Abstract
The investigation of the genetic basis of refractive error and myopia entered a new stage with the introduction of genome-wide association studies (GWAS). Multiple GWAS on many ethnic groups have been published over the years, providing new insight into the genetic architecture and pathophysiology of refractive error. This is a review of the GWAS published to date, the main lessons learned, and future possible directions of genetic studies of myopia and refractive error.
Keywords: refractive error, genetic architecture, GWAS
Introduction
Myopia is the most common eye disorder in the world,1 affecting over a quarter of all adults in Europe and United States2 and three-quarters of urban populations in Southeast Asia.3,4 Myopia is an environmentally driven condition in genetically susceptible individuals. A remarkable rise in the prevalence of myopia worldwide over the last three decades5,6 and associations of refractive error with a host of environmental factors and behavioral characteristics7,8 point to a strong environmental influence on refractive errors. Nevertheless, a strong association of myopia with parental history9 as well as heritability studies10 has consistently indicated that more than half of the variability of refractive error within populations is determined by genetic factors.
Given the prevalence of refractive error and myopia, the economic costs (estimated at an annual US$268 billion worldwide for myopia alone11), and rising numbers of individuals affected (as many as 2.5 billion by 2020),2 there is considerable interest in genetic epidemiological studies as a means of uncovering the mechanisms underlying refractive development in humans. The ultimate goal of these studies is to reduce the burden of refractive errors by identifying potential biological targets for treatment and/or prevention strategies. In addition to these direct benefits, general scientific interest is justifiably high regarding a trait that affects the only visible part of the central nervous system and that is associated at a population level with variations of intelligence quotient12 and academic achievement.13
The full range of genomic tools has been exploited to study refractive error in human populations and families. At least 23 genome-wide linkage studies in related individuals have identified 17 loci across the human genome (a detailed review of linkage studies can be found elsewhere14,15). In addition, a variety of refractive phenotypes have been studied in numerous candidate gene association studies. Candidate regions were chosen based on prior knowledge of gene functionality (often gleaned from animal models of visually induced myopia) or in combination with genetic linkage evidence (see reviews elsewhere15,16). However, these results were generally poorly reproducible, likely because of factors ranging from genetic and phenotypic heterogeneity to methodological issues that are known to reduce the power of genetic association studies.17
Contrary to inherently hypothesis-driven candidate-region genetic studies, genome-wide association studies (GWAS) offer an alternative, hypothesis-free approach that is often more appropriate for the genetic dissection of complex traits, which are affected by numerous genetic variants. They were first reported in the field of myopia research in 2009. This review will provide a brief description of the GWAS of refractive phenotypes published to date, along with the early lessons learned and some conclusions regarding the future of genomics research in the field. For ease of presentation we will divide these GWAS into two groups defined by the traits under consideration and their respective study designs: GWAS associated with high myopia in case–control analyses; and population-based GWAS of ocular refraction treated as a quantitative trait, with myopes in the negative part of the distribution, as quantified by the spherical equivalent refraction in diopters (D).
The first published GWAS for refractive error was designed as a two-stage analysis, using a cohort of 297 cases of pathological myopia (axial length > 26 mm) and 977 controls drawn from the general population18; all participants were of declared Japanese ethnicity. Follow-up association of the 22 most suggestive loci from the discovery stage in 533 cases and 977 controls revealed the strongest association at rs577948 (P = 2.22 × 10−07), located on 11q24.1 approximately 44 kb upstream of the BLID gene. The risk allele conferred a relative increase in the odds of high myopia of 1.37 (95% confidence interval: 1.21–1.54).
Li et al.19 performed another two-stage association analysis of high myopia (≤ −6 D) in Chinese and Japanese subjects. The first stage was a meta-analysis of two cohorts of ethnic Chinese cases: one preadolescent group (65 cases and 238 controls) and the other group comprising young adults (222 cases and 435 controls). After replication in a Japanese cohort (959 cases and 2128 controls, who overlapped the ones used in the previously described association study18), the strongest association was obtained for rs6885224 (Fisher's combined P = 7.84 × 10−06). This variant is an intronic single nucleotide polymorphism (SNP) within the CTNND2 gene on 5p15.2. It should be noted that neither of these initial association signals met the conventional threshold for statistical significance in GWAS (P < 5 × 10−08).
