Abstract
Solid-state nanopores show promise as single-molecule sensors for biomedical applications, but to increase their resolution and efficiency analyte molecules must remain longer in the nanopore sensing volume. Here we demonstrate a novel, facile, and customizable nanopore sensor modification that reduces double stranded DNA translocation velocity by two orders of magnitude or more via interactions outside the nanopore. This is achieved by electrospinning a copolymer nanofiber mesh (NFM) directly onto a solid-state nanopore (NP) chip. The effect of NFMs on dsDNA translocation through a nanopore is highlighted using a set of NFMs of varying mesh composition that reduce translocation speed relative to a bare pore from 1× to >100×. A representative NFM from this set is effective on DNA as long as 20 kbp, improves nanopore resolution, and allows discrimination among different DNA lengths.
The emergence of novel nanomaterials and nanofabrication tools is accelerating the development of single-molecule biosensors to directly detect genomic information, such as single-nucleotide polymorphisms, structural variations, and epigenetic markers. One of the simplest and most versatile biosensors in this class is the solid-state nanopore (NP).1 Similar to protein based NP2, solid-state NPs employ electrophoretic forces to thread and slide electrically charged biopolymers through a nanoscale hole fabricated in ultra-thin, insulating, membrane. A large voltage potential applied across a NP immersed in an electrolyte solution induces an intense ion current, producing a strongly diverging electrical field in the vicinity of the pore. This field gradient efficiently focuses electrically charged biopolymers, such as DNA molecules, allowing the detection of minute sample concentrations.3 The potential ability of a NP to scan and to resolve small, local features along DNA necessitates that the nominal thickness of the membrane be limited to only a few nanometers or even less.4 However, such thin membranes significantly limit the surface of the pore available to interact with DNA and thereby slow its movement through the NP: a conundrum. Typical translocation (or sliding) speeds of double-stranded DNA (dsDNA) in solid-state NP range from tens to hundreds of ns per basepair,1a and cannot be resolved with sufficient accuracy due to the inherent electrical noise in the detection system at these bandwidths.5 Thus, methods capable of slowing (or better yet controlling) translocation speeds are of interest for sensing applications such as DNA and RNA sequencing, gene expression, and genotyping.6
To regulate the translocation speed of DNA molecules we have developed a highly porous and tunable synthetic coating which is applied to either the entry or the exit (cis or trans, respectively) faces of a thin silicon nitride membrane containing the pore, as depicted schematically in Figure 1. The coating is formed by a polymeric nanofiber mesh (NFM) with tunable chemical and physical properties that is electrospun directly onto a surface.7 Previous studies have demonstrated that nanofiber meshes can act as an adsorbent to separate biopolymers.8 We propose that a low-density, high-surface-area NFM proximal to a NP will significantly slow DNA passage by interacting with the biopolymer prior to and during translocation through the NP. Our approach differs from previous strategies for NP modification, which influence translocation by altering the pore surface itself,9 directly tethering and manipulating the DNA,10 or changing properties of the surrounding medium, such as viscosity, pressure, or ionic strength.11 These approaches typically do not decouple improvements in translocation dynamics from other characteristics of the device, such as conductivity, blockage level, or wall charge. This can lead to undesirable consequences, including reduced threading efficiency and ion current stability, smaller signal/noise ratios, and reduced NP hydration efficiency.
Figure 1.
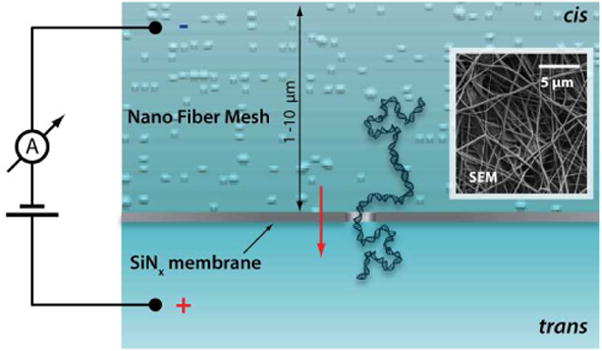
Schematic cross-section of a solid-state nanopore with nanofiber mesh spun on its cis. The DNA interacts with the mesh as it is electrophoretically threaded through the pore (not to scale). Inset: SEM of nanofiber mesh on nanopore chip.
