Abstract
The vertebrate glucocorticoid receptor (GR) is cytoplasmic without hormone and localizes to the nucleus after hormone binding. GR has two nuclear localization signals (NLS): NL1 is similar in sequence to the SV40 NLS; NL2 is poorly defined, residing in the ligand-binding domain. We found that GR displayed similar hormone-regulated compartmentalization in Saccharomyces cerevisiae and required the Sxm1 nuclear import receptor for NL2-mediated import. Two metazoan homologues of Sxm1, importin 7 and importin 8, bound both NL1 and NL2, whereas importin α selectively bound NL1. In an in vitro nuclear import assay, both importin 7 and the importin α-importin β heterodimer could import a GR NL1 fragment. Under these conditions, full-length GR localized to nuclei in the presence but not absence of an unidentified component in cell extracts. Interestingly, importin 7, importin 8, and importin α bound GR even in the absence of hormone; thus, hormonal control of localization is exerted at a step downstream of import receptor binding.
INTRODUCTION
To survive in a complex environment, cells sense external cues and commonly respond by altering gene expression, in part by regulating the intracellular localization and activity of transcriptional regulatory factors. For example, the intracellular localization of the yeast Pho4 and mammalian SREBP factors is altered in response to changes in external phosphate and circulating sterol concentrations, respectively (Kaffman et al., 1998b; Nagoshi et al., 1999). Similarly, changes in blood glucose levels and a wide range of physiological stress signals modulate the circulating level of glucocorticoids, changing the activity and localization of the glucocorticoid receptor (GR). Within minutes of glucocorticoid binding, GR enters the nucleus and activates or represses target gene transcription (Picard and Yamamoto, 1987). However, hormone binding is reversible; after hormone withdrawal, GR locates back to the cytoplasm (t1/2 = 10 h; Hache et al., 1999).
Proteins and RNAs enter and exit the nucleus through the nuclear pore, a 125-MDa complex in the nuclear envelope (Stoffler et al., 1999). Small molecules readily diffuse through the nuclear pore, whereas molecules greater than ∼40 kDa require import and export machinery for transport (Davis, 1995; Pante and Aebi, 1995). The first nuclear localization sequence (NLS) was identified in the SV40 large T-antigen (Kalderon et al., 1984). Development of an in vitro nuclear import assay facilitated the identification of two proteins that import substrates containing SV40 NLS-like domains. The adapter molecule importin α binds to the NLS of the substrate and the importin β transport receptor, creating a trimeric complex that imports the substrate into the nucleus through the nuclear pore (Strom and Weis, 2001).
Based on similarity to importin β, a family of nuclear import and export receptors was identified and shown to transport a wide variety of proteins and RNAs (Gorlich and Kutay, 1999) into and out of the nucleus. After reaching the nucleoplasm, import receptors bind the small GTPase Ran in the GTP-bound form (RanGTP), causing substrate dissociation. In contrast, nuclear export receptors bind substrate in the presence of RanGTP, forming a trimeric export receptor-substrate-RanGTP complex that crosses the nuclear pore and releases the substrate into the cytoplasm upon hydrolysis of RanGTP to RanGDP by the cytoplasmic RanGAP (Melchior and Gerace, 1998; Weis, 1998; Gorlich and Kutay, 1999; Kuersten et al., 2001).
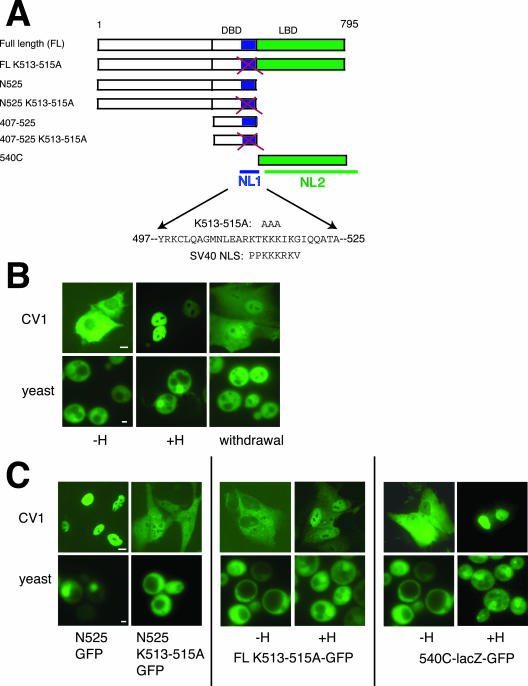
The prevailing view is that protein localization is regulated by modulating the substrate: transport receptor interaction and the accessibility of substrate to the nuclear pore (Kaffman and O'Shea, 1999; Turpin et al., 1999). It is not known how glucocorticoids regulate GR intracellular localization. GR has two nuclear localization signals and one nuclear export signal (Picard and Yamamoto, 1987; Tang et al., 1997; Black et al., 2001). The first nuclear localization signal, NL1 (amino acids 497–524 of rat GR), is located within the DNA-binding domain (DBD) and is similar in amino acid sequence to the SV40 NLS (Figure 1A). NL1 binds importin α2 in yeast two-hybrid and GST pull-down assays (Savory et al., 1999). The second nuclear localization domain, NL2 (amino acids 540–795) is in the ligand binding domain (LBD) of GR (Figure 1A). It fails to interact with importin α by GST pull-down and yeast two-hybrid analysis (Savory et al., 1999) and is otherwise poorly characterized.
Figure 1.
Import of GR is dependent on hormone and two nuclear localization signals (NL1 and NL2) in yeast and mammalian cells. Bar, 10 μM for CV1 cell images, 1 μM for yeast images. (A) Diagram shows full-length GR with positions of the DNA-binding domain (DBD), ligand-binding domain (LBD), NL1, and NL2, other GR constructs used, and sequence similarities between GR and the SV40 large T-antigen. In K513-515A, lysines are mutated to alanines. (B) CV1 cells were transfected with pEGFP-N795. Twenty-four hours after transfection, cells were treated with EtOH (–H) or 1 μM Dex (+H) as indicated for 1 h. For hormone withdrawal, hormone-treated cells were incubated overnight in media lacking hormone. W303-1a yeast were transformed with pAdh-GR-GFP and grown overnight in selective media containing EtOH (–H) or 5 μM DAC (+H) as indicated; yeast were diluted to an OD595 of 0.1 and grown to OD595 of 0.4 before microscopy. For hormone withdrawal, treated cells were incubated in media lacking hormone for 1 h. The kinetics of GR nuclear transport are different in yeast than in mammalian cells; the cause of this kinetic difference has not been investigated. (C) CV1 cells were transfected and yeast cells transformed with noted plasmids and treated with EtOH (–H), 1 μM Dex (CV1 cells +H), or 5 μM DAC (yeast +H) as indicated.
In the simplest model, hormone binding might establish a GR conformation amenable to binding nuclear import receptors (Picard and Yamamoto, 1987; Pratt et al., 1999). It appears, however, that importin α2 can bind to NL1 in vitro in the presence and absence of hormone (Savory et al., 1999), implying that additional factors are required for the hormonal regulation of GR localization. Because the NL2-containing region of GR conveys hormone-dependent localization to heterologous fusion proteins (Picard and Yamamoto, 1987), we hypothesized that hormone regulates GR localization by regulating the interaction of NL2 with its nuclear import receptors. Thus, we screened in Saccharomyces cerevisiae for nuclear import receptors that would bind NL2, tested whether mammalian homologues of those receptors could import GR in vitro, and investigated the role of hormone in regulating the interaction with GR.
MATERIALS AND METHODS
Plasmid Constructs
pEGFP-N795 K513-515A was made from pEGFP-N795 (full-length rat-GR; R. Sitcheran and Keith R. Yamamoto, unpublished results) using Quick Change Mutagenesis (Stratagene, La Jolla, CA). DNA fragments encoding the first N-terminal 525 amino acids of rat GR from pEGFP-N795 and pEGFP-N795 K513-515A were amplified with primers containing XhoI and BglII sites and subcloned into the EcoRI-Kleneaw-BamHI sites of pSG5 (Promega, Madison, WI) making pSG5-N525 and pSG5-N525 K513-515. pEGFP-lacZ-540C is a fusion between β-galactosidase, GFP, and residues 540–795 of rat GR. The lacZ gene was amplified from pCMV-βgal (Spaete and Mocarski, 1985) and cloned into the BglII site of pEGFP-C1 (CLONTECH, Palo Alto, CA) producing pEGFP-lacZ. A DNA fragment encoding amino acids 540–795 of GR was amplified from pEGFP-N795 with primers containing SalI and BglII sites and cloned into the SalI-BamHI sites of pEGFP-lacZ.
pJW248, containing the ADH1 promoter and GFP, was kindly provided by Jonathan Weissman (UCSF). pAdh-N795-GFP, wt or K513-515A, was constructed by excising full-length rat GR from pEGFP-N795 or pEGFP-N795 with BamHI-XhoI and ligating into the SalI-BglII sites of pJW248. pGPD-N795-GFP and pGPD-N795 K513-515A GFP (expressing GR from a GPD promoter) were created by excising GR-GFP from pAdh-N795-GFP or pAdh-N795-GFP-K513-515A with BamHI-BglII and subcloning into the BamHI site of pRS316-GPD-PGK, a kind gift from Anastasia Kralli (Scripps).
