Abstract
Metal hypersensitivity is a common immune disorder. Human immune systems mount the allergic attacks on metal ions through skin contacts, lung inhalation and metal-containing artificial body implants. The consequences can be simple annoyances to life-threatening systemic illness. Allergic hyper-reactivities to nickel (Ni) and beryllium (Be) are the best-studied human metal hypersensitivities. Ni-contact dermatitis affects 10 % of the human population, whereas Be compounds are the culprits of chronic Be disease (CBD). αβ T cells (T cells) play a crucial role in these hypersensitivity reactions. Metal ions work as haptens and bind to the surface of major histocompatibility complex (MHC) and peptide complex. This modifies the binding surface of MHC and triggers the immune response of T cells. Metal-specific αβ T cell receptors (TCRs) are usually MHC restricted, especially MHC class II (MHCII) restricted. Numerous models have been proposed, yet the mechanisms and molecular basis of metal hypersensitivity remain elusive. Recently, we determined the crystal structures of the Ni and Be presenting human MHCII molecules, HLA-DR52c (DRA*0101, DRB3*0301) and HLA-DP2 (DPA1*0103, DPB1*0201). These structures revealed unusual features of MHCII molecules and shed light on how metal ions are recognized by T cells.
Keywords: MHC, TCR, Hypersensitivity, Metal, Structure
Metal and immune systems
Metal ions are essential nutrients in all forms of life. From zinc to magnesium, from iron to manganese, different metals play different roles, from DNA replication to enzymatic catalysis, from electron transfer to signal transduction. Despite their important roles, metals can also be toxic and elicit different kinds of immune responses, especially in humans. Inhalation of Be and cobalt dusts can cause CBD or hard metal disease in lungs. Metal-induced allergic contact dermatitis is expressed in a wide range of cutaneous reactions following dermal and systemic exposure. Apart from the well-known significance of Ni in developing contact dermatitis, other metals such as aluminum, Be, chromium, cobalt, copper, gold (Au), iridium, mercury, palladium, platinum, rhodium and titanium represented emerging causes of skin hypersensitivity [1]. Heavy metals, such as, Au, cadmium and mercury, can induce human autoimmune diseases [2]. Exposure to mercury vapors can result in elevated IgE levels and increased T lymphocyte numbers in humans [3, 4].
Metals are also directly involved in antigen presentation. The involvement of zinc in superantigen activation of T cells has been well established [5]. A zinc-binding site was revealed close to the MHC-binding site on superantigen staphylococcal enterotoxin A (SEA) and several other superantigens [6, 7]. This site interacts with the MHCII molecule via the exposed βHis81 of the β1 domain. This arrangement implies that SEA cross-links two or more MHC molecules on the surface of the antigen-presenting cell (APC) that greatly enhances SEA’s binding, “pre-clustering” and more efficient T cell activation. Recent studies showed that zinc may function as an ionic signaling messenger to regulate T cell activation [8, 9].
Metal ions have also been observed to inhibit antigen presentation to T cells. For example, Griem et al. [65] showed that Au+ interferes with antigen processing by the formation of Au+-peptide chelate complexes. CD4+ T cell hybridomas specific for different antigenic peptides were tested in the presence of disodium aurothiomalate (Au+ salts). In these studies, T cell recognition of peptides possessing two or more cysteine residues could be inhibited by the addition of Au+ salts. The requirement of two or more cysteine residues for the formation of stable Au+ complexes was confirmed by the substitution of cysteine by serine in an immunodominant peptide: The inhibitory effect of Au+ on peptide-specific T cell proliferation was completely eliminated.
Metal hypersensitivity
Allergic reactions to metals are on the rise [10]. Contact hypersensitivity to metal afflicts around of 10–15 % of the human population [11, 12]. Ni represents the most common occupational as well as public contact allergen. In 2008, Ni also named as “Allergen of the Year” by The American Contact Dermatitis Society, because of its rise as a cause of significant contact dermatitis in the North America, particularly among children [13, 14]. Ni-induced contact dermatitis is a skin inflammation resulting in swollen, reddened and itchy skin due to direct contact with an allergen. In most cases, the resulting rash occurs only at the site of contact, though it may be found on other parts of the body as well. Ni sensitivity is acquired through intimate and prolonged skin exposure to Ni metal or Ni salts. Once this sensitization has occurred, exposure to sufficient amounts of Ni ions may result in an allergic response. In addition to Ni, Au, Be and cobalt contacts may also result in immune-mediated contact hypersensitivity. Orthodontic appliances, dental casting alloys and knee arthroplasties are also common sources of Ni and other metal hypersensitivity [15, 16]. Although both CD4+ and CD8+ T cells are major effector cells in Ni hypersensitivity [17], there has been no particular MHC allele or isotype associated with sensitivity to Ni.
A number of groups have isolated T cell clones from patients sensitive to Ni in vitro [18–20]. One of the TCRs (ANi-2.3) was isolated from a Ni-reactive T cell originally isolated from a human patient with Ni-contact hypersensitivity [21]. This TCR (Vα1/Vβ17) recognizes Ni bound to the human MHCII molecule, HLA-DR52c. Previous experiments showed that recognition by this TCR requires the combination of Ni, a particular MHC molecule, HLA-DR52c, and an unknown specific MHC-bound peptide produced in B cells [22]. The paraformaldehyde-fixed APCs could present Ni to ANi-2.3 cells indicating that antigen processing was not required and suggesting that the Ni was presented by a preformed MHC/peptide complex [22]. Ni and the fixed APC interaction assays showed that Ni is reversibly bound to the fixed APC surface via a pH sensitive (pH 3.5–5.0) interaction consistent with coordination by amino acid side chains from His and/or acidic residues.