NFMs were formulated from copolymer blends of poly(ε-caprolactone) (PCL) (70-90 kg/mol, Sigma) and poly(glycerol-monostearate-co-ε-caprolactone) (PGC-C18) (22 kg/mol) (Figure 2a). PGC-C18 is synthesized according to our previously published protocol (see SI).7a,12 Doping PCL with increasing quantities of the hydrophobic PGC-C18 increases the resulting mesh hydrophobicity, as characterized by water droplet contact angle (Figure 2b). These measurements of hydrophobicity indicate changes in chemical composition. Figure 1 inset shows a typical SEM image of a 7:3 PCL:PGC-C18 hydrophobic NFM electrospun onto a NP chip, with fiber diameters ranging from 300-450 nm. In all cases the electrospinning deposition time, voltage, and needle position were adjusted to produce uniform NFM thicknesses with fibers of similar morphology across all polymer blends (see Table S1, Figures S5-S7). The NFM fabrication step is facile, parallel, and fast. For example, coating 50 chips required about a minute and a few milligrams of polymer. The resulting NFM has low volume fraction, high surface area, and is mechanically stable. In this study, we electrospun NFMs onto NP chips using the following PCL:PGC-C18 copolymer blend ratios: 10:0 (PCL only), 9:1, 8:2, 7:3, 6:4, and 5:5.
Figure 2.
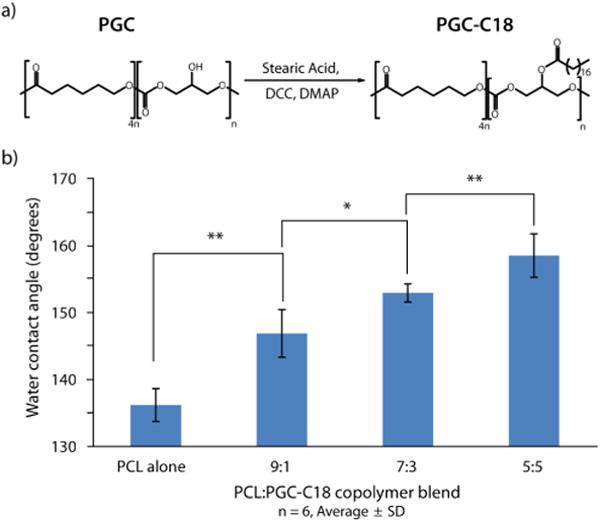
a) Synthesis of PGC-C18. b) Effect of PCL:PGC-C18 copolymer ratio on nanofiber mesh hydrophobicity, measured via contact angle (n=6)(*p<0.05, **p<0.01)
The nanopore-nanofiber mesh (NP-NFM) sensor consists of a small NP, drilled with a tightly focused electron TEM beam in an LPCVD-deposited, low-stress SiNx membrane (25 nm thick; Figure S1).13 NP chips were sealed in a custom-built flow cell permitting a low-noise recording of the ion current flowing through the pore.13 DNA added to the cis side of an unmodified NP (no NFM) under an applied electric potential induces blockades in the ionic current corresponding to translocations of DNA from the grounded cis side to the positively biased trans side of the membrane. Typical translocation events for 1000 bp DNA in an unmodified SiNx bare NP are shown in Figure 3a (blue).
Figure 3.
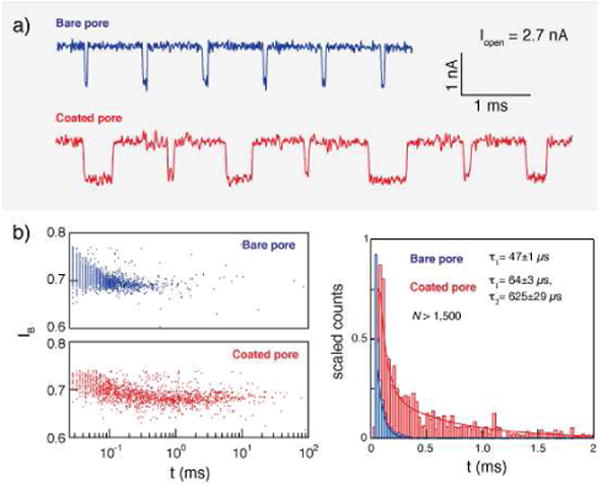
Comparing translocation times through a bare nanopore (NP) and an NP-NFM. a) Representative current traces for 1000 bp dsDNA passing through the same NP, as a bare NP (upper) and as a 7:3 PCL:PGC-C18 NP-NFM (lower), showing similar blockage levels but some significantly slower translocation times for the NP-NFM (V=300 mV, Iopen = 2.7 nA). b) Histogram of event duration for the same uncoated NP (blue) and 7:3 PCL:PGC-C18 NP-NFM (red), showing exponential tail fits (error bar: τ ± 95% confidence interval).
