Abstract
Microvascular adaptation to metabolic stress is important in the maintenance of tissue homeostasis. Nowhere is this more important than in the central nervous system (CNS) where the cellular constituents of the neurovascularture including endothelial cells, pericytes and some astroglia must make fine-tuned autoregulatory modulations that maintain the delicate balance between oxygen availability and metabolic demand. miRNAs have been reported to play an important regulatory role in many cellular functions including cell differentiation, growth and proliferation, lineage determination, and metabolism. In this study, we investigated the possible role of miRNAs in the CNS capillary pericyte response to hypoxic stress. Micro-array analysis was used to examine the expression of 388 rat miRNAs in primary rat cortical pericytes with and without exposure to low oxygen (1%) after 24 or 48 hr. Pericytes subjected to hypoxia showed 27 miRNAs that were higher than control and 31 that were lower. Validation and quantification was performed by Real Time RT-PCR on pericytes subjected to 2 hr, 24 hr, or 48 hr of hypoxia. Hypoxia induced changes included physiological pathways governing the stress response, angiogenesis, migration and cell cycle regulation. miRNAs associated with HIF-1α (miR−322[1], miR−199a [2]), TGF-β1 (miR−140[3], miR−145[4], miR−376b−3p[5]) and VEGF (miR−126a[6], miR−297[7], miR−16[8], miR−17−5p[9]) were differentially regulated. Systematic and integrative analysis of possible gene targets analyzed by DAVID bioinformatics resource (http://david.abcc.ncifcrf.gov) and MetaSearch 2.0 (GeneGo) for some of these miRNAs was conducted to determine possible gene targets and pathways that may be affected by the post-transcriptional changes after hypoxic insult.
Keywords: Brain, hypoxia, microRNA, microvasculature, pericyte, posttranscriptional regulation, translational repression
INTRODUCTION
In response to environmental stress, the cells of the microvasculature undergo a number of endogenous adaptive changes in order to maintain hemodynamic and tissue homeostasis. In the central nervous system (CNS), adaptation to hypoxia is an important homeostatic mechanism, since the balance between delivery of oxygen and glucose must be maintained to meet neuronal metabolic demand. To accomplish this function brain microvessels make fined tuned regulatory adjustments that involve cross-talk between the cellular constituents of the neurovascular unit (endothelial cells, pericytes, glial cells and neurons). One of the immediate responses to hypoxia in the neurovascular unit is transcriptional activation of several genes leading to angiogenesis and metabolic adaptation [10]. A central mediator of this transcriptional activation is the hypoxia-inducable factor (HIF) family of proteins. HIF-1-a/b dimer binds to hypoxia response elements, and activates target genes involved in cellular metabolism, angiogenesis and erythropoiesis [10, 11].
Recent studies indicate that in addition to the canonical HIF-1 pathway, further levels of regulation exist to control hypoxic responses. In particular, it was revealed that microRNAs (miRNA) play important roles in the hypoxic response to stress [12]. microRNAs are a class of small non- protein coding RNAs approximately 21 nucleotides long that regulate gene expression post-transcriptionally. miRNAs have been found to play an important regulatory role in many, if not all, cellular processes, including differentiation, growth and proliferation, lineage determination, and metabolism [13-16]. miRNAs act cytoplasmically to control translation of protein encoding mRNAs but have also been speculated to have a role in influencing translation in other cells by being secreted. Weber and colleagues [17] were able to profile the microRNA spectrum in 12 body fluids including plasma, saliva, breast milk, tears, CSF and others. In some of these fluids, they were able to identify over 450 different miRNAs. These extracellular miRNAs exist either packaged in exosomes or more commonly as vesicle free molecules associated with the highly stable Ago2 protein [18, 19]. This large extracellular distribution would tend to imply that some miRNAs may have functional roles associated with signaling of the surrounding tissue.
In hypoxia, two mechanisms are proposed for miRNA involvement. First, they may act to regulate the expression of genes that are normally required in a normoxic environment but must be inhibited in hypoxic conditions, and secondly, by regulating the expression of genetic factors that provide alternative metabolic pathways or other advantages in hypoxia [20]. miRNAs may be regulated in an HIF-1-dependent or -independent manner, and they may act to repress the expression of HIF-1α, HIF-1β, or a plethora of possible downstream targets to effect hypoxic responses [20, 21]. Most of the studies investigating the relationship be tween hypoxia and miRNAs focus on the endothelial cells and tumor cells. However, pericytes also play an important role in the formation and maintenance of CNS vasculature in both normoxic and hypoxic conditions [10, 22]. Therefore, to elucidate the miRNA changes in pericytes following hypoxia, microarray analysis was performed and the expression level of 388 rat miRNAs in primary rat cortical pericytes exposed to low oxygen (1%) for 24 or 48 hr were evaluated, and compared to control normoxic cultures.
MATERIALS AND METHODS
Microvessel and Pericyte Isolation by Differential Adhesion
Six to eight week old Sprague Dawley male rats (purchased from Harlan Laboratories) were decapitated and the brain tissue immediately removed using sterile technique. Primary microvessels (MV) and pericytes were isolated as previously described [23]. Briefly, brain tissue was removed within minutes of decapitation using sterile technique. Tissue was homogenized in 10 volumes of Dulbecco's Modified Eagle Media (DMEM,Sigma, St. Louis, MO) and 10% Fetal calf serum (FCS, Gibco BRL, Grand Island, NY) (pH 7.4) using a glass-Teflon homogenizer mechanically shaved to leave 0.25 μm between the plunger and the glass surface. After 20-25 up-and-down strokes at 420 rpm, the homogenate was centrifuged and the pellet was resuspended in DMEM with 17% Dextran 70 (Sigma). The soluble portion was harvested and suspension was centrifuged at 5000 rpm for 10 min. The pellet was resuspended and filtered through a sterile 118 μm nylon mesh (Tetco, Braerclif Manor, NJ) to get rid of the larger vessels. The filtrate was then passed through an 80μm nylon mesh. MV were collected from the 80 μm mesh, by washing the mesh vigorously with 10 volumes of DMEM. MV were suspended in DMEM (sigma), 50 Pg/ml penicillin – streptomycin, 2.5 μg/ml nystatin, and 0.1% collagenase type II (Worthington Corp., Freehold, NJ) and incubated at 37°C overnight. Microvessel digests were vigorously pipetted up and down. Cells were pelleted and digests washed 3x then resuspended in DMEM, 10% FCS, and 1% antibiotics. Cells (5×105) were incubated for 4-6 hr to allow the pericytes to adhere to uncoated plastic dishes. Any non-adhered cells and fragments were vigorously washed off. Adherent cells were maintained at 37°C in DMEM +10%FCS. The average yield is 106 pericyte/mg MV or 2-3 × 106 cells total from 15 rats. Non-adherent cells were 95% Factor VIII positive and bound the Griffonia symplicifolia agglutinin (GSA). Upon culture, non-adherent cells displayed the morphological characteristics of EC. Adherent cells were GSA negative and did not express Factor VIII after culture at 37°C. Adherent cells were 95-98% positive for the expression of platelet derived growth factor beta receptor (PDGFBR) and displayed the morphological appearance consistent with that of pericytes (data not shown).