Another study of ethnic Han Chinese participants was conducted by Li et al.20 The genome-wide discovery stage focused on 102 high-grade myopia cases (≤ −8 D) with retinal degeneration and 335 “myopia-free” controls. Replication was attempted in two further stages using 2628 cases and 9485 controls in one stage and 263 cases and 586 controls in the other. The strongest evidence for association was observed for rs10034228 (meta-analysis P = 7.70 × 10−13), a high-frequency variant (minor allele frequency [MAF] = 0.5), located in a gene desert within the MYP1121 myopia linkage locus on 4q25.
Shi et al.22 identified another locus associated with high myopia (≤ −6 D) using a discovery cohort of 419 cases and 669 controls, all of Han Chinese ancestry, and replicating their findings in a combined 2803 cases and 5642 controls. They found the strongest evidence of association at rs9318086 (P = 1.91 × 10−16), an intronic SNP within the MIPEP gene on chromosome 13q12. This variant is common across populations including Asians (MAF = 42% and 46% in Han Chinese and Japanese HapMap samples, respectively).
The defining characteristic of the above studies is that they were aimed at identifying susceptibility variants for high myopia. These studies were all conducted in East Asia, where the prevalence of this condition is highest and has been increasing the most within the last few decades. Their results, however, have not been replicated in other GWAS of similar design, ethnic background, and phenotypic definition (high myopia).
Among Europeans, the prevalence of high-grade myopia is lower than in East Asians, and the main approach has been to study quantitative association with the whole spectrum of refractive error. This approach has the advantage of utilizing all data from population-based samples. The first two GWAS of refractive error in European subjects included 4270 British23 and 5328 Dutch24 individuals from the general population in their respective GWAS discovery cohorts. For replication, both studies used more than 10,000 subjects drawn from each other's main discovery cohorts and a smaller shared pool of replication samples. Associations survived customary GWAS significance thresholds for two separate loci, one near the RASGRF1 gene and the other near GJD2 (P = 2.70 × 10−09 for rs8027411 on 15q25.1 and 2.21 × 10−14 for rs634990 on 15q14).
Over a year later, the only case–control GWAS for high-grade myopia in a population of European origin was published from France.25 Using 192 cases (≤ −6 D) and 1064 controls but no independent replication sample, this study identified suggestive evidence of association for a marker located within the MYP10 linkage locus 3 kb downstream of PPP1R3B26 (P = 6.32 × 10−7 for rs189798). This study did not replicate any of the previously reported loci for refractive phenotypes.
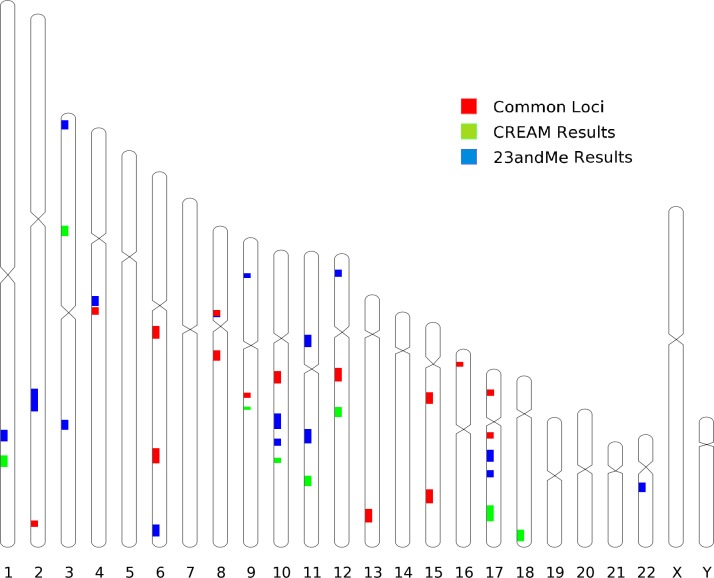
Major developments in myopia genetics occurred in early 2013. Two studies, one from the international Consortium for Refractive Error and Myopia (CREAM)27 and one by the direct-to-consumer genotyping company 23andMe28 (Mountain View, CA, USA), amassed 37,382 and 55,177 subjects of European ancestry, respectively. In addition, the CREAM study reported on 12,332 subjects of various Southeast Asian ancestries. For the first time deploying high statistical power in the field, the two studies followed different study designs and analytical strategies. The CREAM study was a classic meta-analysis of GWAS data from linear regressions on spherical equivalent obtained from 35 participating centers. The 23andMe study involved a GWAS survival analysis on age of onset of myopia (<30 years), obtained from questionnaire data. Both studies replicated associations to the RASGRF1 and GJD2 loci previously discovered in British and Dutch populations.23,24 In addition, the CREAM study identified a total of 24 novel loci in its multiethnic panel while the 23andMe study independently identified 20 novel loci. Very surprisingly for two studies of such different designs, both GWAS independently identified or replicated at near GWAS significance the same 25 genetic loci across the genome29 (Fig. 1). Moreover, despite being measured on different scales (diopters for CREAM and hazard ratios for 23andMe), the direction of the estimated effects was consistent across all significant loci. Even though the effects were generally small, the genetic basis underlying both age of onset of myopia and degree of refractive error was shown to be very similar.
