Abstract
Neurospora crassa has a long history as an excellent model for genetic, cellular, and biochemical research. Although this fungus is known as a saprotroph, it normally appears on burned vegetations or trees after forest fires. However, due to a lack of experimental evidence, the nature of its association with living plants remains enigmatic. Here we report that Scots pine (Pinus sylvestris) is a host plant for N. crassa. The endophytic lifestyle of N. crassa was found in its interaction with Scots pine. Moreover, the fungus can switch to a pathogenic state when its balanced interaction with the host is disrupted. Our data reveal previously unknown lifestyles of N. crassa, which are likely controlled by both environmental and host factors. Switching among the endophytic, pathogenic, and saprotrophic lifestyles confers upon fungi phenotypic plasticity in adapting to changing environments and drives the evolution of fungi and associated plants.
The filamentous fungal species, Neurospora crassa has become a popular experimental model microbe for genetic, cellular, and biochemical research in the latter half of the 20th century1,2,3. N. crassa is commonly found on carbohydrate-rich foodstuffs and residues of sugar-cane processing1. Most N. crassa strains collected for studies of geographical distribution have been from tropical and subtropical regions4. However, N. crassa and N. discreta can also be found as far north as Montana and Alaska, respectively5, a natural habitat that contains coniferous trees. Although the lifestyle of N. crassa in the wild is unknown, it normally appears on burned vegetations or trees after forest fires5,6. It has been suggested that heat from forest fires stimulates the germination of ascospores in the soil and provides a sterile, nutrient-containing environment that stimulates growth5. Neurospora species has also been isolated from the artificial plantation of Acer ginnala (Amur Maple) in northeastern China and proposed to be an endophyte7; however, no strong experimental evidence of its association with living plants is available. Global distribution and comprehensive collection of isolates of Neurospora species offer a new platform to decipher its ecology and evolution in nature4,5. Fungi have been traditionally categorized as parasites, symbionts, or saprotrophs based on their strategies for nutrient acquisition. However, accumulated evidence suggests that fungal lifestyles are plastic in relation to their hosts and environments, rather than rigidly dictated only by their genetic makeup8,9,10,11. Fungal endophytes exhibit a broad range of lifestyles (e.g., latent saprotrophs or pathogens, and mutualists), which are determined by the fitness benefits conferred on their hosts, the production of secondary metabolites, and/or their colonization strategies8,12,13,14. Mycorrhizal fungi can also act as endophytes, necrotrophic pathogens, and antagonists of host or non-host plants8,9. Here, we performed a series of experiments to reveal the alternative lifestyles of N. crassa and provide the first evidence of endophytic and pathogenic N. crassa lifestyles in Scots pine (Pinus sylvestris).
Results
Association of N. crassa with Scots pine
In order to investigate the alternative lifestyles of N. crassa, Scots pine seedlings grown in microcosm were inoculated with conidial suspension (105 conidia/ml) and the colonization patterns were documented over a period of 5 months by fluorescence and confocal microscopy (Supplementary Fig. S1). Most of seedlings looked healthy and were indistinguishable from those without inoculation. Surprisingly, however, fungal hyphae expressing GFP were observed from inside of inoculated seedlings, but not from uninoculated ones. During its growth in the cells of Scots pine seedlings, N. crassa was found to proliferate and survive for up to 5 months without causing any disease symptoms, suggesting its endophytic lifestyle. Inside the roots, fungal growth was confined mostly to the root epidermis and cortex layers (Supplementary Fig. S1e). More compelling evidence of endophytic lifestyle is being described in the later section. To further decipher the innate association between N. crassa and Scots pine, we performed a series of inoculation experiments on Scots pine seedlings grown on water agar.
Can N. crassa be a plant pathogen on Scots pine?
