS2031), luxOD47E, ΔtsrA (YS2032), and ΔluxO, ΔtsrA (YS2033), luxOD47E, Δqrr1–4, ΔtsrA (YS2037), and Δqrr1–4, ΔtsrA (YS2039) strains in the absence and presence of plasmid-encoded Qrr4 (pSLS155).
The Qrr sRNAs repress T6SS in a HapR-independent manner
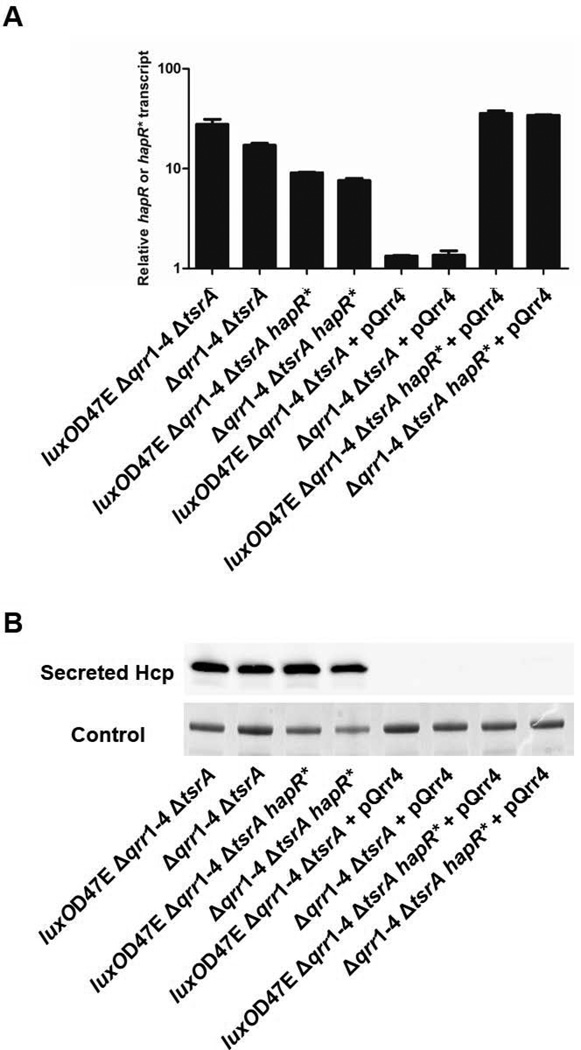
There are two possible interpretations for the above results: First, the Qrr sRNAs repress T6SS exclusively through repression of HapR, which is required to activate T6SS and for secretion of Hcp. Second, the Qrr sRNAs act through HapR and they also repress T6SS in a HapR-independent manner. To determine whether there is HapR-independent regulation of T6SS, we engineered a mutation in the 5’ UTR region of hapR (called hapR*) that renders it insensitive to Qrr control (Figure S1 and S2). The data in Figure 3A provide proof that hapR* is not subject to Qrr control. Specifically, introduction of a plasmid expressing Qrr4 repressed hapR in the luxOD47E, Δqrr1–4, ΔtsrA strain and the Δqrr1–4, ΔtsrA strain, but not in the strains when they carried hapR*. We next tested the effect of the hapR* mutation on T6SS. Secretion of Hcp occurred in strains carrying both WT hapR and the hapR* mutation and, importantly, Qrr4 functioned equally well to repress Hcp secretion in both strains (Figure 3B). These results show that, in addition to functioning through HapR, the Qrr sRNAs also repress T6SS via a HapR-independent mechanism.
Figure 3. The Qrr sRNAs repress T6SS in a HapR-independent manner.
(A) hapR or hapR* mRNA and (B) Secreted Hcp protein in V. cholerae O1 El Tor C6706 luxOD47E, Δqrr1–4, ΔtsrA (YS2037), Δqrr1–4, ΔtsrA (YS2039), luxOD47E, Δqrr1–4, ΔtsrA, hapR* (YS2047), and Δqrr1–4, ΔtsrA, hapR* (YS2048) strains in the absence and presence of plasmid-encoded Qrr4 (pSLS155). mRNA levels and secreted protein levels were determined using Western blot and qRT-PCR, respectively. hapR* has a mutation that renders it insensitive to Qrr sRNA control (Figure S2).