Figure 1.
Location of the loci associated with refractive error in the CREAM meta-analysis (green), with myopia age of onset in the 23andMe study (blue), or in both (red). The size of each locus is chosen for better visualization and does not correspond to any of its properties.
Lessons From GWAS
Genome-wide association studies in largely European populations have provided great insight into the genetics of refractive variation. They represent important first steps in what is bound to become an essential path in the study of myopia genetics in human populations: aggregation of a large number of subjects using methodology (GWAS) that critically depends on large numbers to achieve robust results. As large GWAS have the statistical power to yield highly reliable results, they provide the firm foundations needed to draw the first lessons from genetic analysis of myopia and offer insights into what lies ahead. Some of the lessons that can be drawn from these studies are listed below.
First, genome-wide association studies have identified genetic loci for refractive error and myopia that have been independently replicated; the simultaneous discovery of more than 20 novel loci for refractive phenotypes by two independent groups leaves little doubt as to the validity of their findings. Scientific rigor from study design to analytical protocol30 is often rewarded in sufficiently powered GWAS. As already noted, the two largest GWAS of refractive error and myopia age at onset yielded almost identical results. There was a high correlation between effect sizes (regression coefficient and hazard ratios) in these two studies.29 Interestingly, the CREAM study offers credible evidence that the genetic architecture of refractive error may be largely the same in at least two continental populations (European and Asian). Genome-wide association results published from high-grade myopia studies were not replicated by the meta-analysis of the CREAM cohorts, which may be due to phenotypic or genetic heterogeneity and power needed to reach statistically significant conclusions from these studies.17,31 However, underpowered studies are more likely to produce spurious results, and any findings (be they from GWAS of other study designs) must be interpreted in light of their statistical power.
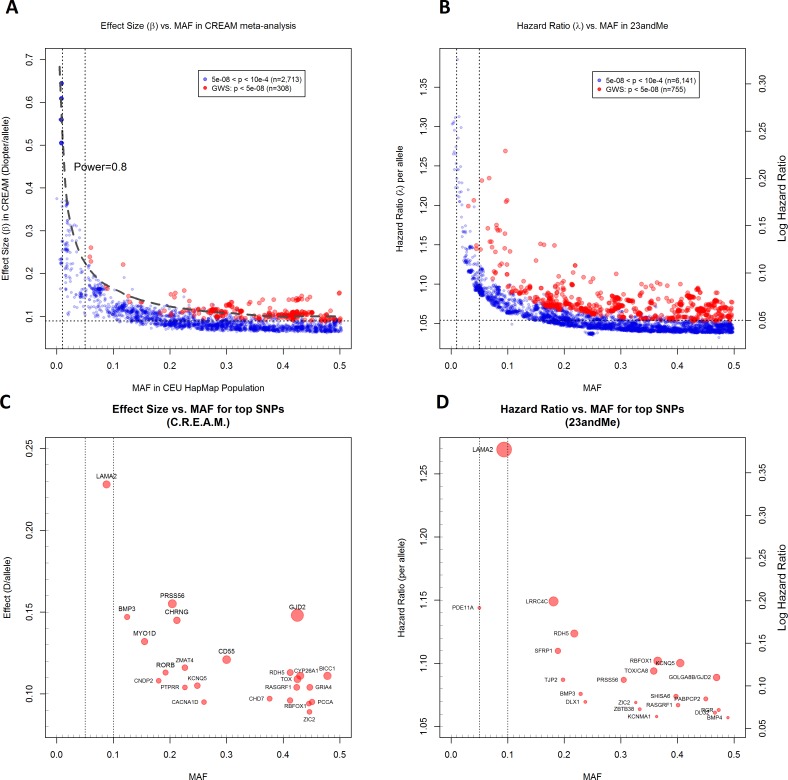
Second, the allelic spectrum of the identified refractive error loci confirms the polygenic nature of myopia. A comparison of findings from the CREAM and 23andMe studies shows that loci associated with population variations in refractive error are generally of small effect. Interestingly, most of the significantly associated alleles in each study were of high frequency. A plot of the effect sizes observed versus the MAF at these loci (Fig. 2) shows that the effect size of the discovered genetic variants is inversely proportional to their allele frequency and that variants with strongest effects tended to have lower allele frequencies. This observation reflects the fact that there may be very few common loci with large effect sizes, and the limited power of the current GWAS precludes identification of rare variants with lesser effects.