Our most remarkable finding is that N. crassa can act not only as an endophyte but also as a pathogen on Scots pine, when the host plant was grown on water agar or under controlled environments in the greenhouse. Infection with N. crassa incited typical disease symptoms, eventually causing the death of Scots pine seedlings. The mortality rate reached to 83% (90 out of 108 seedlings) at 5 weeks post inoculation (wpi) (Fig. 1a). The abiosis of Scots pine caused by N. crassa takes 4–5 weeks, whereas its well-adapted fungal pathogen Heterobasidion annosum exerts a similar effect in only 3–4 weeks (96% mortality rate) (Fig. 1b). During the initial stage of infection, N. crassa conidia germinated, formed a hyphopodia-like structure (Supplementary Fig. S2c), penetrated into plant tissues, and grew intra- or intercellularly between adjacent cells (Fig. 1d–i; Supplementary Fig. S2d–f; Supplementary Fig. S3). Invasive growth continued from root cortical cells (Fig 1d–e) to the core area (Fig. 1f), and they were found in almost 50% of infected root cells at 5 wpi (Supplementary Fig. S4). At the end of the infection stage, N. crassa hyphae could grow out from the stomata on the stem of infected Scots pine seedling (Supplementary Fig. S5a–f) and develop conidiophores with conidia (Supplementary Fig. S5g). These observations clearly demonstrate that N. crassa can complete its life cycle in association with Scots pine and further support the hypothesis that N. crassa has a pathogenic lifestyle. Moreover, culture filtrate of N. crassa induced similar cell death in Scots pine seedlings (Supplementary Fig. S6b), suggesting that N. crassa may produce phytotoxic compounds and function as a necrotrophic plant pathogen on Scots pine. To understand if N. crass can incite disease symptom not only in seedlings grown on water agar, but also grown trees, 3-year-old Scots pine trees were inoculated with wood dowels pre-colonized by N. crassa in the greenhouse. N. crassa could incite clear necrosis on 3-year-old trees 6 wpi. The necrosis areas by N. crassa and H. annosum were 42.5% and 67.2%, respectively, when measured by the ratio of the white (healthy) and brown area (necrosis) (Fig. 2a–c). These infected trees were sampled and heat-treated at 121°C for 10 min to understand whether N. crassa inside of tree could survive under harsh conditions such as forest fire. Surprisingly, N. crassa was grown out and was the sole surviving fungal taxon on wood trunks after heat treatment when incubated for 2 weeks (Fig. 2d). This further supports our previous finding and suggests that N. crassa within host cells can survive as a pathogen or an endophyte and grow out from the burned tree as a saprotroph.
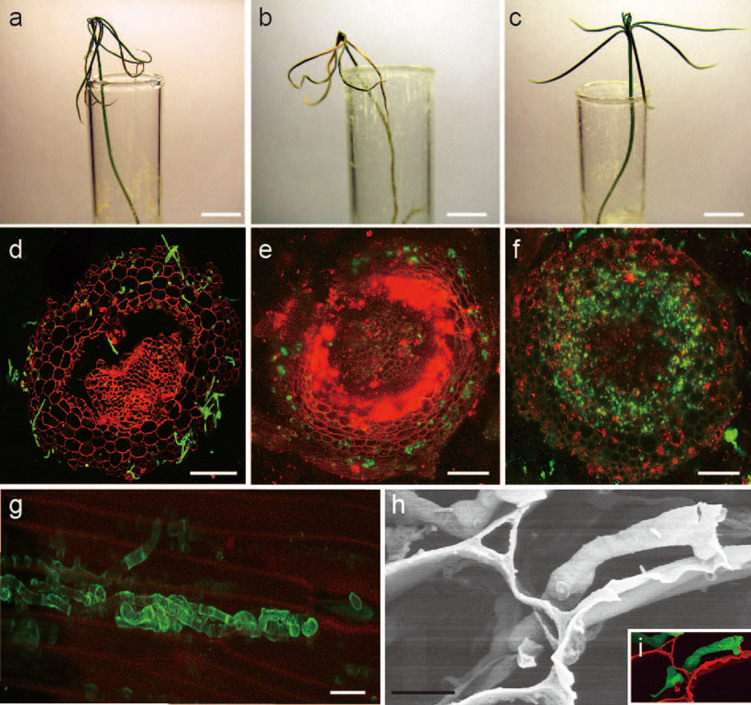
Figure 1. Pathogenic interactions between N. crassa and Scots pine seedlings.
Scots pine seedlings were inoculated with N. crassa (a), H. annosum (b), and control (c). (d) Transverse section of Scots pine root inoculated with N. crassa. Plant cell walls were stained with PI, and fungal hyphae were labeled with WGA. (e and f) Transverse sections of Scots pine roots inoculated with N. crassa FGSC 10589. GFP images were obtained by staining with FM4-64 at different stages of infection from 3 (e) to 5 (f) weeks post inoculation (wpi). (g) Image of N. crassa hyphae stained with WGA within host plant cells. (h) SEM image of N. crassa hyphae growing from one plant cell to another. (i) Colored SEM image, red and green indicate plant cell wall and N. crassa hyphae, respectively. Bars = 1 cm (a–c); 100 μm (d, e, and f); 10 μm (g); 5 μm (h). N. crassa strains used in a, d, g, h, and i was FGSC 2489.
