Abstract
This paper describes a new class of salt-responsive poly(ethylene glycol) (PEG) self-assembled monolayers (SAMs) on top of polyelectrolyte multilayer (PEMs) films. PEM surfaces with poly(diallyldimethylammonium chloride) as the topmost layer are chemically patterned by microcontact printing (μCP) oligomeric PEG molecules with an activated carboxylic acid terminal group (m-dPEG acid). The resistive m-d-poly(ethylene glycol) (m-dPEG) acid molecules on the PEMs films were subsequently removed from the PEM surface with salt treatment, thus converting the nonadhesive surfaces into adhesive surfaces. The resistive PEG patterns facilitate the directed deposition of various macromolecules such as polymers, dyes, colloidal particles, proteins, liposomes, and nucleic acids. Further, these PEG patterns act as a universal resist for different types of cells (e.g., primary cells, cell lines), thus permitting more flexibility in attaching a wide variety of cells to material surfaces. The patterned films were characterized by optical microscopy and atomic force microscopy (AFM). The PEG patterns were removed from the PEM surface at certain salt conditions without affecting the PEM films underneath the SAMs. Removal of the PEG SAMs and the stability of the PEM films underneath it were characterized with ellipsometry and optical microscopy. Such salt- and pH-responsive surfaces could lead to significant advances in the fields of tissue engineering, targeted drug delivery, materials science, and biology.
Introduction
The development of new tunable and structured surfaces capable of assembling two or more biological elements such as proteins, liposomes, nucleic acids, and cells onto a surface resulting in arrays of biological molecules has generated tremendous interest in the past years. Here, we describe a new class of salt-responsive poly(ethylene glycol) (PEG) self-assembled monolayers (SAMs) on top of polyelectrolyte multilayer (PEMs) films. Current approaches to engineer tunable surfaces are based on sophisticated methods that use light-, laser-, and UV-induced and electrochemical surface modifications, which tend to affect the morphology and properties of the underlying surfaces and are not compatible when extended to biological systems involving cells and proteins.1–3 The PEG patterns developed in this study are tunable at certain salt conditions, unveiling active regions of the film while leaving the attached biomolecules on the PEM surface undisturbed. The resistive PEG patterns facilitate the directed deposition of various macromolecules such as polymers, dyes, colloidal particles, proteins, liposomes, and nucleic acids. Further, these PEG patterns act as a universal resist for different types of cells (e.g., primary cells, cell lines), thus permitting more flexibility in attaching a wide variety of cells to material surfaces. Such salt- and pH-responsive surfaces could lead to significant advances in the fields of tissue engineering, targeted drug delivery, materials science, and biology.4–6
The ability to control cell adhesion in vitro may lead to advances in diverse fields, ranging from cell biology to tissue engineering. A number of fabrication strategies such as photolithography, microcontact printing, micromolding, inkjet printing, and dippen spotting7–10 have been applied to create micropatterned surfaces for manipulating the cell environment. In these approaches, the cells have been localized to adhesive regions on a substrate, thus limiting their use to one cell type. Most cell-patterning studies that engineered patterned co-cultures have involved selective