pYN525-GFP and pYN525 K513-515A GFP were created by amplifying a DNA fragment encoding amino acids 1–525 from pEGFP-N795 and pEGFP-N795 K513-515A using primers containing XhoI and BamHI restriction sites. The resulting fragment was subcloned into the SalI-BglII sites of pJW248. pY540C-lacZ-GFP is a fusion between amino acids 540–795 of GR, β-galactosidase, and GFP. Fragments encoding residues 540–795 of rat-GR (540C) and the lacZ gene were amplified as described above and ligated into the SalI-BglII and BglII sites of pJW248, respectively. The pL2G3z GR lacZ reporter plasmid (Bohen and Yamamoto, 1993) EB0347 expressing Pho4-GFP (Kaffman et al., 1998b), pyxLhp1gfp expressing a fusion between Lhp1p-GFP (Rosenblum et al., 1997), pPS1574 expressing Sxm1p, and pPS1117 expressing Sxm1p-GFP (Seedorf and Silver, 1997) have been described.
pGST-imp7 and pGST-imp8 were constructed by amplifying the inserts from pQE70 importin 7 and zz60 importin 8 (Gorlich et al., 1997) and subcloning into pGEX4t-1 (Amersham, Piscataway, NJ). Plasmids encoding GST-tagged mouse importin α2 GST-PTAC 58 (Imamoto et al., 1995) and GST alone, pGEX 4t-1, have been described. For in vitro transcription, GR fragments were subcloned into pSG5, which has an integrated T7 promoter. pSG5-N795 (rGR; Darimont et al., 1998) has been described. pSG5–540C was constructed by amplifying the DNA encoding amino acids 540–795 of pEGFP-N795 using primers containing SalI and BglII and cloning the resulting fragment into the EcoRI/Klenow-BglII sites of pSG5.
For nuclear import substrates, a series of GFP-GST–tagged import domains were constructed. To create pGST-GFP, the gene encoding GFP was amplified from pEGFP-C1 (CLONTECH) and subcloned into the BglII site of pGEX4t-1. Human SRP19a cDNA was amplified from HeLa cell total RNA using the Superscript One-Step RT-PCR kit (Invitrogen, Carlsbad, CA) and subcloned into the EcoRI-XhoI sites of pGST-GFP generating pGST-GFP-SRP19a. A DNA fragment encoding residues 407–525 of rat GR was amplified from pEGFP-GR and subcloned into the EcoRI-XhoI sites of pGST-GFP creating pGST-GFP-407-525. Plasmid pGST-GFP-407-525 K513-515A was made from pGST-GFP-407-525 by Quick Change Mutagenesis. pGST-GFP-BIB was constructed by amplifying the DNA encoding the BIB domain from plasmid 4zL123a-cys (Jakel and Gorlich, 1998) using primers containing EcoRI and XhoI sites. Plasmid pGEX-2t-GFP-NLS was generously provided by Mary S. Moore (Baylor College of Medicine, Houston, TX). For nuclear import assays, pQt Ran (pKW356), pQtRanQ69L (pKW592; Nachury and Weis, 1999), and GST-PTAC97 (importin β; Kose et al., 1997) have been described. Klenow polymerase and all restriction enzymes were obtained from New England Biolabs (Beverly, MA). All constructs were verified by DNA sequencing.
Mammalian Cell Culture, Transfections, and In Vivo Localization Studies
CV1, HeLa, and A549 cells were obtained from ATCC (Manassas, VA) and cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 5% fetal bovine serum (FBS; Hyclone, Logan, UT). For transient transfections, CV1 cells were split into 24-well plates (5 × 104 cells per well) on poly-dl-lysine (Sigma, St. Louis, MO) precoated glass coverslips in phenol red-free DMEM (Invitrogen) supplemented with 5% charcoal stripped FBS (Picard and Yamamoto, 1987; Freeman and Yamamoto, 2001), ∼16 h before transfection. Cells were transfected with Lipfectamine 2000 (Invitrogen). Twenty-four to 48 h after transfection, dexamethasone (Dex; Sigma) was added to 1 μM for 1 h. The cells were fixed in freshly prepared 4% paraformaldehyde (Sigma) in phosphate-buffered saline (PBS) for 20 min and mounted onto glass slides in mounting medium (Citiflouor, Ted Pella Inc., Irvine, CA) For hormone withdrawal studies, cells were washed extensively in PBS and incubated in phenol red–free DMEM with charcoal stripped FBS for 24 h before fixation.
For GFP-tagged proteins, localization was assessed by fluorescence microscopy with a Zeiss Axiovert S100 inverted microscope (Thornwood, NY), 40× objective, and Spot digital camera. For immunofluorescence, fixed cells were permeabilized with PBS-0.1% Triton for 20 min and stained with the BuGR2 antibody (Gametchu and Harrison, 1984) in PBS/5% bovine serum albumen (BSA) to detect rat GR. Coverslips were washed three times with PBS, incubated with a goat anti-mouse rhodamine-linked secondary antibody (Molecular Probes, Eugene, OR) in PBS/5% BSA for 1 h, and mounted on glass slides for microscopy. Within experiments, all images were taken using identical camera and microscope settings. All subcellular localization experiments were performed a minimum of four times.
Yeast Strains, Handling, Transformations, and Localization Studies
For localization studies, W303–1a (Thomas and Rothstein, 1989) yeast were grown in YEPD or minimal SD media supplemented with appropriate amino acids and 2% glucose at 30°C unless otherwise noted. Yeast were transformed by the lithium acetate method (Gietz and Woods, 2002). For localization studies, overnight cultures in the appropriate SD medium in the presence or absence of 5 μM deacylcortivazol (Dac; Thompson et al., 1981) were diluted to an OD595 of 0.1 and grown to an OD595 of 0.4 in the identical medium. Two microliters of this suspension was spotted on a slide beneath a coverslip, and protein localization was assessed by fluorescence microscopy with a 100× oil immersion lens on the Zeis microscope described above. For hormone withdrawal experiments, yeast were washed three times in SD medium lacking hormone and incubated at 30°C for 1 h.
The localization of GR-GFP was monitored in 14 yeast strains each deficient in a different nuclear transport receptor: CRMI (Stade et al., 1997), CSEI (Xiao et al., 1993), Kap95 (Iovine et al., 1995), Kap104 (Aitchison et al., 1996), Kap123 (Seedorf and Silver, 1997), LOS1 (Hopper et al., 1980), MSN5 (Kaffman et al., 1998a), MTR10 (Kadowaki et al., 1994), NMD5 (He and Jacobson, 1995), PSEI (Seedorf and Silver, 1997), SRP1 (Yano et al., 1992), SXM1 (Seedorf and Silver, 1997), Kap114 (Morehouse et al., 1999; Pemberton et al., 1999), and PDR6 (Titov and Blobel, 1999). Yeast strains PSY580, pse1-1, PSY1200, and PSY902 have been described (Seedorf and Silver, 1997). For temperature-sensitive strains, suspension cultures were grown overnight at permissive temperature, diluted to an OD595 of 0.1, and placed at the restrictive temperature. Pho4-gfp localization was performed as described (Kaffman et al., 1998b).
Yeast reporter assays were performed as follows: Yeast were transformed with pGPD-N795-GFP (URA) or pGPD-N795-GFP K513-515A (URA), the pL2G3z (LEU) reporter plasmid, and the pPS1117 SXM1-GFP (TRP1) rescue construct. Colonies were grown to saturation at 25°C, diluted 1/40 into media with 5 μM Dac and grown to an OD595 between 0.2 and 0.3 for analysis on a Molecular Devices Therma Max plate reader (Menlo Park, CA).
Protein Expression and Purification
GST, GST-imp7, GST-imp8, GST-PTAC 58, GST-PTAC97, GST-GFP, GST-GFP-BIB, GST-GFP-SRP19, GST-GFP-NLS, GST-GFP-407-525, and GST-GFP-407-525 K513-515A were expressed in BL21-Codon Plus (DE3)-RIL Escherichia coli (Stratagene). Saturated overnight cultures in LB/Carb were diluted 1/100 to 1 liter and grown to an OD595 of 0.7 at 37°C. Cultures were then placed at 25°C for 20 min, induced with 0.4 mM IPTG for 7 h, and harvested by centrifugation. Cell pellets were frozen, thawed, and resuspended in lysis buffer: PBS with 2 mM EDTA, 2 mM DTT, and protease inhibitors mix (1 μg/ml leupeptin, pepstatin, aprotinin, and 1 mM PMSF; Sigma). Lysates were incubated with 1 mg/ml lysozyme (Sigma) for 30 min, sonicated, and clarified at 40,000 × g for 30 min at 4°C. For nuclear import substrates, GST-GFP, GST-GFP-Bib, GST-GFP-SRP19, GST-GFP-NLS, GST-GFP-407-525, and GST-GFP-407-525 K513-515A, cell pellets were resuspended in the above lysis buffer with 1% Triton X-100. Proteins were purified according to Moore and Schwoebel (2000). For import assays, GST-PTAC 58 and GST-PTAC97 were purified according to Kurisaki et al. (2001) with the following variations: importin α2 or importin β were cleaved from GST using 10 U thrombin (Amersham) overnight at 4°C, and eluate was passed over p-aminobenzamidine resin (Sigma) to remove thrombin. The soluble protein was passed over a PD10 column (Amersham) equilibrated with 20 mM HEPES, pH 7.3, 110 potassium acetate, 0.5 mM EGTA, 2 mM DTT, and protease inhibitors and concentrated on a Centricon 30 (Millipore, Beverly, MA).