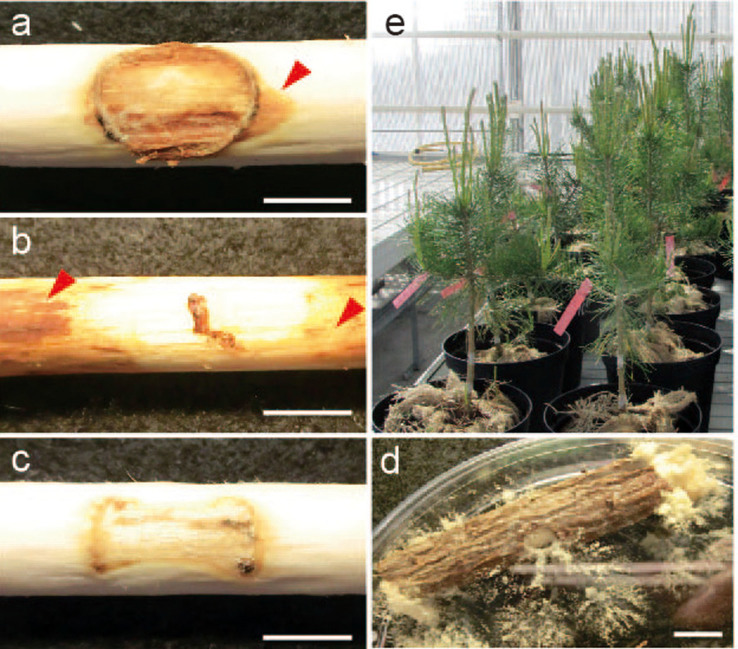
Figure 2. Infection of 3-year-old Scots pine trees by N. crassa.
Disease symptoms caused by N. crassa (a), H. annosum (b), and control (c). Disease symptom was measured 6 wpi. Red arrows indicate typical necrosis symptoms. (d) Survival of N. crassa in plant tissues after heat treatment. Three-year-old N. crassa-infected trees were heat-treated to 121°C at 100 kPa for 10 min, followed by incubation at 24°C for 2 weeks. (e) Three-year-old Scots pines used for infection experiments in the greenhouse. Bar = 1 cm.
Biochemical and molecular mechanisms underlying N. crassa and Scots pine interactions
To determine whether N. crassa infection elicits a defense response in Scots pine seedlings, host-defense-related reactions were observed. Plant cells at infection sites killed by N. crassa were observed by staining with Evans blue (Supplementary Fig. S6a). Furthermore, callose deposition and the accumulation of ROS around infection sites on stem were evident following staining with aniline fluorochrome blue (Fig. 3a) and diaminobenzidine (DAB) (Fig. 3b), respectively. These data indicate that the interaction between N. crassa and Scots pine represents a typical host–pathogen interaction.
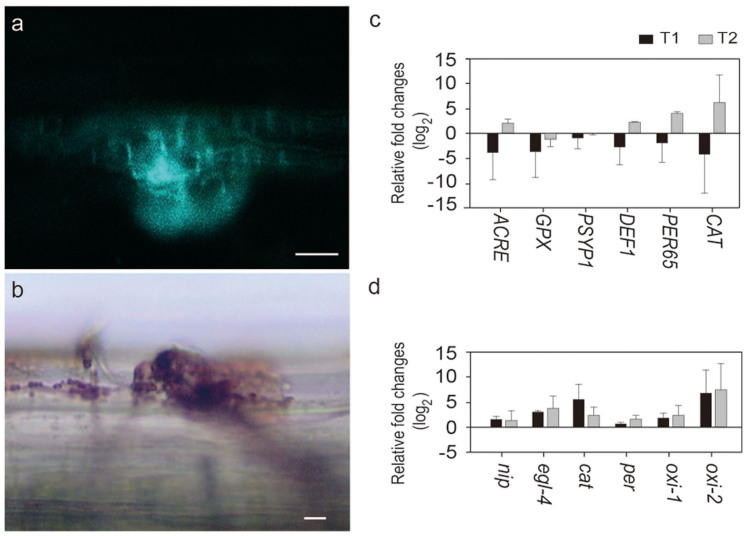
Figure 3. Host response and gene expression during interaction with N. crassa.
(a) Callose deposition around the infection site on stem, stained with aniline fluorochrome. (b) Accumulation of reactive oxygen species (ROS) at the infection site on stem, stained with diaminobenzidine (DAB). Bars = 20 μm. (c) Expression profiles of defense-related genes in the roots of Scots pine after N. crassa inoculation: ACRE, Avr9/Cf-9 rapidly elicited defense-related gene; PER65, peroxidase 65; PSYP1, class III peroxidase; GPX, glutathione peroxidase; DEF1, defensin; and CAT, catalase. (d) Expression profiles of N. crassa genes during its interaction with Scots pine's roots. nip, Necrosis-inducing protein; cat, catalase-1; per, dyp-type peroxidase; oxi-1 and oxi-2, two oxidoreductases. The level of expression was measured at one (T1) and two (T2) weeks after inoculation. Fold changes are relative to uninfected Scots pine seedlings (c) and N. crassa grown on Vogel's medium (d). The bars indicate standard deviation among 3 biological replicates.