adhesion of one cell type over another. For example, studies to design co-cultures with primary hepatocytes and fibroblasts required adhesion of primary hepatocytes to the surface prior to attaching the second cell type, i.e., fibroblast. This is because fibroblast typically can attach to any surface, whereas primary hepatocytes are more selective in their attachment to surfaces. Thus, capitalizing upon this fact permits the design of pattern co-cultures of primary hepatocytes and fibroblasts. Due to the lack of a tunable universal surface resistant to all cell types,11,12 there is a need to develop surfaces with the ability to dynamically and locally switch substrate adhesiveness to different types of cells and thus more easily facilitate the patterning of two or more cell types in spatially defined cocultures.
Engineering of micrometer- and nanoscale protein arrays is important for a wide range of applications such as drug delivery, biosensors, and basis cell studies.7,13,14 Most of the studies developed have focused on forming arrays of single proteins.7,8,15–20 However, few studies have reported engineering of multiple protein arrays on surfaces and have various limitations including pattern resolution,21,22 protein degradation,23 and exposure to harsh chemicals.24
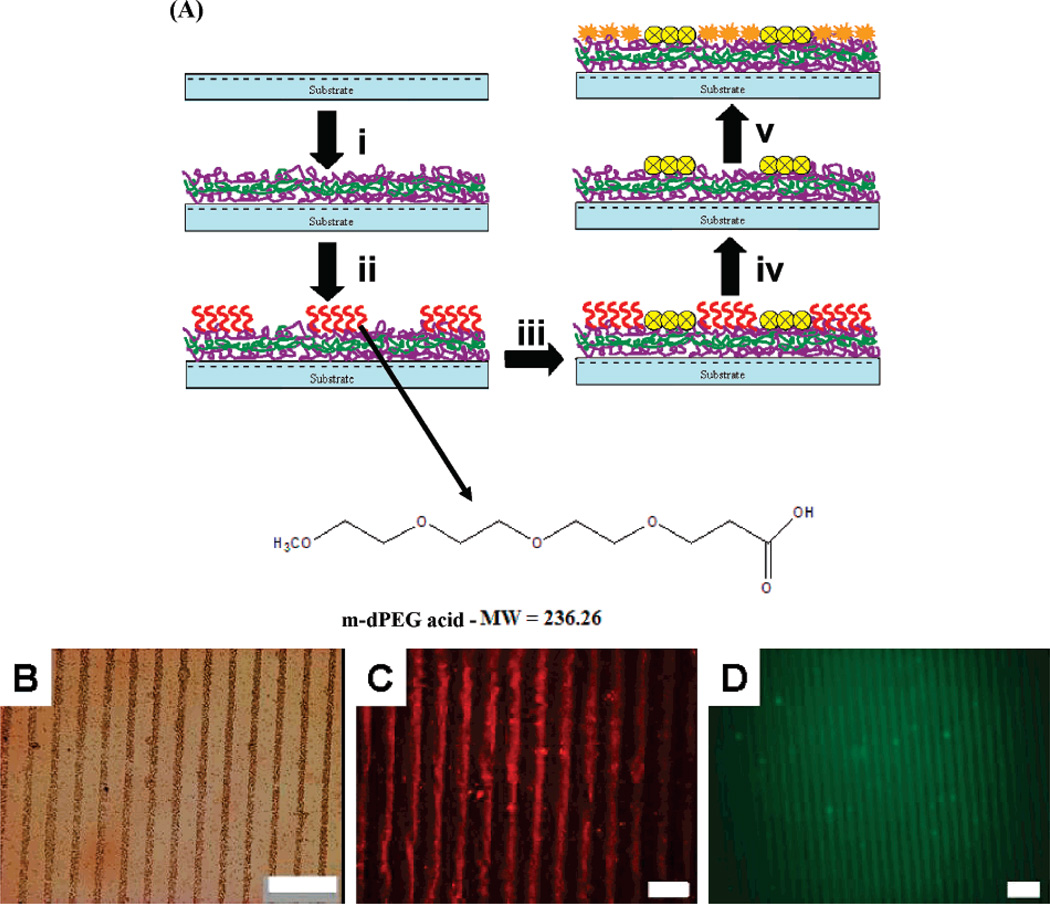
In this study, we engineered a novel salt tunable resistive m-dPEG acid SAM patterns on PEM surfaces which provides a template to design numerous sorted surfaces that can be used in a wide variety of applications. We capitalized upon the ionic interactions to deposit thin, uniform SAM patterns of resistive m-dPEG acid molecules atop the PEM films using μCP (Figure 1A).24 The PEG molecule has a degree of polymerization of four and an activated carboxylate functional group at the end. This carboxylate functional group in the PEG molecule ionically binds to the topmost positive surface of the PEM surfaces, and the other end of the PEG molecule resists the deposit of subsequent macromolecules including polyelectrolyte layers. The resistive nature of the PEG patterns was used to achieve formation of complex polyelectrolyte multilayer structures25–27 and directed assembly of a wide range of macromolecules such as colloid particles, proteins, as well as cells (as shown in Figure 1B–D and Figure 3). The PEG patterns were tuned under mild conditions (salt concentration = 0.25 M) to reveal active regions that can be used to create multicomponent systems. Hammond and co-workers developed surface-directed templates capable of creating multicomponent systems of colloidal particles and polymers.24 The current study extends the tunable PEG surfaces to engineer multicomponent systems of macromolecules with similar physical and chemical properties.
Figure 1.
(A) Diagram illustrating the formation of salt tunable m-dPEG acid SAMs on a PDAC/SPS multilayer platform. (i) PEMs (PDAC/SPS)10.5 build on top of the substrates. (ii) Patterned PEG SAMs on PEMs. (iii) Directed assembly of molecules due to the presence of resistive PEG SAMs. (iv) PEG SAMs are removed by treating it with salt, giving rise to new active regions. (v) The new active regions are filled with a new set of molecules. The chemical structure of the m-dPEG acid molecule. (B–D) Optical microscope images of directed deposition of macromolecules on PEG patterns: (B) 0.5 µm colloid particles (brown lines), scale bar = 25 µm; (C) Alexa Fluoro tagged sADH, scale bar = 25 µm; (D) FITC tagged nucleic acid, scale bar = 50 µm. The dark lines represent the m-dPEG acid regions.
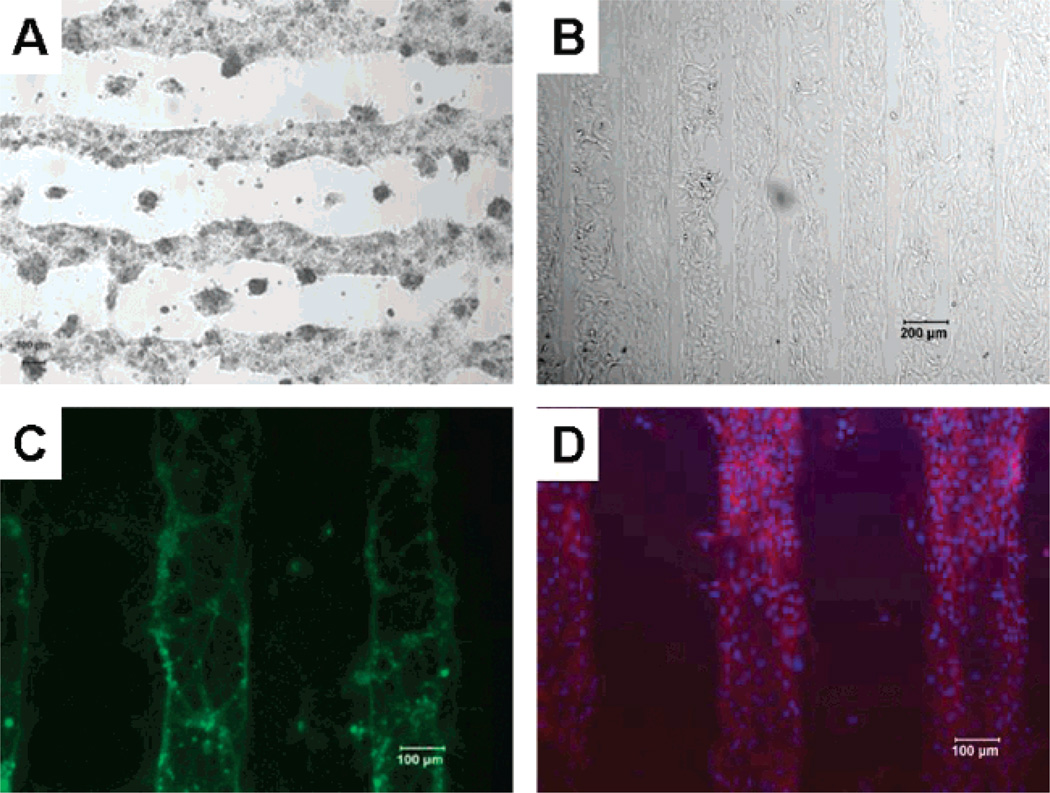
Figure 3.