A 7:3 PCL:PGC-C18 polymer solution was electrospun onto the very same NP. The NP-NFM device was readily hydrated and permitted both buffer and sample exchange (see SI Figure S8). Translocations of 1,000 bp DNA through this NP-NFM (Figure 3) revealed two important characteristics of the modified NP: First, and most noticeably, we observe a broader spread in the DNA translocation time. Specifically, the dwell-time of a large fraction of the events falls between 0.5 ms and 10 ms (Figure 3b); a range which exceeds the typical translocation time of the same uncoated pore by roughly an order of magnitude. Second, the presence of the NFM does not substantially affect the open pore current (the ion current prior to DNA entry into the pore), the blocked current level, or the noise in the NP (Figure S9). A closer evaluation of the translocation events (see sample events in Figure 3a and the dwell time histograms in Figure 3b) suggests that instead of a uniform shift of the entire dwell time histogram towards longer timescales, the NFM induces a bimodal distribution containing populations of “normal” and “long” events. Indeed, a mono-exponential tail fit fails to represent the dwell time histogram of the NFM coated pore as accurately as does a double exponential fit. The shorter timescale, τ1, is close to the typical timescale for the uncoated pore, while the longer timescale, τ2, is nearly 10× longer. A detailed analysis of the fits and errors obtained for all data sets are presented in the SI (Table S2).
To further characterize the nature of the slowed DNA translocations induced by the presence of a tunable NFM, we fabricated and tested a set of NP-NFMs representing six different PCL:PGC-C18 copolymer blends spun onto 4-4.5 nm pores. Similar to our observations for the 7:3 copolymer blend, the relative blockage level (IB = Iblock/Iopen) and the conductance measured are nearly the same across all of these compositions (Figures S10, S11). These results suggest that ion mobility near the NP are similar to that of a bare NP, and consistent with a highly porous NFM structure.
In contrast to the ion current levels, dsDNA translocation dynamics are highly dependent on the NFM composition. We measured the characteristic translocation times of 1000 bp dsDNA using different NFM copolymer blend coatings, once again tail-fitting the resulting dwell-time distributions to double exponential functions (see SI Figures S14 and S15 for all fits). Other metrics for analyzing the translocation time were explored (e.g., histogram peaks (tP) for logarithmic binning of the data), but these approaches generally failed to capture both the short and long translocation event populations (see SI Table S2 and SI Figure S13). We defined the relative τ as the ratio of the timescale for the “long” event population (τ2) at each coating normalized by the characteristic timescale of translocation in the bare pore. We repeated these measurements at two applied voltages: 300 mV and 500 mV. Our results are summarized in Figure 4 (error bars show corresponding 95% confidence intervals for fits). For reference we also show the relative short τ values using the normal translocation population (τ1) where available, generally showing a value around unity.
Figure 4.
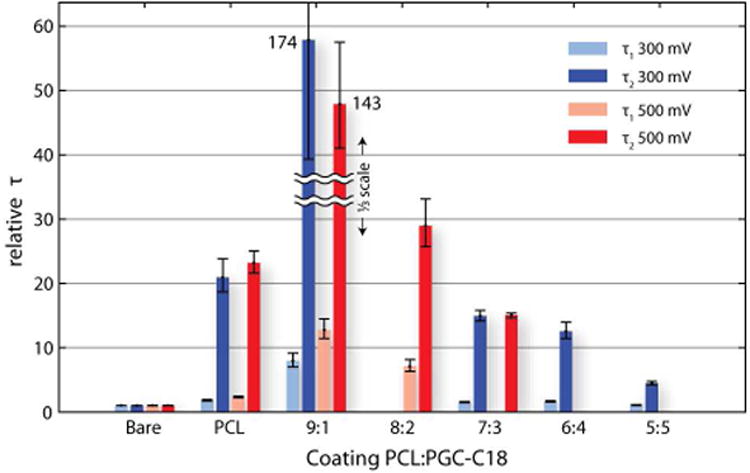
Slowing factor τrelative (where τrelative = τcoated/τbare) for various coatings: Bare pore (τrelative = 1), PCL only, 9:1, 8:2, 7:3, 6:4, and 5:5 PCL:PGC-C18 blends. All data are for 1000 bp dsDNA in 4-4.5 nm nanopores, 300 mV (blue) and 500 mV (red) (error bar: τ ± 95% fit confidence interval of exponential tail fits; 9:1 PCL:PGC-C18 blend at both 300 and 500 mV is shown at 1/3 scale for clarity). τrelative values calculated using either τ1 or τ2 are shown (light and dark colors, respectively).