Importin 7 was purified as in Jakel and Gorlich (1998), and importin 8 was purified as in Dean et al. (2001) with variations. Proteins were expressed in XL1-Blue cells (Stratagene), induced with 4 mM IPTG for 8 h at 16°C at an OD595 of 0.8. After sonication, lysates were clarified at 40,000 × g for 30 min and purified over nickel agarose (QIAGEN, Chatsworth, CA) in 50 mM Tris, pH 7.5, 200 mM NaCl, 5 mM MgCl2. Eluate was passed over a Superdex 200 gel filtration column (Amersham) and concentrated to 20 μM by a Centricon 30 concentrator. Ran-GDP and Ran-GTP Q69L were expressed in SG13 cells and purified according to Nachury and Weis (1999). Highly purified rat GR (N795) has been described (Perlmann and Wrange, 1988). Human importin β (Kutay et al., 1997) was also induced in XL1-Blue cells as above. After clarification, importin β lysate was flash-frozen in liquid nitrogen.
Binding Assays
Lysates containing GST, GST-GRIP1, GST-importin 7, GST-importin 8, GST-PTAC 58 (importin α2), GST-GFP-SV40 NLS, GST-GFP-BIB, GST-GFP-407-525, and GST-GFP-407-525 K514-515A were thawed and incubated with 15 μl glutathione-agarose beads (Sigma) for >1 h at 4°C in binding buffer: 20 mM Tris, pH 8.1, 150 mM NaCl, 0.1% NP40, 14 mM β-mercaptoethanol (β-ME), and protease inhibitors. GST beads were then washed three times in binding buffer containing 2 M NaCl.
For GST pull-downs with purified full-length GR, 15 μl glutathione beads bearing 4 μg bound GST fusion proteins were incubated with 150 ng purified rat GR overnight at 4°C in pull-down buffer: 20 mM Tris, pH 8.0, 100 mM NaCl, 10% glycerol, protease inhibitors, 20 μg/ml BSA, 2 mM DTT, 1 mM EDTA, and 0.01% NP40. Beads were washed, and bound proteins were eluted as described previously (Darimont et al., 1998) and immunoblotted with the BuGR2 antibody. For GST pull-downs with in vitro transcribed and translated proteins, GR and truncation derivatives in the pSG5 vector were transcribed and translated (TNT) using the Promega TNT kit as described (Darimont et al., 1998). Immobilized import receptors were incubated with 12 ng GR (as computed from the amount of incorporated 35S-labeled methionine) in binding buffer for 1 h at 23°C. Bound proteins were washed four times with binding buffer containing 500 mM NaCl, eluted with SDS loading buffer, resolved by SDS-PAGE, and analyzed on a PhosphorImager (Storm 820; Molecular Dynamics).
For GST pull-downs with immobilized nuclear localization domains, 4 μg immobilized import domain was incubated with 4 μg purified importin 7 or 72 μg HeLa extract overnight at 4°C in binding buffer. Bound proteins were eluted and resolved as described above and immunoblotted with a polyclonal importin 7 antibody (Gorlich et al., 1997).
For binding experiments with recombinant RanQ69L, immobilized import receptors were preincubated with buffer or 40 μM RanQ69L for 1 h at 4°C, before addition of 12 ng full-length GR for an additional 2 h at 4°C, washed, and resolved as described (Darimont et al., 1998).
For binding experiments with importin β, bacterial lysates containing 4 μg GST, GST-importin α, or GST-importin 7 were incubated with 15 μl glutathione-agarose beads overnight in pull-down buffer at 4°C in the presence or absence of lysate containing 4 μg human importin β. Beads were washed three times in pull-down buffer and then incubated with 4 μg of GR fragment EX556 (Freedman et al., 1989) containing NL1 for 3 h at 4°C. Bound proteins were resolved and immunoblotted with the BuGR2 antibody.
For hormone-dependent binding assays, immobilized import receptors or GRIP were mixed with 12 ng IVT full-length GR that had been translated in the presence or absence of hormone, as described (Darimont et al., 1998). Bound proteins were resolved by SDS-PAGE and analyzed by PhosphorImager.
A549 extracts were made as described (Yamamoto et al., 1974) with slight modifications. A549 cells were grown in phenol red–free DMEM supplemented with 10% charcoal-stripped FBS for 3 days. Cells from six confluent 15-cm plates were resuspended in an equal volume of lysis buffer 10 mM Tris, pH 8.1, 1 mM EDTA, 10% vol/vol glycerol, 50 mM NaCl, 1 mM β-ME, 100 μg/ml BSA, and protease inhibitors. Cells were lysed with 30 stokes of a Teflon homogenizer (Wheaton, Millville, NJ) and clarified at 20,000 × g for 10 min at 4°C. Typical extract concentration was 6–8 mg/ml. Immobilized importin 7, importin 8, importin α, or GST beads were incubated with 240 μg extract in the presence or absence of 100 nM dexamethasone for 30 min at room temperature. Bound proteins were washed and resolved as described (Darimont et al., 1998). Gels were immunoblotted with GR-specific polyclonal antibody 283 (a kind gift of Michael Garabedian, NYU).
Nuclear Import Assay
HeLa extract was prepared from 4 liters of HeLa cells grown in suspension according to Takizawa et al. (1999) with modifications. The cell pellet was washed twice with PBS and once with 10 mM HEPES, pH 7.3, 110 mM KOAc, 2 mM MgOAc, and 2 mM DTT with protease inhibitors. Cell pellets were resuspended in 1.5 volumes 5 mM HEPES, pH 7.3, 10 mM KOAc, 2 mM MgOAc, 2 mM DTT, and protease inhibitors and incubated on ice for 10 min. Cells were lyzed by Dounce homogenization with a type A pestle (20 strokes). After addition of 1/30 volume of 3 M KOAc, homogenate was clarified at 23,500 × g for 20 min and centrifuged at 100,000 × g for 1 h. Supernatant was dialyzed into transport buffer: 20 mM HEPES, pH 7.4, 110 mM KOAc, 2 mM MgOAc, 0.5 mM EGTA, protease inhibitors, and 2 mM DTT and flash-frozen in liquid nitrogen.
Nuclear import assays were performed as described (Adam et al., 1990; Nachury and Weis, 1999; Takizawa et al., 1999; Moore and Schwoebel, 2000). Cells were grown on poly-dl-lysine–coated glass coverslips for 48 h. Cells were washed twice with transport buffer, incubated with 40 μg/ml digitonin in transport buffer for 6 min on ice, washed twice in transport buffer, and kept on ice for 10 min. Coverslips containing permeabilized cells were incubated for 30 min at 30 or 4°C where noted on top of a 15-μl nuclear import reaction, which contained: 0.5–2 μM GFP-tagged import substrate or 30 nM purified rat GR, 2 mg/ml HeLa extract, and an ATP-regenerating system (1 mM ATP, 5 mM creatine phosphate, 10 U/ml creatine kinase). For ATP depletion, 2 U apyrase (Sigma) was added instead of the ATP regenerating system. After the import reaction, cells were washed twice in transport buffer, fixed in formaldehyde, and analyzed by immunofluorescence or direct fluorescence. For import assays with purified components, reactions contained 0.5–2 μM GFP-tagged substrate or 30 nM purified rat GR, 3 μM RanGDP, 2 μM RanBP7 or RanBP8 or 4 μM importin α2 + 1 μM importin β. For extract dilution experiments, HeLa extract was diluted to the designated concentration with transfer buffer and supplemented as noted with 2 μM RanBP7 or 4 μM importin α2. RanQ69L-GTP was added to the import reactions to 6 μM as noted. All import experiments were performed a minimum of four times.
RESULTS
NL1 and NL2 Are Functional in Yeast
GR is a transcriptional regulatory factor whose intracellular localization is controlled by its hormonal ligand (Figure 1B). We sought to identify nuclear import receptors that confer this redistribution. It has been previously shown that mammalian GR is functional when expressed in the budding yeast S. cerevisiae (Schena and Yamamoto, 1988). Similar to GR localization in mammalian cells, full-length GR was cytoplasmic in the absence of hormone, nuclear in the presence of hormone, and redistributed to the cytoplasm during hormone withdrawal in yeast (Figure 1B).