Optical microscope images of directed deposition of cells on PEG patterns: (A) primary hepatocytes, (B) HeLa cells, (C) primary neurons, and (D) fibroblast.
Experimental Details
1. Materials
Sulfonated polystyrene (SPS) (Mw = 70 000), poly-(diallyldimethylammonium chloride) (PDAC) (Mw = 100 000–200 000) as a 20wt%solution, and sodium chloride were purchased from Aldrich Chemical, Milwaukee, WI. The m-dPEG acid molecule (Mw = 236) was obtained from Quanta Biodesign. Poly(dimethylsiloxane) (PDMS) from the Sylgard 184 silicone elastomer kit (Dow Corning, Midland, MI) was used to prepare stamps used in microcontact printing. Carboxyfluorescein (6-CF), fluorescence dye, was purchased and used as received from Sigma. Carboxylated polystyrene latex particles (4 µm diameter), purchased from Polysciences, were used for a colloidal adsorption study on m-dPEG self-assembled monolayer patterned polyelectrolyte templates. Dulbecco’s Modified Eagle Medium (DMEM) with 4.5 g/L glucose, 10×DMEM,fetal bovine serum (FBS), penicillin, and streptomycin were purchased from Life Technologies (Gaithersburg, MD). Insulin and glucagon were purchased from Eli Lilly and Co. (Indianapolis, IN); epidermal growth factor was purchased from Sigma Chemical (St. Louis, MO). Adult female Sprague-Dawley rats were obtained from Charles River Laboratories (Boston, MA). Actin cytoskeleton and focal adhesion staining kit was purchased from Chemicon (Temecula, CA).
2. Preparation of Polyelectrolyte Multilayers
Films were prepared as described in our earlier study.24 Briefly, 10.5 bilayers of PEMs were built using a Carl Zeiss slide stainer equipped with a custom-designed ultrasonic bath connected to a computer to perform layer-by-layer assembly. The stamping conditions were varied to optimize the microcontact printing of them-dPEG acid. The optimized conditions, as determined by us in our previous work, were used for making PEG patterns.28 The stamped regions were designed to act as resists to adsorption as the oligoethylene glycol graft chains of PEG did. In the procedure of creating complex 3-D microstructures, m-dPEG acid was stamped onto the PEM surface (PDAC surface) followed by a sequential adsorption layer-by-layer deposition process to build additional patterned polyelectrolyte multilayers outside the stamped region.
3. Ellipsometry
Ellipsometric measurements were obtained with a rotating analyzer ellipsometer (model M-44; J. A. Woollam) using WVASE32 software. Substrate parameters (n and k) were measured after absorption of MPA. This ensures that any changes in substrate reflectivity due to Au–S bonds will not affect subsequent measurements. The angle of incidence was 75° for all experiments. The thickness values for PEM films were determined using 44 wavelengths between 414.0 and 736.1 nm.
4. Cell Culture
Hepatocyte Isolation
Primary rat hepatocytes were isolated from 2 month old adult female Sprague-Dawley rats (Charles River Laboratories, Boston, MA) according to a two-step collagenase perfusion technique described by Seglen29 and modified by Dunn.27 The liver isolations yielded 150–300 × 106 hepatocytes. Using trypan blue exclusion the viability ranged from 90% to 98%. Primary hepatocyte culture medium consisted of DMEM supplemented with 10% FBS, 14 ng/mL glucagon, 20 ng/mL epidermal growth factor, 7.5 µg/mL hydrocortisone, 200 µg/mL streptomycin (10 000 µg/mL)-penicillin (10 000 U/mL) solution, and 0.5 U/mL insulin.
Hepatocyte Culture
The cells were seeded under sterile tissue culture hoods and maintained at 37 °C in a humidified air/CO2 incubator (90/10 vol %). Primary hepatocytes were cultured on PEM-coated 6-well TCPS coating PEG patterns. The multiplayer-coated TCPS plates were sterilized by spraying with 70% ethanol and exposing them to UV light before seeding the cells onto these surfaces. The cell culture experiments were performed on PEM surfaces without adhesive proteins. Collagen-coated TCPS and uncoated TCPS were used as controls in these studies. A collagen gel solution was prepared by mixing 9 parts of the 1.2 mg/mL collagen suspension in 1 mM HCl with 1 part of concentrated (10×) DMEM at 4 °C. The control wells were coated with 0.5 mL of this collagen gel solution, and the coated plates were incubated at 37 °C for 1 h. Freshly isolated hepatocytes were seeded at a concentration of 2 × 105 cells per well, and 2 mL was added to all surfaces studied. One milliliter of fresh medium was supplied daily to the cultures after removal of the supernatant. Samples were kept in a temperature-and humidity-controlled incubator.
