Abstract
Estrogen-related receptor α (ERRα) is one of the first orphan nuclear receptors to be identified, yet its physiological functions are still unclear. We show here that ERRα is an effector of the transcriptional coactivator PGC-1α [peroxisome proliferator-activated receptor γ (PPARγ) coactivator 1α], and that it regulates the expression of genes involved in oxidative phosphorylation and mitochondrial biogenesis. Inhibition of ERRα compromises the ability of PGC-1α to induce the expression of genes encoding mitochondrial proteins and to increase mitochondrial DNA content. A constitutively active form of ERRα is sufficient to elicit both responses. ERRα binding sites are present in the transcriptional control regions of ERRα/PGC-1α-induced genes and contribute to the transcriptional response to PGC-1α. The ERRα-regulated genes described here have been reported to be expressed at reduced levels in humans that are insulin-resistant. Thus, changes in ERRα activity could be linked to pathological changes in metabolic disease, such as diabetes.
Estrogen-related receptor α (ERRα, NR3B1) was identified on the basis of its sequence similarity to classical, hormoneregulated steroid receptors (1). Based on its ability to recognize similar DNA sequences as the estrogen receptors, ERRα has been proposed to modulate estrogen signaling (2–5). ERRα may also regulate bone formation, given that it is highly expressed at ossification sites, promotes osteoblast differentiation in vitro, and activates the promoter of the bone matrix protein osteopontin (6, 7). Finally, ERRα may regulate fatty acid oxidation. Consistent with this function, ERRα is prominently expressed in tissues with high capacity for β-oxidation of fatty acids, such as brown fat, heart, muscle, and kidney, and induces the expression of the medium-chain acyl-CoA dehydrogenase gene (8, 9).
A better understanding of the transcriptional programs and cellular pathways that depend on ERRα has been hampered by the lack of tools to regulate the activity of this receptor. Despite the high similarity between ERRα and other ligand-dependent nuclear receptors, it is not clear whether ERRα activity is regulated by small lipophilic ligands. Compounds that inhibit ERRα-dependent transcription, such as toxaphene, chlordane, and diethylstilbestrol, have been described (10, 11). However, these compounds are not specific enough for ERRα to facilitate studies of its cellular function. Recently, we demonstrated that the transcriptional coactivator peroxisome proliferator-activated receptor γ (PPARγ) coactivator 1α (PGC-1α) regulates ERRα function (12). PGC-1α induces the expression of ERRα and interacts physically with ERRα, enabling it to activate transcription (12, 13). These findings suggest that PGC-1α can be used as a protein “ligand” to regulate ERRα-dependent transcription and study ERRα function.
PGC-1α has been identified as a tissue-specific coactivator of nuclear receptors (14–16). The expression of PGC-1α is most prominent in tissues with high energy demands, similar to the expression pattern of ERRα (12, 16). PGC-1α mRNA levels are induced in response to signals that relay metabolic needs, such as exposure to cold, fasting, and physical exercise (reviewed in refs. 15 and 16). Strikingly, increases in PGC-1α levels seem sufficient to induce cellular pathways important for energy metabolism, including adaptive thermogenesis, mitochondrial biogenesis, and fatty acid oxidation (14, 17–19). This is accomplished by the interaction of PGC-1α with transcription factors, which recruit PGC-1α to target DNA regulatory sequences and enable the induction of genes important in energy metabolism pathways. Transcription factors that guide PGC-1α action to specific genes include nuclear receptors, members of other transcription factor families, such as nuclear respiratory factor (NRF)-1, which controls the expression of mitochondrial proteins, and myocyte enhancer factors 2C/2D (15, 16).
The recent identification of ERRα as a protein that is coexpressed with and induced and activated by PGC-1α suggests that ERRα plays a role in some of the known PGC-1α-regulated pathways. Consistent with this hypothesis, we show here that ERRα and PGC-1α cooperate to induce mitochondrial biogenesis.
Materials and Methods
Adenoviruses and Plasmids. Adenoviral vectors expressing GFP, PGC-1α, small interfering RNA (siRNA) for ERRα, and control AdSUPER have been described (12). Adenoviruses expressing ERRα or ERRα fused to the VP16 activation domain (VP16-ERRα) were constructed by using the insert of pSG5-mERRα or pSG5-ΔA/BmERRα, respectively (6). For the reporter plasmids, human genomic DNA and gene-specific oligonucleotides (Table 1, which is published as supporting information on the PNAS web site) were used to amplify the sequences -385 to +90 and -686 to +55 (relative to transcription initiation site) of the ATP synthase β (ATPsynβ) and cytochrome c, somatic (Cyt c) genes, respectively. The PCR products were cloned upstream of the luciferase coding sequences of pGL3-Basic (Promega). Mutations and deletions were introduced by fusion PCR (20). The ERR response elements (ERR Es) at AT Psynβ/-338 (CCAAGGACA), Cyt c/-596 (ACAAGGTCA), and Cyt c/-9 (CCAAGGACA) were changed to CCAgatctt, ACAgatctA and CCAgatctA, respectively. The NRF-2 binding sites of ATPsynβ were deleted by removing sequences -300 to -270; the NRF-1 binding site of Cyt c (CCAGCATGCGCG) was changed to CCAGgATcCaac.