The Qrr sRNAs repress T6SS through base pairing to the 5’ UTR of the mRNA
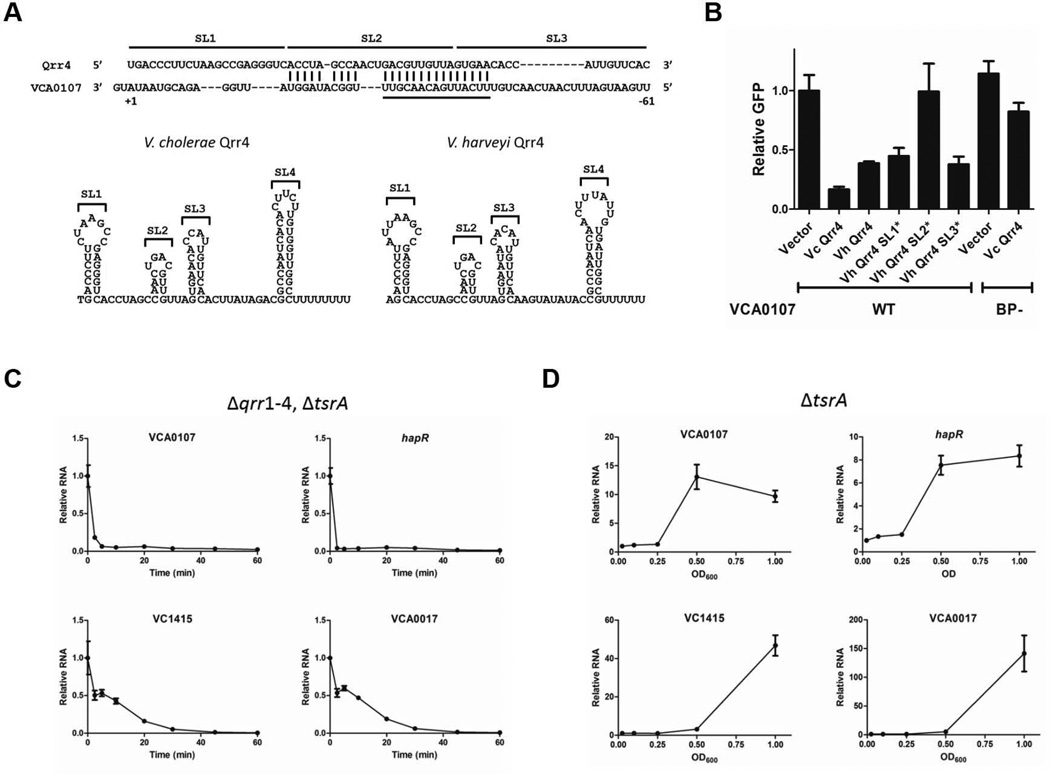
To define the mechanism underpinning HapR-independent regulation of T6SS by the Qrr sRNAs, we tested a direct base pairing mechanism. In V. cholerae, the T6SS proteins are encoded in three gene clusters, one large cluster (VCA0107–VCA0124) and two small clusters (VC1415–VC1421 and VCA0017–VCA0021) (Pukatzki et al., 2006; Zheng et al., 2011) (Figure S3). Sequence analysis identified a potential Qrr sRNA binding site in the 5’ UTR of the large T6SS cluster, but not in the 5’ UTR of the two small clusters (Figure 4A). To examine this, we generated a VCA0107-GFP translational fusion (−250 to +30 with +1 as the translational start site) and introduced it into E. coli. We also introduced a plasmid encoding inducible V. cholerae qrr4. Our rationale was that, if the VCA0107-GFP fusion could be controlled by V. cholerae Qrr4 in a heterologous system, it would suggest that the Qrr sRNAs act directly to repress T6SS. Indeed, V. cholerae Qrr4 repressed VCA0107-GFP (~5-fold) in E. coli, suggesting that the Qrr sRNAs repress the large T6SS cluster through base pairing to the 5’ UTR of the mRNA (Figure 4B). By contrast, V. cholerae Qrr4 did not repress translation fusions to VC1415 and VCA0017 (−64 to +30 with +1 as the translational start site), suggesting that the Qrr sRNAs do not regulate the small T6SS clusters via direct base pairing (Figure S4). The Qrr sRNAs in V. cholerae and the related bacterium V. harveyi are highly conserved at both the primary sequence level and the secondary structural level (Figure 4A and Figure S5). Our previous analyses of the Qrr sRNAs in V. harveyi defined the second stem-loop (SL2) region as critical for base pairing with and regulating target genes (Shao et al., 2013). Figure 4B shows that similar to V. cholerae Qrr4, V. harveyi Qrr4 represses VCA0107-GFP (~2.5-fold) in E. coli. Thus, we reasoned we could use our existing set of V. harveyi Qrr4 stem-loop variants to test the roles of different portions of the Qrr sRNAs in control of the T6SS large cluster. Inversion of SL2 but not SL1 or SL3 in V. harveyi Qrr4 eliminated T6SS repression as judged by the GFP reporter (Figure 4B; for SL2 p > 0.05). Quantification of the corresponding VCA0107 mRNA levels show the mRNA tracks with the GFP reporter output (Figure S6). This effect is not due to expression level differences since the stem-loop inversion constructs are as stable as WT Qrr4 (Shao et al., 2013). This result suggests that SL2 of the Qrr sRNA is critical for regulation. Indeed, when the predicted base pairing region in the 5’ UTR of the large T6SS cluster is mutated to its complementary sequence (denoted BP-in Figure 4B), regulation by Qrr4 is lost. These results confirm the predicted base pairing pattern (Figure 4A).