Metal hypersensitivity in human lungs causes chronic CBD, cobalt-induced hard metal lung disease. CBD is a granulomatous disorder that predominantly affects the lung, although the lymphatic system, skin, liver and spleen may also be affected [23, 24]. This disorder is estimated to occur in up to 18 % of exposed workers, depending on the nature of the exposure and the individual’s genetic susceptibility [25–28]. Be exposure in the form of metal, metal oxide, or metal alloy continues to be a major public health concern with approximately 1,000,000 individuals currently at risk for disease development [29, 30]. Most evidence indicates that CD4+ T cells are critically involved in the immunopathogenesis of CBD [18, 31–33]. Genetic susceptibility to CBD is associated with certain alleles of the MHCII molecule, HLA-DP, especially HLA-DPB1*0201 and other alleles that contain a glutamic acid residue at position 69 of the β-chain (βGlu69). After more than a decade of research, the reason for the genetic linkage between CBD and particular HLA-DP alleles has been a mystery [34].
The three-dimensional structures of HLA-DR52c and HLA-DP2
In spite of extensive studies on metal hypersensitivity, the structural basis for major MHC-restricted metal recognition is still unclear [35]. We determined the structure of HLA-DR52c bound to a self-peptide derived from the Tu elongation factor (pTu) [36]. Although HLA-DR52c/Tu peptide could not present Ni [22], a structure of HLA-DR52c would be important to understand the HLA-DR52c-restricted Ni presentation and to design the peptide screening libraries. HLA-DR52c has unique peptide-binding pockets. The nature of P4 pocket explains the strong selection for Asn (or Asp) among peptides known to bind to HLA-DR52c [36]. Asn is coordinated by an intricate hydrogen bond network involving three water molecules and Gln α9, Glu β28, Lys β72 and Gln β74 of HLA-DR52c [37]. These hydrogen bonds lock the position of the pTu peptide Asn side chain. A Pro at P6 sits in a pocket with both hydrophilic and hydrophobic properties that could accept virtually any aliphatic or short polar amino acid side chain. The Ile at P9 sits at the top of a deep mostly hydrophobic tunnel at the bottom of which lies Glu β9. This pocket is large enough to accept virtually any hydrophobic amino acid, such as Phe or Tyr. In a manner similar to the mouse MHCII molecule, IEk, this pocket might also accept a Lys with its ε-amino group salt bridged to the carboxylate of Glu β9 [38].
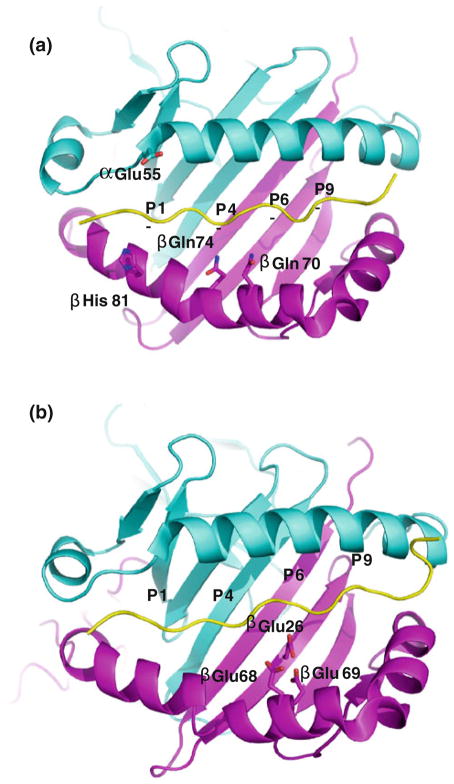
The potential Ni-binding site was not apparent on the TCR interacting surface of HLA-DR52c, because the natural Ni-presenting peptide was still unknown [37]. Ni is normally coordinated by at least four ligands. The common Ni ligands are His and Glu (or Asp), although Cys, Ser and Gln (or Asn) ligands were also occasionally found [39]. Since HLA-DR52c and specific peptide are involved in Ni binding and the types of Ni-binding amino acids are limited, we could identify four possible HLA-DR52c residues for the coordination of the Ni, based on the distance between HLA-DR52c surface residues and the peptide (Fig. 1a). It was suggested that the βHis81 of the HLA-DR52c and/or its bound peptide could coordinate Ni for the TCR recognition [22]. Based on HLA-DR52c structure, βGln70, βGln74 and αAsp55 could also potentially coordinate the Ni. However, none of these residues alone are sufficient to keep the Ni. This is in agreement with previous data that a specific peptide is needed to form the Ni/HLA-DR52c/peptide complex [22]. The HLA-DR52c structure was determined at 1.8 Å. At this resolution, we can use B-factor to define the flexible regions in the molecule with confidence (Fig. 2). The higher B-factor between β62 and β73 of HLA-DR52c shows this region is less rigid. Both molecular dynamics simulations’ modeling and conformationally sensitive monoclonal antibody probing showed this region (residues β53–67) may be flexible [40, 41]. Normally, the Ni ion-binding sites are in flexible parts of proteins. Thus, it is tempting to speculate the Ni binding around βGln70 and βGln74 of HLA-DR52c β1 domain.
Fig. 1.
Ribbon representations of the MHCII/peptide complexes. View is from the top looking down the α1 helix and β1 helix of MHCII (the potential metal-binding residues are shown with coloring: oxygen, red; nitrogen, blue). The α1 domain is shown in cyan and the β1 domain in magenta. The peptide is colored in yellow. The figures are prepared with program, Pymol. The amino acid names and the peptide anchoring residues are labeled. The four peptide anchoring pockets are labeled with P1, P4, P6 and P9 in all figures a The DR52c/Tu peptide complex, b The DP2/DRA peptide complex
Fig. 2.