Figure 2.
Effect size versus minor allele frequency (MAF) for all SNPs showing associations at P < 0.0001 (blue circles) and P < 5 × 10−08 (red circles) with refractive error in the CREAM European (A, C) and with age of onset of myopia in 23andMe (B, D) analyses. All SNPs meeting these significance thresholds are shown; SNPs within loci are not independent because of linkage disequilibrium. Effect sizes shown are for the allele associated with a positive linear regression coefficient (CREAM) or hazard ratio > 1 (23andMe). The dashed line shows the power of 0.8 in a GWAS at a type 1 error (α) of 5 × 10−08 for the given effect size and MAF, assuming a sample size of 40,000 individuals, an additive genetic model, and a standard deviation = 2.2 D for refraction. Vertical dotted lines show MAF cutoffs of 0.01 and 0.05. Horizontal dotted lines show the empirical asymptotic limit of detection in CREAM (0.09 D/allele) and 23andMe (hazard ratio = 1.05/allele) at genome-wide significance (P < 5 × 10−08). Effect size versus minor allele frequencies are plotted for the top SNPs at each genome-wide significant locus for CREAM (C) and 23andMe (D). The closest genes (within 100 kb from the strongest associated SNP) at each locus are labeled. Circle diameters are scaled according to the –log10 P value; larger circles represent greater statistical significance within each study.
As the vast majority of the variants in the human genome have population frequencies of less than 5%, it is thus possible to be optimistic about what may still be hidden among them that future GWAS may find. As ever-larger consortia are being formed, they will have the power to detect effects of uncommon variants. Because of their low frequency, individually these variants are unlikely to account for a significant proportion of the population variance in refractive error or to a high population-attributable risk of myopia. Nor is the potential of larger consortia to discover even rarer variants of strong effect inexhaustible. Evidence from simulation studies32 shows that at relative risks (RR) above 2.0, the power of the conventional linkage studies starts to exceed that of association studies of equal sample size. Although some associations with myopia for very rare and highly penetrant variants will almost certainly be identified from genotyping or sequencing of large cohorts, most of the phenotypic variability at the population level will be explained by common variants as suggested for other traits.33
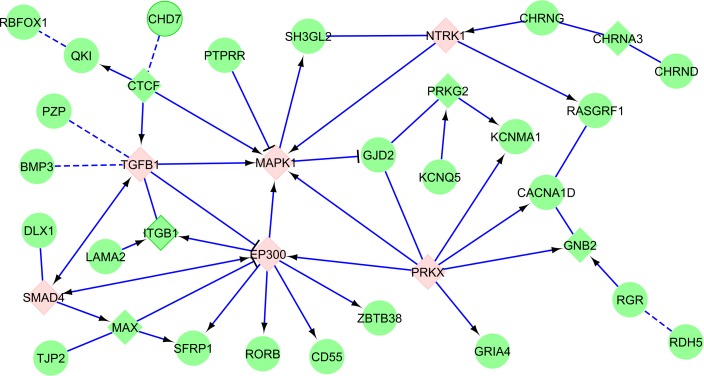
Third, refractive error and myopia are caused by genes acting along a finite number of potentially interacting physiologic pathways. The signature of specific functional pathways (or ontological classifications) is very strong and reproducible across many studies.34 Most of the variants identified lie outside transcript coding regions, and the mechanisms by which they control ocular growth are uncertain. Inferences about their functionality have at this stage been made judging only by their proximity to protein-coding genes, although this criterion does not necessarily account for factors that can decrease the accuracy of annotation, such as long-ranging linkage disequilibrium. Genes in the proximity of loci associated with refractive error are enriched for certain functional annotations such as neurotransmitter functions (GJD2, RASGRF1, GRIA4, and so on), retinoic acid metabolism (RDH5, RGR, RORB), and ion channel activity (KCNQ5, KCNJ2, KCNMA1, CACNA1D) or are involved in ocular and central nervous system development (SIX6, CHD7, ZIC2, and PRSS56). Although, at first sight, these gene networks appear to have little in common, the effect of these genes and their protein products may be highly coordinated. Based on existing knowledge on protein–protein interactions involving products of genes identified through the two large-scale GWAS shows that many of the genes are related to cell cycle and growth pathways such as the MAPK and TGF-beta/SMAD pathways (Fig. 3). This is consistent with the view that molecular mechanisms underlying refractive error and eye growth overlap. Further study of the topology of gene networks involved in refraction will provide insight into the mechanistic architecture of refractive development and yield potential molecular avenues for intervention.