Cell Culture and Transfections. Cells were cultured in DMEM supplemented with 9% charcoal-stripped FCS. SAOS2 (SAOS2-GR(+) in ref. 12) cells were infected with adenoviruses at a multiplicity of infection (moi) of 20–100. COS7 cells were transfected by calcium phosphate precipitation and analyzed as described (21). The amounts of plasmids per transfection were 100 ng of the reporters pCytc/-686Luc or pATPsynβ/-385Luc, 100 ng pcDNA3/HA-PGC-1α (21), and 50 ng pcDNA3/ERRα (22).
cRNA Preparation and Array Hybridization. Total RNA (10 μg) was reverse transcribed with the SuperScript Choice system (Invitrogen). The cDNA (1 μg) was in vitro transcribed by using the Enzo BioArray High Yield RNA system (Enzo Diagnostics). The cRNA (10 μg) was fragmented and hybridized to a HG-U133A GeneChip (Affymetrix, Santa Clara, CA) by using standard procedure (45°C, 16 h). Washing and staining were performed in a Fluidics Station 400 (Affymetrix) by using the protocol EukGE-WS2v4 and scanned in an Affymetrix GeneChip 2500 scanner.
Microarray Analysis. Data from three experiments were analyzed with microarray suite 5 software (Affymetrix) and gene-spring 5.1 (Silicon Genetics, Redwood City, CA). Changes in gene expression were assessed by looking for concordant changes between replicates by using a signed Wilcoxon rank test. The “change” P-value threshold was <0.003. Genes whose detection P-value was >0.05 in all experimental conditions were excluded from the analysis. Genes that reproducibly changed in the same direction were subjected to a one-way ANOVA test (P < 0.05) with a Benjamini and Hochberg multiple testing correction. Classification into genes encoding mitochondrial proteins was based on annotations of the Affymetrix NetAffx Analysis Center, SOURCE and the National Center for Biotechnology Information PubMed, and the OXPHOS and human_mitoDB_6_2002 lists curated at the Whitehead Institute Center for Genome Research (23).
DNA Isolation and Quantification. Total DNA was prepared according to standard procedures and digested with 100 μg/ml RNase A for 30 min at 37°C. The relative copy numbers of mitochondrial and nuclear DNA were determined by real-time PCR with primers specific to the COX2 (mitochondrial) and β actin (nuclear) genes (Table 1), 2 ng DNA, and the Light Cycler system (Roche Diagnostics). Serial dilutions of DNA from uninfected cells were analyzed in parallel to establish a standard curve. Quantification was as described (24).
RNA Analysis. Isolation of RNA, conversion to cDNA, and quantification of transcripts by real-time PCR by using the Light Cycler system (Roche Diagnostics) and gene-specific primers (Table 1) have been described (24).
Western Analysis. Cell lysates were subjected to Western analysis by using antibodies against PGC-1α (12) and ERRα (3).
Labeling of Mitochondria and Flow Cytometry. Cells were incubated, first with 500 nM CM-H2XRos or 500 nM MitoFluor Red 594 (Molecular Probes) in culture medium for 30 min and, second, in fresh, dye-free medium for 30 min at 37°C. CM-H2XRos-labeled mitochondria were visualized by fluorescence microscopy. MitoFluor Red 594-labeled cells were analyzed by flow cytometry (FACSCalibur, Becton Dickinson), by using the software winmdi 2.8.
In Silico Analysis for ERREs. Thirty-five sequences reported to bind ERRα (3, 4, 8, 25) were aligned by using clustalw and used to compile a position-weighted nucleotide distribution matrix. Crossvalidation of the matrix revealed a mean and median score for the 35 sequences of 0.915 and 0.946, with a maximum at 0.994 and a minimum of 0.695, the best possible score being 1. For candidate genes, 5 kb of 5′ upstream region sequence were searched for matches to the matrix, by using a variant of the nubiscan algorithm (26).