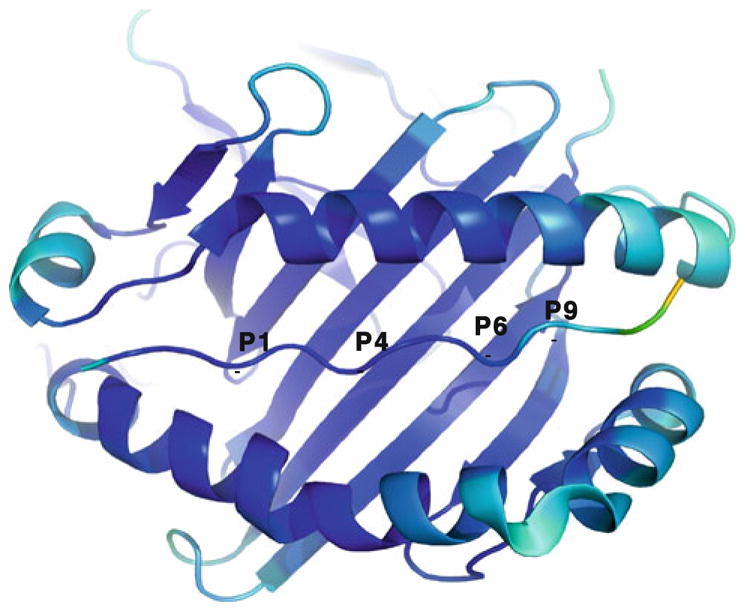
Ribbon representation of the HLA-DR52c/Tu peptide is viewed as Fig. 1. The model is color coded by B-factor. Blue indicates a low B-factor through to yellow indicating a high B-factor
The HLA-DP2 structure is critical for the understanding of Be presentation in CBD. Genetic susceptibility to CBD has been associated with certain alleles of the MHCII molecule, HLA-DP, especially HLA-DPB1*0201 and other alleles that contain a glutamic acid residue at position 69 of the β-chain (βGlu69) [34]. The structure of Be presenting HLA-DP2 with a bound self-peptide derived from the HLA-DR α chain (pDRA) was solved to a resolution of 3.25 Å [42]. Properties of the HLA-DP2 peptide side chain-binding pockets are different from other HLA alleles [43]. The very hydrophobic P1 pocket is similar to that found in a number of MHCII molecules, such as HLA-DR1 and HLA-DR4, that have a Gly at β86 [44]. This deep and very hydrophobic pocket can accommodate virtually all hydrophobic residues including large aromatic amino acids. It is occupied by a Phe of pDRA. In many other MHCII molecules, including most alleles of HLA-DR and HLA-DQ, the P6 peptide-binding pocket heavily selects for small, often charged amino acids. However, the P6 pocket of HLA-DP2 is large and very deep. It is lined with hydrophobic and polar residues: αTyr11, αAla14, αPhe25, αAsn65, αLeu69, βPhe9, βGly11, βGln13 and βTyr30. Its deepness is due to the amino acids αAla14 and βGly11, whose small side chains open up the bottom of the pocket. The P6 pocket is occupied by a Phe in the HLA-DP2-pDRA structure. The P1 and P6 positions of peptides appear to be the main peptide anchor residues for binding to DP2. The structure is in agreement with previous analysis of peptides naturally found bound to DP2 and mutational studies [42, 45]. The unique binding groove of the HLA-DP2 suggested the Be-binding peptides might have an unusual binding motif.
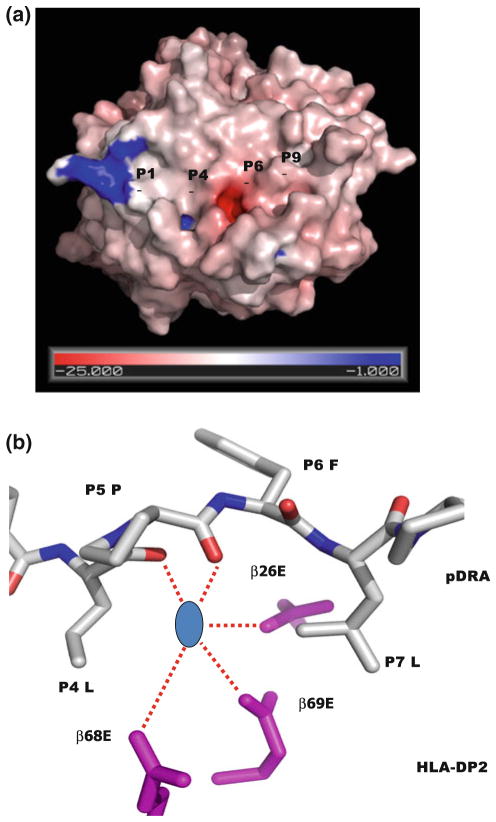
The unusual feature of the DP2 is the position of the β-chain α-helix in relation to the peptide and the rest of the molecule [43]. The P4-peptide-binding pocket of DP2 is large enough to accept positively charged amino acids or hydrophobic amino acids. A Leu is located at p4 position of pDRA. However, the backbone of pDRA has risen up in the binding groove at the P4 position, and the side chain of p4Leu of pDRA has moved up to the surface of the DP2-pDRA complex. The other unique property of the DP2 structure is the exceptionally wide peptide binding groove compared to other MHCII molecules, mostly due to the movement of the DP2 β-chain α-helix away from the peptide in the region between β69Glu of the DP2 β-chain and the p5Pro of the peptide [43]. The distance between the peptide P5 Cα atom and the DP2 β69 Cα atom is 11.2 Å, the widest gap seen in any MHCII structure so far [43]. As a result of this structural change, an acidic pocket opposite the P5 amino acid is formed, between the pDRA p4Leu and p7Leu (Fig. 3a). The acidity of this pocket is due to the presence of three DP2 β-chain amino acids: β68Glu and β69Glu from the β-chain α-helix and β26Glu from the floor of the peptide binding groove (Fig. 3b). In the crystal structure, the guanidinium group of an Arg from symmetry-related molecules filled into this pocket [43]. This positively charged moiety forms an extensive H-bond/salt bridge network with the three glutamic acids, offering a possible model for how Be compounds might occupy this site.