Figure 3.
Network connections of genes associated with refractive error in two GWAS.28,38 The genes directly identified in these GWAS are shown in round green nodes, linker elements in square nodes. Key MAPK, TGF-beta/SMAD pathway elements are highlighted in pink. Solid blue edges symbolize known protein–protein interactions; dashed blue edges symbolize corregulation relationship. The network was constructed using Reactome database.39,40
Fourth, genes associated with refractive error have pleiotropic effects on a number of nonocular systems. Specific SNPs associated with refractive error (CREAM) or age of onset of myopia (23andMe) are generally not indexed in the database of GWAS SNPs as associated with other diseases.35 However, cross referencing genes near markers associated with refractive error or onset of myopia reveals that a number of these genes have been found in association with a number of other phenotypes in past GWAS (Table).
Table.
Genome-Wide Association Studies Results for the Refractive Error– and Myopia-Associated Genes in the GWAS Database as of August 1, 2013
|
|
Gene |
Trait |
PMID |
| Stature | BMP2 | Height | 18391951, 20881960 |
| BMP3 | Height | 18391951, 20881960 | |
| KCNJ2 | Height | 20881960 | |
| PDE11A | Height | 20881960 | |
| SIX6 | Height | 20881960 | |
| ZBTB38 | Height | 21998595, 18193045, 18391950, 18391951, 18391952, 19343178, 19396169, 19893584, 20189936, 20881960 | |
| Diabetes, insulin, and glucose metabolism | CACNA1D | Insulin resistance/response | 21901158 |
| TCF7L2 | Fasting glucose-related traits | 20081858, 22581228, 21873549, 20081857, 17293876, 17460697, 17463246, 17463248, 17463249, 17554300, 17668382, 18372903, 19056611, 19401414, 20581827, 22101970, 22693455, 19734900 | |
| ZMAT4 | Fasting plasma glucose | 17903298, 17903298 | |
| Obesity, fat metabolism | BMP2 | Body mass index | 19079261 |
| CYP26A1 | Triglycerides | 20686565 | |
| KCNMA1 | Obesity | 21708048 | |
| LAMA2 | Body mass index | 20397748 | |
| RORB | Adipose tissue ratio | 22589738 | |
| TJP2 | Renal sinus fat | 22044751 | |
| TOX | Adipose tissue ratio | 22589738 | |
| Liver disease | EHBP1L1 | Nonalcoholic fatty liver disease | 20708005 |
| PZP | Nonalcoholic fatty liver disease | 20708005 | |
| Neuropsychiatric | BMP4 | Parkinson's disease | 19915575 |
| CA8 | Response to amphetamines | 22952603 | |
| DLG2 | Parkinson's disease | 17052657 | |
| KCNJ2 | Hypokalemic periodic paralysis | 22863731, 22399142 | |
| KCNJ2 | Thought disorder in schizophrenia | 22648509 | |
| LRRC4C | Temperament (bipolar disorder) | 22365631 | |
| SH3GL2 | Cognitive performance | 19734545 | |
| SH3GL2 | Parkinson's disease | 22451204 | |
| TOX | Cognitive performance | 19734545 | |
| Electrocardiographic | CACNA1D | Ventricular conduction | 21076409 |
| KCNJ2 | QT interval | 19305409 | |
| RASGRF1 | RR interval (heart rate) | 20031603 | |
| Cardiovascular | CA8 | Cardiac hypertrophy | 21348951 |
| KCNJ2 | Cardiac repolarization | 22342860 | |
| KCNMA1 | Mortality in heart failure patients | 20400778 | |
| MYO1D | Hypertension risk | 22322875 | |
| SH3GL2 | Heart failure | 20445134 | |
| TCF7L2 | Coronary heart disease | 21347282 | |
| Developmental | BMP2 | Sagittal craniosynostosis | 23160099 |
| KCNJ2 | Primary tooth development | 20195514 | |
| Glaucoma | 66 | Glaucoma (primary open angle) | 22570617, 22419738 |
| Cancer | BMP4 | Colorectal cancer | 19011631 |
| DLG2 | Wilm's tumor | 22544364 | |
| MYO1D | Pancreatic cancer | 20686608 | |
| ZBTB38 | Prostate cancer | 21743467 | |
| Hematologic | LRFN5 | Hemostasis | 22443383 |
| TCF7L2 | Glycated hemoglobin levels | 20849430 | |
| Immune | ANTXR2 | Ankylosing spondylitis | 21743469, 20062062 |
| BMP4 | Immune response to smallpox | 22610502 | |
| QKI | Response to TNF antagonist | 18615156 | |
| SH3GL2 | Multiple sclerosis | 19010793 | |
| Pulmonary | CHD7 | Pulmonary function decline | 22424883 |
| LRRC4C | Osteoporosis | 20548944 | |
| Miscellaneous | DLG2 | Phospholipid levels (plasma) | 22359512 |
| DLG2 | Protein quantitative trait loci | 18464913 | |
| NPLOC4 | Eye color traits | 20463881 | |
| TCF7L2 | Metabolic syndrome | 20694148 | |
| ZBTB38 | Prion disease | 22210626 |