In mammalian cells, nuclear localization depends on two domains, NL1 and NL2 (Figure 1A). We found that truncated GR containing NL1 but not NL2 (N525) was nuclear in yeast even in the absence of hormone, consistent with the hormone-independent localization of this fragment in mammalian cells (Figure 1C). A fusion between NL2 (540C), GFP, and lacZ displayed hormone-dependent localization in both mammalian and yeast cells (Figure 1C). We replaced three of the conserved lysines of NL1 with alanines (Figure 1A) by site-directed mutagenesis. Mutations in these residues have been shown to abolish NL1 activity in mammalian cells (Savory et al., 1999). As predicted, these mutations rendered the N525 K513-515A GFP fusion protein largely cytoplasmic (Figure 1C). If the concentration of GR in the nucleus were significantly reduced, we would expect that the expression of a GR target gene would be reduced as well. Indeed the mutations abolished the transcriptional activity of this fragment in both mammalian and yeast transcription assays (our unpublished results). However, corresponding mutations had only mild effects on the intracellular distribution of full-length GR (Figure 1C). As previously observed in mammalian cells (Savory et al., 1999), the transcriptional activity of full-length GR K513-515A in the presence of hormone was modestly reduced (about twofold) relative to wild-type receptor (Figure 2C). We conclude that GR import requires NL1 and NL2 in yeast.
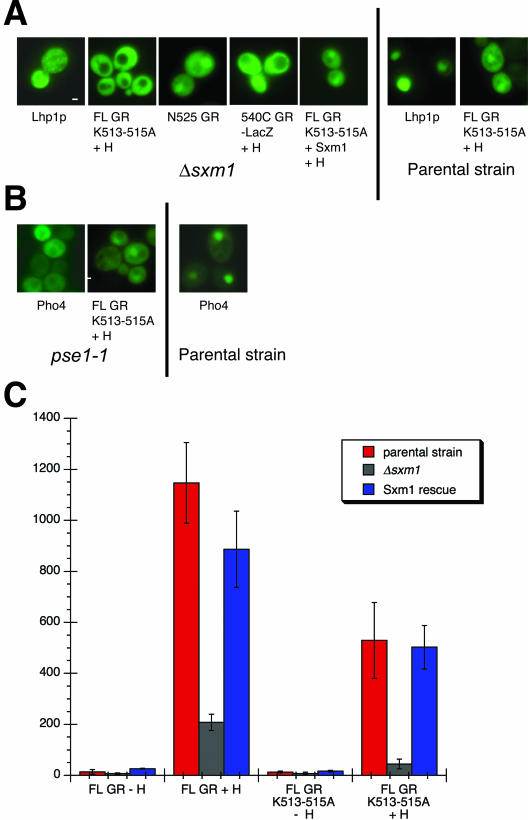
Figure 2.
NL2-mediated import is altered in a Δsxm1 yeast strain. Bar, 1 μM. (A) Plasmids encoding FL GR K513-515A-GFP, N525-GFP, 540C-lacZ-GFP, and Lhp1 were transformed into PSY1200 (Δsxm1 strain) or PSY902 (parental control); as noted. Plasmid pPS1574 was cotransformed to ectopically express Sxm1p and yeast treated with either EtOH (–H) or 5 μM DAC (+H). (B) pAdh-N795 K513-515A-GFP and Pho4-GFP (EB0347) were transformed into pse1-1ts or the control psy580 strain. (C) PSY1200 (red bars) or PSY902 (gray bars) were transformed with pAdh-N795-GFP or pAdh-N795-K513-515A-GFP and the pL2g3z GR reporter plasmid. For ectopic Sxm1p expression (blue bars), PSY1200 yeast were transformed with above plasmids and pPPS1117 expressing Sxm1p. Data represent the average values of six independent assays. Note: N795 K513-515A localization was unaffected in an Δsxm1 strain constructed in the Blobel lab (Rosenblum et al., 1997) in a different strain background than PSY1200 (Seedorf and Silver, 1997). These results suggest the presence of a genetic modifier, implying the existence of multiple import receptors for NL2 in some yeast strains.
Sxm1 Is a Nuclear Import Receptor for GR in Yeast
We screened a panel of 14 yeast strains with either null or temperature-sensitive alleles of nuclear transport receptors, for those strains in which full-length GR K513-515A displayed altered localization (MATERIALS AND METHODS). We found one strain, Δsxm1, in which full-length GR K513-515A and NL2 (540C)-lacZ failed to accumulate in the nucleus in the presence of hormone (Figure 2A). In addition, ectopic expression of Sxm1p was able to rescue this localization defect (Figure 2A). As expected, a known substrate for Sxm1, Lhp1p (Rosenblum et al., 1997), also failed to accumulate in the nucleus of Δsxm1 yeast (Figure 2A). Finally, N525 was nuclear in the Δsxm1 strain (Figure 2A), demonstrating that NL1-mediated import is not substantially affected in these cells.
GR nuclear accumulation was not significantly altered in any of the other 13 strains (our unpublished results). For example, GR localization was unaffected in a pse1-1 temperature-sensitive strain, in contrast to the altered localization of Pho4, a known Pse1 substrate (Kaffman et al., 1998b; Figure 2B). Finally, the transcriptional activity of a GR-responsive reporter driving lacZ expression was reduced in the Δsxm1 strain, and this decrease in transcriptional activity could be rescued by ectopically expressed Sxm1p (Figure 2C). Thus, Sxm1 is nuclear import receptor for GR in yeast.
Importin 7 and Importin 8 Bind Directly to GR
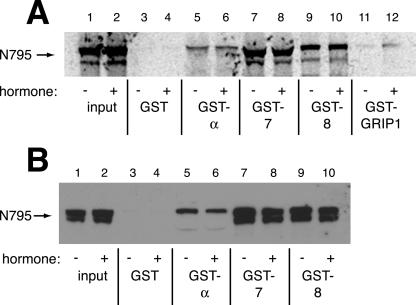
The yeast Sxm1 protein has two mammalian homologues, importin 7 and importin 8, each of which have 23% identity and 43% similarity to Sxm1p (Gorlich et al., 1997). We constructed fusions of both proteins to GST and tested whether they would directly bind to hormone-bound GR purified from rat liver (Figure 3A). As expected, GST alone failed to bind GR under the conditions tested (lane 2); however, GST-importin α, previously shown to interact with NL1, bound purified GR (lane 5). GST-importin 7 and GST-importin 8 also directly bound GR (lanes 3 and 4), suggesting that they may function as GR import receptors in mammalian cells.
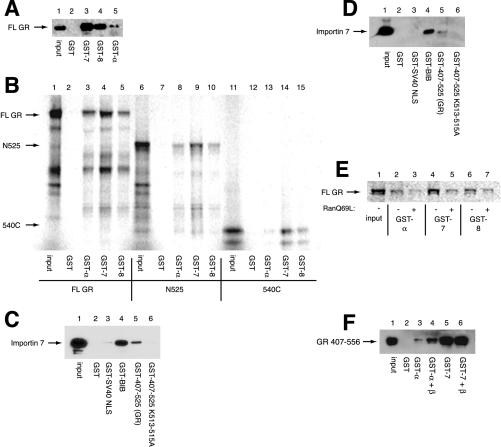
Figure 3.
GR binds to importin 7 and importin 8. Input lane corresponds to 20% input. (A) GST, GST-importin 7 (7), GST-importin 8 (8), or GST-importin α (α) (4 μg) were immobilized on glutathione resin and incubated with 150 ng of purified rat GR for 2 h at 4°C. Bound proteins were resolved by SDS-PAGE and immunoblotted with rat GR-specific BuGR2 antibody. (B) Immobilized GST proteins in A were incubated with 12 ng in vitro transcribed and translated N795, N525, or 540C for 1 h at RT. Bound proteins were resolved by SDS-PAGE and detected by PhosphorImager. (C) Immobilized GST-BIB, GST NLS, GST-407-525, GST-407-525 K513-515A, or GST alone (4 μg) were incubated with 4 μg of purified importin 7 overnight at 4°C. Bound proteins were resolved by SDS-PAGE and immunoblotted with a polyclonal antibody to importin 7. (D) Substrates described in C were incubated with 72 μg of HeLa extract overnight at 4°C and immunoblotted as above. (E) Immobilized import receptors (4 μg) were preincubated with 40 μM RanQ69L for 1 h at 4°C and then incubated with 12 ng in vitro transcribed and translated full-length GR for 2 h at 4°C. (F) GST, GST-α, and GST-7 (4 μg) were immobilized on glutathione resin in the presence or absence of importin β overnight at 4°C. Bound import receptor complexes were incubated with 4 μg of a purified GR fragment (407–565) containing NL1 for 3 h at 4°C.