Electrophoretic Mobility Shift Assay. In vitro translated ERRα (0.5 μl) [T7 Coupled Reticulocyte system (Promega)] or unprogrammed lysate was incubated in 20 μl of buffer (10 mM Hepes, pH 7.5/2.5 mM MgCl2/50 mM EDTA/1 mM DTT/6% glycerol) with 1 ng of 32P-end-labeled oligonucleotide probe and 1 μg of poly dI:dC, in the absence or presence of 100 ng of unlabeled oligonucleotide competitor (Table 1). Complexes were resolved in 6% native polyacrylamide gels.
Results
PGC-1α Induces Mitochondrial Biogenesis in SAOS2 Cells Through a Pathway That Requires Interaction with Nuclear Receptors. To identify the cellular programs that are regulated by PGC-1α in SAOS2 cells and where ERRα could play a role, we used high-density oligonucleotide arrays and compared the RNA profiles of cells expressing PGC-1α to those of control cells. Seventeen hours after infection with a PGC-1α expressing adenovirus, 151 of the up-regulated transcripts were classified as nuclear genes encoding mitochondrial proteins (Table 2, which is published as supporting information on the PNAS web site). These genes define “mitochondrial functions” that are upregulated in the early phase of the PGC-1α-induced response (≈12 h after PGC-1α protein becomes detectable), and encode proteins with roles in many facets of mitochondrial biogenesis and function, including mitochondrial protein synthesis (20 genes), transport across the mitochondrial membrane (17 genes), fatty acid oxidation (8 genes), the tricarboxylic acid cycle (17 genes), and oxidative phosphorylation (55 genes) (Table 2). An additional 23 of the up-regulated transcripts represent genes that do not encode known mitochondrial proteins but have been reported as coregulated with “mitochondrial genes” and proposed to carry functions relevant to mitochondrial biology (27) (Table 2). PGC-1α also induced the expression of mitochondrial transcription and translation factor A (mtTFA). Interestingly, PGC-1α did not affect the expression of NRF-1 or NRF-2 (Table 2), the transcription factors that regulate the expression of many nuclear genes encoding mitochondrial proteins and that are induced by PGC-1α in C2C12 cells (17). We concluded that PGC-1α induces the gene expression program of mitochondrial biogenesis in SAOS2 cells in a manner that differs from the NRF-1 pathway described in C2C12 cells (17).
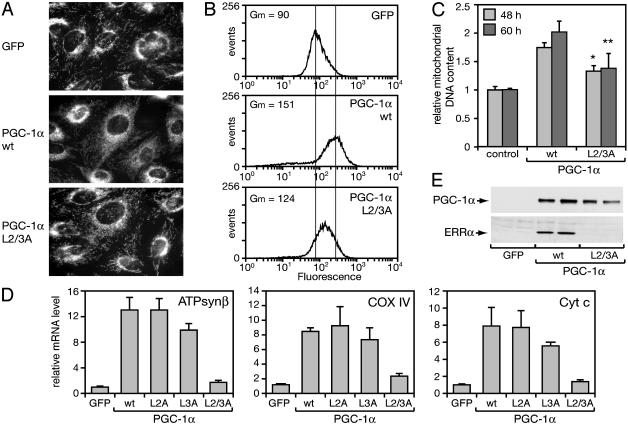
To determine whether the PGC-1α-mediated induction of mitochondrial proteins led to an increase in mitochondrial content, SAOS2 cells were stained with MitoTracker, a dye that accumulates specifically in respiring mitochondria. Mitochondria in control cells infected with a GFP-expressing adenovirus had a characteristic tubular appearance and were concentrated around the nuclei, similar to mitochondria in noninfected cells (Fig. 1A and data not shown). Expression of PGC-1α led to a distinct mitochondrial reticulum, which filled the cytoplasm. The increased mean fluorescence intensity in PGC-1α expressing cells (geometric mean = 151) compared with control cells (geometric mean = 90) was consistent with an increase in mitochondrial content (Fig. 1B). To measure mitochondrial DNA directly, we isolated total DNA and determined the relative copy number of mitochondrial DNA by quantitative PCR. PGC-1α expression led to an increase in mitochondrial DNA content per cell, by 1.7- and 2-fold, at 48 and 60 h, respectively (Fig. 1C).
Fig. 1.