In addition to biochemical response, expression patterns of ROS- and defense-related genes, such as those encoding peroxidases15 (peroxidase 65 [PER65], class III peroxidase [PSYP1], and glutathione peroxidase [GPX]), Avr9/Cf-9 rapidly elicited defense-related gene (ACRE)16, defensin (DEF1)17, and catalase (CAT)15 were monitored by qRT- PCR. All genes were differentially expressed in Scots pine's roots at 2 wpi with N. crassa (Fig. 3c). Expression of genes encoding catalase and peroxidases was most highly up-regulated. Expression of ACRE and DEF1 was also up-regulated in the roots (Fig. 3c) and stems (Supplementary Fig. S7a)after N. crassa infection. We also monitored the expression profiles of pathogenicity-related genes in N. crassa, including those encoding necrosis-inducing protein (nip)18, endoglucanase IV (egl-4)19, catalase (cat), peroxidase (per), and two oxidoreductases (oxi-1 and oxi-2).
Expression of egl-4 was highly up-regulated in roots (Fig. 3d and Supplementary Fig. S7b). Similar expression pattern of egl-4 was observed in H. irregulare during infection of Scots pine19. Expression of the genes encoding the two oxidoreductases and catalase were also highly up-regulated during interaction with Scots pine seedling (Fig. 3d). Together, expressions of genes responsible for ROS modulation were highly up-regulated in both the host and the pathogen. ROS is known to play key roles in maintaining the balance between endophytic and pathogenic fungal lifestyles with host plants20. Reducing ROS levels in the host can stimulate latent pathogens to cause disease20,21. Our gene expression data suggest that the pathogen and the host use opposite strategies to manipulate the level of ROS to maintain their relationships. Scots pine appears to reduce the expression of catalase (leading to accumulation of ROS) (Fig. 3c), resulting in a toxic response to the pathogen22 in the roots. In N. crassa, on the other hand, expression of catalase gene was highly up-regulated at an early stage of root infection (Fig. 3d), reducing the plant-derived ROS level. Later, the host catalase begins to be produced at high levels (Fig. 3c), likely due to excess ROS caused by the presence of the pathogen. Catalase expression of N. crassa was down-regulated, maintaining the virulence (Fig. 3d) of the fungus to the host, which can lead to an ectopic oxidative burst and cell death (accumulation of ROS)23. At the same time, both the pathogen and the host were preparing for the next phase of interactions in the other part of plant such as stem cells (Supplementary Fig. S7a,b). Since expression of a gene encoding a flavoprotein oxidoreductase (NCU06061)24 was highly up-regulated during infection (Fig. 3d), two deletion mutants (Δoxi-1 and Δoxi-2) were tested for their virulence. Indeed, deletion mutants showed significantly lower virulence on Scots pine seedlings (p = 0.02 and 0.07 in Δoxi-1 and Δoxi-2, respectively) (Supplementary Fig. S8), although they did not have any defect on mycological characters including mycelial growth, colony morphology and pigmentation. These combined biochemical and gene expression data suggest that the association between N. crassa and Scots pine is a typical intimate host–pathogen interaction.
Discussion
Understanding of lifestyle switching in fungi is critical for deciphering the evolution of host–microbe interactions and carbon/nitrogen cycling in the ecosystem. Our data and previous reports8,9,10,11 suggest that fungal lifestyles are not stable but dynamic, and are likely influenced by the genetic makeup of the fungal species, host factors, and changing environments. The endophytic stage represents a balanced interaction between the fungus and its host. However, endophytic fungal species can become pathogens or saprotrophs when this balance is disturbed or the host dies, respectively. In contrast, saprotrophic wood decomposers can colonize Scots pine's roots as mycorrhizal fungal species25. Opportunistic fungal pathogens, including Aspergillus fumigatus26,27 and Candida albicans28 are pathogenic only in immunocompromised humans26,27,28. Therefore, many fungi have likely evolved to switch their lifestyles among the Endophyte–Pathogen–Saprotroph as a circle (EPS Ring; Fig. 4) to adapt to various hosts and changing environmental conditions. However, the mechanisms underlying the appearance of N. crassa as the first fungal colonizer after forest fires in nature remain unknown. Although ascospores rather than conidia in the soil or on the tree were proposed as a source of Neurospora after a forest fire6,29, this is contradicted by the fact that conidia are observed most frequently in the field5. It has also been reported that desiccated conidia can survive after treatment at >100°C30 and our data also provides the evidence that N. crassa can survive on wood trunks after heat-treated (Fig. 2d). However, we do not rule out the possibility that ascospores could also survive such extreme temperature. These further suggest that N. crassa within host cells can survive and grow out from the burned tree in which it resides as a pathogen or an endophyte. To better understand the ecology of N. crassa and provide more ecological relevance of our findings, we attempted to detect N. crassa from forest soil samples collected from tropical (Indonesia) to Nordic (Finland) areas including post-forest fire sites (Supplementary Table S1). We were unable to amplify N. crassa ITS region by PCR from these soils. Even when soil DNA samples from post-forest fire sites were analyzed by pyrosequencing, no sequence signature was found as Neurospora species. These results would support our hypothesis that N. crassa may not be living as a saprotroph in forest soils but as an endophyte or a pathogen in their natural host, Scots pine. Taken together, our study will provide a new paradigm to understand fungal lifestyles in nature and coevolution with its associated plants.