Figure 4. The Qrr sRNAs repress T6SS through direct base pairing.
(A) Putative base pairing between V. cholerae Qrr4 and the VCA0107 5’ UTR was predicted by RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/). The VCA0107 translational start site is denoted +1. Sequences encoding stem-loops are shown with over-lines. The mutated base pairing region is shown by the underline. Predicted secondary structures of V. cholerae Qrr4 and V. harveyi Qrr4. (B) Fluorescence from E. coli carrying a plasmid with an IPTG inducible translational GFP fusion to VCA0107 (WT, pYS254) was measured in the presence of an empty vector (pLF253), a vector expressing tetracycline inducible V. cholerae qrr4 (Vc Qrr4, pYS245), V. harveyi qrr4 (Vh Qrr4, pLF127), V. harveyi qrr4 stem-loop 1 inversion (Vh Qrr4 SL1*, pYS230), V. harveyi qrr4 stem-loop 2 inversion (Vh Qrr4 SL2*, pYS231), or V. harveyi qrr4 stem-loop 3 inversion (Vh Qrr4 SL3*, pYS232). Fluorescence from a GFP fusion to VCA0107 which harbors a 15 nucleotide mutation UUCAUUGACAACGUU to AAGUAACUGUUGCAA in the predicted base pairing region (BP-, pYS350) was measured in the presence of an empty vector (pLF253) or a vector expressing tetracycline inducible V. cholerae qrr4 (Vc Qrr4, pYS245), GFP production from three independent cultures was measured and the means and SEMs are shown, with all measurements normalized to the mean of the vector controls. (C) mRNA levels of VCA0107, hapR, VC1415, and VCA0017 in V. cholerae O1 El Tor C6706 Δqrr1–4, ΔtsrA (YS2039) were measured by qRT-PCR following pulse induction of V. cholerae Qrr4 (pYS249). Samples were collected at the designated time points, and means and SEMs of triplicate samples are shown. (D) mRNA levels of VCA0107, hapR, VC1415, and VCA0017 were measured by qRT-PCR in V. cholerae O1 El Tor C6706 ΔtsrA (YS2031) from LCD to HCD. Samples were collected at the designated OD600, and means and SEMs of triplicate samples are shown.
When vibrios launch their quorum-sensing programs, target genes that are directly regulated by the Qrr sRNAs respond more rapidly than target genes that are controlled indirectly via Qrr-regulated transcription factors (e.g. AphA and HapR) (Shao et al., 2013). Our above analyses suggest that the V. cholerae large T6SS cluster is directly controlled by the Qrr sRNAs. If so, we expect this cluster to respond rapidly to a change in quorum-sensing mode. We used pulse induction of Qrr4 in the V. cholerae Δqrr1–4, ΔtsrA strain to probe the timing of quorum-sensing-controlled T6SS gene expression. Figure 4C shows that the mRNA encoding the large T6SS cluster VCA0107–VCA0124 decreases within 10 min to its minimum level following pulse induction of Qrr4. This timing is similar to that established for targets directly controlled by the Qrr sRNAs such as hapR (Figure 4C) (Shao et al., 2013). Our findings are consistent with the idea that the Qrr sRNAs regulate the T6SS large cluster directly. By contrast, the mRNAs encoding the two small clusters, VC1415–VC1421 and VCA0017–VCA0021, showed slower responses to pulse induction of Qrr4 as the mRNAs decreased to minimum levels only after 30 min (Figure 4C). We suspect that this delay in timing occurs because, once the Qrr sRNAs repress translation of HapR and the large T6SS cluster that encodes VasH, an activator of the two small T6SS clusters, it takes time for existing HapR and VasH to decay. Only after HapR and VasH levels decrease, does a reduction in the mRNAs encoding the two small T6SS clusters occur. A similar rapid response of the large cluster and a correspondingly slower response of the small clusters occurred in the Δqrr1–4 strain containing an intact tsrA gene (Figure S7). This result indicates that the mechanism of TsrA repression of T6SS does not involve altering the timing of mRNA production. Based on these results, we expect that, in response to cell population density changes, activation of expression of the large T6SS cluster should occur prior to the activation of expression of the two small clusters. To explore this idea, we monitored expression of type VI secretion genes from LCD to HCD in the V. cholerae ΔtsrA strain. Indeed, expression of the large cluster increased between OD600 0.25 and 0.5, whereas the increase in expression of the two small clusters occurred later, between OD600 0.5 and 1.0 (Figure 4D). These results suggest that, during the LCD to HCD transition, the VasH and HapR regulators are produced prior to the secreted effectors encoded by the two small clusters.