Next, we mapped the regions of GR that interact with importin 7 and importin 8 by expressing 35S-methionine–labeled fragments of GR in reticulocyte lysate and testing for their interaction with GST-tagged import receptors by coprecipitation (Figure 3B). As expected, full-length GR bound importin α, importin 7, and importin 8 (lanes 3–5). Importin α also interacted with N525 (containing NL1; lane 8), but bound NL2 poorly (540C; lane 13). These results are consistent with importin α serving as an import receptor for NL1 but not NL2. Surprisingly, GST-importin 7 and GST-importin 8 bound both NL1 and NL2 (lanes 9, 10, 14, and 15), suggesting that there are at least two binding sites on GR for these import receptors.
We immobilized import substrates and tested their binding to purified importin 7. As a positive control, we used the beta-like importin binding (BIB) domain from L123a that has previously been shown to bind importin 7 (Jakel and Gorlich, 1998). As expected, GST-BIB was able to bind purified importin 7 (Figure 3C, lane 4) and importin 7 only weakly bound to the SV40 NLS (Figure 3C, lane 3). In contrast, the GR 407-525 (containing NL1) bound purified importin 7 (Figure 3c, lane 5), and 407-525 K513-515A failed to interact (Figure 3C, lane 6), consistent with a requirement of NL1 for binding. Similarly, the BIB domain and the GR 407-525 fragment of GR bound to importin 7 in a HeLa cell extract (Figure 3D, lanes 4 and 5), and mutation of GR 407-525 K513-515A again abrogated this interaction (Figure 3D, lane 6). We conclude that GR can bind importin 7 and importin 8.
Finally, consistent with other importin 7 and importin 8 substrates (Jakel and Gorlich, 1998; Dean et al., 2001; Jakel et al., 2002), the presence of a GTPase-deficient Ran mutant, RanQ69L, reduced the affinity of these import receptors for in vitro–transcribed and translated full-length GR (Figure 3E, lanes 5 and 7), suggesting that these importins may import GR. In addition, RanQ69L reduced the binding observed between GST-importin α and in vitro–transcribed and translated full-length GR (Figure 3E, lane 3). Importin α does not bind Ran directly (Rexach and Blobel, 1995), suggesting that another factor in reticulocyte lysate contributes to the observed binding between GR and importin α in the absence of RanQ69L. Because importin β binds importin α and the importin α/importin β/SV40 NLS complex can be dissociated by RanQ69L (Rexach and Blobel, 1995; Gorlich et al., 1996), we investigated the role of importin β in the binding of importin α to the NL1 of GR. The presence of recombinant importin β potentiated the binding of importin α but not importin 7 to a purified GR fragment (407–556) containing NL1 (Figure 3F, compare lanes 3–4 and 5–6), suggesting that importin β plays an important role in importin α but not importin 7 mediated GR nuclear import.
Reconstitution of GR Nuclear Import In Vitro
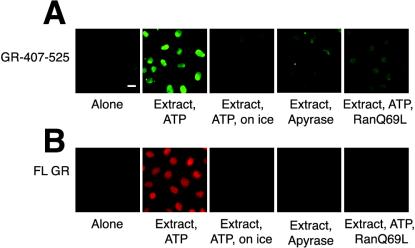
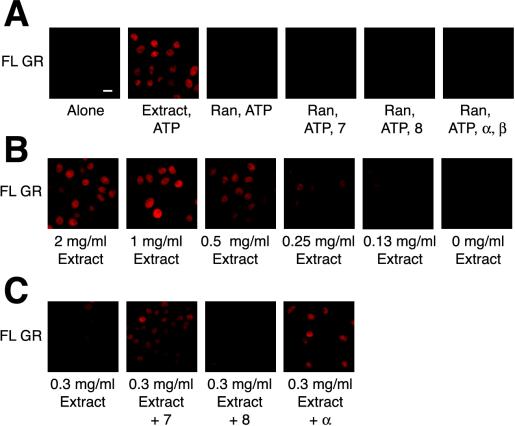
To determine if importin α, importin 7, and importin 8 can import GR, we used the digitonin-permeabilized cell assay (Adam et al., 1990). We first assessed nuclear import of a GR NL1-containing fusion protein, 407-525-GST-GFP, expressed and purified from bacteria. We found that nuclear import required HeLa cell extract and failed to occur on ice or in the presence of apyrase (Figure 4A), suggesting that energy is required for import. Next, full-length GR purified from rat liver was tested in the import assay. Because this protein is not tagged, we detected nuclear accumulation by indirect immunofluoresence. Import of full-length GR similarly required HeLa extract and ATP and was not observed in the presence of apyrase or at 0–4°C (Figure 4B). Import of both GR 407-525 and full-length GR was abolished in the presence of RanQ69L, suggesting that Ran plays a role in the import of both the GR 407-525 fragment (Figure 4A) and full-length GR (Figure 4B).
Figure 4.
Reconstitution of GR nuclear import in vitro. Bar, 10 μM. (A) GFP-GST-407-525 (0.5 μM) was incubated with permeabilized cells, 2 mg/ml HeLa extract, ATP mix, 6 μM RanQ69L, or apyrase for 30 min at 30°C or 4°C, as indicated. (B) Purified GR (30 nM) was incubated with digitonin-permeabilized cells, 2 mg/ml HeLa extract, ATP mix, 6 μM RanQ69L, or apyrase for 30 min at 30°Cor4°C, as indicated.
Importin 7 and Importin α/β Can Import GR In Vitro
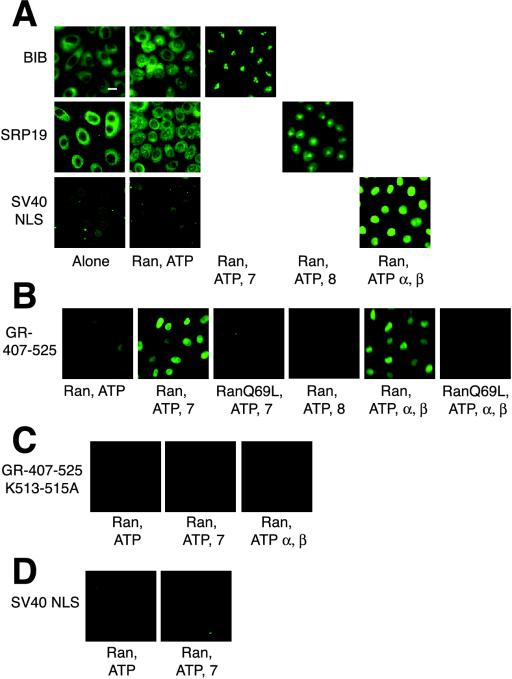
Next, we attempted to reconstitute 407-525-GST-GFP import with purified components. As positive controls for our assay, we used the SV40 NLS, which is imported by importin α and β (Nigg, 1997), the BIB domain, which is imported by importin 7 (Jakel and Gorlich, 1998), and SRP19, which is imported by importin 8 (Dean et al., 2001). Each was purified from bacteria as a protein fusion with GST and GFP. As observed previously by others, the import of each substrate required HeLa extract and ATP for import (our unpublished results). In addition, as reported, a low level of import was observed in the presence of the small GTPase Ran-GDP and ATP (Jakel and Gorlich, 1998; Figure 5A), due presumably to the presence of residual import receptors in digitonin-permeabilzed cells. However, the import of the BIB domain was significantly stimulated by importin 7, SRP19 by importin 8, and SV40 NLS by the importin α-importin β heterodimer (Figure 5A). As shown previously, the import of these substrates by purified import receptors was abolished in the presence of the Ran mutant RanQ69L (our unpublished results).
Figure 5.
Importin 7 and the importin α-importin β heterodimer import GR 407-525 in vitro. Bar, 10 μM. (A) BIB, SRP19, and SV40 import substrates (0.5–1 μM) and an ATP mix, 3 μM RanGDP, 6 μM RanQ69L, 2 μM importin 7, 2 μM importin 8, 4 μM importin α,or1 μM importin β, as indicated, were incubated with permeabilized cells as above. (B) Import assays were performed with 0.5 μM GFP-GST-407-525 and indicated import machinery. (C) Import assays were performed as above with 0.5 μM GFP-GST-407-525 K513-515A. (D) Import assays were performed with 1 μM SV40 NLS and importin 7.
The import of the GR 407-525 was also stimulated by Ran-GDP and ATP (Figure 5B). Addition of purified importin 7 or the importin α-importin β heterodimer significantly increased GR 407-525 nuclear import (Figure 5B). Importin 8 failed to import GR 407-525 (Figure 5B), although, it was able to import the SRP19 substrate (Figure 5A). As before, addition of RanQ69L abolished the nuclear import observed with purified import receptors (Figure 5B), again implicating Ran in the import of GR 407-525.
NL1 is strongly homologous to the SV40 NLS but shares little apparent similarity to either histone H1(o) or the BIB domain, previously identified as substrates for importin 7 (Jakel and Gorlich, 1998; Jakel et al., 1999). We tested whether the residues required for import by importin α were also required for the import of GR 407-525 by importin 7. Mutation of three conserved NL1 lysine residues (407-525 K513-515A) abolished the ability of both the importin α-importin β heterodimer and importin 7 to import GR407-525 into the nucleus (Figure 5C), suggesting that both classes of import receptors utilize NL1. Importin 7 failed to import the SV40 NLS in our assay, suggesting that although similar in sequence to the SV40 NLS, NL1 can be imported by additional import receptors (Figure 5D).