When the NFMs are ranked in order of increasing hydrophobicity according to contact angle measurement, we observed non-monotonic changes in relative τ: The most- and least-hydrophobic NFMs, respectively, had relatively little effect on translocation speed. PCL alone slowed translocations by more than 20× at both 300 mV and 500 mV. The superhydrophobic 6:4 and 5:5 PCL:PGC-C18 meshes only slowed DNA by 12× and 4×, respectively, at the lower driving force of 300 mV. For intermediate copolymer blends, the data collected at both 300 and 500 mV clearly showed a more pronounced slowing effect than the most- and least-hydrophobic meshes. In particular, the 9:1 PCL:PGC-C18 NFM slowed translocations by more than 140× at 500 mV, and more than 170× at 300 mV. At 300 mV, nearly 20% of events for this mesh were longer than 10 ms. For comparison, <0.2% of events in the bare pore at 300 mV are longer than 10 ms.
The variation of relative τ with mesh composition suggests that the translocating DNA interacts with the NFM as it approaches and threads through the NP, and that the strength of these interactions changes with mesh chemical composition. Moreover, the fact that the values obtained for τ1 are close to the bare pore translocation times suggests that only a fraction of the DNA molecules interact with the NFM. This observation is consistent with the presence of very sparse NFMs. The interactions of strong polyelectrolytes, such as DNA, with dielectric surfaces are governed by a complex interplay between electrostatic and hydrophobic forces, which depend not only upon chemical composition, but also on the material's structure, texture, and other steric considerations.14 The most hydrophobic NFMs produce relatively small retardation effects, while the set of NFMs characterized by intermediate hydrophobicity levels creates maximum drag on the DNA. To account for this complex behavior, a detailed model describing the DNA-NFM interactions must be developed. Yet, from a practical standpoint this interplay provides flexibility in tuning the material properties of the NFMs.
Finally, we collected translocation events using the 7:3 PCL:PGC-C18 copolymer blend NFM, at 500 mV, for five different dsDNA lengths ranging from 0.5 kbp to 20 kbp to determine if longer biopolymers interacted more strongly with the NFM than shorter biopolymers. One might expect that the number of contact points between the mesh and DNA would increase with biopolymer length affording a more stable overall interaction. To maintain consistency across the samples, all measurements were performed sequentially in a single 6 nm diameter pore (with the same NFM coating), where some data sets were collected twice at different time points to ensure reproducibility. The characteristic ion current level and dwell time of each event was extracted and plotted on an ‘event diagram’ (Figure 5). Events that displayed a folded DNA translocation pattern15 were excluded in the analysis to simplify interpretation of the results.
Figure 5.
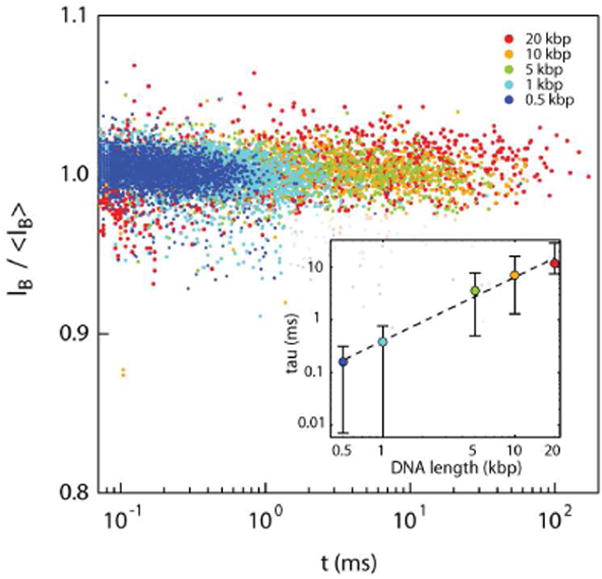
Event diagram for five lengths of dsDNA translocating through a 6 nm nanopore coated with 7:3 PCL:PGC-C18. Ib is normalized for clarity. Inset: Characteristic translocation time τ for each DNA length (error bar: τ ± 95% fit confidence interval). Dotted line is a guide to the eye.