In Vitro Exposure to Low Oxygen
A complete GasPak 100 system consisting of hydrogen and carbon dioxide generating envelops, a polycarbonate anaerobic jar, disposable methylene blue indicator, and catalyst chambers containing palladium catalyst pellets was used to generate the hypoxic environment (Becton, Dickinson and company, BD, Franklin Lakes, NJ). Pericytes were seeded as discussed above. Cells were plated in standard culture medium and incubated in the hypoxia chambers 2 hr, 24 hr or 48 hr as indicated in the experimental results. Time zero was defined at that time when the methylene blue indicator changed color. Following exposure to low oxygen the cells were removed from the chambers and harvested, spun down and fast frozen before isolation of microRNA as detailed below. Analysis of the hypoxic environment has shown that the GasPak system generates an oxygen level of 1-5% oxygen within the culture medium (24). Oxygen levels reached in the culture medium are comparable to those reached in brain tumors and high altitude. Control cells were cultured as above and maintained in normoxic (20 % oxygen) conditions until harvested.
| GasPAK System | |
|---|---|
| Lid Temp. | 30°C |
| % O2 (air 3 hours) | 0.5-1 |
| (medium,3 hours) | 1-5 |
| Discoloration of methylene blue | 7 hours |
| pressure | 2 lb/in2 |
| Condensation | small amount |
| CO2 | Not done |
MicroRNA ARRAY
Total RNA was extracted from pericyte cultures using mirVana™ MiRNA isolation kit from Ambion® being careful to include isolation of small RNAs. 5 μg of control normoxic or 24 hr and 48 hr hypoxic samples were sent to be analyzed by LC Sciences (Houston, Tx, USA) using their μParaflo microfluidic chips containing 388 unique rat miRNA transcripts listed in Sanger miRBase Release 14.0. Each array was hybridized with 1 of the samples labeled with Cy3. For the sample to be counted, the signal intensity had to exceed 3X background standard deviation and then normalized using a LOWESS filter (Locally-Weighted Regression). All samples with signals greater than 32 were measured. In depth data analysis was provided by LC Sciences.
RT-PCR
Changes in the expression of some of the most interesting miRNAs were confirmed by quantitative real time RT-PCR. 20 ng of total RNA from normoxic control, 2 hr, 24 hr, and 48 hr hypoxic dishes (n=3 per group) was reverse transcribed using specific primers to the miRNA of interest (TaqMan® MicroRNA Assay Reverse Transcription Primers, Applied Biosystems, Foster City, CA, USA) followed by realtime PCR with TaqMan® MicroRNA assay primers or by reverse transcribing with random primers and amplification with miRNA specific LNA primers from Exiqon (Woburn, MA). The relative expression of miR−140−5p, miR−145, miR−24, miR−345−5p, miR−126, miR−376b−3p and miR−150 were analyzed using the standard 2–ΔΔCt method [24]. Results are shown as fold change verses normoxic control cultures. U87 was used as the endogenous control for all samples. U87 (also known as SNORD87) is a small non-coding nucleolar RNA that is highly conserved and abundant which functions in the modification of other small nuclear RNAs.
Identification of Gene Targets and Biological Significance: Bioinformatics
Systematic and integrative analysis of possible gene targets was analyzed by DAVID (the database for annotation, visualization and integrated discovery) bioinformatics resource (http://david.abcc.ncifcrf.gov) [25, 26]. This free online resource allows one to extract biological features/meaning associated with large gene lists. It systematically maps a large number of interesting genes in a list to the associated biological annotation (ontology terms) and then statistically highlights the most overrepresented biological annotation. miRNAs of greatest interest from the microarray results were first screened using the miRDB search tool (http://mirdb.org/miRDB) and microrna.org (http://www.microrna.org/microrna) to identify possible gene targets of each miRNA. These gene lists were then loaded into DAVID for analysis. The functional groups and clusters with the highest significance are reported here.
microRNAs of interest were also analyzed by Meta-Search 2.0 (GeneGo Inc.) to determine what known gene targets have been identified previously for each miRNA of interest and what genes are known to influence the transcription of these miRNAs (Table 4).
Table 4.
Known genes regulating miRNAs or regulated by hypoxia sensitive miRNAs. genes that activate or inhibit transcription of listed miRNAs are shown in the first 2 columns. genes that are activated or inhibited by miRNAs are listed in the last 2 columns.
| miRNA | Influenced by | Effect | Target Gene | Effect |
|---|---|---|---|---|
| miR-24 | BMP2 | activation | KHSRP | activation |
| HIF1A | activation | FEN1 | inhibition | |
| p53 | activation | FOXP3 | inhibition | |
| HTLV1 | activation | Furin | inhibition | |
| TGF-β | activation | GATA-2 | inhibition | |
| AML-1 | activation | Histone H2 | inhibition | |
| E2F1 | activation | HNF4-α | inhibition | |
| E2F3 | activation | NET1 | inhibition | |
| ESR2 | activation | NIPK | inhibition | |
| RelA | activation | p120GAP | inhibition | |
| AML1 | inhibition | PAK4 | inhibition | |
| ESR1 | inhibition | STING | inhibition | |
| miR-126 | G-CSF | activation | VEGF-A | inhibition |
| TLR4 | activation | CYP2A13 | inhibition | |
| VEGF-A | activation | EGFL7 | inhibition | |