We next performed the import assay with purified components and full-length GR. We observed no significant import of full-length GR with Ran-GDP, an energy regeneration system and either importin 7, importin 8, or the importin α-importin β heterodimer (Figure 6A). Furthermore, no import was observed when importin α, importin β, and importin 7 were added in combination to the digitonin permeabilzed cells (our unpublished results). To test if another factor in the extract was required for import of full-length GR, we supplemented import reactions with diluted HeLa extract. Using a concentration of HeLa extract that stimulated intermediate levels of import (Figure 6B), we added recombinant import receptors and found that importin 7 and importin α, but not importin 8 potentiated the import of full-length GR (Figure 6C). This result suggests the presence of additional factor(s) in HeLa extract required for the import of full-length GR.
Figure 6.
Importin 7 and the importin α-importin β heterodimer potentiate the import of full-length rat GR. Bar, 10 μM. (A) GR (30 nM), ATP mix, 3 μM RanGDP, and 2 μM importin 7, 2 μM importin 8, 4 μM importin α,or1 μM importin β, as indicated, were incubated with digitonin permeabilized cells for 30 min at 30°C. (B) GR (30 nM) was incubated with permeabilized cells, ATP mix, and the indicated concentration of HeLa cell extract. (C) GR (30 nM), ATP mix, 0.3 mg/ml HeLa extract and 2 μM importin 7 or 4 μM importin α were incubated with digitonin permeablized cells as above.
Hormone Is Not Required for GR to Bind to Import Receptors
Finally, we tested the effect of hormone on the interaction of GR with importin 7, importin 8, and importin α. We could not readily test whether the interactions of purified full-length GR with nuclear import receptors were hormone dependent, because hormone is required for the purification of GR from rat liver (Perlmann and Wrange, 1988). Therefore, we examined these interactions using in vitro transcribed and translated GR in the presence and absence of hormone. As shown in Figure 7A, hormone did not affect the binding of importin α, importin 7, or importin 8 to full-length GR (lanes 5–10). As a positive control, we used GR-interacting protein (GRIP1), a transcriptional cofactor previously determined to bind GR in a hormone-dependent manner (Darimont et al., 1998). Indeed, hormone substantially stimulated the binding of GRIP1 to full-length GR (Figure 7A, lanes 11 and 12). Consistent with these results, hormone did not affect the interaction of endogenous full-length GR from human A549 cells with GST-tagged importin α, importin 7, and importin 8 (Figure 7B, lanes 5–10). These results suggest that hormone does not regulate the interaction of GR to its import receptor and thus must control GR intracellular localization in another way.
Figure 7.
Interaction of importin 7, importin 8, and importin α with GR is hormone independent. Input lane corresponds to 20% input. (A) GST, GST-importin 7 (7), GST-importin 8 (8), GST-importin α (α) were immobilized on glutathione-agarose and incubated with 12 ng of 35S-labeled GR, which had been in vitro transcribed and translated in the presence or absence of 10 μM Dex. Bound proteins were resolved by SDS-PAGE and analyzed by PhosphorImager. (B) Immobilized import receptors were incubated with 240 μg of A549 cytosol in the presence or absence of 100 nM Dex for 30 min at room temperature. Bound proteins were resolved by SDS-PAGE and immunoblotted with the GR 283 polyclonal antibody.
DISCUSSION
It has long been established that hormonal ligands direct the intracellular distribution of GR (Picard and Yamamoto, 1987). The GR LBD, which also houses NL2, conveys hormone-dependent nuclear localization when fused to heterologous substrates (Picard and Yamamoto, 1987). However, technical problems with purifying functional recombinant GR have precluded biochemical identification of nuclear import receptors for NL2. We therefore took a genetic approach, screening a panel of S. cerevisiae strains to infer nuclear import receptors for GR NL2 in yeast and then testing their vertebrate homologues. Indeed, this may be an efficient general strategy for identifying candidate import receptors for mammalian proteins.
Both NL1 and NL2 nuclear localization signals are functional in yeast, and localization of various GR truncations was similar in yeast and mammalian cells (Figure 1). In yeast, a family of nuclear transport receptors homologous to importin β has been identified, and a set of strains was constructed with individual disruptions in these genes (Kaffman et al., 1998a, 1998b). We tested NL2-mediated import in these strains and found that import was abolished in strains defective in Sxm1p (Figure 2A).
Surprisingly, the mammalian homologues of Sxm1p, importin 7 and importin 8, bound both NL1 and NL2 of GR. Although NL2 has not been precisely mapped, the NL2 region lacks sequences homologous to NL1 or to any of the identified importin 7 and importin 8 substrates. NL1 is closely related to the well-characterized NLS sequences of SV40 T-antigen and nucleoplasmin (Figure 1; Picard and Yamamoto, 1987; Tang et al., 1997). NL1 binds importin α (Savory et al., 1999; and Figure 3), which together with importin β confer nuclear localization in digitonin permeablized cells (Figure 5B). In parallel experiments, NL1 also bound to importin 7 and importin 8 (Figure 3) and was imported by importin 7 (Figure 5B). Whether importin α, importin 7, and importin 8 recognize distinct or identical residues within NL1 remains to be determined. Furthermore, we observed that importin β potentiates the binding of importin α to GR (Figure 3F). Similar results have been observed for the SV40 NLS (Rexach and Blobel, 1995; Gorlich et al., 1996). It will be interesting to determine the role that importin β plays in this interaction, whether it simply stabilizes the importin α-GR NL1 interaction or can bind to NL1 directly.
Gorlich and colleagues (Jakel et al., 2002) have suggested that importin 7 and importin 8, among other import receptors, may bind exposed basic regions and act as cytoplasmic chaperones to prevent aggregation; conceivably, importin 7 and importin 8 might play a role in maintaining GR solubility during nuclear transport. It will be interesting to determine if importin 7 can import other proteins that carry NLSs similar to those on NL1 and the SV40 T-antigen (Figure 1A), which are considered substrates of importin α. Notably, however, importin 7 failed to bind (Figure 3D) or import (Figure 5D) the SV40 NLS itself, demonstrating previously unrecognized selectivity within this class of nuclear localization motifs. Moreover, NL2 appears to be charge-neutral rather than basic yet bound importin 7 and importin 8 (Figure 3B). How importin 7 and importin 8 recognize apparently distinct motifs and whether they can simultaneously bind the same GR molecule remains to be determined. It is apparent, though, that multiple classes of nuclear import receptors can act on NL1.
Importin 7 alone and the importin α-importin β heterodimer were each competent to import an NL1-containing fragment of GR in an in vitro import assay in the presence of RanGDP and ATP (Figure 5B), whereas they failed to import purified full-length GR (Figure 6A). Addition of diluted HeLa cell extract to importin 7 or importin α permitted import of full-length GR (Figure 6C), suggesting that components in the extract absent from the highly purified import system are required to reconstitute import. It is unlikely that the diluted extract is simply providing additional importin 7 to the import reaction, for the amount of recombinant importin 7 in the reactions was >103-fold greater than that found in the diluted extract (our unpublished results). The observation that two separate import pathways could each import GR 407-525 but not full-length GR suggests that our biochemical import system lacks one or more general components required for GR import. Possible candidates include nuclear import machinery and molecular chaperones. Molecular chaperone complexes interact with GR and other intracellular receptors to facilitate hormone binding (Dittmar and Pratt, 1997) and the disassembly of GR-containing transcriptional regulatory complexes (Freeman et al., 2000).
Importin 7 and importin 8 display 64% sequence similarity, and each is 23% identical to Sxm1p. We found that although both importin 7 and importin 8 could bind GR, only importin 7 could import GR. Such substrate specificity is reminiscent of SRP19 and RPL123a, which bind both import receptors but are imported only by importin 8 and importin 7, respectively (Jakel and Gorlich, 1998; Dean et al., 2001). However, we cannot exclude the possibility that importin 8 might import GR under certain in vitro or in vivo conditions.
It is apparent from our work that multiple import receptors can transport GR to the nucleus. Most other transcriptional regulatory factors examined seem to enter the nucleus via a single import pathway (Kaffman and O'Shea, 1999), although certain nontranscription factor substrates for importin 7 and importin 8 can also be imported by other import receptors (Jakel and Gorlich, 1998; Jakal et al., 1999; Dean et al., 2001). Perhaps tissue-specific import-receptor expression modulates the relative rates or efficiencies of GR import in different target tissues, contributing to tissue specific responsiveness. It is also conceivable that different import receptors might be engaged when GR is activated by particular signals or combinations of signals.