PGC-1α induces mitochondrial biogenesis in SAOS2 cells, dependent on interaction with nuclear receptors. Cells were infected with GFP- or (WT or mutant L2/3A) PGC-1α-expressing adenoviruses at an moi of 40. (A) Mitochondria in cells labeled with CM-H2Xros were imaged 48 h after infection. (B) Accumulation of MitoFluor Red 594 in cells was measured by flow cytometry 48 h after infection. Gm represents the geometric mean fluorescence intensity of 20,000 cells. (C) Mitochondrial (COX2) DNA levels normalized to nuclear (β actin) DNA levels are expressed relative to levels in control cells expressing GFP, which were set to 1, at 48 and 60 h after infection. Data are the mean ± SEM of three experiments performed in duplicates. *, P < 0.0001 versus WT PGC-1α at 48 h; **, P < 0.001 versus WT PGC-1α at 60 h, as determined by the Student's t test. (D) mRNA levels of ATPsynβ, Cyt c, and COX4 at 48 h after infection were determined by quantitative RT-PCR, normalized to the mRNA levels of 36B4, and expressed relative to levels in GFP-infected cells. (E) Protein levels of PGC-1α and ERRα were determined by Western analysis at 48 h after infection.
PGC-1α interacts with nuclear receptors through two leucinerich motifs. Leucine motif 2 (L2) mediates interaction with most nuclear receptors, including ERRα, whereas motif 3 (L3) recognizes specifically ERRα and the related receptors ERRβ and ERRγ (12, 13, 28). Mutation of L2 and L3 (mutation L2/3A) disrupts interactions with nuclear receptors, without affecting the interaction domains for other factors, like NRF-1 and myocyte enhancer factor 2C (15, 16). To determine the role of nuclear receptors in PGC-1α-induced mitochondrial biogenesis, we tested the effect of the L2/3A mutation. As seen in Fig. 1 A–C, the PGC-1α variant L2/3A showed a reduced ability to induce mitochondria when compared with WT PGC-1α. The L2/3A PGC-1α was also deficient in inducing the expression of nuclear genes encoding mitochondrial proteins and of ERRα (Fig. 1 D and E). The single L2A mutant, which is defective for interactions with peroxisome proliferator-activated receptors, the glucocorticoid receptor, and thyroid hormone receptors (TR), but retains interactions with ERRα (12, 13, 18, 29, 30), was as active as WT PGC-1α in inducing the expression of target genes (Fig. 1D and data not shown). We concluded that interactions of PGC-1α with nuclear receptors and potentially ERRα are important for PGC-1α to induce the program of mitochondrial biogenesis.
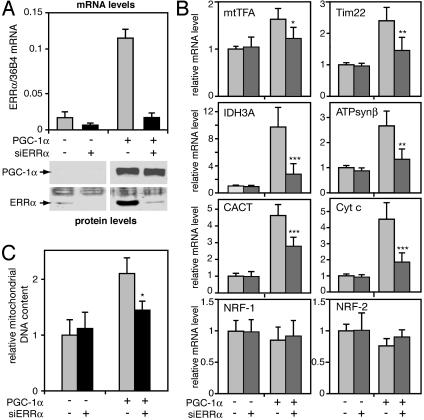
ERRα Expression Is Required for the PGC-1α-Induced Mitochondrial Biogenesis. To address the involvement of ERRα specifically, we compared the ability of PGC-1α to induce genes encoding mitochondrial proteins in cells that express endogenous ERRα and in cells in which ERRα expression was inhibited by siRNA. As seen in Fig. 2A, siRNA specific for ERRα abolished the basal expression of ERRα in the absence of PGC-1α and reduced strongly the induction of ERRα by PGC-1α. Under these conditions, we determined the mRNA levels of PGC-1α up-regulated genes that carry important roles in different aspects of mitochondrial biogenesis and function: mtTFA (mitochondrial DNA replication and transcription), Tim22 (protein import into mitochondria), isocitrate dehydrogenase α (IDH3A; tricarboxylic acid cycle), carnitine/acylcarnitine translocase (fatty acid oxidation), and Cyt c and ATPsynβ (oxidative phosphorylation). For all six genes, PGC-1α expression led to increases in their mRNA levels when endogenous ERRα levels were not perturbed, confirming results from the arrays. Inhibition of ERRα expression by siRNA reduced significantly the ability of PGC-1α to induce these genes, without affecting basal levels in the absence of PGC-1α (Fig. 2B). Because the siRNA diminished but did not abolish ERRα expression, the remaining induction by PGC-1α could still be mediated by the low levels of ERRα (Fig. 2 A) as well as by other pathways. Inhibition of ERRα did not prevent PGC-1α from inducing glucocorticoid receptor targets, such as p21 (data not shown), or affect the mRNA levels of the transcription factors NRF-1 and NRF-2 (Fig. 2B).
Fig. 2.