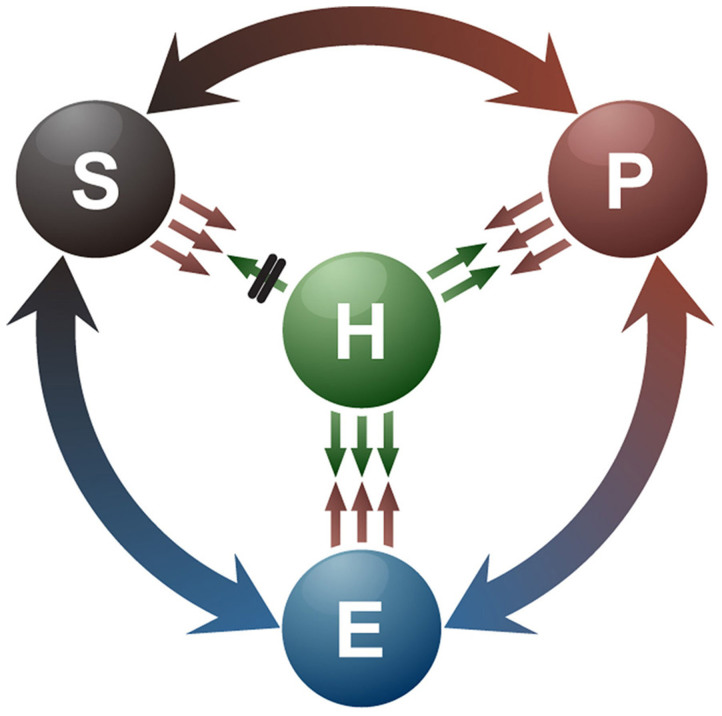
Figure 4. Dynamic relationship between a fungus and its host plant as a function of the fungal lifestyles, Endophyte–Pathogen–Saprotroph, as a circle (EPS Ring).
The endophytic stage represents a balanced interaction between fungal virulence and host defense factors. When this balance is disturbed or the host dies, endophytes may become pathogens or saprotrophs, respectively. Saprotrophs and pathogens may switch their lifestyles to endophytes/pathogens and endophytes/saprotrophs, respectively, in the presence of appropriate environmental factors. H, host; E, endophyte; P, pathogen; S, saprotroph.
Methods
Plant and fungal materials
Neurospora crassa strains (Supplementary Table S2) used in this study were obtained from the Fungal Genetic Stock Center, Missouri, USA, and grown on Vogel's medium (pH 6.5)2. Heterobasidion annosum P-type (isolate 03012 provided by Kari Korhonen, METLA, Finland) was maintained and grown on MEG medium (0.5% malt extract, 0.5% glucose, 2% agar). Scots pine (Pinus sylvestris L.) seeds were obtained from Svenska Skogsplantor (Saleby FP-45), Finland and sown on 2% water agar plates (or test tubes) or a mixture of sterilized Kekkilä White 420-W peat for microcosm assay. Prior to use, 100 seeds were sterilized by soaking in 30 ml of 33% Hydrogen peroxide for 15 min, and then washing with 2 L sterilized water31. Experiments on water agar or in microcosm were performed in growth chamber or in plant growth room, respectively. The controlled conditions were a 12-h light/12-h dark photoperiod at 24°C with 80% RH in growth chamber and a 16-h light/8-h dark period at 18°C in plant growth room.