In the absence of a bound agonist, the GR aporeceptor (apoGR) resides in a complex with molecular chaperones, competent to bind hormone, but inactive with respect to its other functions, such as DNA binding, regulatory cofactor recruitment, dimerization, and nuclear localization (Yamamoto et al., 1974; Picard and Yamamoto, 1987; Dahlman-Wright et al., 1990; Darimont et al., 1998). Hence, in the simplest view, the maintenance of apoGR in the cytoplasm may reflect a failure of the aporeceptor to associate with import receptors. Indeed, localization of the yeast transcriptional regulator Pho4 is governed at least in part by phosphorylation-dependent binding to the import receptor PseI (Kaffman et al., 1998b); the binding of Smad3 to importin β is regulated by phosphorylation (Kurisaki et al., 2001). However, we found that the interaction of GR with importin α, importin 7, or importin 8 is hormone independent, suggesting that the aporeceptor complex includes import receptors. Thus, the hormone-regulated step for nuclear localization appears to be downstream of import receptor binding. For example, GR has been proposed to be rapidly exported from the nucleus in the absence of hormone, although evidence for this model remains controversial (Hache et al., 1999; Savory et al., 1999; Liu and DeFranco, 2000; Holaska et al., 2001). A nuclear export sequence has been identified in GR (Black et al., 2001), but its role in regulating GR localization in the absence of hormone, remains to be determined.
An alternative possibility is that GR may be tethered to a cytoplasmic factor or complex in the absence of hormone. Pratt and colleagues have demonstrated that components of the apoGR complex bind cytoskeletal factors (Galigniana et al., 2001). Alternatively, Kino et al. (2003) have suggested that binding to the 14-3-3σ protein helps tether apoGR to the cytoplasmic compartment. Given these observations, it would be interesting to examine the intracellular distribution of GR in the absence of hormone by electron microscopy.
Our findings are also consistent with a third model, in which nuclear accumulation of GR may require the hormone-dependent binding of a novel factor, perhaps corresponding to the putative component in the diluted HeLa cell extract required for the in vitro import of full-length GR. In the future, it should be possible to purify this factor by fractionating the extract and monitoring GR import activity.
It is worth noting that GR is a member of a 48-gene family of intracellular receptors that display a wide spectrum of localization phenotypes. For example, nuclear accumulation of GR depends fully on ligand binding, whereas localization of the estrogen receptor is hormone stimulated but not dependent, and the thyroid hormone and retinoid receptors are constitutively nuclear. It will be interesting to determine whether other members of the intracellular receptor family serve as importin 7 substrates, if they require the putative factor used by GR for nuclear import, and the hormonal requirements for their interaction and import. A detailed understanding of the molecular features governing the nuclear localization of these receptors would provide insight into the physiological rationale for the observed differences in localization.
Acknowledgments
We thank M. Garabedian, D. Gorlich, A. Hopper, A. Kralli, A. Kurisaki, J. Li, M. S. Moore, E. O'Shea, L. Pemberton, T. Pieler, P. Silver, R. Sitcheran, K. Weis, J. Weissman, O. Wrange, and Y. Yoneda for their kind gifts of reagents. We also thank I. Herskowitz, J. Li, E. O'Shea, C. Takizawa, and members of the Yamamoto Laboratory for advice and discussions and E. Bolton, M. Diamond, H. Luecke, S. Meijsing, I. Rogatsky, W. Wang, K. Weis, and K. Witkin for thoughtful critiques of the manuscript. This work was supported in part by an American Heart Association Predoctoral fellowship to N.D.F. and by grants from the National Institutes of Health and National Science Foundation to K.R.Y.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–11–0839. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03–11–0839.
References
- Adam, S.A., Marr, R.S., and Gerace, L. (1990). Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111, 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison, J.D., Blobel, G., and Rout, M.P. (1996). Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science 274, 624–627. [DOI] [PubMed] [Google Scholar]
- Black, B.E., Holaska, J.M., Rastinejad, F., and Paschal, B.M. (2001). DNA binding domains in diverse nuclear receptors function as nuclear export signals. Curr. Biol. 11, 1749–1758. [DOI] [PubMed] [Google Scholar]
- Bohen, S.P., and Yamamoto, K.R. (1993). Isolation of Hsp90 mutants by screening for decreased steroid receptor function. Proc. Natl. Acad. Sci. USA 90, 11424–11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlman-Wright, K., Siltala-Roos, H., Carlstedt-Duke, J., and Gustafsson, J.A. (1990). Protein-protein interactions facilitate DNA binding by the glucocorticoid receptor DNA-binding domain. J. Biol. Chem. 265, 14030–14035. [PubMed] [Google Scholar]
- Darimont, B.D., Wagner, R.L., Apriletti, J.W., Stallcup, M.R., Kushner, P.J., Baxter, J.D., Fletterick, R.J., and Yamamoto, K.R. (1998). Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 12, 3343–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, L.I. (1995). The nuclear pore complex. Annu. Rev. Biochem. 64, 865–896. [DOI] [PubMed] [Google Scholar]
- Dean, K.A., von Ahsen, O., Gorlich, D., and Fried, H.M. (2001). Signal recognition particle protein 19 is imported into the nucleus by importin 8 (RanBP8) and transportin. J. Cell Sci. 114, 3479–3485. [DOI] [PubMed] [Google Scholar]
- Dittmar, K.D., and Pratt, W.B. (1997). Folding of the glucocorticoid receptor by the reconstituted Hsp90-based chaperone machinery. The initial hsp90.p60.hsp70-dependent step is sufficient for creating the steroid binding conformation. J. Biol. Chem. 272, 13047–13054. [DOI] [PubMed] [Google Scholar]
- Freedman, L.P., Yoshinaga, S.K., Vanderbilt, J.N., and Yamamoto, K.R. (1989). In vitro transcription enhancement by purified derivatives of the glucocorticoid receptor. Science 245, 298–301. [DOI] [PubMed] [Google Scholar]
- Freeman, B.C., Felts, S.J., Toft, D.O., and Yamamoto, K.R. (2000). The p23 molecular chaperones act at a late step in intracellular receptor action to differentially affect ligand efficacies. Genes Dev. 14, 422–434. [PMC free article] [PubMed] [Google Scholar]
- Freeman, B.C., and Yamamoto, K.R. (2001). Continuous recycling: a mechanism for modulatory signal transduction. Trends Biochem. Sci. 26, 285–290. [DOI] [PubMed] [Google Scholar]
- Galigniana, M.D., Radanyi, C., Renoir, J.M., Housley, P.R., and Pratt, W.B. (2001). Evidence that the peptidylprolyl isomerase domain of the hsp90-binding immunophilin FKBP52 is involved in both dynein interaction and glucocorticoid receptor movement to the nucleus. J. Biol. Chem. 276, 14884–14889. [DOI] [PubMed] [Google Scholar]
- Gametchu, B., and Harrison, R.W. (1984). Characterization of a monoclonal antibody to the rat liver glucocorticoid receptor. Endocrinology 114, 274–279. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., and Woods, R.A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87–96. [DOI] [PubMed] [Google Scholar]
- Gorlich, D., Dabrowski, M., Bischoff, F.R., Kutay, U., Bork, P., Hartmann, E., Prehn, S., and Izaurralde, E. (1997). A novel class of RanGTP binding proteins. J. Cell Biol. 138, 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich, D., and Kutay, U. (1999). Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Gorlich, D., Pante, N., Kutay, U., Aebi, U., and Bischoff, F.R. (1996). Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 15, 5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Hache, R.J., Tse, R., Reich, T., Savory, J.G., and Lefebvre, Y.A. (1999). Nucleocytoplasmic trafficking of steroid-free glucocorticoid receptor. J. Biol. Chem. 274, 1432–1439. [DOI] [PubMed] [Google Scholar]
- He, F., and Jacobson, A. (1995). Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 9, 437–454. [DOI] [PubMed] [Google Scholar]
- Holaska, J.M., Black, B.E., Love, D.C., Hanover, J.A., Leszyk, J., and Paschal, B.M. (2001). Calreticulin is a receptor for nuclear export. J. Cell Biol. 152, 127–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper, A.K., Schultz, L.D., and Shapiro, R.A. (1980). Processing of intervening sequences: a new yeast mutant which fails to excise intervening sequences from precursor tRNAs. Cell 19, 741–751. [DOI] [PubMed] [Google Scholar]