The Qrr sRNAs repress T6SS in non-pandemic V. cholerae strains
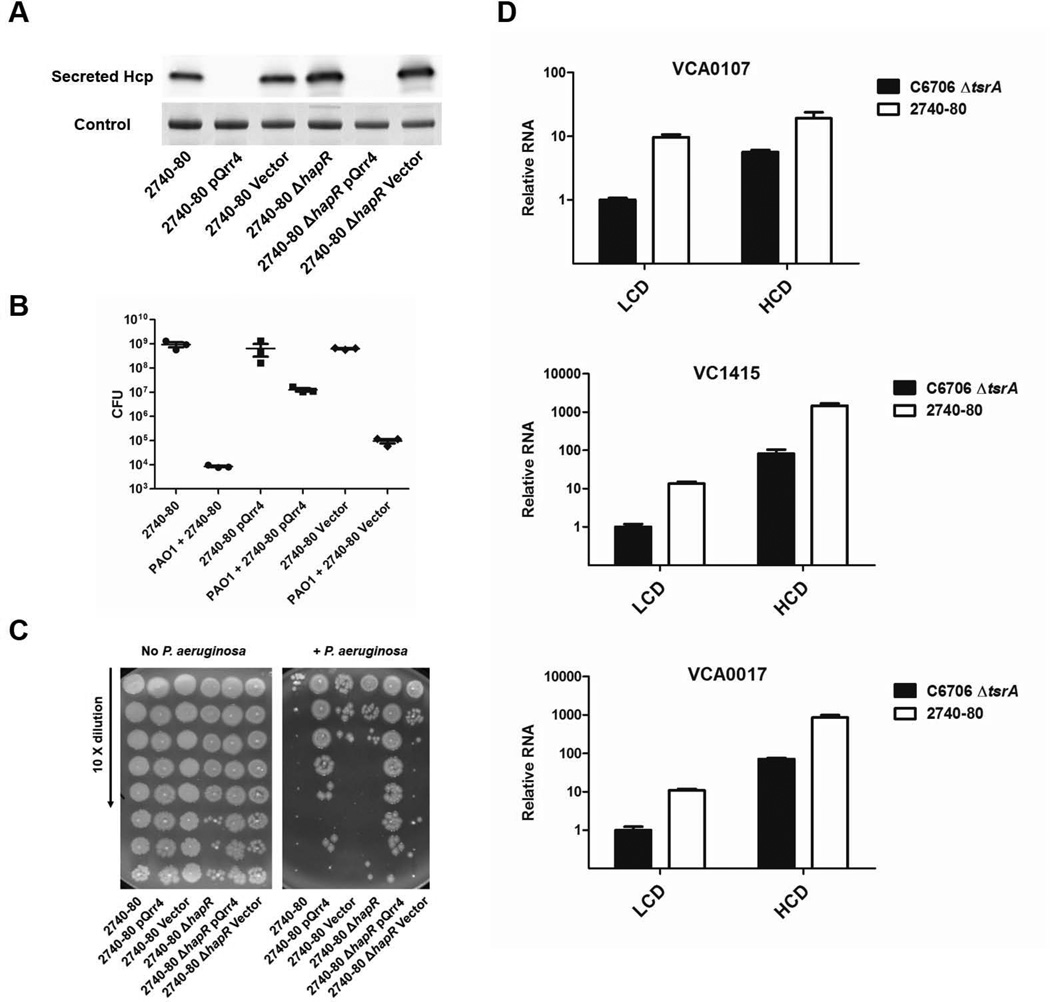
Unlike in pandemic V. cholerae strains in which T6SS expression is regulated, in non-pandemic strains, under laboratory conditions, expression of T6SS is constitutive (Pukatzki et al., 2006; Zheng et al., 2010). This feature makes non-pandemic V. cholerae strains such as strain V52 and strain 2740-80 especially useful for exploring the biological function of T6SS. Such analyses have led to the notion that T6SS is used as a defense mechanism to battle other T6SS-containing bacterial species. For example, attack by one T6SS+ species induces counter-attack by other T6SS+ strains in killing assays (Basler et al., 2013). Considering that the quorum-sensing circuits as well as the three T6SS clusters are conserved between pandemic and non-pandemic V. cholerae strains, we wondered whether Qrr sRNA-directed repression of T6SS is likewise conserved. To test this, we introduced our Qrr4-expressing plasmid into the non-pandemic V. cholerae strain 2740-80. We found that Hcp secretion dramatically decreased (Figure 5A). We performed a T6SS killing assay to measure the regulatory effect of the Qrr sRNA on the counter-attack process. V. cholerae 2740-80 containing the Qrr4-expressing plasmid showed ~100 fold higher survival from P. aeruginosa T6SS attack than the strain harboring the control plasmid (Figure 5B). Presumably, the Qrr sRNA repressed T6SS expression in V. cholerae 2740-80 so P. aeruginosa did not attack as frequently. We also investigated the function of HapR in control of T6SS gene expression in non-pandemic V. cholerae strains. In contrast to the pandemic strain O1 El Tor C6706 in which HapR is required for Hcp secretion, HapR is dispensable for Hcp secretion in V. cholerae strains 2740-80 and V52 (Figure S1) (Ishikawa et al., 2009; Tsou et al., 2009; Zheng et al., 2010). When we introduced our plasmid expressing Qrr4 into the non-pandemic V. cholerae 2740-80 and 2740-80 ΔhapR strains, Hcp secretion was fully repressed (Figure 5A). Consistent with this, V. cholerae 2740-80 and 2740-80 ΔhapR containing the Qrr4-expressing plasmid showed higher survival in the killing assay than the strains harboring the control plasmid (Figure 5C). Similar results were obtained in V. cholerae V52 and V52 ΔhapR stains (data not shown). Thus, the Qrr sRNAs are capable of repressing T6SS in both pandemic and non-pandemic V. cholerae strains, and furthermore, in non–pandemic strains, HapR appears to be irrelevant in terms of T6SS control. Type VI secretion gene expression increases in both the pandemic V. cholerae ΔtsrA strain and the non-pandemic V. cholerae 2740-80 strain as the cells transition from LCD to HCD. However, basal expression is ~10-fold higher at LCD in the non-pandemic strain than in the pandemic strain. This finding could explain the constitutive Hcp secretion phenotype of the non-pandemic strain (Figure 5D) as well as the overall higher Hcp secretion that occurs in non-pandemic strains compared to the pandemic strain (Figure S1). These results cannot be not due to differential expression of either hapR or the qrr genes since their expression levels are indistinguishable between pandemic and non-pandemic strains (data not shown).