Fig. 3.
The unusual features of HLA-DP2 a The DP2/DRa peptide complex is viewed as Fig. 1. The water accessible surface of the DP2 molecule (without bound pDRA) is shown. The HLA-DP2 β-chain α-helix colored by the relative charge of the surface atoms (red, negative and blue, positive). The acidic pocket is located in the area between p5Pro and the DP2 β-chain α-helix. b A hypothetic Be compound is modeled in the acidic pocket. View of the acidic pocket looking down the peptide binding groove from the top. Wireframe representations of the side chains of β26Glu, β68Glu and β69Glu are colored in magenta. A wireframe representation of P4 to P6 of pDRA is shown with white carbon, red oxygen and blue nitrogen. Also, shown is a blue circle as a hypothetic Be compound
The fact that β69Glu is strongly associated with genetic susceptibility to CBD is in agreement that solved structures of other proteins associated with Be compounds generally show coordination of the compound by acidic amino acids [46] and also suggest that this acidic pocket is the site of binding of Be compounds to HLA-DP2. β26Glu and β68Glu are invariant among DP alleles. The model suggests that the Be antigen-presentation complex consists of MHCII HLA-DP with Be bound to carboxylate groups in the acidic pocket that is formed by a select group of peptides that give Be an appropriate amount of room to enter the acidic pocket. Mutation analysis of the acidic amino acids in the binding grove show that the βGlu69 position is necessary and sufficient to bind Be in a manner that allows T cell activation. Interestingly, there are several more acidic residues around the pocket, for example, Glu 67, Asp26. The roles of these acidic residues were not tested. Probably, they serve as charge attractants to help Be to find the pocket.
How the T cells recognize metal ions complexes with the MHC/peptide
TCRs commonly recognize linear peptides of antigenic proteins bound to MHC molecules. However, MHC molecules can also present components other than, or in addition to, peptides to T cells. Small chemical haptens presented by MHCI and MHCII to T cells have long been known [47]. Organic haptens such as dinitrophenol, trinitrophenol and fluorescein have long been used experimentally to study T cell responses and hypersensitivity [48–50]. T cell-mediated hypersensitivity to natural chemically active haptens, such as the catechols of poison ivy and poison oak, is common. T cells are also involved in allergic reactions to drugs, such as, β-lactam antibiotics and sulphamethoxazole [51]. Ni, Be, Au, cobalt and other metals ions form a unique group of chemicals for which T cell hypersensitivity has been demonstrated two decades ago [20, 34, 52]. While hypersensitivity to Be has been associated with particular alleles of HLA-MHCII alleles, especially HLA-DP2 [53, 54], there has been no particular MHC allele or isotype associated with sensitivity to Ni or Au [55, 56]. However, most often these clones require autologous APC for response to the metal ion, suggesting a role for either a specific peptide or particular MHC polymorphic amino acids in either the metal binding or the interaction with the TCR [57].
Conventionally, the TCRs dock diagonally on top of the surface of the MHC/peptide. The 6 CDR loops contact MHC/peptide, usually with CDR1α and CDR2α over the β helix of MHCII or the α2 helix of MHCI, and CDR1β and 2β over the α1 helix of MHCII or of MHCI. CDR3α and CDR3β are usually focused on amino acids of the peptide [58, 59]. Recent ternary complex structures of the mouse Vβ8 TCR and MHC/peptide demonstrate that some amino acids in the CDR1 and CDR2 regions of TCR Vα’s and Vβ’s may be evolutionarily conserved for interaction with particular conserved sites on MHC molecules and contribute to the bias of TCRs toward MHC ligands [60–62]. This heavy germline bias of the TCR for MHC may account for the TCR diagonal docking mode on MHC [60, 61].
How metal ions influence the docking orientations of TCRs on MHCs is unknown. There are no structural data for how metal ions become an essential part of the TCR ligand. It has been proposed that metals may bind on the surface of MHC molecules with or without a contribution from the bound peptide, such that they become part of the interface during TCR engagement [52]. Depending on what degree these metals dominate the TCR/MHC interface, TCR interaction with the MHC molecule and its bound peptide may also contribute to the overall specificity of the interaction. A study of effects of noble metal complexes on MHCII peptide binding has raised the possibility of another way that metals may create new epitopes [63]. In these experiments, some platinum, palladium and Au3+ complexes with square planar coordination were shown to destabilize peptide binding to MHCII creating stable molecules with the properties of “empty” MHCII, perhaps by binding to the MHCII in such a way as to stabilize an open conformation. Although not tested in the study, one could envision that these empty MHCII molecules could be recognized by T cells as neoantigens, accounting for hypersensitivity in some cases. Based on current biochemical data, hypothetically the metal ions can incorporate into MHC and become the ligands of T cell at least in four different ways (Fig. 4).
Fig. 4.