Infection experiments and sample preparations
Five microliters of conidial suspension (105 conidia per mL−1 in water) were placed near the base of stems (in test tubes) or on roots (on Petri dish) of 3-week-old Scots pine seedlings on water agar. For microcosm assay, six Scots pine seeds were sown in each cup and 100 μl conidial suspensions were then placed on the peat near the roots. The plants were maintained in the plant growth room. The fungal colonization patterns were monitored over a period of 5 weeks and 5 months for the samples on water agar and in microcosm, respectively, by fluorescence and confocal microscopy. To make fungal infections in fully developed trees, 3-year-old Scots pines were used. The trees were obtained from the field in Ruotsinkylä (field station of the Finnish Forest Research Institute, Finland) and grown in natural peat (Kekkilä Oy, Finland) in a greenhouse (22°C during the day and 18°C at night). The average height and diameter of the trees were 60 cm and 15 mm, respectively. The wood dowels (autoclaved Scots pine wood, 1 cm height × 1 cm diameter) pre-colonized (4 weeks) by N. crassa and H. annosum were used as inocula. Disease symptom was measured at 6 weeks after inoculation from 9 trees for each experimental group.
For cytotoxicity assay of culture filtrate, N. crassa was grown in Vogel's liquid medium for 5 days and filtered with 0.22 μm filter to remove fungal mycelia and spores. Prior to use, three-week-old Scots pine seedlings were exposed to light for 30 min.
Microscopic lesions and fungal hyphae were visualized by staining infected plant tissues. The tissues were cleared in alcoholic lactophenol, consisting of 1 volume of lactophenol (phenol: glycerol: lactic acid: water, 1:1:1:1, v/v) and 2 volumes of ethanol, followed by rinsing with lactophenol. The tissues were then transferred into diluted trypan blue solution (250 μg mL−1 trypan blue in lactophenol) and boiled for 1 min. Destaining was performed by replacing the staining solution with chloral hydrate (250 g/100 ml), followed by a wash in 50% glycerol; and tissues were then mounted on glass slides for microscopic observation. Cell death was detected by staining with Evans blue solution (0.25% [w/v] in 0.1 mM CaCl2, pH 5.6; Sigma)17.
Diaminobenzidine (DAB) was used to detect accumulated H2O232. Aniline blue staining was used to reveal callose formation33. Propidium iodide (PI)34,35 and wheat germ agglutinin (WGA) from Triticum vulgaris conjugated to fluorescein isothiocyanate (FITC) were used to stain plant and fungal cell walls, respectively.
Wide-field fluorescence microscopy and light microscopy
Wide-field fluorescence microscopy was performed using a Leitz Laborlux S with a Leitz NPL Fluotar 100×/1.32 oil 160/0.17 objective. A fluorescein filter set with excitation at 395 nm and emission at 495 nm was used. Light microscopy (Olympus CX31) was performed with a PlanC N 10×/0.25 or a PlanC N 40×/0.65 objective.
Confocal laser scanning microscopy
Confocal laser scanning microscopy was performed using a Leica TCS SP5II HCS A inverted microscope with an HCX PL APO 20× (0.7 NA) CS (air) objective and an HCX PL APO 63× (1.2 NA) W Corr/0.17 CS (water) objective. GFP-, FITC- and FM4-64-labeled images were captured simultaneously using 488 nm excitation with an argon laser and fluorescence detection at 500–520 nm (for GFP and FITC) and >600 nm for FM4-6436. The fluorescence excitation maximum for PI was 535 nm and the emission maximum was 617 nm. Image analysis was performed using Imaris ver. 7.5.2 (Bitplane Scientific Software, Zurich).
Imaging by scanning electron microscope
Specimens were dissected into 0.5 mm with a scalpel, dried on the sterilized and uncoated cellophane placed on water solid medium for 24 h at room temperature. The cellophane with specimens was cut into 1 cm- by-1 cm square and fixed with fixing solution (2% glutaraldehyde, 2% formaldehyde, 0.1% tannic acid, 4.5% sucrose in 70 mM sodium phosphate buffer (pH 7.4) and immersed overnight at 4°C. Specimens were subsequently dehydrated with serial concentrations of ethanol (50–100%), critical point drying using Bal-Tec CPD 030 device, mounted to aluminum specimen stubs, and coated with platinum using Platinum sputter for 20 sec. The specimen was examined at 5 kV beam voltage, spot size 4.5, and 60 Pascal pressure. Digital images were captured by FEI Quanta 250 Field Emission Gun Scanning Electron Microscope (FEI Co., Eindhoven, The Netherlands) using a Large Field Low vacuum secondary electron detector (LFD).