| ETS1 | activation | RGS16 | inhibition | |
| ETS2 | activation | SDF-1 | inhibition | |
| p53 | activation | SLC45A3 | inhibition | |
| FGF2 | inhibition | ADAM9 | inhibition | |
| IL-3 | inhibition | CRK | inhibition | |
| NGF | inhibition | DNMT1 | inhibition | |
| IBP2 | inhibition | |||
| IRS-1 | inhibition | |||
| K-RAS | inhibition | |||
| MER | inhibition | |||
| p85-β | inhibition | |||
| PITPNC1 | inhibition | |||
| PLK2 | inhibition | |||
| PTP-MEG2 | inhibition | |||
| SDF-1 | inhibition | |||
| SHMT2 | inhibition | |||
| SLC7A5 | inhibition | |||
| SOX2 | inhibition | |||
| TOM1 | inhibition | |||
| VCAM1 | inhibition | |||
| HOXA9 | inhibition | |||
| SPRED1 | inhibition | |||
| miR-140 | p53 | activation | SFD-1 | inhibition |
| SOX5 | activation | dynamin-1 | inhibition | |
| SOX6 | activation | GLUT3 | inhibition | |
| SOX9 | activation | RIP140 | inhibition | |
| IFN-γ | inhibition | situin1 | inhibition | |
| IL-1α | inhibition | aggrecanase-2 | inhibition | |
| IL-1β | inhibition | BMP2 | inhibition | |
| TNF-α | inhibition | dnpep | inhibition | |
| EGR2 | inhibition | |||
| HDAC4 | inhibition | |||
| IBP5 | inhibition | |||
| SMAD3 | inhibition | |||
| SP1 | inhibition | |||
| ULBP1 | inhibition | |||
| Mir-145 | BMP4 | activation | VEGF-A | inhibition |
| IFN-β | activation | N-cadherin | inhibition | |
| IFN-γ | activation | Oct-3/4 | inhibition | |
| MKL1 | activation | p70 S6 | inhibition | |
| TGF-β | activation | PAI1 | inhibition | |
| CSX | activation | Rhotekin | inhibition | |
| FKHR | activation | RLI | inhibition | |
| FOXO3A | activation | RREB1 | inhibition | |
| KLF2 | activation | situin1 | inhibition | |
| p53 | activation | SMAD3 | inhibition | |
| SMAD3 | activation | SOX2 | inhibition | |
| SMAD4 | activation | SOX9 | inhibition | |
| SRF | activation | SWAP-70 | inhibition | |
| NOTCH-1 | activation | PLK-1 | inhibition | |
| myocardin | activation | |||
| EGFR | inhibition | |||
| C/EBP-β | inhibition | |||
| EWS | inhibition | |||
| Oct-3/4 | inhibition | |||
| PRDM5 | inhibition | |||
| RREB1 | inhibition | |||
| miR-150 | c-MPL | activation | IL-10 | activation |
| IFN-α | activation | AKT2 | inhibition | |
| IFN-β | activation | c-Myb | inhibition | |
| SDF-1 | activation | CXCR4 | inhibition | |
| TLR3 | activation | NAP57 | inhibition | |
| FOXP1 | inhibition | |||
| Huntington | inhibition | |||
| IL-18 | inhibition | |||
| Mucin4 | inhibition | |||
| NOTCH3 | inhibition | |||
| P2X7 | inhibition | |||
| p53 | inhibition | |||
| PDGF-B | inhibition | |||
| Survivin | inhibition | |||
| Utrophin | inhibition | |||
| TNF-α | inhibition | |||
| miR-345 | none | ABCC1 | inhibition | |
| BAG-3 | inhibition | |||
| p21 | inhibition | |||
| miR-376b | MEF2 | activation | ATG4C | inhibition |
| NF-κB | inhibition | BDNF | inhibition | |
| Beclin1 | inhibition |
RESULTS
Exposure of CNS Microvessel Pericytes to Hypoxia
Cultured primary CNS pericytes (Fig. 1) were exposed to low oxygen using an in vitro hypoxia system (GasPak100) as detailed in methods. During the experimental exposure period ranging from 2 to 24 hrs, pericytes remained viable and showed no obvious morphological changes even after 48 hrs. These results were similar to those previously published [22].
Fig. (1).
Primary CNS microvascular pericytes were subcultured from pure microvessel preparations. Microvessels were 2-3% pre-capillary arterioles, 2-3% post capillary venules and 92-96% capillaries.
The Effect of Hypoxia on Mirna Expression in Pericytes
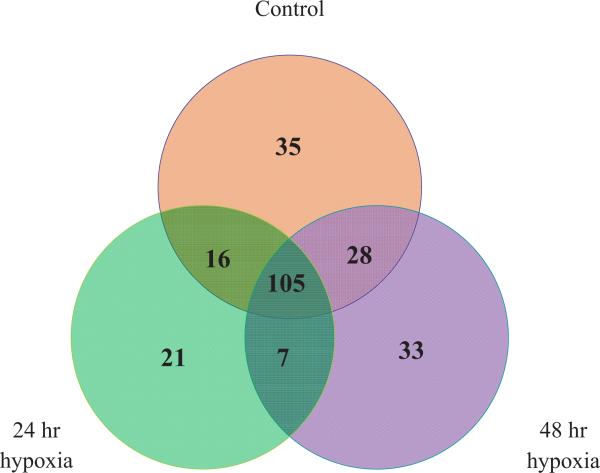
Previously, we have shown that pericytes are highly responsive to low O2 levels [10, 22, 27]. As early as one hour following exposure to low O2, pericytes undergo changes in eicosanoid repertoire [22] and mount a HIF-1 response [22]. In this study, the miRNA expression profiles of pericytes were determined following 24 hr or 48 hr of low O2 (1%) exposure, and compared to control pericytes. The number of genes detected in each group is presented as a Venn diagram (Fig. 2). In the control pericytes, we were able to detect 184 miRNAs. The 24 hr hypoxia animals had 149, and the 48 hr hypoxia group had 173 detectable miRNAs. In the 24 hr group, there were 21 miRNAs that were not detected in either control or 48 hr cells (Fig. 2, Table 1). 33 miRNAs were detected at the 48 hr time point only. There were 7 miRNAs that were not detected in control but were reported following both hypoxia time periods (24+48 hr column in Table 1). Interestingly, there were constitutive miRNAs present in control cells that were down regulated to the point where they could not be detected in the hypoxia groups. These included miR−1, miR−100, and miR−127.
Fig. (2).
Venn diagram showing the numbers of miRNAs that were expressed in rat pericytes constitutively (orange circle) or following 24 hr (green circle) and 48 hr (purple circle) hypoxia. A total of 105 miRNAs were present in all 3 groups while only 7 were present in the hypoxic groups but not in the control group 35 miRNAs were present in the control group but not detectable in the hypoxic groups.
Table 1. Differential expression of miRNAs at 24 and 48 Hours.
miRNAs detected by arrays with differential expression after 24h or 48 hr Hypoxia. miRNAs from venn diagram that were expressed either only at one time point after hypoxia (24 or 48 h), only after hypoxia (Both 24h and 48h), or in control and one hypoxia time point only.