Figure 5. The Qrr sRNAs repress T6SS in non-pandemic V. cholerae strains.
(A) Western blot of secreted Hcp in V. cholerae 2740-80 and 2740-80 ΔhapR (YS2029), and in V. cholerae 2740-80 and 2740-80 ΔhapR (YS2029) harboring plasmid-encoded Qrr4 (pSLS155) or a vector control (pEVS143). (B) T6SS Killing assay showing numbers of surviving colonies of V. cholerae 2740-80, V. cholerae 2740-80 in the presence of plasmid-encoded Qrr4 (pSLS155), and V. cholerae 2740-80 carrying the vector control (pEVS143) without and with challenge by P. aeruginosa PAO1. (C) Survival of V. cholerae 2740-80 and 2740-80 ΔhapR (YS2029) in the absence and presence of plasmid-encoded Qrr4 (pSLS155) or the vector control (pEVS143). The left and right panels show the killing assay performed in the absence and presence of P. aeruginosa PAO1 challenge, respectively. (D) mRNA levels of VCA0107, VC1415, and VCA0017 in V. cholerae O1 El Tor C6706 ΔtsrA (YS2031) and in V. cholerae 2740-80 were measured by qRT-PCR Samples were collected at OD600 ~0.03 (LCD) and OD600 ~1.0 (HCD), and means and SEMs of triplicate samples are shown.
Discussion
Quorum sensing is a cell-cell communication process that bacteria use to control group behaviors including bioluminescence, biofilm formation, and virulence factor production (Ng and Bassler, 2009). In the case of quorum sensing in V. cholerae, the Qrr sRNAs function at the heart of the quorum-sensing circuit. The Qrr sRNAs indirectly control production of the canonical V. cholerae virulence factors TCP and CTX through regulation of the quorum-sensing LCD master regulator AphA and the HCD master regulator HapR (Rutherford et al., 2011). Recently, it was discovered that T6SS is crucial for V. cholerae virulence against both eukaryotic cells and other proteobacteria, and a connection to quorum sensing through LuxO and HapR has been established (Zheng et al., 2010). Here we find that the Qrr sRNAs are responsible for repression of T6SS at LCD, which further links virulence to quorum-sensing control. We define two mechanisms by which the Qrr sRNAs control T6SS: 1) they repress the HCD master regulator HapR which is an activator of T6SS gene expression; 2) they repress T6SS directly through base pairing to the mRNA encoding the large T6SS cluster (Figure 1). We also show that Qrr-mediated regulation of T6SS is conserved in both pandemic and non-pandemic V. cholerae strains. It should be pointed out that the role of HapR in control of T6SS gene expression differs in pandemic and non-pandemic V. cholerae strains: HapR is required for Hcp secretion in the pandemic V. cholerae strain O1 El Tor C6706 but dispensable in the non-pandemic V. cholerae strain V52 and strain 2740-80. We note that our findings in this respect differ from those in a previous study in which deletion of hapR decreased Hcp secretion in V. cholerae V52 (Zheng et al., 2010). Finally, unlike what has been reported for V. cholerae O1 strain A1552, no induction of Hcp secretion occurred in V. cholerae O1 strain El Tor C6706 under high-osmolarity and low temperature conditions. We presume this discrepancy is due to strain differences (Ishikawa et al., 2012).