Inhibition of ERRα expression impairs the induction of mitochondrial biogenesis by PGC-1α. SAOS2 cells were infected with control-(AdSUPER) or an adenovirus-expressing siRNA for ERRα (siERRα) at a moi of 100. Two days later, cells were infected with GFP- or PGC-1α-expressing adenoviruses at a moi of 20 (A and B) or 40 (C). Cells were harvested 24 h (A and B) or 48 h (C) later. (A) ERRα mRNA levels were determined by quantitative RT-PCR and normalized to 36B4 levels. Data shown are the mean ± SEM of three experiments performed in duplicates. (B) mRNA levels for mtTFA, Tim22, IDH3A, ATPsynβ, carnitine/acylcarnitine translocase (CACT), Cyt c, NRF-1, and NRF-2, were determined by quantitative RT-PCR, normalized to the mRNA levels of 36B4, and expressed relative to levels in AdSUPER/GFP infected cells. Data are the mean ± SEM of three experiments performed in duplicates. *, P < 0.02; **, P < 0.003; ***, P < 0.0005 versus PGC-1α-expressing cells in the absence of siERRα. (C) Mitochondrial (COX2) DNA levels were normalized to nuclear (β actin) DNA levels and expressed relative to levels in control-infected (AdSU-PER and GFP) cells, which were set to 1. Data are the mean ± SEM of two experiments performed in duplicates. *, P < 0.008 versus PGC-1α-expressing cells in the absence of siERRα.
The requirement of ERRα for the induction of genes, such as mtTFA and Tim22, suggests that ERRα is required for PGC-1α-dependent mitochondrial biogenesis. Indeed, inhibition of ERRα expression significantly diminished the ability of PGC-1α to increase mitochondrial DNA content (Fig. 2C). Inhibition of ERRα had no effect on mitochondrial DNA in the absence of PGC-1α, leading us to conclude that ERRα contributes to the PGC-1α-mediated induction but not the basal expression of genes important in mitochondrial biogenesis.
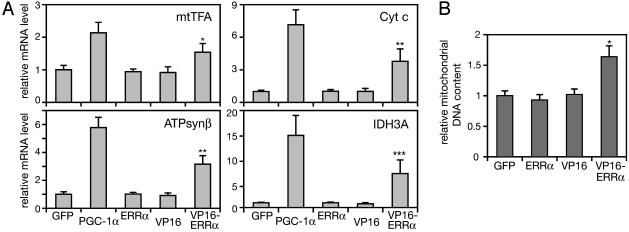
A Constitutive Form of ERRα Induces Mitochondrial Biogenesis in the Absence of PGC-1α. The lack of effect of ERRα on the basal expression of genes encoding mitochondrial proteins could reflect the low levels of ERRα in the absence of PGC-1α (Fig. 2 A), the low transcriptional activity of ERRα in the absence of PGC-1α, and/or the requirement for other PGC-1α-dependent pathways that enable the induction of these genes. To address these possibilities, we determined the effect of overexpression of ERRα or of ERRα endowed with a heterologous strong transcriptional activation domain in the absence of PGC-1α. ERRα, VP16-ERRα, and, as control, the VP16 activation domain alone, were expressed in SAOS2 cells by using adenoviral vectors. As seen in Fig. 3, neither ERRα nor VP16 by itself induced the expression of mtTFA, ATPsynβ, Cyt c, or IDH3A. In contrast, VP16-ERRα induced all four genes to ≈50% of the PGC-1α-induced levels (Fig. 3A). VP16-ERRα also led to a significant increase in the amount of cellular mitochondrial DNA (Fig. 3B), indicating that ERRα is capable of inducing mitochondrial biogenesis in the absence of PGC-1α, if activated by other means.
Fig. 3.
A constitutively active ERRα induces mitochondrial biogenesis. SAOS2 cells were infected with adenoviruses expressing GFP, PGC-1α, ERRα, VP16, or VP16-ERRα (moi 40) and analyzed 24 h (A) or 60 h (B) later. (A) mRNA levels for the indicated genes were determined by quantitative RT-PCR, as in Fig. 1D. Data are the mean ± SEM of three experiments performed in duplicates. *, P < 0.002; **, P ≤ 0.0001; ***, P < 0.0004 versus GFP-infected cells. (B) Mitochondrial DNA content was determined as in Fig. 1C. Data are the mean ± SEM of two experiments performed in triplicates. *, P < 0.0001 versus GFP-infected cells.