| 24 hr Only | 24 hr + 48 hr | 24 hr + Control | 48 hr Only | 48 hr + Control |
|---|---|---|---|---|
| miR-105 | miR-330* | miR-106b | miR-217 | miR-129 |
| miR-139-3p | miR-343 | miR-107 | miR-218* | miR-133a |
| miR-201 | miR-345-3p | miR-138* | miR-27a* | miR-15b |
| miR-25* | miR-490 | miR-139-5p | miR-291-5p | miR-17-5p |
| miR-26b* | miR-493 | miR-147 | miR-292-3p | miR-181a |
| miR-29b-2* | miR-532-3p | miR-151* | miR-292-5p | miR-196b |
| Mir-301a | miR-708 | miR-19b | miR-294 | miR-215 |
| miR-301b | miR-200b | miR-297 | miR-218 | |
| miR-32 | miR-204 | miR-29a* | miR-224 | |
| miR-339-5p | miR-300-3p | miR-29b-1* | miR-23a* | |
| miR-376a* | miR-339-3p | miR-30c-2* | miR-28* | |
| miR-463 | miR-450a | miR-325-3p | miR-295 | |
| miR-532-5p | miR-628 | miR-325-5p | miR-298 | |
| miR-540 | miR-760-3p | miR-33 | miR-29b | |
| miR-542-5p | miR-873 | miR-331 | miR-30a* | |
| miR-615 | miR-99a | miR-335 | miR-30d* | |
| miR-632 | miR-337 | miR-30e* | ||
| miR-652 | miR-349 | miR-322 | ||
| miR-653 | miR-34a | miR-324-5p | ||
| miR-99a* | miR-376-5p | miR-326 | ||
| Let-7e* | miR-377 | miR-338* | ||
| miR-379 | miR-347 | |||
| miR-381 | miR-350 | |||
| miR-423 | miR-351 | |||
| miR-487b | miR-378* | |||
| miR-488 | miR-382 | |||
| miR-495 | miR-7a | |||
| miR-496 | miR-93 | |||
| miR-501 | ||||
| miR-542-3p | ||||
| miR-592 | ||||
| miR-598-3p | ||||
| miR-678 |
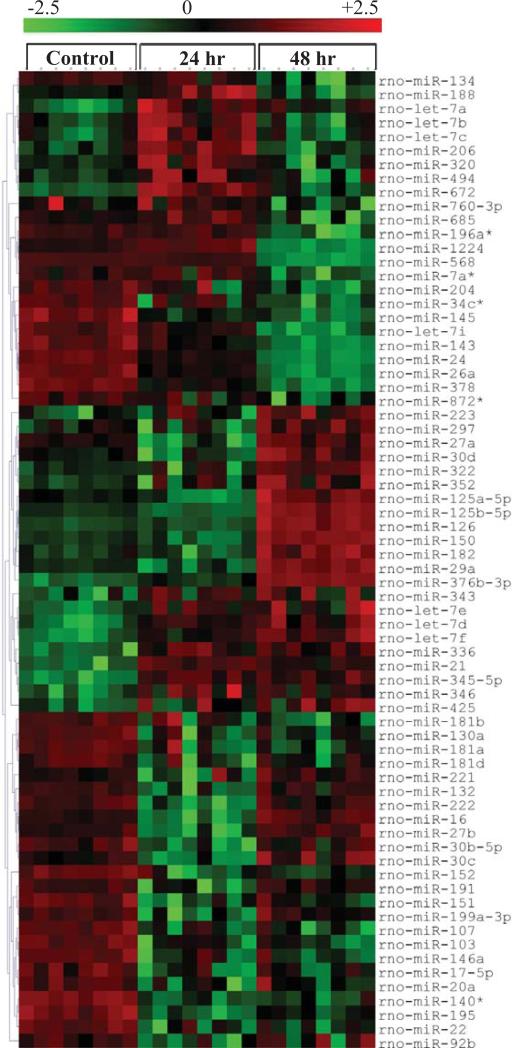
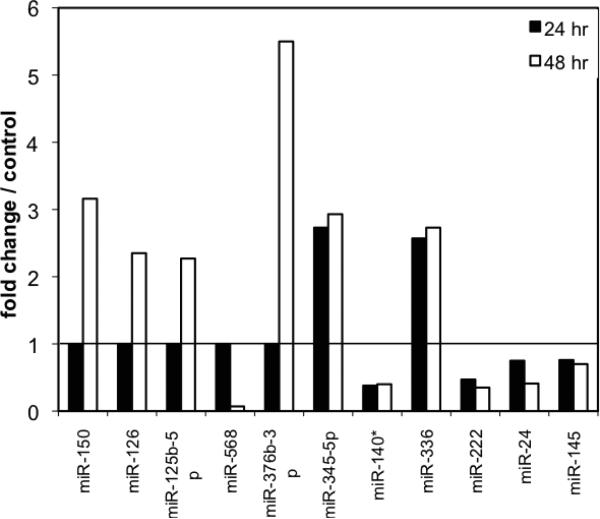
When we evaluated the heat map of array results, approximately 70 miRNAs were significantly (p<0.01) changed, 39 of which were suppressed while 31 miRNAs were upregulated following either 24 or 48 hr of hypoxia compared to the control group (Fig. 3). In Fig. (3), red indicates increased expression and green indicates down regulation. Among those, the most over expressed miRNAs at both hypoxia time points were miR-345-5p (2.73 and 2.93 fold increase respectively) and miR−336 (2.57 and 2.73 fold respectively). The most downregulated miRNAs at both time points were miR−140* (0.38 and 0.40 fold respectively) and miR−222 (0.47 and 0.35 fold). Interestingly, there were several miRNAs that were not changed at 24 hours, but significantly different at 48 hr from control. MiR-150 (3.16 fold), miR-126 (2.35 fold), miR−125b−5p (2.27 fold), miR−322 (2.9 fold), and miR−376b−3p (5.5 fold) were increased after 48 hr of hypoxia while miR−568 was decreased down to 7% of con trol levels by 48 hr (Figs. 3 and 4). On the other hand, miR−140*, miR−222, miR−24, miR−145 were down-regulated after both 24 and 48 hr hypoxia, while miR−345−5p and miR−336 were increased at both time points after hypoxia (Fig. 4).
Fig. (3).
Heat Maps of miRNAs that showed significant (p<0.01) differences between control, 24h hypoxia, and 48h hypoxia groups. Red indicates upregulation over control groups and green indicates downregulation.
Fig. (4).
Based on the array results, the miRNAs most affected after hypoxia were graphed. Relative fold change in expression was given in comparison to control normoxic pericytes, which was set at 1. Black bars represent the 24h hypoxia group and the white bars represent the 48h hypoxia group. Most miRNAs were down-regulated after 24h hypoxia. The greatest up-regulation occurred after 48 h hypoxia.
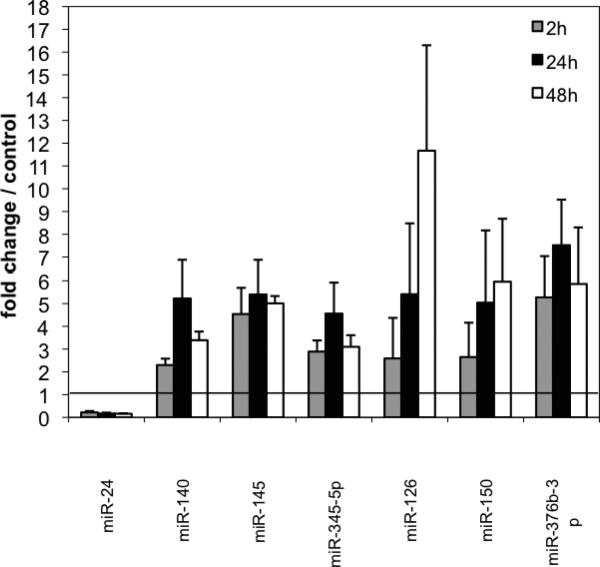
Validation of miRNA Changes in Response to Hypoxia
To validate the changes observed in the array, we analyzed the expression level of specific microRNAs by real time RT-PCR. In addition to the 24 hr and 48 hr timepoints used for the arrays, we also evaluated a 2 hr hypoxia group to determine early changes as well. The miRNAs we vali dated by qRT-PCR were chosen because: 1) they showed the greatest changes (miR−345−5p. miR−140*); 2) they were known to be expressed in pericytes (miR−145) or endothelial cells (miR−24); or 3) showed the most differences in normoxic versus hypoxic conditions. In general there was good concordance between the results. miR−24, which is known to be associated with the microvasculature and is present in endothelial cells, was downregulated by hypoxia to 22% of the control normoxia levels by 2hrs, 17% of control normoxia levels at 24hr and 16% of control normoxia levels at 48hr (Fig. 5). In concordance with the array result, miR−376b−3p was upregulated by hypoxia at all time points (5.3 fold at 2hr, 7.5 fold at 24hr and 5.9 fold at 48hr) compared to control. Similarly, miR345−5p which is involved in cell pro liferation and targets anti-apoptotic BAG-3 [28] was increased at 2hr, 24hr, and 48 hr by 2.89, 4.56 and 3.1 folds respectively.