4. NIH 3T3, HeLa Cell Culture
NIH 3T3 fibroblast and HeLa cell lines were purchased from American Tissue Type Collection. Cells grown to 70% confluency were trypsinized in 0.01% trypsin (ICN Biomedicals) solution in PBS for 10 min and re-suspended in 25 mL of media. Approximately 10% of the cells were seeded into a fresh tissue culture flask, and the rest of the cells were used for the co-culture experiments. Fibroblast medium consisted of DMEM with high glucose, supplemented with 10% bovine calf serum, 200U/mL penicillin, and 200µg/mL streptomycin.
5. Characterization
A Nikon Eclipse ME 600 optical microscope (Nikon, Melville, NY) was used to obtain dark-field images of the m-dPEG acid patterns and the additional microfabricated PEMs. A Nikon Eclipse E 400 microscope was used to obtain the fluorescence images. The 6-carboxyfluorescein (6-CF) dye was used to visualize the m-dPEG SAM patterns on PEM following the stamping and rinsing processes. The dye was dissolved directly in 0.1 M NaOH; samples were imaged by dipping the substrates into the dye solution. The dye, which is negatively charged, preferentially stained the positively charged PDAC surface. The dyed regions appear green when viewed with the fluorescence optical microscope using a FITC filter. Images were captured with a digital camera and processed on a Pentium computer running camera software.
Results and Discussion
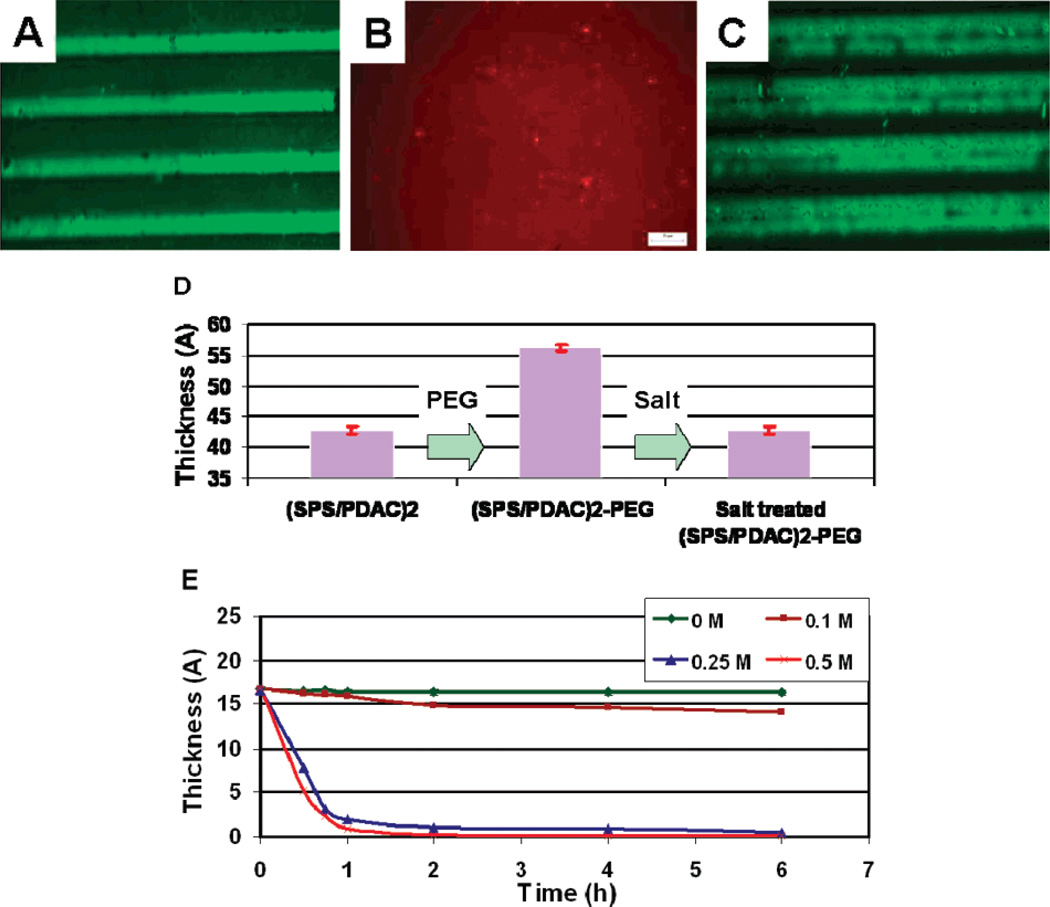
To evaluate the effect of salt on the m-dPEG acid patterns, the PEG patterns were treated with salt and the presence of PEG patterns was analyzed with fluorescence microscopy using a dye specific to positive surfaces. The dye attached selectively to the positive PDAC surface and resulted in patterns of green (PDAC) and dark (PEG) regions, thereby indicating the resistive property of the PEG patterns (Figure 2A). When this surface was exposed to a salt solution, the PEG patterns were removed from the PEM surface to expose the active PDAC surfaces underneath the SAMs as evident by the red dye specific for PDAC surfaces attaching homogeneously to the PEG free PEM films (Figure 2B). As proof-of-concept of the resistive nature of the PEG films, a layer of SPS was added on top of the PEG patterns; due to the presence of the PEG patterns, the SPS preferentially formed a layer on the non-PEG, PDAC surfaces, resulting in alternating regions of PEG and SPS surfaces. Treating the surface with salt removed the PEG patterns, resulting in alternating regions of PDAC (green) and SPS (dark) (Figure 2C). Further evidence of the salt-induced surface tunability of the PEG patterns was established using ellipsometry (Figure 2D). Results showed an increase of 13–15 Å in thickness when PEG SAMs were attached on the PEM films which upon exposure to salt decreased the film thickness to the original thickness, suggesting that the PEG coating was removed and the underlying PEM film was left intact. The effect of salt concentration was studied by monitoring the thickness of the PEG films by varying the ionic strengths over time (Figure 2E). The rate of PEG removal varied with the ionic strength. At lower salt concentrations (0 and 0.1 M) no significant change in the PEG thickness was observed over time, whereas at higher salt concentrations (0.25 and 0.5 M) an appreciable change in the PEG thickness was observed with 85% of the PEG removed upon exposure to salt for 1 h. The salt concentrations used included physiological levels (i.e., 0.25 M) and thus should not affect the biological molecules that may be attached to the PEMs.