Figure 5 shows a clear pattern of longer translocation times with larger DNA molecules. While we did not make an attempt to discriminate collision events (fast events that involve unsuccessful threading of the DNA into the pore) from true translocations, the overall trend of the translocation time is clear and consistent for all lengths. As before, we numerically characterized the translocation dwell-time distributions using exponential tail fits (see SI Figures S16 and S17). These results are shown in the inset of Figure 5, indicating mean translocation speeds of roughly 0.4-0.7 μs/base, which is 20-35× slower than for an uncoated pore under the same conditions (see SI Figure S18). A monotonic growth in the characteristic translocation time as a function of length is observed for DNA in the presence of the NFM coating. We also observed that in the range from 1 kbp to 10 kbp, the slowing factor relative to a bare pore increased slightly (from 20× to 35×, see SI Figure S18, inset). While this is consistent with our original hypothesis, the trend of increased slowing for longer DNA was far less pronounced than expected, and barely significant given the associated fit error. Although this observation partly contradicts our a priori expectation that the longest DNA would be slowed much more than shorter DNA, there are still a number of possible explanations for this behavior. First, some of the events in the 20 kbp sample and even the 10 kbp sample exceeded the acquisition capability of our experimental system (∼250 ms); thus the overall tail fit may reflect shorter timescales than expected. Second, a fully stretched 20 kbp DNA may extend beyond the width (even locally) of the NFM fibers used in this experiment. It is thus reasonable to predict that the retardation factor may stay constant or even become smaller for very long DNAs. Nevertheless, a clear relationship exists between the characteristic translocation times and DNA length.
In summary, the effect of NFM coatings on dsDNA translocation dynamics in solid-state NPs is reported. The NFMs increase DNA translocation time by up to two orders of magnitude or more without altering the ion current levels. This effect is sustained for DNA, up to 20 kbp in length, enabling greater temporal resolution for the longest strands of DNA. NFM composition, as characterized by hydrophobicity, affects translocation times. This observation is consistent with our view that at an intermediate hydrophobicity the DNA interacts strongly with the NFM, the disruption of which is facilitated by the electrophoretic forces applied on the DNA. The process of electrospinning a NFM coating onto a NP is facile, high-throughput, parallel, and compatible for use with a number of chemically diverse polymers. Thus, this method and the resulting device compositions can be readily adjusted for many DNA sensing applications benefiting from control over biopolymer translocation rates. Future work will focus on these and other NFMs to create a NP-based class of biosensors with a broad range of customizable translocation properties.
Supplementary Material
Acknowledgments
We acknowledge support from the Center for Nanoscale Systems at Harvard University, and financial support from NIH awards NIBIB R21-EB017377. (AM & MWG) and NHGRI R01-HG005871 (AM).