Fig. (5).
Relative expression of miRNAs after 2hrs (gray bars) 24hrs (black bars) or 48h (white bars) (of hypoxia was determined by qRTPCR, and normalized to normoxic control samples (set equal to1). N+3 per group. miR−24 was down-regulated at all time points. The others tested were up-regulated.
MiR−126, which stimulates angiogenesis and vascular remodeling and targets VEGF-A, was increased at 2hr by 2.6 fold, at 24hr by 5.5 fold and at 48hr by 11.9 fold over control levels. miR−150, which is packaged and secreted and also targets VEGF-A, went up by 2.65 fold at 2hr, increased to 5.0 fold at 24hr and continued to increase to 6.0 fold over control at 48hr (Fig. 5). miR-126 and miR-150 have both been shown to be downregulated by hyperoxia [29].
Surprisingly, two miRNAs mir−145 and miR−140, that showed down-regulation on the arrays, were seen to increase after hypoxia by quantitative RT-PCR. miR−145 went up 4 ½ to 5 fold at all time points. Likewise, miR−140 was found to be upregulated more than 5 fold at 24hr. The discrepancy between the two methods underscores the need to validate non-quantitative array methods by highly quantitative PCR. In general, arrays detect both mature and pre-miRNAs whereas qRT-PCR targets only the mature form. In addition, the arrays were performed with an n=1, while the PCR was done on groups of 3-4, providing the ability to run statistical analysis and negating the effects of non representative data groups. The other miRNA that was downregulated by hypoxia was miR−24 (Fig. 5).
Ontological and Target Gene Analysis of miRNAs
To associate these miRNA changes with the possible gene targets, we analyzed the most affected miRNAs by DAVID bioinformatic resource [25, 30]. Lists of potential target genes based on sequence homology were generated using both microRNA.org (which incorporates both miRanda and mirSVR algorithms) and miRDB (which uses MirTarget 2 alogorithm). These potential gene target lists were loaded into the DAVID website and clusters of ontologically related genes were generated including enrichment scores. The number of predicted gene targets and ontological clusters for selected miRNAs are listed in Table 2. In addition, the most significant clusters of genes by ontological annotation are listed in Table 3. The enrichment score is used to rank overall importance (enrichment) of gene groups. The enrichment score is based on the score for how relevant each search term is for the gene cluster. A higher score for a group indicates that the gene members in the group are involved in more important (enriched) terms in a given study, therefore, more significance was given to the ones scoring 1.3 and more [30]. Further information can be obtained through the website (http://david.abcc.ncifcrf.gov/). This analysis indicated that many of the target genes for miR−24 clustered to groups associated with membranes and intracellular lumen. Decreased amounts of miR- 24 may lead to a breakdown of cellular integrity and the induction of apoptosis. On the other hand, miR−150 potentially targeted many genes involved in vascular development. Increases in this miRNA, as seen after hypoxia, could play a role in the angiogenic capabilities of pericytes following hypoxic stress.
Table 2.
Ontological gene clustering of predicted targets for miRNAs that changed after hypoxia treatment. Genes are clustered based on enriched biological themes, particularly GO terms and enriched functional-related gene groups.
| MiRNA | Gene Targets | Ontological Clusters |
|---|---|---|
| miR-150 | 157 | 37 |
| miR-24 | 137 | 34 |
| miR-376b-3p | 120 | 34 |
| miR-140 | 119 | 34 |
| miR-145 | 215 | 64 |
| miR-345-5p | 58 | 7 |
| miR-126 | 1 | 1 |
Table 3.
Ontological clustering of predicted gene targets. The enrichment score, number of genes, and annotation is listed for the 3 most enriched clusters for each mirna listed indicating the most likely biological processes affected by these miRNAs after hypoxia.
| miRNA | Cluster | Enrichment Score | # Gene Targets | Functional Annotation |
|---|---|---|---|---|
| miR-150 | 1 | 1.68 | 13 | Protein homodimerization |
| 2 | 1.17 | 7 | Negative regulator of DNA binding | |
| 3 | 1.06 | 7 | Vascular development | |
| miR-24 | 1 | 1.34 | 15 | Intracellular lumen |
| 2 | 1.18 | 3 | Ligase activity | |
| 3 | 1.08 | 5 | Membrane associate | |
| 4 | 0.9 | 4 | Cell development and differentiation | |
| miR-376-3p | 1 | 1.56 | 9 | Ubiquitin and catabolic related |
| 2 | 1.25 | 9 | Mesoderm formation | |
| 3 | 1.15 | 12 | Neuron development | |
| miR-140 | 1 | 1.93 | 8 | Skeletal system, osteoblast |
| 2 | 1.51 | 26 | Metal and ion/cation binding | |
| 3 | 1.48 | 11 | Protein localization and transport | |
| miR-145 | 1 | 1.96 | 12 | GTPase binding |
| 2 | 1.38 | 14 | Embryonic development | |
| 3 | 1.37 | 8 | ER stress response | |
| miR-345-5p | 1 | 0.55 | 18 | Membrane associated |
| 2 | 0.53 | 4 | vesicles | |
| 3 | 0.52 | 6 | Protein transport | |
| miR-126 | 1 | 1 | p-55 |
Some of the miRNAs of interest from our study were also searched using the GeneGo application MetaSearch where known interactions between miRNAs and their gene targets have been reported. Validation methods for each known interaction, whether it be activation/inhibition of a miRNA by another gene or activation/inhibition of a gene by a miRNA is listed with supporting documentation on the GeneGo website. These include sequence verification, luciferase reporter activation, PicTar, Targetscan, miRDB searches, or real time RT-PCR. The miRNAs with previously reported known interactions between genes and miRNAs are summarized in Table 4.
DISCUSSION
The pericyte is a complex adaptive regulatory cell and an integral member of the neurovascular unit [23, 31]. The pericyte is actively involved in the maintenance of tissue and vascular homeostasis [10, 32]. In this study, we defined the changes involved in the pericyte microRNA gene response to low oxygen levels. Our findings indicate that the pericyte miRNA transcriptome is subject to significant but selective alterations in response to hypoxic stress. Also, the duration of exposure to hypoxia fundamentally altered the makeup of miRNA changes. The hypoxia-induced changes in the miRNA transcriptome involved gene targets associated with proliferation, differentiation, and regulation of angiogenesis consistent with the established role of pericytes in the vascular stress response [22].
In pericytes, several miRNAS such as miR−322, miR−345−5p, miR−145, miR−150, miR−140, miR−126, miR−376b−3p and to a lesser degree miR−222 were significantly induced by the hypoxic stress. In contrast other miRNAs such as miR−24, miR−140 and miR−199a were suppressed following exposure to hypoxia. HIF-1α is a transcription factor that is acutely upregulated in response to hypoxia. Recently, miR−322 (rodent homolog of hu-miR−424), miR−20b and miR−199a were identified and named hypoxamirs (induced by hypoxia) [33]. Hypoxamirs have also been reported to affect the expression of HIF-1α [21]. In this study, we show that among those hypoxamirs, miR−322 was upregulated in pericytes, while miR−199a was downregulated following 48 hr of low O2 exposure (Fig 3). Additionally, miR−24 which is transcriptionally activated by HIF-1α , was downregulated by hypoxia. Our results therefore suggest that hypoxic conditions influence hypoxamirs differentially, which might represent a compensation mechanism in primary pericytes isolated from normal tissues. miRNAs are thought to be intimately involved in the fine tuning of protein levels and as such, several miRNAs may act in opposing manners. Previously published experiments [33] were conducted under varying hypoxic conditions and used an heterogeneous cell population including pericytes. Thus, it is not too surprising to see several discrepancies between the present results and published data.