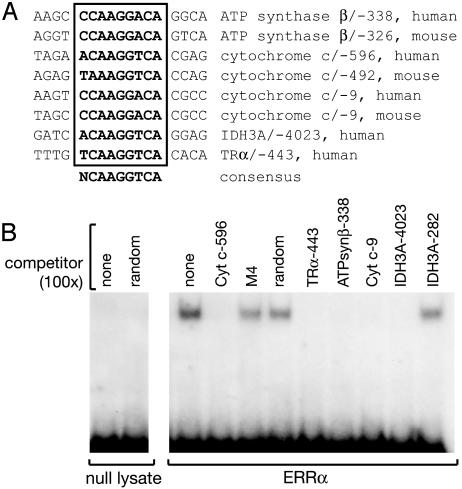
ERRα Binds to Regulatory Sites in the Promoters of ATPsynβ and Cyt c. We next asked whether ERRα acts directly at the promoters of genes encoding mitochondrial proteins. ERRα binds to DNA sites with the consensus sequence TCAAGGTCA, termed ERREs (3, 4, 6, 8). Analysis of the promoter and upstream regulatory sequences of 18 genes that are induced by PGC-1α indicated the presence of putative ERREs in most of them (data not shown). We focused on ATPsynβ and Cyt c, whose promoters have been studied (31–33) and where the putative ERREs are within 1 kb of the characterized transcription initiation sites (Fig. 4A). First, we tested whether ERRα binds to these sites in a gel mobility shift assay. In vitro-translated ERRα formed a specific complex with an oligonucleotide representing the putative ERRE at -596 bp of the Cyt c promoter (Fig. 4B). The complex was inhibited by a 100-fold excess of an oligonucleotide bearing a known ERRE from the TRα promoter (25) and oligonucleotides representing the candidate ERREs from the Cyt c/-9, ATPsynβ/-338, and IDH3A/-4,023 but not by oligonucleotides harboring a mutated TRα ERRE (M4), a random sequence, or another site of the IDH3A gene.
Fig. 4.
ERRα recognizes sites in ATPsynβ and Cyt c regulatory sequences. (A) Sequences of candidate ERREs identified by in silico analysis. The TRα/-443 ERRE has been described (25). (B) Electrophoretic mobility shift assay. ERRα was incubated with a 32P-labeled oligonucleotide containing the ERRE of Cyt c/-596 in the presence of unlabeled oligonucleotides as indicated. M4 oligonucleotide has the TRα/-443 sequence with a 2-bp substitution in the core ERRE (25).
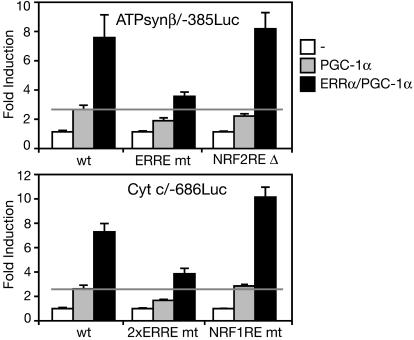
To test the significance of the ERRα binding sites for the induction of ATPsynβ and Cyt c, we measured the response of these two promoters to PGC-1α/ERRα in COS7 cells. PGC-1α induced the ATPsynβ and Cyt c promoters, driving the expression of luciferase by 2.7- and 2.6-fold (Fig. 5). Coexpression of ERRα enhanced further the induction, to 7.6- and 7.3-fold. Mutations in the ERREs decreased the response to PGC-1α and PGC-1α/ERRα by 40–50% (Fig. 4C) without affecting the basal levels of expression in the absence of PGC-1α (data not shown). The ATPsynβ and Cyt c promoters harbor also binding sites for NRF-2 and NRF-1, respectively (32, 33). Deletion of the NRF-2 site in the ATPsynβ promoter caused a drop in basal expression levels (by 40%) and a small decrease in the response to PGC-1α but did not affect the response to ERRα. Mutations in the NRF-1 site of Cyt c also reduced basal levels of expression by 40% but did not decrease the response to PGC-1α or PGC-1α/ERRα (Fig. 5). Taken together, our results indicate that the promoters of the two genes are responsive to ERRα and that the identified ERREs contribute to, but are not solely responsible for, the induction by PGC-1α and ERRα.
Fig. 5.
The ERREs of ATPsynβ and Cyt c contribute to the transcriptional response to PGC-1α. COS7 cells were transfected with reporters pATPsynβ/-385Luc or pCytc/-686Luc, [WT and bearing mutations (mt) or deletions (Δ) at the ERREs and NRF-1/NRF-2 binding sites] and control vector (-), PGC-1α-, and/or ERRα-expressing plasmids as indicated. Data are expressed as fold activation by PGC-1α or PGC-1α/ERRα, with the basal activity of each construct (white bars) set to 1, and are the mean ± SEM of at least three experiments performed in duplicates.