Figure 2.
Fluorescent images of PEG patterns (A) before and (B,C) after salt treatment. (D) Ellipsometric data on the PEG patterns before and after salt treatment. (E) Thickness of the PEG patterns at varying salt concentration.
Directed Assembly of Cells
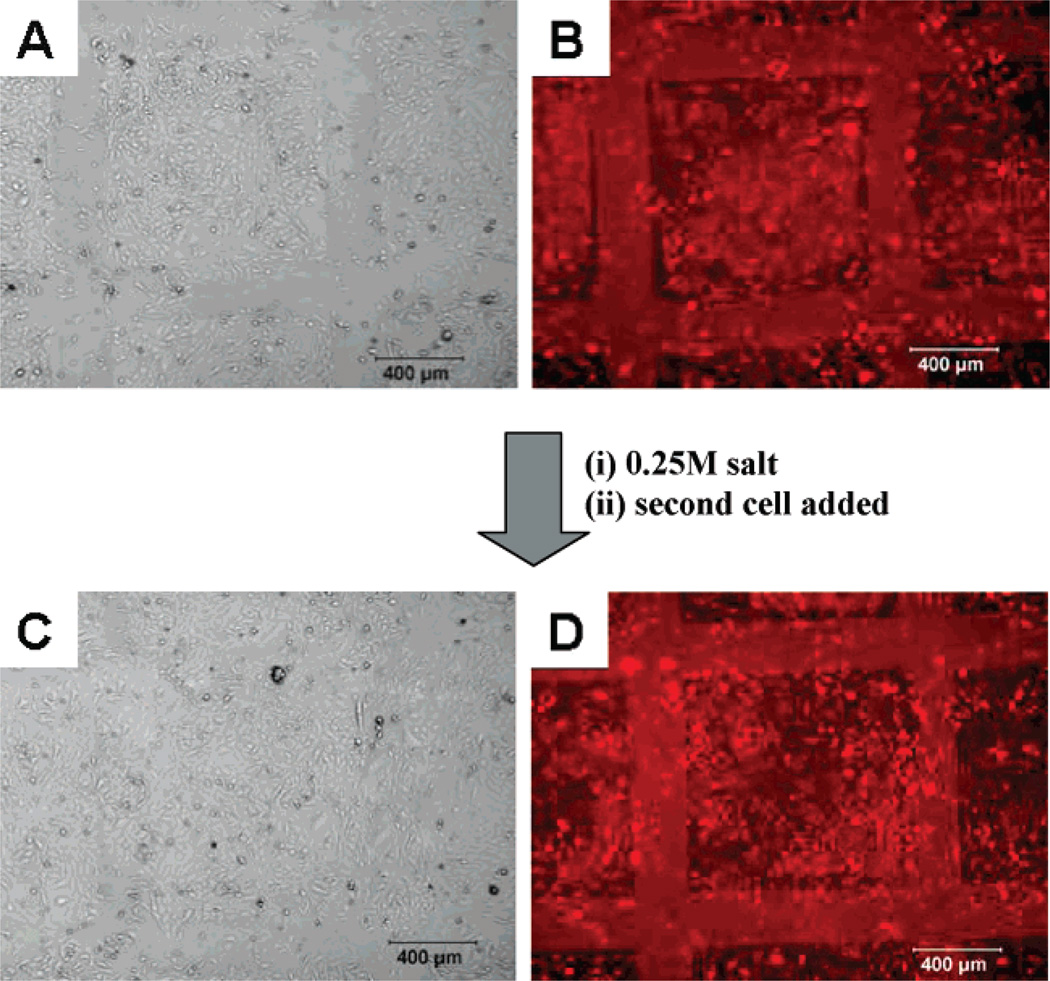
To illustrate the advantage of the salt tunable PEG patterns, we used the PEG patterns to control the adhesion of different types of cells including primary hepatocytes, primary neurons, HeLa cells, and fibroblasts (Figure 3). The removable PEG SAMs developed in this study provide surfaces that can be readily switched from cell repulsive to cell adhesive using cell-friendly conditions. This approach is advantageous since they can be used to form patterned co-cultures irrespective of the types of cell or the order of seeding of the different types of cells. As shown in Figure 3, HeLa cells attached onto the PEG patterns before and after salt treatment, indicating that the salt treatment did not affect the cells attached to the PEM surface. The same cell type was used to illustrate the advantages of this method and its ability to control the adhesion of similar cell types. This approach can be extended to different combinations of cell types for co-culturing (e.g., adherent versus nonadherent, eukaryotic versus prokaryotic). Figure 4A,B shows the phase contrast and fluorescence images of the directed assembly of FITC tagged HeLa cells on top of the PEG patterns, respectively, before salt treatment. When the cell-deposited surfaces were treated with salt, the cells remained attached and intact while the PEG SAMs were removed to expose the underlying active PDAC surface. Next, a second batch of HeLa cells without the fluorescent tag was cultured over the exposed surfaces to form patterned co-culture of HeLa cells (Figure 4C,D). When the cells were imaged, the HeLa cells seeded before salt treatment are visible while the cells added after salt treatment were not fluorescently tagged and thus are not observed under the fluorescence microscope.