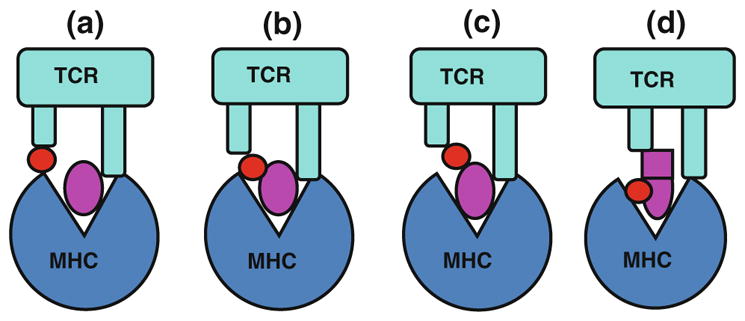
Models of metal presentation to CD4+ T cells in the context of MHCII molecules. Four models of metal presentation have been hypothesized. The schematic drawings of TCR and MHCII are colored as cyan (top) and blue (bottom). A metal ion is showed as a red circle. A peptide is showed as a magenta ellipse. The conformationally changed peptide is showed as a square and a ellipse, a shows the schematic model for the direct binding of a metal to the MHCII molecule only, b depicts the model of interaction where the metal binds to the antigenic peptide and the MHC molecule, c shows that the metal only interacts with peptide, d shows metal induced the change of peptide or MHC as neoantigens
Ni-reactive ANi-2.3 T cell requires DR52c, not other HLA alleles, as the Ni-presenting molecule [22]. ANi-2.3 TCR seems to lie on a diagonal above the face of HLA-DR52c/peptide. Vollmer and his works showed that the specific combinations of α and β chains were required for Ni recognition [17, 21]. More strikingly, an Arg95 and Asp96 dyad of CDRβ3s are absolutely required for the activation of ANi-2.3 T cells. This implied the dyad may be directly ligated to Ni ion or contact HLA-DR52c surface residues. Our recent structure of Ani-2.3 and DR52c/mimotope complex confirmed ANi-2.3 TCR docks on DR52c/peptide in a conventional way (unpublished data). Gamerdinger et al. reported that Ni forms a bridge between HLA-DR molecules and the TCR of a human Ni-reactive T cell clone, SE9 (Vα22/Vβ17), without the requirement for a specific MHC-bound peptide [57]. TCR SE9 is also MHC promiscuous and can be activated by multiple alleles of HLA-DRs [64]. TCR SE9 β chain is not related for TCR SE9 reactivity. Ni activation of these T cells only requires the specific α chain. The His81 of β1 domain of DR molecule and Tyr29 and Tyr94 of Vα domain of SE9 TCR may coordinate the Ni ion [57]. This suggested TCR SE9 has unusual binding topology. The TCR SE9 may greatly tilt toward the β1 domain HLA-DR molecule, causing no interactions between its α chain and the α1 domain of HLA-DR molecule. There are several ways of Ni presentation. One cannot rule out the possibility that metals may change the processing of self-peptide and trigger T cells activation with cryptic self-peptides [57, 65].
Several HLA-DP2-restricted, Be-reactive T cell clones have been identified by Fontenot’s group [66]. A thorough alanine scanning study of both DP2 and TCR surface residues was performed. The results suggested that, like Ni-reactive SE9 T cells, these TCRs bind Be, DP2 and peptide complex in an unexpected docking orientation [66]. None of the alanine mutations in the CDR1α and CDR2α of these TCRs had any effect on Be reactivity. In contrast, the mutational analysis of the TCR β-chains resulted in either reduced or abolished T cell hybridoma responses compared with the wild-type TCR. Overall, these data show that the Vβ domain of TCRs plays the major role in Be recognition, with no contribution from the germline-encoded parts of α chain. However, the Tyr at position 95 of the CDR3α is important for Be presentation. The binding topology of the TCR-peptide-MHC complex can modulate TCR signaling [67]. Until now, the unusual TCR footprints on MHC and peptide have been observed only on autoimmune TCRs [68–70]. These abnormal TCR binding modes may allow a subset of autoreactive T cells to escape thymic selection, causing the autoimmune disease [71, 72]. Likewise, metal ions may induce the unusual docking of TCR on MHC/peptide, triggering hypersensitivity.
Although the structural studies of metal presenting MHC molecules are essential to understand the mechanisms of T cell-mediated metal hypersensitivity, a complete understanding of metal recognition will come from ternary structures of the complete complexes: TCR, MHCII, peptide and metal. The crystallization and structure determination of the complexes of metal ion, MHC, peptide and TCR will be the definitive proof of the role of metal and relative contribution of the MHC molecule, peptide and TCR.
Acknowledgments
This work is supported by NIH KL2 TR000156 and the Boettcher Foundation.
References
- 1.Budinger L, Hertl M. Immunologic mechanisms in hypersensitivity reactions to metal ions: an overview. Allergy. 2000;55(2):108–15. doi: 10.1034/j.1398-9995.2000.00107.x. [DOI] [PubMed] [Google Scholar]
- 2.Schiraldi M, Monestier M. How can a chemical element elicit complex immunopathology? Lessons from mercury-induced autoimmunity. Trends Immunol. 2009;30(10):502–9. doi: 10.1016/j.it.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Vas J, Monestier M. Immunology of mercury. Ann N Y Acad Sci. 2008;1143:240–67. doi: 10.1196/annals.1443.022. [DOI] [PubMed] [Google Scholar]
- 4.Rowley B, Monestier M. Mechanisms of heavy metal-induced autoimmunity. Mol Immunol. 2005;42(7):833–8. doi: 10.1016/j.molimm.2004.07.050. [DOI] [PubMed] [Google Scholar]
- 5.Li H, Llera A, Malchiodi EL, Mariuzza RA. The structural basis of T cell activation by superantigens. Annu Rev Immunol. 1999;17:435–66. doi: 10.1146/annurev.immunol.17.1.435. [DOI] [PubMed] [Google Scholar]
- 6.Papageorgiou AC, Baker MD, McLeod JD, Goda SK, Manzotti CN, Sansom DM, et al. Identification of a secondary zinc-binding site in staphylococcal enterotoxin C2. Implications for superantigen recognition. J Biol Chem. 2004;279(2):1297–303. doi: 10.1074/jbc.M307333200. [DOI] [PubMed] [Google Scholar]