Among the miRNAs that were significantly induced following hypoxia in our studies, miR-345-5p was the most prominent with more than a three-fold increase (Fig. 5). The transient expression of this miRNA has been associated with ischemic injury [34] and viral infection [35]. It is known that miR−345−5p targets Waf1, which regulates cell cycle progression [36], and may thus be important in the pluripotent capacity of pericytes [37, 38]. In addition, it has recently been reported that the BcL12-associated athanogene 3 (BAG3) is an anti-apoptotic protein target of miR−345−5p [28]. Increased BAG3 function is sufficient to suppress colon cancer cell proliferation and invasiveness in vitro [39]. Therefore miR−345 may suppress apoptosis via targeting anti-apoptotic proteins [40, 41].
miR−24, a pro-apoptotic component [42, 43, 44] was found to be down-regulated in pericytes in response to low oxygen. Interestingly, Fiedler and coworkers [43] have shown that the down-regulation of miR−24 led to increased vascularization and prevention of endothelial apoptosis in a myocardial infarction model. The up-regulation of miR−345−5p (anti-apoptotic) and the down-regulation of miR−24 (proapoptotic) suggest that under specific pathological conditions, reduced apoptosis is an hypoxia-induced adaptive mechanism that promotes cell survival.
It is well established that TGF-β is important in pericyte-mediated regulation of angiogenesis. Mice deficient for TGF-β1 signaling components show dilated and irregularly shaped microvessels [45]. Also TGF-β1 appears to orchestrate the de novo induction of pericytes by regulating the differentiation of pericyte progenitors [46]. This function may be related to the effect of TGF-β1 on miR−145, since there is evidence that TGF-β1 treatment leads to the sustained up-regulation of miR-145 in mesenchymal stem cell cultures. Similar to miR−145, miR−140, which was also induced in pericytes following hypoxia was reported to be in- volved in cell differentiation [47]. In contrast, there is a negative interaction between miR−140 and TGF-β. miR−140 acts to regulate the TGF-β pathway by directly repressing Smad3, a key transcription factor [48], and TGF-β suppresses the accumulation of miR−140, forming a double negative feedback loop [48]. The consequences of hypoxia-induced miR−140 and miR−145 in pericytes are currently not known. However, the reported data showing the interaction between those miRNAs and TGF-β1 suggest that mechanisms involved in cell differentiation are finely tuned in pericytes under hypoxic conditions.
miR−376b−5p which was also demonstrated to be highly up-regulated following hypoxia, may be another miRNA participating in the differentiation of pericytes and development of new vessels. In this regard, miR−376b−5p has been reported to be regulated in developing muscles in neonates [49] and involved in pancreatic islet development [50]. miR−376b−3p also directly inhibits ATG4C and Beclin1, both of which are autophagy-related genes potentially necessary for cell remodeling and differentiation. Thus, it is intriguing to speculate that hypoxia stimulates selective miRNAs involved in the regulation of pericyte stem cell activity. Obviously additional studies are needed to confirm this hypothesis.
A potential hypoxia–induced microRNA target of particular importance is vascular endothelial growth factor (VEGF). VEGF is synthesized in response to hypoxia via a HIF-1 depended transcriptional process [51]. VEGF directly induces the proliferation and migration of pericytes under hypoxic conditions and also indirectly regulates pericyte recruitment via endothelial cell production of nitric oxide (NO) [46]. Pericytes constitutively express VEGF mRNA but transcripts are not translated until after the hypoxic stimulus [10, 27]. For this gene, a group of candidate regulatory microRNAs have been identified recently in tumor cells [12, 52] and human monocytes [53, 54]. The levels of miR−15b, −16, −20a, and −20b are decreased following exposure to hypoxia and have been implicated in the down-regulation of VEGF-A expression [52]. Thus, suppression of miR−15b, −16, −20a, and −20b must involve the de-repression of the VEGF gene allowing its induction in pericytes following exposure to hypoxia [10, 27]. Additionally, we found that miRNAs miR−150 and miR−126 which are known to target VEGF and stimulate angiogenesis were induced while miR−16, miR−20a, miR−107, miR−17−5p were suppressed in hypoxia exposed pericytes (Fig. 3), suggesting that any subsequent functional outcomes of the miRNA responses are likely the result of a balance between the up-regulation and down-regulation of this gene.
Beside their regulatory role in the cell where they are formed, miRNAs have been demonstrated to circulate in highly stable, cell-free forms in various body fluids, including serum, and plasma, saliva, urine, cerebral spinal fluid (CSF) and milk [17, 55, 56]. miR−150, found in various body fluids [56, 57], was shown to be upregulated 600 % in pericytes after 48 hr of hypoxia in our study. This miRNA has been shown to be packaged into microvesicles and is actively secreted from cells and enhances human microvascular endothelial cell and monocyte migration [57]. Currently, it is unclear whether hypoxia-induced miR125b−5p or miR−150 are secreted from pericytes following exposure to hypoxia. However, it is intriguing to speculate that pericyte-generated miRNAs may be involved in the recruitment of leukocytes, inflammation-induced cell migration and the acute stress response [10, 58-60].
Analysis of ontological annotations by the DAVID algorithm for genes that were potential targets of these miRNAs showed very broad functional implications. Many of these miRNAs had multiple targets that fell into diverse ontological clusters. For example miR-145, which had 215 potential gene targets clustered into 64 different categories. Direct query of known interactions between genes that regulate miRNA transcription and gene targets of miRNA activation or inhibition by MetaSearch also revealed interesting interactions with some positive and negative feedback loops. Many of the known target genes were involved in angiogenisis, cell differentiation and apoptosis.
Comparing the lists of known activators/inhibitors of miRNA transcription and the known target genes of these miRNAs also showed some interesting similarities (Table 4). P53, a tumor suppressor, activates the transcription of miR−126, miR−140 and miR−145. TGF-β activates transcription of both miR−24 and miR−145. SOX2 is inhibited by both miR−126 and miR−145. We have shown that pericytes are multi-potent and can form neurons in culture [37, 61, 62]. Karow and colleagues [63] recently showed that the co-expression of SOX2 and Mash1 in human adult cerebral cortex pericytes can reprogram them into neuronal cells. Pericytes are also known to differentiate along the mesenchymal lineage (as reviewed Dore-Duffy 2008) and Sox2 is required for pericyte differentiation into osteoblasts. Both miR−126 and miR−145 inhibit VEGF-A expression. In some cases there is a negative feedback loop such that the target gene also acts upon the miRNA. For example, Oct3/4 inhibits expression of miR−145, which in turn inhibits translation of Oct3/4. Oct3/4 is a stem cell marker and involved with maintaining the pluripotency of the cell [64]. Taken together, the results suggest that miRNA may be involved in the functional regulation of pericyte potency.