Discussion
PGC-1α has been shown previously to induce mitochondrial biogenesis and oxidative metabolism in muscle cells, adipocytes, and cardiomyocytes (14, 17, 19). These studies also provided evidence that the transcription factors NRF-1 and NRF-2 mediate the effects of PGC-1α on the expression of nuclear genes encoding mitochondrial proteins (17). We now show that PGC-1α expression in SAOS2 cells, osteoblast progenitors with adipocyte differentiation capacity (34), also induces mitochondrial biogenesis. Interestingly, this PGC-1α-driven program depends on the induction and activation of the orphan nuclear receptor ERRα. Moreover, in the absence of PGC-1α, a constitutively active ERRα induces mitochondrial biogenesis and the expression of genes essential for oxidative phosphorylation. Our findings demonstrate a role for ERRα in the control of mitochondrial biogenesis and function and suggest that, depending on the cell type, ERRα activity is necessary and sufficient to induce mitochondrial biogenesis. Consistent with these findings, RNA profiling studies have recently shown a tight correlation of the expression of ERRα with that of genes encoding mitochondrial proteins (27).
Mitochondrial abundance and oxidative capacity are cell type-specific and regulated by energy demand. For example, physical exercise and chronic exposure to cold lead to the biogenesis of mitochondria in muscle and brown fat, respectively (35, 36). This adaptive response requires the coordinated induction of a large set of nuclear genes, accomplished, at least in part, by PGC-1α and the transcription factors NRF-1 and NRF-2 (35, 37). Because not all genes encoding mitochondrial proteins have binding sites for NRF-1 and NRF-2, additional factors must contribute to the response (37, 38). Possibly, the different factors contribute selectively to mitochondrial biogenesis in different cellular contexts; e.g., the levels of NRF-1 are induced during PGC-1α-mediated mitochondrial biogenesis in muscle but decreased when PGC-1α and mitochondria levels rise during brown fat development (39, 40). NRF-1, NRF-2, and ERRα may act synergistically in some cell types and operate independently in others. The presence of multiple factors may serve to integrate diverse signals into mitochondrial biogenesis. Furthermore, the different factors may enhance differentially the expression of specific genes, thereby enabling the newly made mitochondria to be selectively endowed with cell type- or signal-specific functions. Interestingly, ERRα alone (i.e., in the absence of PGC-1α or other activating signals) had no effect on “mitochondrial genes,” suggesting a function in the tissue-specific or signal-dependent regulation, rather than basal expression of genes encoding mitochondrial proteins.
Consistent with our findings, expression of Cyt c is downregulated in mice that carry a targeted null mutation in the ERRα gene (41). Further studies will be necessary to define mitochondrial defects and to determine whether other factors may partially compensate for the loss of ERRα function in these mice. One such candidate factor is the related receptor ERRγ, which is not expressed in the SAOS2 cells used in our study (data not shown). The ERRα-null mice display also altered expression of many genes involved in lipid metabolism (41). Together with our findings that ERRα is important for the PGC-1α-driven induction of the carnitine/acylcarnitine translocase and medium-chain acyl-CoA dehydrogenase genes (12), these observations suggest that ERRα function contributes to other PGC-1α-induced pathways, such as fatty acid β-oxidation (8, 9, 18). Finally, while our study demonstrates a role for ERRα as an important effector of PGC-1α, it is still possible that ERRα carries additional roles in regulating PGC-1α activity, as previously suggested (28).
Mitochondrial dysfunction and, in particular, decreases in oxidative capacity have been linked to insulin resistance and type 2 diabetes (42, 43). Recent studies also suggest that decreases in the levels of PGC-1α and the related coactivator PGC-1β contribute to the reduced oxidative capacity in diabetic subjects (23, 44). Supporting this notion, polymorphisms in the PGC-1α gene have been associated to an increased risk of diabetes (45, 46), whereas mice overexpressing PGC-1β show increased levels of ERRα and resistance to high-fat induced obesity (47). Strategies aimed at enhancing ERRα activity may thus have therapeutic applications in diseases associated with reduced mitochondrial function, such as diabetes.
Supplementary Material
Acknowledgments
We thank D. Kressler, M. Meyer, P. Coward, J.-M. Vanacker, and J. Mertz for plasmids and antibodies; A. McLachlan and U. Mueller for discussions; and the Swiss National Science Foundation, the University of Basel, the Max Cloëtta Foundation, and the Roche Research Foundation for their support.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ERRα, estrogen-related receptor α; ERRE, ERR response element; PGC-1α, peroxisome proliferator-activated receptor γ (PPARγ) coactivator 1α; NRF, nuclear respiratory factor; siRNA, small interfering RNA; L2/3, leucine motifs 2 and 3; Cyt c, cytochrome c, somatic; ATPsynβ, ATP synthase β; IDH3A, isocitrate dehydrogenase α; TR, thyroid hormone receptor; moi, multiplicity of infection; mtTFA, mitochondrial transcription factor A.