- 7.Li H, Zhao Y, Guo Y, Li Z, Eisele L, Mourad W. Zinc induces dimerization of the class II major histocompatibility complex molecule that leads to cooperative binding to a superantigen. J Biol Chem. 2007;282(9):5991–6000. doi: 10.1074/jbc.M608482200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu M, Lee WW, Tomar D, Pryshchep S, Czesnikiewicz-Guzik M, Lamar DL, et al. Regulation of T cell receptor signaling by activation-induced zinc influx. J Exp Med. 2011;208(4):775–85. doi: 10.1084/jem.20100031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bordon Y. T cell signalling: heavy metal rocks T cells. Nat Rev Immunol. 2011;11(5):300–1. doi: 10.1038/nri2977. [DOI] [PubMed] [Google Scholar]
- 10.Anthony TJ, Goon CLG. Metal allergy in Singapore. Contact Dermatitis. 2005;52(3):130–2. doi: 10.1111/j.0105-1873.2005.00518.x. [DOI] [PubMed] [Google Scholar]
- 11.Loh J, Fraser J. Metal-derivatized major histocompatibility complex: zeroing in on contact hypersensitivity. J Exp Med. 2003;197(5):549–52. doi: 10.1084/jem.20022180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Büdinger MH, Hertl M. Immunologic mechanisms in hypersensitivity reactions to metal ions: an overview. Allergy. 2000;55(2):108–15. doi: 10.1034/j.1398-9995.2000.00107.x. [DOI] [PubMed] [Google Scholar]
- 13.Militello G, Jacob SE, Crawford GH. Allergic contact dermatitis in children. Curr Opin Pediatr. 2006;18(4):385–90. doi: 10.1097/01.mop.0000236387.56709.6d. [DOI] [PubMed] [Google Scholar]
- 14.Hogeling M, Pratt M. Allergic contact dermatitis in children: the Ottawa hospital patch-testing clinic experience, 1996 to 2006. Dermatitis. 2008;19(2):86–9. [PubMed] [Google Scholar]
- 15.McGinley EL, Moran GP, Fleming GJ. Base-metal dental casting alloy biocompatibility assessment using a human-derived three-dimensional oral mucosal model. Acta Biomater. 2012;8(1):432–8. doi: 10.1016/j.actbio.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 16.Fors R, Persson M, Bergstrom E, Stenlund H, Stymne B, Stenberg B. Lifestyle and nickel allergy in a Swedish adolescent population: effects of piercing, tattooing and orthodontic appliances. Acta Derm Venereol. 2012 doi: 10.2340/00015555-1305. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 17.Vollmer J, Weltzien HU, Gamerdinger K, Lang S, Choleva Y, Moulon C. Antigen contacts by Ni-reactive TCR: typical alphass chain cooperation versus alpha chain-dominated specificity. Int Immunol. 2000;12(12):1723–31. doi: 10.1093/intimm/12.12.1723. [DOI] [PubMed] [Google Scholar]
- 18.Fontenot AP, Falta MT, Freed BM, Newman LS, Kotzin BL. Identification of pathogenic T cells in patients with beryllium-induced lung disease. J Immunol. 1999;163(2):1019–26. [PubMed] [Google Scholar]
- 19.Moulon C, Vollmer J, Weltzien HU. Characterization of processing requirements and metal cross-reactivities in T cell clones from patients with allergic contact dermatitis to nickel. Eur J Immunol. 1995;25(12):3308–15. doi: 10.1002/eji.1830251216. [DOI] [PubMed] [Google Scholar]
- 20.Romagnoli P, Labhardt AM, Sinigaglia F. Selective interaction of Ni with an MHC-bound peptide. EMBO J. 1991;10(6):1303–6. doi: 10.1002/j.1460-2075.1991.tb07648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vollmer J, Weltzien HU, Moulon C. TCR reactivity in human nickel allergy indicates contacts with complementarity-determining region 3 but excludes superantigen-like recognition. J Immunol. 1999;163(5):2723–31. [PubMed] [Google Scholar]
- 22.Lu L, Vollmer J, Moulon C, Weltzien HU, Marrack P, Kappler J. Components of the ligand for a Ni++ reactive human T cell clone. J Exp Med. 2003;197(5):567–74. doi: 10.1084/jem.20021762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freiman DG, Hardy HL. Beryllium disease: the relation of pulmonary pathology to the clinical course and prognosis based on a study of 130 cases from the U.S. Beryllium case registry. Hum Pathol. 1970;1:25–44. doi: 10.1016/s0046-8177(70)80003-x. [DOI] [PubMed] [Google Scholar]
- 24.Kriebel D, Brain JD, Sprince NL, Kazemi H. The pulmonary toxicity of beryllium. Am Rev Respir Dis. 1988;137:464–73. doi: 10.1164/ajrccm/137.2.464. [DOI] [PubMed] [Google Scholar]
- 25.Kreiss K, Mroz MM, Newman LS, Martyny J, Zhen B. Machining risk of beryllium disease and sensitization with median exposures below 2 μg/m3. Am J Ind Med. 1996;30:16–25. doi: 10.1002/(SICI)1097-0274(199607)30:1<16::AID-AJIM3>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 26.Kreiss K, Mroz MM, Zhen B, Martyny JW, Newman LS. Epidemiology of beryllium sensitization and disease in nuclear workers. Am Rev Respir Dis. 1993;148:985–91. doi: 10.1164/ajrccm/148.4_Pt_1.985. [DOI] [PubMed] [Google Scholar]
- 27.Kreiss K, Wasserman S, Mroz MM, Newman LS. Beryllium disease screening in the ceramics industry: blood test performance and exposure-disease relations. J Occup Med. 1993;35:267–74. [PubMed] [Google Scholar]