RNA isolation, cDNA synthesis, and qRT-PCR
RNA extraction from inoculated Scots pine seedlings was performed using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. There are two sets of control, RNAs from uninoculated plants and Neurospora crassa grown in the culture medium. Reverse transcription was performed with Moloney murine leukemia virus reverse transcriptase (M-MuLV RT) (Thermo Fisher Scientific). qRT-PCR was performed using a LightCycler® 480 Instrument II with SYBR Green I Master (Roche). The thermal profile for qRT-PCR (primers listed in Supplementary Tables S3 and 4) was an initial 95°C for 5 min, denaturation at 94°C for 10 sec (4.8°C/s), annealing at 59°C for 10 sec (2.5°C/s), extension at 72°C for 10 sec (4.8°C/s), 40 cycles of amplification, and a final extension at 72°C for 3 min. Ct values were calculated using the LightCycler 480 software.á-tubulin (TUBA) and 40 S ribosome were used as reference genes to normalize the data for the host plant and the fungus, respectively. The relative fold changes were calculated by the equation (1)
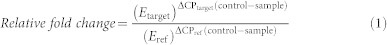 |
DNA extraction and PCR from forest soil samples
A total of 115 forest soil samples collected from tropical (Indonesia) to Nordic areas (Finland) including post-forest fire sites were obtained from the courtesy of Kajar Koster, Department of Forest Sciences, University of Helsinki. Details on collection sites and year, and soil types are described in Supplementary Table S1. Soil DNA was extracted by using cetyltrimethyl ammonium bromide (CTAB) buffer37, and used to detect N. crassa by PCR with ITS primer pair of NcITS-F (AAAACTCCCACAAACCATCG) and NcITS-R (CCGCCACTGATTTTGAGG).
Author Contributions
Conceived and designed the experiments: H.C.K., Y.H.L. Performed the experiments: H.C.K. Analyzed the data: H.C.K., S.H., J.C., F.O.A., J.P.T.V., Y.H.L. Wrote the paper: H.C.K., Y.H.L.
Supplementary Material
Supplementary information-Secret lifestyles of Neurospora crassa
Acknowledgments
This work was supported by the Finland Distinguished Professor Programme (FiDiPro # 138116) from Academy of Finland, and grants from the National Research Foundation of Korea (2012-0001149 and 2012-0000141) to YHL.
Additional information: Reprints and permissions information is available at www.nature. com/reprints.
References
- Davis R. H. & Perkins D. D. Neurospora: a model of model microbes. Nat. Rev. Genet. 3, 397–403 (2002). [DOI] [PubMed] [Google Scholar]
- Davis R. H. Neurospora: contributions of a model organism. (Oxford University Press, New York, 2000). [Google Scholar]
- Seiler S. & Plamann M. The genetic basis of cellular morphogenesis in the filamentous fungus Neurospora crassa. Mol. Biol. Cell 14, 4352–4364 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B. C., Perkins D. D. & Fairfield A. 2001. Neurospora from natural populations: a global study. Fungal Genet. Biol. 32, 67–92 (2001). [DOI] [PubMed] [Google Scholar]
- Jacobson D. J. et al. Neurospora in temperate forests of western North America. Mycologia 96, 66–74 (2004). [PubMed] [Google Scholar]
- Powell A. J., Jacobson D. J., Salter L. & Natvig D. O. Variation among natural isolates of Neurospora on small spatial scales. Mycologia 95, 809–819 (2003). [PubMed] [Google Scholar]
- Qi F., Jing T. & Zhan Y. Characterization of endophytic fungi from Acer ginnala maxim. in an artificial plantation: media effect and tissue-dependent variation. PLoS ONE 7, e46785 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide R. T., Sharda J. N., Herr J. R. & Malcolm G. M. Ectomycorrhizal fungi and the biotrophy-saprotrophy continuum. New Phytol. 178, 230–233 (2008). [DOI] [PubMed] [Google Scholar]
- Hibbett D. S., Gilbert L. B. & Donoghue M. J. Evolutionary instability of ectomycorrhizal symbioses in basidiomycetes. Nature 407, 506–508 (2000). [DOI] [PubMed] [Google Scholar]
- Eaton C. J. et al. Disruption of signaling in a fungal-grass symbiosis leads to pathogenesis. Plant Physiol. 153, 1780–1794 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R. J., White J. F. Jr, Arnold A. E. & Redman R. S. Fungal endophytes: diversity and functional roles. New Phytol. 182, 314–330 (2009). [DOI] [PubMed] [Google Scholar]
- Redman R. S., Dunigan D. D. & Rodriguez R. J. Fungal symbiosis from mutualism to parasitism: who controls the outcome, host or invader? New Phytol. 151, 705–716 (2001). [DOI] [PubMed] [Google Scholar]
- Johnson N. C., Graham J. H. & Smith F. A. Functioning of mycorrhizal associations along the mutualism-parasitism continuum. New Phytol. 135, 575–585 (1997). [Google Scholar]
- Müller C. B. & Krauss J. Symbiosis between grasses and asexual fungal endophytes. Curr. Opin. Plant Biol. 8, 450–456 (2005). [DOI] [PubMed] [Google Scholar]