Figure 4.
Optical microscope images of HeLa cells on PEG patterns before and after salt treatment. Phase contrast (A) and fluorescent images (B) of HeLa cells labeled red on the m-dPEG acid patterns before salt treatment. Phase contrast (C) and fluorescent images (D) of HeLa cells seeded onto the surface after salt treatment.
Directed Assembly of Colloidal Particles
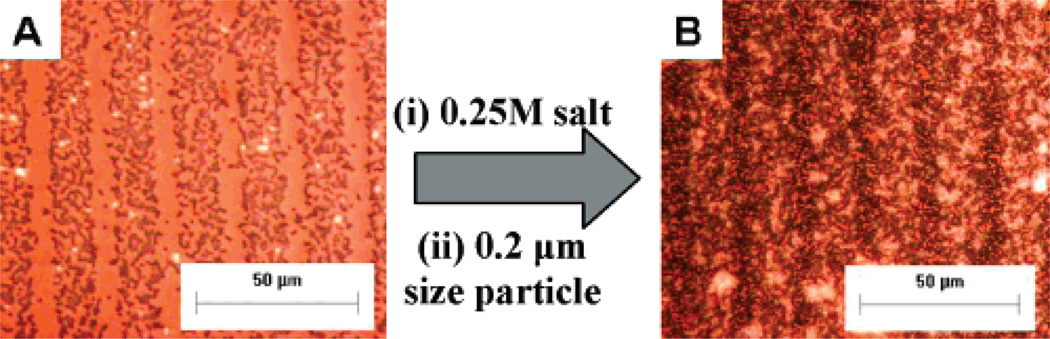
m-dPEG acid molecules were stamped on top of the (PDAC/SPS)10.5 and the negatively charged carboxylated polystyrene PS particles (Poly-sciences, diameter = 0.5 µm) deposited selectively over the positive (PDAC/SPS)10.5 surface but not on the m-dPEG self-assembled monolayer regions (Figure 5A). When the particle-deposited surfaces were treated with salt, the particles remained attached and intact while the PEG SAMs were removed, exposing the underlying active PDAC surface. Next, particles of 0.2 µm diameter were deposited over the exposed surfaces, resulting in a two-particle system (Figure 5B). In our study, both particles have similar chemistry but different sizes were used to illustrate the presence of two different sized particles on the surface. This approach is more universal, convenient, and nonspecific than the previous one by the Hammond group. This effective method provides a flexible and versatile approach to the fabrication of composite colloidal structures irrespective of their properties such as silica and metal-doped particles of varying size and surface functionality and using functionalized spheres modified with PEMs. This approach has potential applications in electronic and optical devices, direct colloid assembly, plastic electronics, and thin film power devices.
Figure 5.
Phase contrast images of colloidal particles on PEG patterns before and after salt treatment: (A) particles (D = 0.5 µm) on the m-dPEG acid patterns before salt treatment, (B) particles (D = 0.2 µm) added onto surface A after salt treatment.
Directed Assembly of Proteins
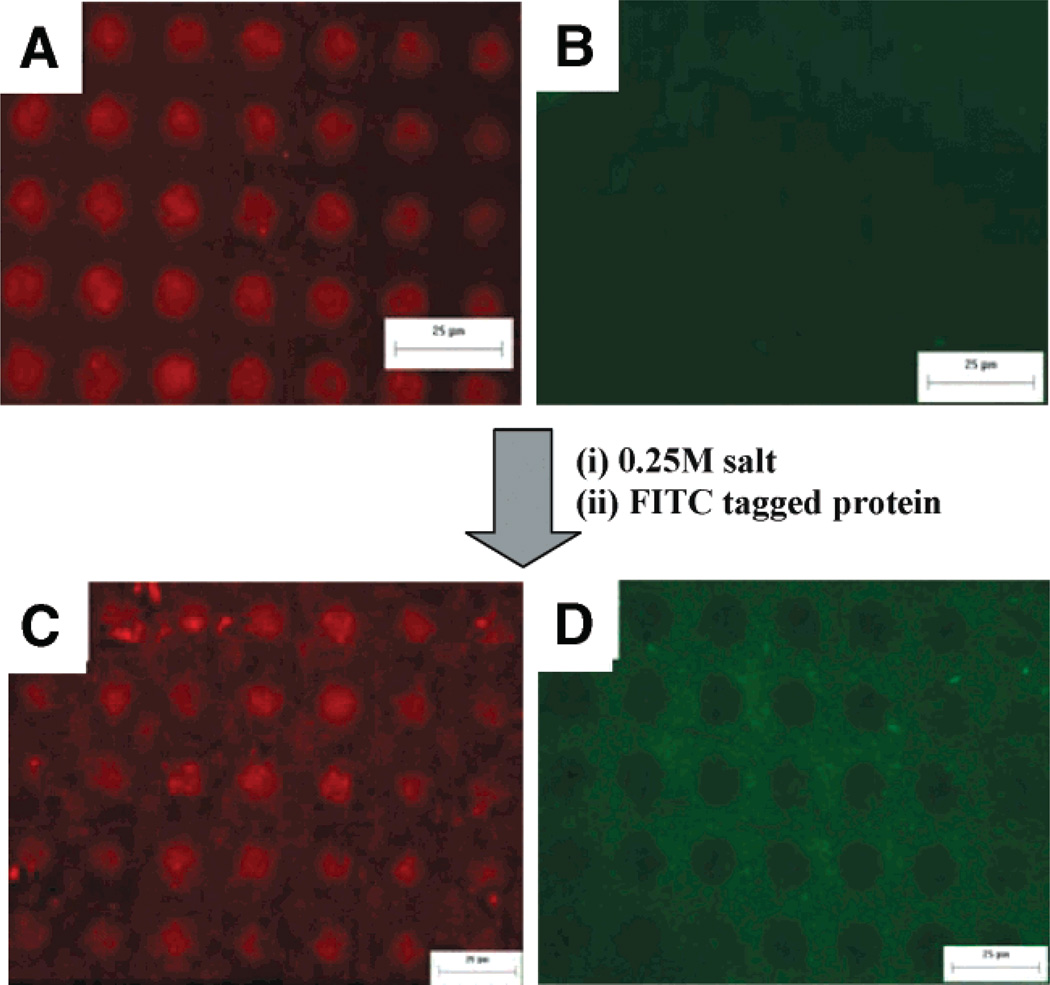
The removable PEG surfaces developed in this study provide a template for designing multiple regions of different proteins onto defined regions of a surface without exposing the proteins to irradiation, organic solvents, or dehydration. As shown in Figure 6A–D, the same protein, secondary alcohol dehydrogenase (sADH), with different fluorescent tags (FITC and Alexa Fluoro) was attached onto the PEG patterns before and after salt treatment, indicating that the salt treatment did not affect attachment of the proteins to the PEM surface. Figure 6A,B shows fluorescence images of the directed assembly of proteins on top of PEG patterns in both red and green channels before salt treatment. The red regions demonstrate the directed attachment of the alexa-fluoro tagged secondary alcohol dehydrogenase (sADH) proteins to the PEMs [(PDAC/SPS)10.5] and away from the resistive m-dPEG monolayer regions (black regions). No proteins were observed when imaged using the green channel. When the protein-deposited surfaces were treated with salt, the proteins remained attached and intact while the PEG SAMs were removed to expose the underlying active PDAC surface. Next FITC-tagged sADH were deposited over the exposed surfaces, resulting in a two-protein array (Figure 6C,D).
Figure 6.
Fluorescent images of sADH protein attachment on PEG patterns before and after salt treatment: (A, B) sADH tagged with Alexa-Fluoro on the m-dPEG acid patterns before salt treatment, and (C, D) sADH tagged with FITC added onto surface A after salt treatment. A and C are pictures taken using the red channel, while B and D were taken using the green channel.