- 28.Van Dyke MV, Martyny JW, Mroz MM, Silveira LJ, Strand M, Cragle DL, et al. Exposure and genetics increase risk of beryllium sensitisation and chronic beryllium disease in the nuclear weapons industry. Occup Environ Med. 2011;68(11):842–8. doi: 10.1136/oem.2010.064220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amicosante M, Fontenot AP. T cell recognition in chronic beryllium disease. Clin Immunol. 2006;121(2):134–43. doi: 10.1016/j.clim.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 30.Fontenot AP, Maier LA. Genetic susceptibility and immune-mediated destruction in beryllium-induced disease. Trends Immunol. 2005;26(10):543–9. doi: 10.1016/j.it.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 31.Fontenot AP, Kotzin BL, Comment CE, Newman LS. Expansions of T-cell subsets expressing particular T-cell receptor variable regions in chronic beryllium disease. Am J Respir Cell Mol Biol. 1998;18(4):581–9. doi: 10.1165/ajrcmb.18.4.2981. [DOI] [PubMed] [Google Scholar]
- 32.Fontenot AP, Palmer BE, Sullivan AK, Joslin FG, Wilson CC, Maier LA, et al. Frequency of beryllium-specific, central memory CD4+ T cells in blood determines proliferative response. J Clin Invest. 2005;115(10):2886–93. doi: 10.1172/JCI24908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saltini C, Winestock K, Kirby M, Pinkston P, Crystal RG. Maintenance of alveolitis in patients with chronic beryllium disease by beryllium-specific helper T cells. N Engl J Med. 1989;320:1103–9. doi: 10.1056/NEJM198904273201702. [DOI] [PubMed] [Google Scholar]
- 34.Richeldi L, Sorrentino R, Saltini C. HLA-DPB1 glutamate 69: a genetic marker of beryllium disease. Science. 1993;262:242–4. doi: 10.1126/science.8105536. [DOI] [PubMed] [Google Scholar]
- 35.Sawyer RT, Maier LA. Chronic beryllium disease: an updated model interaction between innate and acquired immunity. Bio-metals. 2011;24(1):1–17. doi: 10.1007/s10534-010-9376-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verreck FAW, van de Poel A, Drijfhout JW, Amons R, Coligan JE, Koning F. Natural peptides isolated from Gly86/Val86-containing variants of HLA-DR1, -DR11, -DR13, and -DR52. Immunogenetics. 1996;43(6):392–7. doi: 10.1007/BF02199809. [DOI] [PubMed] [Google Scholar]
- 37.Dai S, Crawford F, Marrack P, Kappler JW. The structure of HLA-DR52c: comparison to other HLA-DRB3 alleles. Proc Natl Acad Sci U S A. 2008;105(33):11893–7. doi: 10.1073/pnas.0805810105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fremont DH, Dai S, Chiang H, Crawford F, Marrack P, Kappler J. Structural basis of cytochrome c presentation by IE(k) J Exp Med. 2002;195(8):1043–52. doi: 10.1084/jem.20011971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carrington PE, Chivers PT, Al-Mjeni F, Sauer RT, Maroney MJ. Nickel coordination is regulated by the DNA-bound state of NikR. Nat Struct Biol. 2003;10(2):126–30. doi: 10.1038/nsb890. [DOI] [PubMed] [Google Scholar]
- 40.Painter CA, Cruz A, Lopez GE, Stern LJ, Zavala-Ruiz Z. Model for the peptide-free conformation of class II MHC proteins. PLoS ONE. 2008;3(6):e2403. doi: 10.1371/journal.pone.0002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carven GJ, Chitta S, Hilgert I, Rushe MM, Baggio RF, Palmer M, et al. Monoclonal antibodies specific for the empty conformation of HLA-DR1 reveal aspects of the conformational change associated with peptide binding. J Biol Chem. 2004;279(16):16561–70. doi: 10.1074/jbc.M314315200. [DOI] [PubMed] [Google Scholar]
- 42.Diaz G, Canas B, Vazquez J, Nombela C, Arroyo J. Characterization of natural peptide ligands from HLA-DP2: new insights into HLA-DP peptide-binding motifs. Immunogenetics. 2005;56(10):754–9. doi: 10.1007/s00251-004-0735-5. [DOI] [PubMed] [Google Scholar]
- 43.Dai S, Murphy GA, Crawford F, Mack DG, Falta MT, Marrack P, et al. Crystal structure of HLA-DP2 and implications for chronic beryllium disease. Proc Natl Acad Sci U S A. 2010;107(16):7425–30. doi: 10.1073/pnas.1001772107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, et al. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368(6468):215–21. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 45.Berretta F, Butler RH, Diaz G, Sanarico N, Arroyo J, Fraziano M, et al. Detailed analysis of the effects of Glu/Lys beta69 human leukocyte antigen-DP polymorphism on peptide-binding specificity. Tissue Antigens. 2003;62(6):459–71. doi: 10.1046/j.1399-0039.2003.00131.x. [DOI] [PubMed] [Google Scholar]
- 46.Cho H, Wang W, Kim R, Yokota H, Damo S, Kim SH, et al. BeF3− acts as a phosphate analog in proteins phosphorylated on aspartate: structure of a BeF3− complex with phosphoserine phosphatase. Proc Natl Acad Sci U S A. 2001;98(15):8525–30. doi: 10.1073/pnas.131213698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kapsenberg ML, Bos JD, Wierenga EA. T cells in allergic responses to haptens and proteins. Springer Semin Immunopathol. 1992;13(3–4):303–14. doi: 10.1007/BF00200530. [DOI] [PubMed] [Google Scholar]
- 48.Ortmann B, Martin S, von Bonin A, Schiltz E, Hoschutzky H, Weltzien HU. Synthetic peptides anchor T cell-specific TNP epitopes to MHC antigens. J Immunol. 1992;148(5):1445–50. [PubMed] [Google Scholar]
- 49.Preckel T, Breloer M, Kohler H, von Bonin A, Weltzien HU. Partial agonism and independent modulation of T cell receptor and CD8 in hapten-specific cytotoxic T cells. Eur J Immunol. 1998;28(11):3706–18. doi: 10.1002/(SICI)1521-4141(199811)28:11<3706::AID-IMMU3706>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 50.Hughes EA, Kinnel G, Wickerham C, Atkeson B, Owen JA. Fine specificity analysis indicates that the primary and secondary fluorescein-specific cytotoxic T cell receptor repertoires are indistinguishable. Immunol Cell Biol. 1995;73(2):153–7. doi: 10.1038/icb.1995.24. [DOI] [PubMed] [Google Scholar]