In conclusion, we report that many pericyte miRNAs responded to an induced hypoxic insult in pericytes. Most of the miRNAs that were found in pericytes following hypoxia potentially target genes involved in protein transport, vesicle formation, cell differentiation, migration and angiogenesis. miRNAs are known to act on multiple genes and the verification of target genes for the individual miRNAs must be conducted. Further studies are needed to delineate the functional consequences of miRNA changes observed in pericytes after hypoxia. Importantly, our results suggest that miRNAs are finely tuned and may play an essential role in pericyte function and adaptation to hypoxia.
ACKNOWLEDGEMENTS
None declared.
Footnotes
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
REFERENCES
- 1.Ghosh G, Subramanian IV, Adhikari N, et al. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-alpha isoforms and promotes angiogenesis. J Clin Invest. 120(11):4141–54. doi: 10.1172/JCI42980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gu S, Chan WY. Flexible and Versatile as a Chameleon-Sophisticated Functions of microRNA-199a. Int J Mol Sci. 13(7):8449–66. doi: 10.3390/ijms13078449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pais H, Nicolas FE, Soond SM, et al. Analyzing mRNA expression identifies Smad3 as a microRNA-140 target regulated only at protein level. RNA. 16(3):489–94. doi: 10.1261/rna.1701210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis-Dusenbery BN, Chan MC, Reno KE, et al. down-regulation of Kruppel-like factor-4 (KLF4) by microRNA-143/145 is critical for modulation of vascular smooth muscle cell phenotype by transforming growth factor-beta and bone morphogenetic protein 4. J Biol Chem. 286(32):28097–110. doi: 10.1074/jbc.M111.236950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakhshandeh B, Soleimani M, Paylakhi SH, Ghaemi N. A microRNA signature associated with chondrogenic lineage commitment. J Genet. 91(2):171–82. doi: 10.1007/s12041-012-0168-0. [DOI] [PubMed] [Google Scholar]
- 6.Zhu N, Zhang D, Xie H, et al. Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2. Mol Cell Biochem. 351(1-2):157–64. doi: 10.1007/s11010-011-0723-7. [DOI] [PubMed] [Google Scholar]
- 7.Jafarifar F, Yao P, Eswarappa SM, Fox PL. Repression of VEGFA by CA-rich element-binding microRNAs is modulated by hnRNP L. EMBO J. 30(7):1324–34. doi: 10.1038/emboj.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chamorro-Jorganes A, Araldi E, Penalva LO, Sandhu D, Fernandez-Hernando C, Suarez Y. MicroRNA-16 and microRNA-424 regulate cell-autonomous angiogenic functions in endothelial cells via targeting vascular endothelial growth factor receptor-2 and fibroblast growth factor receptor-1. Arterioscler Thromb Vasc Biol. 31(11):2595–606. doi: 10.1161/ATVBAHA.111.236521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lichner Z, Mejia-Guerrero S, Ignacak M, et al. Pleiotropic action of renal cell carcinoma-dysregulated miRNAs on hypoxia-related signaling pathways. Am J Pathol. 180(4):1675–87. doi: 10.1016/j.ajpath.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 10.Dore-Duffy P, LaManna JC. Physiologic angiodynamics in the brain. Antioxidants & redox signaling. 2007;9(9):1363–71. doi: 10.1089/ars.2007.1713. [DOI] [PubMed] [Google Scholar]
- 11.Wenger RH, Stiehl DP, Camenisch G. Integration of Oxygen Signaling at the Consensus HRE. Sci STKE. 2005;2005(306):re12. doi: 10.1126/stke.3062005re12. [DOI] [PubMed] [Google Scholar]
- 12.Kulshreshtha R, Davuluri RV, Calin GA, Ivan M. A microRNA component of the hypoxic response. Cell Death Differ. 2008;15(4):667–71. doi: 10.1038/sj.cdd.4402310. [DOI] [PubMed] [Google Scholar]
- 13.Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Ann Rev Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- 14.Ivey KN, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7(1):36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 15.Harris A, Krams SM, Martinez OM. MicroRNAs as immune regulators: implications for transplantation. Am J Transplant. 2010;10(4):713–9. doi: 10.1111/j.1600-6143.2010.03032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee YS, Dutta A. MicroRNAs: small but potent oncogenes or tumor suppressors. Curr Opin Investig Drugs. 2006;7(6):560–4. [PubMed] [Google Scholar]
- 17.Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56(11):1733–41. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 108(12):5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic acids Res. 2011;39(16):7223–33. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pocock R. Invited review: decoding the microRNA response to hypoxia. Pflugers Archiv : Eur J Physiol. 2011;461(3):307–15. doi: 10.1007/s00424-010-0910-5. [DOI] [PubMed] [Google Scholar]
- 21.Loscalzo J. The cellular response to hypoxia: tuning the system with microRNAs. J Clin Invest. 2010;120(11):3815–7. doi: 10.1172/JCI45105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dore-Duffy P, Balabanov R, Beaumont T, Katar M. The CNS pericyte response to low oxygen: early synthesis of cyclopentenone prostaglandins of the J-series. Microvasc Res. 2005;69(1-2):79–88. doi: 10.1016/j.mvr.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Dore-Duffy P. Isolation and characterization of cerebral microvascular pericytes. Methods Mol Med. 2003;89:375–82. doi: 10.1385/1-59259-419-0:375. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif. Dec. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Prot. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Dennis G, Jr., Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 27.Dore-Duffy P, Wang X, Mehedi A, Kreipke CW, Rafols JA. Differential expression of capillary VEGF isoforms following traumatic brain injury. Neurol Res. 2007;29(4):395–403. doi: 10.1179/016164107X204729. [DOI] [PubMed] [Google Scholar]
- 28.Tang JT, Wang JL, Du W, et al. MicroRNA 345, a methylation-sensitive microRNA is involved in cell proliferation and invasion in human colorectal cancer. Carcinogenesis. 32(8):1207–15. doi: 10.1093/carcin/bgr114. [DOI] [PubMed] [Google Scholar]
- 29.Bhaskaran M, Xi D, Wang Y, et al. Identification of microRNAs changed in the neonatal lungs in response to hyperoxia exposure. Physiol Genomics. 2012;44:970–80. doi: 10.1152/physiolgenomics.00145.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang da W, Sherman BT, Stephens R, et al. DAVID gene ID conversion tool. Bioinformation. 2008;2(10):428–30. doi: 10.6026/97320630002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakagawa S, Deli MA, Nakao S, et al. Pericytes from brain microvessels strengthen the barrier integrity in primary cultures of rat brain endothelial cells. Cell Mol Neurobiol. 2007;27(6):687–94. doi: 10.1007/s10571-007-9195-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dore-Duffy P, Cleary K. Morphology and properties of pericytes. Methods Mol Biol. 2011;686:49–68. doi: 10.1007/978-1-60761-938-3_2. [DOI] [PubMed] [Google Scholar]