The V. cholerae T6SS machine is encoded by three gene clusters: one large cluster VCA0107–VCA0124, and two small clusters VC1415–VC1421 and VCA0017–VCA0021 (Figure S3) (Pukatzki et al., 2006; Zheng et al., 2011). Based on bioinformatic analyses and previous studies of their roles in Hcp expression, Hcp secretion, and virulence towards E. coli and Dictyostelium discoideum, T6SS proteins are divided into four groups: 1) components of the structural apparatus; 2) regulators; 3) secreted effectors and immunity proteins; 4) proteins of unknown function. Hcp, which forms the inner tubular structure, and VgrG, which assembles into a trimeric complex at the tip of the Hcp tube, are encoded by the two small clusters. However, most components of the structural apparatus and regulators (such as VasH, which activates the transcription of the two small clusters) are encoded by the large cluster (Figure S3) (Pukatzki et al., 2006; Zheng et al., 2011). Previous studies have identified several T6SS activators in pandemic V. cholerae strains including HapR, VasH, and RpoN that directly bind to the promoter regions of the two small clusters (Ishikawa et al., 2009; Tsou et al., 2009; Bernard et al., 2011; Kitaoka et al., 2011; Dong and Mekalanos, 2012). Our finding that the Qrr sRNAs directly repress the large T6SS gene cluster adds a new layer of regulation. Since VasH, the activator of the two small clusters, is encoded in the large cluster, our results suggest that the Qrr sRNAs function at the top of the regulatory hierarchy (Figure 1). Specifically, during the LCD to HCD transition, quorum sensing represses the Qrr sRNAs. The absence of the Qrr sRNAs enables the initial launch of the T6SS structural apparatus via relief of repression of the large cluster. Subsequently, when the bacteria switch into quorum-sensing mode, HapR is made. It, together with VasH, activates the expression of the T6SS small clusters encoding the secreted effectors. This coupled direct followed by indirect regulation by the Qrr sRNAs ensures the logically timed progression of assembly: machine first and secreted components second.
Among the over 200 serogroups of V. cholerae, the O1 and O139 strains are responsible for cholera pandemics, whereas the non-O1 and non-O139 strains cause sporadic outbreaks of gastroenteritis or extraintestinal infections (Rahman et al., 2008). Although T6SS gene clusters are conserved between pandemic and non-pandemic V. cholerae strains, and we show that the Qrr sRNAs can repress T6SS expression in each case, major differences exist in terms of endogenous T6SS expression. Under standard laboratory conditions, the T6SS apparatus is constitutively expressed in non-pandemic V. cholerae strains. This finding is consistent with higher basal level T6SS expression at LCD, however, T6SS expression is strictly regulated in pandemic strains. This difference could be a consequence of evolutionary pressures driven by the different lifestyles of the two types of V. cholerae strains. Non-pandemic V. cholerae strains primarily reside in aquatic environmental reservoirs, whereas pandemic strains colonize humans. Thus, non-pandemic strains might require a constitutive T6SS to successfully compete with other bacterial species in the environment and to survive predation by unicellular eukaryotes such as amoebae. By contrast, pandemic V. cholerae strains are transmitted into the gastrointestinal tracts of the host during ingestion of contaminated water or food. At early stages of infection, pandemic V. cholerae colonizes the host via attachment through the TCP and biofilm formation, and the cells produce CTX. TCP and CTX act as the primary virulence factors. T6SS is likely not required for initial colonization so it is repressed. At late stages of infection, V. cholerae terminates production of the primary virulence factors, and simultaneously activates production of proteases that enable exit from the host. The finding that a T6SS constitutively-active pandemic V. cholerae mutant causes diarrhea in infant rabbits indicates that T6SS could contribute to exit from the host during late stages of infection (Zheng et al., 2010). Increased levels of T6SS during late stages of infection might be a mechanism that primes exiting V. cholerae for life in the environment in between hosts. Additionally, new results showing that T6SS promotes the killing of other bacterial cells during infection suggests yet another advantage for induction of T6SS in pandemic V. cholerae while in the host (Fu et al., 2013).
Experimental Procedures
Bacterial strains and growth conditions
P. aeruginosa PAO1 (Holloway, 1955), V. cholerae 2740-80 (Basler et al., 2013), V52 (Pukatzki et al., 2006), C6706 (Thelin and Taylor, 1996) and derivatives were grown aerobically in Luria-Murine (LB) medium at 37°C. E. coli strains S17-1λpir (de Lorenzo and Timmis, 1994) and BW-RI (Levine et al., 2007) were grown aerobically in LB medium or M9 medium (0.5% glucose) at 37°C. Strains used in this study are described in Table S1. Antibiotics (Sigma-Aldrich) were used at the following concentrations: 50 U mL−1 polymyxin B (Pb), 100 µg mL−1 ampicillin (Amp), 100 µg mL−1 kanamycin (Kan), 10 µg mL−1 chloramphenicol (Cm), 60 µg mL−1 spectinomycin (Spec), and 100 µg mL−1 Streptomycin (Strep). Plasmids were introduced into electrocompetent E. coli S17-1λpir and BW-RI using 0.1 cm gap cuvettes (USA Scientific) and a MicroPulser Electroporator (Bio-Rad).