- 51.Pichler WJ, Yawalkar N. Allergic reactions to drugs: involvement of T cells. Thorax. 2000;55(Suppl 2):S61–5. doi: 10.1136/thorax.55.suppl_2.S61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sinigaglia F. The molecular basis of metal recognition by T cells. J Invest Dermatol. 1994;102(4):398–401. doi: 10.1111/1523-1747.ep12372149. [DOI] [PubMed] [Google Scholar]
- 53.Maier LA, McGrath DS, Sato H, Lympany P, Welsh K, Du Bois R, et al. Influence of MHC class II in susceptibility to beryllium sensitization and chronic beryllium disease. J Immunol. 2003;171(12):6910–8. doi: 10.4049/jimmunol.171.12.6910. [DOI] [PubMed] [Google Scholar]
- 54.Fontenot AP, Torres M, Marshall WH, Newman LS, Kotzin BL. Beryllium presentation to CD4+T cells underlies disease-susceptibility HLA-DP alleles in chronic beryllium disease. Proc Natl Acad Sci U S A. 2000;97(23):12717–22. doi: 10.1073/pnas.220430797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hashizume H, Seo N, Ito T, Takigawa M, Yagi H. Promiscuous interaction between gold-specific T cells and APCs in gold allergy. J Immunol. 2008;181(11):8096–102. doi: 10.4049/jimmunol.181.11.8096. [DOI] [PubMed] [Google Scholar]
- 56.Emtestam L, Zetterquist H, Olerup O. HLA-DR, -DQ and -DP alleles in nickel, chromium, and/or cobalt-sensitive individuals: genomic analysis based on restriction fragment length polymorphisms. J Invest Dermatol. 1993;100(3):271–4. doi: 10.1111/1523-1747.ep12469732. [DOI] [PubMed] [Google Scholar]
- 57.Thierse HJ, Gamerdinger K, Junkes C, Guerreiro N, Weltzien HU. T cell receptor (TCR) interaction with haptens: metal ions as non-classical haptens. Toxicology. 2005;209(2):101–7. doi: 10.1016/j.tox.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 58.Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–66. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 60.Yin L, Huseby E, Scott-Browne J, Rubtsova K, Pinilla C, Crawford F, et al. A single T cell receptor bound to major his-tocompatibility complex class I and class II glycoproteins reveals switchable TCR conformers. Immunity. 2011;35(1):23–33. doi: 10.1016/j.immuni.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dai S, Huseby ES, Rubtsova K, Scott-Browne J, Crawford F, Macdonald WA, et al. Crossreactive T Cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules. Immunity. 2008;28(3):324–34. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histo-compatibility complex interaction ‘codon’. Nat Immunol. 2007;8(9):975–83. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 63.De Wall SL, Painter C, Stone JD, Bandaranayake R, Wiley DC, Mitchison TJ, et al. Noble metals strip peptides from class II MHC proteins. Nat Chem Biol. 2006;2(4):197–201. doi: 10.1038/nchembio773. [DOI] [PubMed] [Google Scholar]
- 64.Gamerdinger K, Moulon C, Karp DR, Van Bergen J, Koning F, Wild D, et al. A new type of metal recognition by human T cells: contact residues for peptide-independent bridging of T cell receptor and major histocompatibility complex by nickel. J Exp Med. 2003;197(10):1345–53. doi: 10.1084/jem.20030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Griem P, Panthel K, Kalbacher H, Gleichmann E. Alteration of a model antigen by Au(III) leads to T cell sensitization to cryptic peptides. Eur J Immunol. 1996;26(2):279–87. doi: 10.1002/eji.1830260202. [DOI] [PubMed] [Google Scholar]
- 66.Bowerman NA, Falta MT, Mack DG, Kappler JW, Fontenot AP. Mutagenesis of beryllium-specific TCRs suggests an unusual binding topology for antigen recognition. J Immunol. 2011;187(7):3694–703. doi: 10.4049/jimmunol.1101872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adams JJ, Narayanan S, Liu B, Birnbaum ME, Kruse AC, Bowerman NA, et al. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity. 2011;35(5):681–93. doi: 10.1016/j.immuni.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maynard J, Petersson K, Wilson DH, Adams EJ, Blondelle SE, Boulanger MJ, et al. Structure of an autoimmune T cell receptor complexed with class II peptide-MHC: insights into MHC bias and antigen specificity. Immunity. 2005;22(1):81–92. doi: 10.1016/j.immuni.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 69.Li Y, Huang Y, Lue J, Quandt JA, Martin R, Mariuzza RA. Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule. EMBO J. 2005;24(17):2968–79. doi: 10.1038/sj.emboj.7600771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hahn M, Nicholson MJ, Pyrdol J, Wucherpfennig KW. Unconventional topology of self peptide-major histocompatibility complex binding by a human autoimmune T cell receptor. Nat Immunol. 2005;6(5):490–6. doi: 10.1038/ni1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nicholson MJ, Hahn M, Wucherpfennig KW. Unusual features of self-peptide/MHC binding by autoimmune T cell receptors. Immunity. 2005;23(4):351–60. doi: 10.1016/j.immuni.2005.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deng L, Mariuzza RA. Recognition of self-peptide-MHC complexes by autoimmune T-cell receptors. Trends Biochem Sci. 2007;32(11):500–8. doi: 10.1016/j.tibs.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]