- 33.Loscalzo J. The cellular response to hypoxia: tuning the system with microRNAs. J Clin Invest. 120(11):3815–7. doi: 10.1172/JCI45105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu DZ, Tian Y, Ander BP, et al. Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J Cerebral Blood flow Metab. 2010;30(1):92–101. doi: 10.1038/jcbfm.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glazov EA, Kongsuwan K, Assavalapsakul W, Horwood PF, Mitter N, Mahony TJ. Repertoire of Bovine miRNA and miRNA-Like Small Regulatory RNAs Expressed upon Viral Infection. PloS one. 2009;4(7):e6349. doi: 10.1371/journal.pone.0006349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu S, Huang S, Ding J, et al. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene. 29(15):2302–8. doi: 10.1038/onc.2010.34. [DOI] [PubMed] [Google Scholar]
- 37.Dore-Duffy P. Pericytes: pluripotent cells of the blood brain barrier. Curr Pharm Des. 2008;14(16):1581–93. doi: 10.2174/138161208784705469. [DOI] [PubMed] [Google Scholar]
- 38.Dore-Duffy P, Mehedi A, Wang X, Bradley M, Trotter R, Gow A. Immortalized CNS pericytes are quiescent smooth muscle actin-negative and pluripotent. Microvascular Res. 2011;82(1):18–27. doi: 10.1016/j.mvr.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang J-T, Wang J-L, Du W, et al. MicroRNA 345, a methylation-sensitive microRNA is involved in cell proliferation and invasion in human colorectal cancer. Carcinogenesis. 2011;32(8):1207–15. doi: 10.1093/carcin/bgr114. [DOI] [PubMed] [Google Scholar]
- 40.Malhotra R, Lin Z, Vincenz C, Brosius FC., 3rd Hypoxia induces apoptosis via two independent pathways in Jurkat cells: differential regulation by glucose. Am J Physiol Cell Physiol. 2001;281(5):C1596–603. doi: 10.1152/ajpcell.2001.281.5.C1596. [DOI] [PubMed] [Google Scholar]
- 41.Weinmann M, Jendrossek V, Handrick R, Guner D, Goecke B, Belka C. Molecular ordering of hypoxia-induced apoptosis: critical involvement of the mitochondrial death pathway in a FADD/caspase-8 independent manner. Oncogene. 2004;23(21):3757–69. doi: 10.1038/sj.onc.1207481. [DOI] [PubMed] [Google Scholar]
- 42.Srivastava N, Manvati S, Srivastava A, et al. miR-24-2 controls H2AFX expression regardless of gene copy number alteration and induces apoptosis by targeting antiapoptotic gene BCL-2: a potential for therapeutic intervention. Breast Cancer Res. 2011;13(2):R39. doi: 10.1186/bcr2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fiedler J, Jazbutyte V, Kirchmaier BC, et al. MicroRNA-24 regulates vascularity after myocardial infarction. Circulation. 2011;124(6):720–30. doi: 10.1161/CIRCULATIONAHA.111.039008. [DOI] [PubMed] [Google Scholar]
- 44.Xie Y, Tobin LA, Camps J, et al. MicroRNA-24 regulates XIAP to reduce the apoptosis threshold in cancer cells. Oncogene. 2012 doi: 10.1038/onc.2012.258. [E-pub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lebrin F, Deckers M, Bertolino P, Ten Dijke P. TGF-beta receptor function in the endothelium. Cardiovas Res. 2005;65(3):599–608. doi: 10.1016/j.cardiores.2004.10.036. [DOI] [PubMed] [Google Scholar]
- 46.Ribatti D, Nico B, Crivellato E. The role of pericytes in angiogenesis. Int J Develop Biol. 2011;55(3):261–8. doi: 10.1387/ijdb.103167dr. [DOI] [PubMed] [Google Scholar]
- 47.Yang J, Qin S, Yi C, et al. MiR-140 is co-expressed with Wwp2-C transcript and activated by Sox9 to target Sp1 in maintaining the chondrocyte proliferation. FEBS lett. 2011;585(19):2992–7. doi: 10.1016/j.febslet.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 48.Pais H, Nicolas FE, Soond SM, et al. Analyzing mRNA expression identifies Smad3 as a microRNA-140 target regulated only at protein level. RNA (New York, NY. 2010;16(3):489–94. doi: 10.1261/rna.1701210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McDaneld TG, Smith TP, Doumit ME, et al. MicroRNA transcriptome profiles during swine skeletal muscle development. BMC genomics. 2009;10:77. doi: 10.1186/1471-2164-10-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joglekar MV, Joglekar VM, Hardikar AA. Expression of islet-specific microRNAs during human pancreatic development. Gene Expr Patterns. 2009;9(2):109–13. doi: 10.1016/j.gep.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 51.Liu LX, Lu H, Luo Y, et al. Stabilization of Vascular Endothelial Growth Factor mRNA by Hypoxia-Inducible Factor 1. Biochem Biophys Res Commun. 2002;291(4):908–14. doi: 10.1006/bbrc.2002.6551. [DOI] [PubMed] [Google Scholar]
- 52.Hua Z, Lv Q, Ye W, et al. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PloS One. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jafarifar F, Yao P, Eswarappa SM, Fox PL. Repression of VEGFA by CA-rich element-binding microRNAs is modulated by hnRNP L. EMBO J. 2011;30(7):1324–34. doi: 10.1038/emboj.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karaa ZS, Iacovoni JS, Bastide A, Lacazette E, Touriol C, Prats H. The VEGF IRESes are differentially susceptible to translation inhibition by miR-16. RNA (New York, NY. 2009;15(2):249–54. doi: 10.1261/rna.1301109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meinl E, Meister G. MicroRNAs in the CSF: Macro-advance in MS? Neurology. 2012;79:2162–3. doi: 10.1212/WNL.0b013e31827597d1. [DOI] [PubMed] [Google Scholar]
- 56.Chen X, Liang H, Zhang J, Zen K, Zhang C-Y. Secreted microRNAs: a new form of intercellular communication. Trends in Cell Biol. 2012;22:125–32. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Y, Liu D, Chen X, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Molecular cell. 2010;39(1):133–44. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 58.Bonkowski D, Katyshev V, Balabanov RD, Borisov A, Dore-Duffy P. The CNS microvascular pericyte: pericyte-astrocyte crosstalk in the regulation of tissue survival. Fluids Barriers CNS. 8(1):8. doi: 10.1186/2045-8118-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiao C, Calado DP, Galler G, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131(1):146–59. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 60.Zhao H, Kalota A, Jin S, Gewirtz AM. The c-myb proto-oncogene and microRNA-15a comprise an active autoregulatory feedback loop in human hematopoietic cells. Blood. 2009;113(3):505–16. doi: 10.1182/blood-2008-01-136218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dore-Duffy P, Katychev A, Wang X, Van Buren E. CNS microvascular pericytes exhibit multipotential stem cell activity. J Cereb Blood Flow Metab. 2006;26(5):613–24. doi: 10.1038/sj.jcbfm.9600272. [DOI] [PubMed] [Google Scholar]
- 62.Dore-Duffy P, Mehedi A, Wang X, Bradley M, Trotter R, Gow A. Immortalized CNS pericytes are quiescent smooth muscle actin-negative and pluripotent. Microvasc Res. 82(1):18–27. doi: 10.1016/j.mvr.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]