- Vornam B., Arkhipov A. & Finkeldey R. Nucleotide diversity and gene expression of catalase and glutathione peroxidase in irradiated Scots pine (Pinus sylvestris L.) from the Chernobyl exclusion zone. J. Environ. Radioactiv. 106, 20–26 (2012). [DOI] [PubMed] [Google Scholar]
- Rowland O. et al. Functional analysis of Avr9/Cf-9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf-9–dependent disease resistance in tomato. Plant Cell 17, 295–310 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. Y., Jin J., Lee Y. W., Kang S. & Lee Y. H. Rice blast fungus (Magnaporthe oryzae) infects Arabidopsis via a mechanism distinct from that required for the infection of rice. Plant Physiol. 149, 474–486 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R. A. et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434, 980–986 (2005). [DOI] [PubMed] [Google Scholar]
- Olson Å. et al. Insight into trade-off between wood decay and parasitism from the genome of a fungal forest pathogen. New Phytol. 194, 1001–1013 (2012). [DOI] [PubMed] [Google Scholar]
- Tanaka A., Christensen M. J., Takemoto D., Park P. & Scotta B. Reactive oxygen species play a role in regulating a fungus-perennial ryegrass mutualistic interaction. Plant Cell 18, 1052–1066 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogel K. H., Franken P. & Hückelhoven R. Endophyte or parasite-what decides? Curr. Opin. Plant Biol. 9, 358–363 (2006). [DOI] [PubMed] [Google Scholar]
- Mittler R. et al. Transgenic tobacco plants with reduced capability to detoxify reactive oxygen intermediates are hyperresponsive to pathogen infection. Proc. Natl. Acad. Sci. U.S.A. 96, 14165–14170 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamnongpol S. et al. Defense activation and enhanced pathogen tolerance induced by H2O2 in transgenic tobacco. Proc. Natl. Acad. Sci. U.S.A. 95, 5818–5823 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Xu L. G., Li X., Zhai Z. & Shu H. B. AMID, an apoptosis-inducing factor-homologous mitochondrion-associated protein, induces caspase-independent apoptosis. J. Biol. Chem. 277, 25617–25623 (2002). [DOI] [PubMed] [Google Scholar]
- Vasiliauskas R., Menkis A., Finlay R. D. & Stenlid J. Wood-decay fungi in fine living roots of conifer seedlings. New Phytologist 174, 441–446 (2007). [DOI] [PubMed] [Google Scholar]
- Mullins J., Harvey R. & Seaton A. Sources and incidence of airborne Aspergillus fumigatus (Fres). Clin. Allergy 6, 209–217 (1976). [DOI] [PubMed] [Google Scholar]
- Latgé J. P. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12, 310–350 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller M. A. & Diekema D. J. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20, 133–163 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. Asexual and sexual developments of Neurospora crassa on natural substrata. Fungal Ecol. 5, 223–229 (2012). [Google Scholar]
- Lindegren C. C. The genetics of Neurospora I. The inheritance of response to heat-treatment. Bull. Torrey Bot. Club 59, 85–102 (1932). [Google Scholar]
- Jaber E., Xiao C. & Asiegbu F. O. Comparative pathobiology of Heterobasidion annosum during challenge on Pinus sylvestris and Arabidopsis roots: an analysis of defensin gene expression in two pathosystems. Planta 10.1007/s00425-013-2012-z (2013) [DOI] [PubMed] [Google Scholar]
- Rusterucci C., Aviv D. H., Holt B. F. III, Dangl J. L. & Parker J. E. The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13, 2211–2224 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J. M., Heard J. E., Asai T. & Ausubel F. M. Simulation of fungal-mediated cell death by fumonisin B1 and selection of fumonisin B1-resistant (fbr) Arabidopsis mutants. Plant Cell 12, 1811–1822 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treitschke S., Doehlemann D., Schuster M. & Steinberg G. The myosin motor domain of fungal chitin synthase V is dispensable for vesicle motility but required for virulence of the maize pathogen Ustilago maydis. Plant Cell 22, 2476–2494 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E. & Haseloff J. A simple way to identify non-viable cells within living plant tissue using confocal microscopy. Plant Methods 4, 15 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag M., Hickey P. C., Raju N. B., Selker E. U. & Read N. D. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 41, 897–910 (2004). [DOI] [PubMed] [Google Scholar]
- Jasalavich C. A., Ostrofsky A. & Jellison J. Detection and identification of decay fungi in spruce wood by restriction fragment length polymorphism analysis of amplified genes encoding rRNA. Appl. Environ. Microbiol. 66, 4725–4734 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information-Secret lifestyles of Neurospora crassa