DNA manipulations and mutant construction
DNA manipulations were performed as in (Sambrook et al., 1989). E. coli S17-1λpir was used for cloning. iProof DNA polymerase (Bio-Rad) was used for regular PCR reactions, and PfuUltra DNA polymerase (Agilent) was used for constructing point mutations. Restriction enzymes, T4 polynucleotide kinase, Antarctic phosphatase, and T4 DNA ligase were purchased from New England Biolabs (NEB). Plasmids used in this study are described in Table S2. Primers from Integrated DNA Technologies (IDT) are listed in Table S3. All plasmids were confirmed by sequencing at Genewiz. Arabinose inducible qrr4, anhydrotetracycline inducible qrr4, and target-GFP translational fusions were constructed as described (Shao et al., 2013). V. cholerae mutants were constructed as described (Skorupski and Taylor, 1996).
Western blot analysis
Cell-free culture fluids from 5 ml cultures grown to OD600 ~1.0 were collected by centrifugation at 5000 g for 15 min followed by filtration through 0.22 µm membranes (Millipore). Secreted proteins were precipitated with 20% Trichloroacetic acid (TCA) and resuspended in TE buffer (10 mM Tris-HCl, pH 8.0 and 1 mM EDTA) with 0.5% SDS. 100 µg bovine serum albumin (BSA) was added prior to precipitation as a control. Protein samples were analyzed by SDS polyacrylamide gel electrophoresis (SDS-PAGE) on 4–20% gradient gels (Bio-Rad). Proteins were wet transferred to nitrocellulose membranes at 100 volts for 1 h. Membranes were subsequently blocked in TBS-T with 5% milk overnight at 4°C, incubated with Hcp antibody in TBS-T with 5% milk at a concentration of 1:2500 for 1 h, washed in TBS-T three times for 10 min each, incubated with secondary antibody in TBS-T with 5% milk at a concentration of 1:10 000 for 1 h, and again washed in TBS-T three times for 10 min each. Proteins were visualized using the ECL Western Blotting Detection Reagents (GE Healthcare). HRP conjugated anti-rabbit IgG was used as the secondary antibody (Promega).
RNA isolation and qRT-PCR
RNA used for quantitative RT-PCR (qRT-PCR) was isolated from V. cholerae cultures using the RNeasy Mini Kit (Qiagen) followed by DNase treatment (Ambion). In experiments requiring pulse induction of V. cholerae Qrr4, 0.2% arabinose was added to cultures at OD600 ~0.5. cDNA was generated with SuperScript III reverse transcriptase (Invitrogen) using ~3 µg of total RNA. qRT-PCR was performed on an ABI Prism 7900HT Sequence Detection System using Sybr Green PCR master mix (ABI). Triplicate samples were measured and analyzed by a comparative CT method (Applied Biosystems) in which the relative amount of target RNA was normalized to an internal control RNA (hfq) first, and subsequently, to each other. Statistical analyses were performed using the unpaired t test, with a two-tailed p value < 0.05 considered significant.
GFP reporter assay
Overnight cultures of E. coli reporter strains were diluted 1:1000 in triplicate into M9 medium (0.5% glucose). Upon dilution, 100 ng mL−1 anhydrotetracycline (Clontech) was added to induce qrr4 expression. Target-GFP translational fusions were induced by 0.5 mM IPTG. GFP fluorescence was measured after 8~10 h of growth using FACS (BD Biosciences FACSAria III cell sorter). Statistical analyses were performed using the unpaired t test, with a two-tailed p value < 0.05 considered significant.
T6SS killing assay
Overnight cultures of V. cholerae and of P. aeruginosa were diluted into fresh medium and grown to OD600 ~1.0. Cells from 5 ml of each culture were resuspended in 500 µl of medium (to OD600 ~10) and combined at a 10:1 ratio of P. aeruginosa:V. cholerae. 5 µl of the mixture was spotted on a dry LB agar plate. After ~4 h incubation at 37°C, the bacterial spots were excised and the cells were resuspended in 1 ml LB. The suspension was serially diluted in LB, and 100 µl of the suspensions were plated onto streptomycin-containing plates to select for V. cholerae. Colonies were counted after overnight incubation at 37°C.
Supplementary Material
Acknowledgements
We thank Dr. John Mekalanos for generously providing us the Hcp antibody and the non-pandemic V. cholerae strains 2740-80 and V52. We thank members of the Bassler laboratory for insightful discussions. This work was supported by the Howard Hughes Medical Institute, National Institutes of Health (NIH) Grant 5R01GM065859 and National Science Foundation (NSF) Grant MCB-0343821 to B.L.B.