Footnotes
Supporting Information: Experimental procedures, synthesis, characterization, statistics, and supporting figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.a) Venkatesan BM, Bashir R. Nat Nanotechnol. 2011;6:615. doi: 10.1038/nnano.2011.129. [DOI] [PubMed] [Google Scholar]; b) Miles BN, Ivanov AP, Wilson KA, Doğan F, Japrung D, Edel JB. Chem Soc Rev. 2013;42:15. doi: 10.1039/c2cs35286a. [DOI] [PubMed] [Google Scholar]
- 2.a) Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Proc Natl Acad Sci. 1996;93:13770. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Biophys J. 1999;77:3227. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Proc Natl Acad Sci. 2000;97:1079. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wanunu M, Morrison W, Rabin Y, Grosberg AY, Meller A. Nature Nanotech. 2010;5:160. doi: 10.1038/nnano.2009.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Singer A, Rapireddy S, Ly DH, Meller A. Nano Lett. 2012;12:1722. doi: 10.1021/nl300372a. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wanunu M, Cohen-Karni D, Johnson RR, Fields L, Benner J, Peterman N, Zheng Y, Klein ML, Drndić M. J Am Chem Soc. 2011;133:486. doi: 10.1021/ja107836t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smeets RM, Keyser UF, Dekker NH, Dekker C. Proc Natl Acad Sci U S A. 2008;105:417. doi: 10.1073/pnas.0705349105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branton D, Deamer D, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, Jovanovich SB, Krstic PS, Lindsay S, Ling XS, Mastrangelo CH, Meller A, Oliver JS, Pershin YV, Ramsey M, Riehn R, Soni GV, Tabard-Cossa V, Wanunu M, Wiggin M, Schloss JA. Nature Biotechnol. 2008;26:1146. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Yohe ST, Colson YL, Grinstaff MW. J Am Chem Soc. 2012;134:2016. doi: 10.1021/ja211148a. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yohe ST, Freedman JD, Falde EJ, Colson YL, Grinstaff MW. Adv Funct Mater. 2013 doi: 10.1002/adfm.201203111. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Bhardwaj N, Kundu SC. Biotechnol Adv. 2010;28:325. doi: 10.1016/j.biotechadv.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Teeters MA, Conrardy SE, Thomas BL, Root TW, Lightfoot EN. J Chromatogr A. 2003;989:165. doi: 10.1016/s0021-9673(03)00027-x. [DOI] [PubMed] [Google Scholar]
- 9.a) Yusko EC, Johnson JM, Majd S, Prangkio P, Rollings RC, Li J, Yang J, Mayer M. Nat Nanotechnol. 2011;6:253. doi: 10.1038/nnano.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Anderson BN, Muthukumar M, Meller A. ACS Nano. 2013;7:1408. doi: 10.1021/nn3051677. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Iqbal SM, Akin D, Bashir R. Nat Nanotechnol. 2007;2:243. doi: 10.1038/nnano.2007.78. [DOI] [PubMed] [Google Scholar]; d) Hall AR, Scott A, Rotem D, Mehta KK, Bayley H, Dekker C. Nat Nanotechnol. 2010;5:874. doi: 10.1038/nnano.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Wanunu M, Meller A. Nano Lett. 2007;7:1580. doi: 10.1021/nl070462b. [DOI] [PubMed] [Google Scholar]; f) Wei R, Gatterdam V, Wieneke R, Tampé R, Rant U. Nat Nanotechnol. 2012;7:257. doi: 10.1038/nnano.2012.24. [DOI] [PubMed] [Google Scholar]; g) Yameen B, Ali M, Neumann R, Ensinger W, Knoll W, Azzaroni O. Nano Lett. 2009;9:2788. doi: 10.1021/nl901403u. [DOI] [PubMed] [Google Scholar]
- 10.a) Keyser UF, Koeleman BN, Van Dorp S, Krapf D, Smeets RM, Lemay SG, Dekker NH, Dekker C. Nat Phys. 2006;2:473. [Google Scholar]; b) Hyun C, Kaur H, Rollings R, Xiao M, Li J. ACS Nano. 2013;7:5892. doi: 10.1021/nn4012434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Fologea D, Uplinger J, Thomas B, McNabb DS, Li J. Nano Lett. 2005;5:1734. doi: 10.1021/nl051063o. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kowalczyk SW, Wells DB, Aksimentiev A, Dekker C. Nano Lett. 2012;12:1038. doi: 10.1021/nl204273h. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhang H, Zhao Q, Tang Z, Liu S, Li Q, Fan Z, Yang F, You L, Li X, Zhang J. Small. 2013 doi: 10.1002/smll.201301263. [DOI] [PubMed] [Google Scholar]
- 12.Wolinsky JB, Ray WC, Colson YL, Grinstaff MW. Macromolecules. 2007;40:7065. [Google Scholar]
- 13.Kim MJ, Wanunu M, Bell DC, Meller A. Adv Mater. 2006;18:3149. [Google Scholar]
- 14.Pividori MI, Alegret S. Top Curr Chem. 2005;260:1. [Google Scholar]
- 15.Wanunu M, Sutin J, McNally B, Chow A, Meller A. Biophys J. 2008;95:4716. doi: 10.1529/biophysj.108.140475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