Conclusions
The strategy presented here for preparation of removable resistive SAMs provides a template to design various surfaces that can be used in a wide range of applications. We have shown that these PEG patterns can be removed using physiological salt conditions that do not compromise the underlying polymers, charged particles, and biological molecules, including living cells, deposited on the surface. The salt-responsive PEG SAMs are ideal for optical technologies such as electroluminescent and conducting surfaces, where templates of multicomponent particle arrays on PEMs are required. We have also shown that these removable surfaces can be used to form patterns of multiple proteins and cells, which may be relevant to drug discovery, drug delivery, and tissue engineering applications. This new approach is an environmentally friendly and biocompatible route to designing versatile salt tunable surfaces.
Acknowledgment
This work was funded by the MSU Foundation, Michigan Economic Development Corporation, and in part by the NSF (BES 0222747, BES 0331297, BES 0425821, and CTS 0609164), NIH (1R01GM079688-01), Whitaker Foundation, and Environmental Protection Agency. We thank Lufang Sheng and Yifei Wu for isolating the primary hepatocytes and Deebika for isolating primary neurons. We also thank Srividhya Kidambi for help with ellipsometry and Brian Hassler for help with tagging the proteins.
References
- 1.Huang JY, Dahlgren DA, Hemminger JC. Photopatterning of self-assembled alkanethiolate monolayers on gold—A simple monolayer photoresist utilizing aqueous chemistry. Langmuir. 1994;10(3):626–628. [Google Scholar]
- 2.Kumar A, Biebuyck HA, Whitesides GM. Patterning self-assembled monolayers: Applications in materials science. Langmuir. 1994;10(5):1498–1511. [Google Scholar]
- 3.Bain CD, Whitesides GM. Molecular-level control over surface order in self-assembled monolayer films of thiols on gold. Science. 1988;240(4848):62–63. doi: 10.1126/science.240.4848.62. [DOI] [PubMed] [Google Scholar]
- 4.Langer R, Tirrell DA. Designing materials for biology and medicine. Nature. 2004;428(6982):487–492. doi: 10.1038/nature02388. [DOI] [PubMed] [Google Scholar]
- 5.Griffith LG, Naughton G. Tissue engineering—Current challenges and expanding opportunities. Science. 2002;295(5557):1009–1014. doi: 10.1126/science.1069210. [DOI] [PubMed] [Google Scholar]
- 6.Hubbell JA. Bioactive biomaterials. Curr. Opin. Biotechnol. 1999;10(2):123–129. doi: 10.1016/s0958-1669(99)80021-4. [DOI] [PubMed] [Google Scholar]
- 7.Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Soft lithography in biology and biochemistry. Annu. Rev. Biomed. Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 8.Lee K-B, Park S-J, Mirkin CA, Smith JC, Mrksich M. Protein nanoarrays generated by dip-pen nanolithography. Science. 2002;295(5560):1702–1705. doi: 10.1126/science.1067172. [DOI] [PubMed] [Google Scholar]
- 9.Lemmo AV, Rose DJ, Tisone TC. Inkjet dispensing technology: applications in drug discovery. Curr. Opin. Biotechnol. 1998;9(6):615–617. doi: 10.1016/s0958-1669(98)80139-0. [DOI] [PubMed] [Google Scholar]
- 10.Kidambi S, Lee I, Chan C. Controlling primary hepatocyte adhesion and spreading on protein-free polyelectrolyte multilayer films. J. Am. Chem. Soc. 2004;126(50):16286–16287. doi: 10.1021/ja046188u. [DOI] [PubMed] [Google Scholar]