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- Basler M, Ho BT, Mekalanos JJ. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell. 2013;152:884–894. doi: 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard CS, Brunet YR, Gavioli M, Lloubès R, Cascales E. Regulation of type VI secretion gene clusters by sigma54 and cognate enhancer binding proteins. J Bacteriol. 2011;193:2158–2167. doi: 10.1128/JB.00029-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks TM, Unterweger D, Bachmann V, Kostiuk B, Pukatzki S. Lytic activity of the Vibrio cholerae type VI secretion toxin VgrG-3 is inhibited by the antitoxin TsaB. J Biol Chem. 2013;288:7618–7625. doi: 10.1074/jbc.M112.436725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong TG, Ho BT, Yoder-Himes DR, Mekalanos JJ. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc Natl Acad Sci U S A. 2013;110:2623–2628. doi: 10.1073/pnas.1222783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong TG, Mekalanos JJ. Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic Acids Res. 2012;40:7766–7775. doi: 10.1093/nar/gks567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman Ja, Bassler BL. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol Microbiol. 1999;31:665–677. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- Fu Y, Waldor MK, Mekalanos JJ. Tn-Seq Analysis of Vibrio cholerae Intestinal Colonization Reveals a Role for T6SS-Mediated Antibacterial Activity in the Host. Cell Host Microbe. 2013;14:652–663. doi: 10.1016/j.chom.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- Holloway BW. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol. 1955;13:572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Rompikuntal PK, Lindmark B, Milton DL, Wai SN. Quorum sensing regulation of the two hcp alleles in Vibrio cholerae O1 strains. PLoS One. 2009;4:e6734. doi: 10.1371/journal.pone.0006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Sabharwal D, Bröms J, Milton DL, Sjöstedt A, Uhlin BE, Wai SN. Pathoadaptive conditional regulation of the type VI secretion system in Vibrio cholerae O1 strains. Infect Immun. 2012;80:575–584. doi: 10.1128/IAI.05510-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaoka M, Miyata ST, Brooks TM, Unterweger D, Pukatzki S. VasH is a transcriptional regulator of the type VI secretion system functional in endemic and pandemic Vibrio cholerae. J Bacteriol. 2011;193:6471–6482. doi: 10.1128/JB.05414-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Levine E, Zhang Z, Kuhlman T, Hwa T. Quantitative characteristics of gene regulation by small RNA. PLoS Biol. 2007;5:e229. doi: 10.1371/journal.pbio.0050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V, Timmis KN. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- Ma AT, McAuley S, Pukatzki S, Mekalanos JJ. Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe. 2009;5:234–243. doi: 10.1016/j.chom.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci U S A. 2010;107:19520–19524. doi: 10.1073/pnas.1012931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. Parallel Quorum Sensing Systems Converge to Regulate Virulence in Vibrio cholerae. Cell. 2002;110:303–314. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- Miyata ST, Bachmann V, Pukatzki S. Type VI secretion system regulation as a consequence of evolutionary pressure. J Med Microbiol. 2013;62:663–676. doi: 10.1099/jmm.0.053983-0. [DOI] [PubMed] [Google Scholar]
- Ng W-L, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A. 2007;104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MH, Biswas K, Hossain MA, Sack RB, Mekalanos JJ, Faruque SM. Distribution of genes for virulence and ecological fitness among diverse Vibrio cholerae population in a cholera endemic area: tracking the evolution of pathogenic strains. DNA Cell Biol. 2008;27:347–355. doi: 10.1089/dna.2008.0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, Kessel JCvan, Shao Y, Bassler BL. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev. 2011;25:397–408. doi: 10.1101/gad.2015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Vol. 1. NY: Cold Spring Harbor Laboratory Press; 1989. 2, 3 (1989) [Google Scholar]
- Shao Y, Bassler BL. Quorum-sensing non-coding small RNAs use unique pairing regions to differentially control mRNA targets. Mol Microbiol. 2012;83:599–611. doi: 10.1111/j.1365-2958.2011.07959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Feng L, Rutherford ST, Papenfort K, Bassler BL. Functional determinants of the quorum-sensing non-coding RNAs and their roles in target regulation. EMBO J. 2013;32:2158–2171. doi: 10.1038/emboj.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorupski K, Taylor RK. Positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- Skorupski K, Taylor RK. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol Microbiol. 1999;31:763–771. doi: 10.1046/j.1365-2958.1999.01215.x. [DOI] [PubMed] [Google Scholar]
- Thelin KH, Taylor RK. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect Immun. 1996;64:2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou AM, Cai T, Liu Z, Zhu J, Kulkarni RV. Regulatory targets of quorum sensing in Vibrio cholerae: evidence for two distinct HapR-binding motifs. Nucleic Acids Res. 2009;37:2747–2756. doi: 10.1093/nar/gkp121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Ho B, Mekalanos JJ. Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One. 2011;6:e23876. doi: 10.1371/journal.pone.0023876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Shin OS, Cameron DE, Mekalanos JJ. Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc Natl Acad Sci U S A. 2010;107:21128–21133. doi: 10.1073/pnas.1014998107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci U S A. 2002;99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.