The relationships summarized here are between the nearest gene to a GWAS association and do not necessarily involve the same SNP as those associated with refractive error. References given in the PMID column are the PubMed accession numbers.
This preliminary observation (for all the shortcomings arising from disease classification subjectivity, as well as publication and methodological biases) suggests that the genes associated with refractive error may be significantly enriched for involvement in body height, glucose and insulin metabolism, and neuropsychiatric disorders, further supporting the earlier conjecture that they are related to growth regulation mechanisms, as well as fatty acid metabolism and neurotransmission. However, the crossover of refractive error genes with these other disorders may be a reflection of the publication bias, as diseases causing a major public health burden (and technically straightforward phenotypes) have historically been investigated in more depth and with larger cohort sizes. A systematic analysis of available GWAS data and a more methodical categorization of different traits and disorders would better elucidate the pleiotropy of genes involved in refractive error.
Implications for the Future
Genome-wide association studies have been very valuable instruments that have, within the span of less than a decade, generated significant amounts of new knowledge about the pathophysiology of many complex disorders, including refractive error. Although the quest for translating GWAS findings into prevention strategies or therapies of direct benefit to patients has just begun, persevering with gene mapping for these conditions is likely to pay significant dividends in the future.
There are at least three areas in which further genetic investigation using already available technologies is possible. The first approach is “bigger is better”: using ever-increasing sample sizes, either as part of international collaboration initiatives or through recruitment of larger cohorts. Through improved statistical power, this approach will enable identifying additional common variants of even smaller effect as well as finding variants of intermediate frequency (MAF between 0.05 and 0.15).
The second approach is the more intensive genotyping of existing cohorts by using higher-density SNP chips or high-throughput direct sequencing that is becoming increasingly affordable (such as various next-generation sequencing platforms). There are two reasons this is likely to yield results. The first is that most present-day GWAS are relying on imputation of genotypes for analyses of the majority of (unmeasured) SNPs. This approach tends to be inaccurate at low allele frequencies. As most genetic variation falls into this category, direct genotyping with dense SNP arrays or sequencing will improve the power of the analyses. The second reason is that larger effect sizes tend to be present in low-frequency alleles (Fig. 2), and finer mapping or sequencing will identify variants with very high effect sizes not currently present in commercial SNP arrays or imputation panels. These variants may be rare, but the importance of a gene in the pathophysiology of a disease is unrelated to the frequency of its polymorphisms. Knowledge of rare variants within these genes may offer new insights about the genetic architecture of refractive error and myopia. Next-generation sequencing approaches to gene identification, though still being developed, are promising and have already identified rare variants apparently associated with severe forms of myopia.36,37
The third promising area of investigation is in the rediscovered usefulness of traditional linkage studies, particularly in conjunction with the newly available high-throughput sequencing methods, which capitalize on the sharing of causal loci among affected relatives. Linkage studies are particularly powerful in identifying rare variants of strong effect, such as those segregating in families with Mendelian forms of pathological myopia. Next-generation sequencing technologies currently offer the unprecedented possibility to query a large number of rare variants for cosegregation with refractive errors within affected family members.