- Imamoto, N., Shimamoto, T., Takao, T., Tachibana, T., Kose, S., Matsubae, M., Sekimoto, T., Shimonishi, Y., and Yoneda, Y. (1995). In vivo evidence for involvement of a 58 kDa component of nuclear pore-targeting complex in nuclear protein import. EMBO J. 14, 3617–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine, M.K., Watkins, J.L., and Wente, S.R. (1995). The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J. Cell Biol. 131, 1699–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel, S., Albig, W., Kutay, U., Bischoff, F.R., Schwamborn, K., Doenecke, D., and Gorlich, D. (1999). The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J. 18, 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel, S., and Gorlich, D. (1998). Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 17, 4491–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel, S., Mingot, J.M., Schwarzmaier, P., Hartmann, E., and Gorlich, D. (2002). Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 21, 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki, T., Chen, S., Hitomi, M., Jacobs, E., Kumagai, C., Liang, S., Schneiter, R., Singleton, D., Wisniewska, J., and Tartakoff, A.M. (1994). Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J. Cell Biol. 126, 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman, A., and O'Shea, E.K. (1999). Regulation of nuclear localization: a key to a door. Annu. Rev. Cell Dev. Biol. 15, 291–339. [DOI] [PubMed] [Google Scholar]
- Kaffman, A., Rank, N.M., O'Neill, E.M., Huang, L.S., and O'Shea, E.K. (1998a). The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature 396, 482–486. [DOI] [PubMed] [Google Scholar]
- Kaffman, A., Rank, N.M., and O'Shea, E.K. (1998b). Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 12, 2673–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon, D., Roberts, B.L., Richardson, W.D., and Smith, A.E. (1984). A short amino acid sequence able to specify nuclear location. Cell 39, 499–509. [DOI] [PubMed] [Google Scholar]
- Kino, T., Souvatzoglou, E., De Martino, M.U., Tsopanomihalu, M., Wan, Y., and Chrousos, G.P. (2003). Protein 14-3-3-sigma interacts with and favors cytoplasmic subcellular localization of the glucocorticoid receptor, acting as a negative regulator of the glucocorticoid signaling pathway. J. Biol. Chem. 278, 25651–25656. [DOI] [PubMed] [Google Scholar]
- Kose, S., Imamoto, N., Tachibana, T., Shimamoto, T., and Yoneda, Y. (1997). Ran-unassisted nuclear migration of a 97-kD component of nuclear pore-targeting complex. J. Cell Biol. 139, 841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuersten, S., Ohno, M., and Mattaj, I.W. (2001). Nucleocytoplasmic transport: Ran, beta and beyond. Trends Cell Biol. 11, 497–503. [DOI] [PubMed] [Google Scholar]
- Kurisaki, A., Kose, S., Yoneda, Y., Heldin, C.H., and Moustakas, A. (2001). Transforming growth factor-beta induces nuclear import of Smad3 in an importin-beta1 and Ran-dependent manner. Mol. Biol. Cell 12, 1079–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay, U., Izaurralde, E., Bischoff, F.R., Mattaj, I.W., and Gorlich, D. (1997). Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO J. 16, 1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., and DeFranco, D.B. (2000). Protracted nuclear export of glucocorticoid receptor limits its turnover and does not require the exportin 1/CRM1-directed nuclear export pathway. Mol. Endocrinol. 14, 40–51. [DOI] [PubMed] [Google Scholar]
- Melchior, F., and Gerace, L. (1998). Two-way trafficking with Ran. Trends Cell Biol. 8, 175–179. [DOI] [PubMed] [Google Scholar]
- Moore, M.S., and Schwoebel, E.D. (2000). Nuclear import in digitonin-permeabilized cells. In: Current Protocols in Cell Biology, ed. J.S. Bonifacino, New York: John Wiley, 11.17.11–11.17.17. [DOI] [PubMed]
- Morehouse, H., Buratowski, R.M., Silver, P.A., and Buratowski, S. (1999). The importin/karyopherin Kap114 mediates the nuclear import of TATA-binding protein. Proc. Natl. Acad. Sci. USA 96, 12542–12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury, M.V., and Weis, K. (1999). The direction of transport through the nuclear pore can be inverted. Proc. Natl. Acad. Sci. USA 96, 9622–9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi, E., Imamoto, N., Sato, R., and Yoneda, Y. (1999). Nuclear import of sterol regulatory element-binding protein-2, a basic helix-loop-helix-leucine zipper (bHLH-Zip)-containing transcription factor, occurs through the direct interaction of importin beta with HLH-Zip. Mol. Biol. Cell 10, 2221–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg, E.A. (1997). Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature 386, 779–787. [DOI] [PubMed] [Google Scholar]
- Pante, N., and Aebi, U. (1995). Exploring nuclear pore complex structure and function in molecular detail. J. Cell Sci. Suppl. 19, 1–11. [DOI] [PubMed] [Google Scholar]
- Pemberton, L.F., Rosenblum, J.S., and Blobel, G. (1999). Nuclear import of the TATA-binding protein: mediation by the karyopherin Kap114p and a possible mechanism for intranuclear targeting. J. Cell Biol. 145, 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmann, T., and Wrange, O. (1988). Specific glucocorticoid receptor binding to DNA reconstituted in a nucleosome. EMBO J. 7, 3073–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard, D., and Yamamoto, K.R. (1987). Two signals mediate hormone-dependent nuclear localization of the glucocorticoid receptor. EMBO J. 6, 3333–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt, W.B., Silverstein, A.M., and Galigniana, M.D. (1999). A model for the cytoplasmic trafficking of signalling proteins involving the hsp90-binding immunophilins and p50cdc37. Cell Signal. 11, 839–851. [DOI] [PubMed] [Google Scholar]
- Rexach, M., and Blobel, G. (1995). Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 83, 683–692. [DOI] [PubMed] [Google Scholar]
- Rosenblum, J.S., Pemberton, L.F., and Blobel, G. (1997). A nuclear import pathway for a protein involved in tRNA maturation. J. Cell Biol. 139, 1655–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savory, J.G., Hsu, B., Laquian, I.R., Giffin, W., Reich, T., Hache, R.J., and Lefebvre, Y.A. (1999). Discrimination between NL1- and NL2-mediated nuclear localization of the glucocorticoid receptor. Mol. Cell. Biol. 19, 1025–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena, M., and Yamamoto, K.R. (1988). Mammalian glucocorticoid receptor derivatives enhance transcription in yeast. Science 241, 965–967. [DOI] [PubMed] [Google Scholar]
- Seedorf, M., and Silver, P.A. (1997). Importin/karyopherin protein family members required for mRNA export from the nucleus. Proc. Natl. Acad. Sci. USA 94, 8590–8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaete, R.R., and Mocarski, E.S. (1985). Regulation of cytomegalovirus gene expression: alpha and beta promoters are trans activated by viral functions in permissive human fibroblasts. J. Virol. 56, 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade, K., Ford, C.S., Guthrie, C., and Weis, K. (1997). Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90, 1041–1050. [DOI] [PubMed] [Google Scholar]
- Stoffler, D., Fahrenkrog, B., and Aebi, U. (1999). The nuclear pore complex: from molecular architecture to functional dynamics. Curr. Opin. Cell Biol. 11, 391–401. [DOI] [PubMed] [Google Scholar]
- Strom, A.C., and Weis, K. (2001). Importin-beta-like nuclear transport receptors. Genome Biol. 2, REVIEWS3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa, C.G., Weis, K., and Morgan, D.O. (1999). Ran-independent nuclear import of cyclin B1-Cdc2 by importin beta. Proc. Natl. Acad. Sci. USA 96, 7938–7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Y., Ramakrishnan, C., Thomas, J., and DeFranco, D.B. (1997). A role for HDJ-2/HSDJ in correcting subnuclear trafficking, transactivation, and transrepression defects of a glucocorticoid receptor zinc finger mutant. Mol. Biol. Cell 8, 795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, B.J., and Rothstein, R. (1989). Elevated recombination rates in transcriptionally active DNA. Cell 56, 619–630. [DOI] [PubMed] [Google Scholar]
- Thompson, E.B., Simons, S.S., Jr., and Harmon, J.M. (1981). Deacylcortivazol, a potent glucocorticoid with unusual structure and unusual anti-leukemic cell activity. Adv. Exp. Med. Biol. 138, 315–324. [DOI] [PubMed] [Google Scholar]
- Titov, A.A., and Blobel, G. (1999). The karyopherin Kap122p/Pdr6p imports both subunits of the transcription factor IIA into the nucleus. J. Cell Biol. 147, 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpin, P., Ossareh-Nazari, B., and Dargemont, C. (1999). Nuclear transport and transcriptional regulation. FEBS Lett. 452, 82–86. [DOI] [PubMed] [Google Scholar]
- Weis, K. (1998). Importins and exportins: how to get in and out of the nucleus. Trends Biochem. Sci. 23, 185–189 [published erratum appears in Trends Biochem. Sci. (1998). 23(7), 235]. [DOI] [PubMed] [Google Scholar]
- Xiao, Z., McGrew, J.T., Schroeder, A.J., and Fitzgerald-Hayes, M. (1993). CSE1 and CSE2, two new genes required for accurate mitotic chromosome segregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 13, 4691–4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, K.R., Stampfer, M.R., and Tomkins, G.M. (1974). Receptors from glucocorticoid-sensitive lymphoma cells and two classes of insensitive clones: physical and DNA-binding properties. Proc. Natl. Acad. Sci. USA 71, 3901–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, R., Oakes, M., Yamaghishi, M., Dodd, J.A., and Nomura, M. (1992). Cloning and characterization of SRP1, a suppressor of temperature-sensitive RNA polymerase I mutations, in Saccharomyces cerevisiae. Mol. Cell. Biol. 12, 5640–5651. [DOI] [PMC free article] [PubMed] [Google Scholar]