- 11.Bhatia SN, Yarmush ML, Toner M. Controlling cell interactions by micropatterning in co-cultures: hepatocytes and 3T3 fibroblasts. J. Biomed. Mater. Res. 1997;34(2):189–99. doi: 10.1002/(sici)1097-4636(199702)34:2<189::aid-jbm8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 12.Bhandari R, Riccalton L, Lewis A, Fry J, Hammond A, Tendler S, Shakesheff K. Liver tissue engineering: A role for co-culture systems in modifying hepatocyte function and viability. Tissue Eng. 7. 2001;(3):345–357. doi: 10.1089/10763270152044206. [DOI] [PubMed] [Google Scholar]
- 13.Folch A, Toner M. Microengineering of cellular interactions. Annu. Rev. Biomed. Eng. 2000;2:227–256. doi: 10.1146/annurev.bioeng.2.1.227. [DOI] [PubMed] [Google Scholar]
- 14.Ito Y. Surface micropatterning to regulate cell functions. Biomaterials. 1999;20(23/24):2333–2342. doi: 10.1016/s0142-9612(99)00162-3. [DOI] [PubMed] [Google Scholar]
- 15.Orth RN, Clark TG, Craighead HG. Biosensors: Avidin-biotin micropatterning methods for biosensor applications. Biomed. Microdevices. 2003;5(1):29–34. [Google Scholar]
- 16.Nicolau DV, Taguchi T, Taniguchi H, Yoshikawa S. Microsize protein patterning on diazonaphthoquinone/novolak thin polymeric films. Langmuir. 1998;14(7):1927–1936. [Google Scholar]
- 17.Yang Z, Chilkoti A. Microstamping of a biological ligand onto an activated polymer surface. Adv. Mater. 2000;12(6):413–417. [Google Scholar]
- 18.Doh J, Irvine DJ. Photogenerated polyelectrolyte bilayers from an aqueous-processible photoresist for multicomponent protein patterning. J. Am. Chem. Soc. 2004;126(30):9170–9171. doi: 10.1021/ja048261m. [DOI] [PubMed] [Google Scholar]
- 19.Holden MA, Cremer PS. Light activated patterning of dye-labeled molecules on surfaces. J. Am. Chem. Soc. 2003;125(27):8074–8075. doi: 10.1021/ja035390e. [DOI] [PubMed] [Google Scholar]
- 20.Blawas AS, Oliver TF, Pirrung MC, Reichert WM. Step-and-repeat photopatterning of protein features using caged-biotin-BSA: Characterization and resolution. Langmuir. 1998;14(15):4243–4250. [Google Scholar]
- 21.Lee K-B, Lim J-H, Mirkin CA. Protein nanostructures formed via direct-write dip-pen nanolithography. J. Am. Chem. Soc. 2003;125(19):5588–5589. doi: 10.1021/ja034236p. [DOI] [PubMed] [Google Scholar]
- 22.Tien J, Nelson CM, Chen CS. Fabrication of aligned microstructures with a single elastomeric stamp. Proc. Natl. Acad. Sci. U.S.A. 2002;99(4):1758–1762. doi: 10.1073/pnas.042493399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sorribas H, Padeste C, Tiefenauer L. Photolithographic generation of protein micropatterns for neuron culture applications. Biomaterials. 2001;23(3):893–900. doi: 10.1016/s0142-9612(01)00199-5. [DOI] [PubMed] [Google Scholar]
- 24.Kidambi S, Chan C, Lee I. Selective depositions on polyelectrolyte multilayers: Self-assembled monolayers of m-dPEG acid as molecular templates. J. Am. Chem. Soc. 2004;126(14):4697–4703. doi: 10.1021/ja039359o. [DOI] [PubMed] [Google Scholar]
- 25.Jiang XP, Ortiz C, Hammond PT. Exploring the rules for selective deposition: Interactions of model polyamines on acid and oligoethylene oxide surfaces. Langmuir. 2002;18(4):1131–1143. [Google Scholar]
- 26.Zheng HP, Rubner MF, Hammond PT. Particle assembly on patterned “plus/minus” polyelectrolyte surfaces via polymer-on-polymer stamping. Langmuir. 2002;18(11):4505–4510. [Google Scholar]
- 27.Zheng HP, Lee I, Rubner MF, Hammond PT. Two component particle arrays on patterned polyelectrolyte multilayer templates. Adv. Mater. 2002;14(8):569–572. [Google Scholar]
- 28.Seglen PO. Preparation of isolated rat liver cells. Methods Cell Biol. 1976;13:29–83. doi: 10.1016/s0091-679x(08)61797-5. [DOI] [PubMed] [Google Scholar]
- 29.Dunn JC, Tompkins RG, Yarmush ML. Long-term in vitro function of adult hepatocytes in a collagen sandwich configuration. Biotechnol. Prog. 1991;7(3):237–245. doi: 10.1021/bp00009a007. [DOI] [PubMed] [Google Scholar]