The technology-driven approaches, combined with a wider range of and more highly developed statistical tools (such as better utilization of environmental or lifestyle exposures in the analyses, the construction of hybrid models that incorporate linkage information, and network-based methods) have the potential to change the way we see ocular refraction and myopia as the product of complex interactions between inherited and environmental factors. In time, combining association methods with other sources of “omic” information, such as transcriptomics, would lead to the gradual adaptation of a systems biology approach that has the potential to amplify the individual power of each of these methods.
The use of GWAS has led to great progress in understanding the genetics of myopia and refractive error. However, to date only a fraction of the variation in refractive error (approximately 3.4% in the CREAM dataset38) is explained by the variants identified. A lot more remains to be done, and future GWAS using larger and better genotyped or sequenced cohorts will vastly increase our knowledge of the genetic architecture of refractive error and may suggest better ways to counter it.
Acknowledgments
Supported by a Fight for Sight Early Career Investigator award (PGH) and a National Institutes of Health Mentored Research Career Development Award (NEI:1K08EY022943) (RW).
Disclosure: P.G. Hysi, None; R. Wojciechowski, None; J.S. Rahi, None; C.J. Hammond, None
References
- 1. Pizzarello L, Abiose A, Ffytche T, et al. VISION 2020: The Right to Sight: a global initiative to eliminate avoidable blindness. Arch Ophthalmol. 2004; 122: 615–620 [DOI] [PubMed] [Google Scholar]
- 2. Kempen JH, Mitchell P, Lee KE, et al. The prevalence of refractive errors among adults in the United States, Western Europe, and Australia. Arch Ophthalmol. 2004; 122: 495–505 [DOI] [PubMed] [Google Scholar]
- 3. Lin LL, Shih YF, Hsiao CK, Chen CJ. Prevalence of myopia in Taiwanese schoolchildren: 1983 to 2000. Ann Acad Med Singapore. 2004; 33: 27–33 [PubMed] [Google Scholar]
- 4. Wu HM, Seet B, Yap EP, Saw SM, Lim TH, Chia KS. Does education explain ethnic differences in myopia prevalence? A population-based study of young adult males in Singapore. Optom Vis Sci. 2001; 78: 234–239 [DOI] [PubMed] [Google Scholar]
- 5. Mutti DO, Zadnik K. Age-related decreases in the prevalence of myopia: longitudinal change or cohort effect? Invest Ophthalmol Vis Sci. 2000; 41: 2103–2107 [PubMed] [Google Scholar]
- 6. Vitale S, Sperduto RD, Ferris FL III. Increased prevalence of myopia in the United States between 1971–1972 and 1999–2004. Arch Ophthalmol. 2009; 127: 1632–1639 [DOI] [PubMed] [Google Scholar]
- 7. Wong TY, Foster PJ, Johnson GJ, Seah SK. Education, socioeconomic status, and ocular dimensions in Chinese adults: the Tanjong Pagar Survey. Br J Ophthalmol. 2002; 86: 963–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rahi JS, Cumberland PM, Peckham CS. Myopia over the lifecourse: prevalence and early life influences in the 1958 British birth cohort. Ophthalmology. 2011; 118: 797–804 [DOI] [PubMed] [Google Scholar]
- 9. Jones LA, Sinnott LT, Mutti DO, Mitchell GL, Moeschberger ML, Zadnik K. Parental history of myopia, sports and outdoor activities, and future myopia. Invest Ophthalmol Vis Sci. 2007; 48: 3524–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lopes MC, Andrew T, Carbonaro F, Spector TD, Hammond CJ. Estimating heritability and shared environmental effects for refractive error in twin and family studies. Invest Ophthalmol Vis Sci. 2009; 50: 126–131 [DOI] [PubMed] [Google Scholar]
- 11. Smith TS, Frick KD, Holden BA, Fricke TR, Naidoo KS. Potential lost productivity resulting from the global burden of uncorrected refractive error. Bull World Health Organ. 2009; 87: 431–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saw SM, Tan SB, Fung D, et al. IQ and the association with myopia in children. Invest Ophthalmol Vis Sci. 2004; 45: 2943–2948 [DOI] [PubMed] [Google Scholar]
- 13. Au Eong KG, Tay TH, Lim MK. Education and myopia in 110,236 young Singaporean males. Singapore Med J. 1993; 34: 489–492 [PubMed] [Google Scholar]
- 14. Tang WC, Yap MK, Yip SP. A review of current approaches to identifying human genes involved in myopia. Clin Exp Optom. 2008; 91: 4–22 [DOI] [PubMed] [Google Scholar]
- 15. Hawthorne FA, Young TL. Genetic contributions to myopic refractive error: insights from human studies and supporting evidence from animal models. Exp Eye Res. 2013; 114: 141–149 [DOI] [PubMed] [Google Scholar]
- 16. Wojciechowski R. Nature and nurture: the complex genetics of myopia and refractive error. Clin Genet. 2011; 79: 301–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001; 29: 306–309 [DOI] [PubMed] [Google Scholar]
- 18. Nakanishi H, Yamada R, Gotoh N, et al. A genome-wide association analysis identified a novel susceptible locus for pathological myopia at 11q24.1. PLoS Genet. 2009; 5: e1000660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li YJ, Goh L, Khor CC, et al. Genome-wide association studies reveal genetic variants in CTNND2 for high myopia in Singapore Chinese. Ophthalmology. 2011; 118: 368–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Z, Qu J, Xu X, et al. A genome-wide association study reveals association between common variants in an intergenic region of 4q25 and high-grade myopia in the Chinese Han population. Hum Mol Genet. 2011; 20: 2861–2868 [DOI] [PubMed] [Google Scholar]
- 21. Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmancik JF. A new locus for autosomal dominant high myopia maps to 4q22-q27 between D4S1578 and D4S1612. Mol Vis. 2005; 11: 554–560 [PubMed] [Google Scholar]
- 22. Shi Y, Qu J, Zhang D, et al. Genetic variants at 13q12.12 are associated with high myopia in the Han Chinese population. Am J Hum Genet. 2011; 88: 805–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hysi PG, Young TL, Mackey DA, et al. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat Genet. 2010; 42: 902–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Solouki AM, Verhoeven VJ, van Duijn CM, et al. A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat Genet. 2010; 42: 897–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meng W, Butterworth J, Bradley DT, et al. A genome-wide association study provides evidence for association of chromosome 8p23 (MYP10) and 10q21.1 (MYP15) with high myopia in the French population. Invest Ophthalmol Vis Sci. 2012; 53: 7983–7988 [DOI] [PubMed] [Google Scholar]
- 26. Hammond CJ, Andrew T, Mak YT, Spector TD. A susceptibility locus for myopia in the normal population is linked to the PAX6 gene region on chromosome 11: a genomewide scan of dizygotic twins. Am J Hum Genet. 2004; 75: 294–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Verhoeven VJ, Hysi PG, Saw SM, et al. Large scale international replication and meta-analysis study confirms association of the 15q14 locus with myopia. The CREAM consortium. Hum Genet. 2012; 131: 1467–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kiefer AK, Tung JY, Do CB, et al. Genome-wide analysis points to roles for extracellular matrix remodeling, the visual cycle, and neuronal development in myopia. PLoS Genet. 2013; 9: e1003299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wojciechowski R, Hysi PG. Focusing in on the complex genetics of myopia. PLoS Genet. 2013; 9: e1003442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008; 9: 356–369 [DOI] [PubMed] [Google Scholar]
- 31. Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002; 4: 45–61 [DOI] [PubMed] [Google Scholar]
- 32. Nsengimana J, Bishop DT. Design considerations for genetic linkage and association studies. Methods Mol Biol. 2012; 850: 237–262 [DOI] [PubMed] [Google Scholar]
- 33. Yang J, Benyamin B, McEvoy BP, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010; 42: 565–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hysi PG, Mahroo OA, Cumberland P, et al. Common mechanisms underlying refractive error identified in functional analysis of gene lists from genome-wide association study results in 2 European British cohorts. JAMA Ophthalmol. 2014; 132: 50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hindorff LA, MacArthur J, Morales J, et al. A catalog of published genome-wide association studies. 2013. Available at: www.genome.gov/gwastudies. Accessed August 1, 2013 [Google Scholar]
- 36. Shi Y, Li Y, Zhang D, et al. Exome sequencing identifies ZNF644 mutations in high myopia. PLoS Genet. 2011; 7: e1002084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tran-Viet KN, Powell C, Barathi VA, et al. Mutations in SCO2 are associated with autosomal-dominant high-grade myopia. Am J Hum Genet. 2013; 92: 820–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Verhoeven VJ, Hysi PG, Wojciechowski R, et al. Genome-wide meta-analyses of multiancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat Genet. 2013; 45: 314–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Croft D, O'Kelly G, Wu G, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011; 39: D691–D697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu G, Feng X, Stein L. A human functional protein interaction network and its application to cancer data analysis. Genome Biol. 2010; 11: R53 [DOI] [PMC free article] [PubMed] [Google Scholar]