References
- 1.Giguere, V., Yang, N., Segui, P. & Evans, R. M. (1988) Nature 331, 91-94. [DOI] [PubMed] [Google Scholar]
- 2.Yang, N., Shigeta, H., Shi, H. & Teng, C. T. (1996) J. Biol. Chem. 271, 5795-5804. [DOI] [PubMed] [Google Scholar]
- 3.Johnston, S. D., Liu, X., Zuo, F., Eisenbraun, T. L., Wiley, S. R., Kraus, R. J. & Mertz, J. E. (1997) Mol. Endocrinol. 11, 342-352. [DOI] [PubMed] [Google Scholar]
- 4.Vanacker, J. M., Pettersson, K., Gustafsson, J. A. & Laudet, V. (1999) EMBO J. 18, 4270-4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giguere, V. (2002) Trends Endocrinol. Metab. 13, 220-225. [DOI] [PubMed] [Google Scholar]
- 6.Bonnelye, E., Vanacker, J. M., Dittmar, T., Begue, A., Desbiens, X., Denhardt, D. T., Aubin, J. E., Laudet, V. & Fournier, B. (1997) Mol. Endocrinol. 11, 905-916. [DOI] [PubMed] [Google Scholar]
- 7.Bonnelye, E., Merdad, L., Kung, V. & Aubin, J. E. (2001) J. Cell Biol. 153, 971-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sladek, R., Bader, J. A. & Giguere, V. (1997) Mol. Cell. Biol. 17, 5400-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vega, R. B. & Kelly, D. P. (1997) J. Biol. Chem. 272, 31693-31699. [DOI] [PubMed] [Google Scholar]
- 10.Yang, C. & Chen, S. (1999) Cancer Res. 59, 4519-4524. [PubMed] [Google Scholar]
- 11.Tremblay, G. B., Kunath, T., Bergeron, D., Lapointe, L., Champigny, C., Bader, J. A., Rossant, J. & Giguere, V. (2001) Genes Dev. 15, 833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schreiber, S. N., Knutti, D., Brogli, K., Uhlmann, T. & Kralli, A. (2003) J. Biol. Chem. 278, 9013-9018. [DOI] [PubMed] [Google Scholar]
- 13.Huss, J. M., Kopp, R. P. & Kelly, D. P. (2002) J. Biol. Chem. 277, 40265-40274. [DOI] [PubMed] [Google Scholar]
- 14.Puigserver, P., Wu, Z., Park, C. W., Graves, R., Wright, M. & Spiegelman, B. M. (1998) Cell 92, 829-839. [DOI] [PubMed] [Google Scholar]
- 15.Knutti, D. & Kralli, A. (2001) Trends Endocrinol. Metab. 12, 360-365. [DOI] [PubMed] [Google Scholar]
- 16.Puigserver, P. & Spiegelman, B. M. (2003) Endocr. Rev. 24, 78-90. [DOI] [PubMed] [Google Scholar]
- 17.Wu, Z., Puigserver, P., Andersson, U., Zhang, C., Adelmant, G., Mootha, V., Troy, A., Cinti, S., Lowell, B., Scarpulla, R. C., et al. (1999) Cell 98, 115-124. [DOI] [PubMed] [Google Scholar]
- 18.Vega, R. B., Huss, J. M. & Kelly, D. P. (2000) Mol. Cell. Biol. 20, 1868-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehman, J. J., Barger, P. M., Kovacs, A., Saffitz, J. E., Medeiros, D. M. & Kelly, D. P. (2000) J. Clin. Invest. 106, 847-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knutti, D., Kressler, D. & Kralli, A. (2001) Proc. Natl. Acad. Sci. USA 98, 9713-9718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knutti, D., Kaul, A. & Kralli, A. (2000) Mol. Cell. Biol. 20, 2411-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coward, P., Lee, D., Hull, M. V. & Lehmann, J. M. (2001) Proc. Natl. Acad. Sci. USA 98, 8880-8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mootha, V. K., Lindgren, C. M., Eriksson, K. F., Subramanian, A., Sihag, S., Lehar, J., Puigserver, P., Carlsson, E., Ridderstrale, M., Laurila, E., et al. (2003) Nat. Genet. 34, 267-273. [DOI] [PubMed] [Google Scholar]
- 24.Kressler, D., Schreiber, S. N., Knutti, D. & Kralli, A. (2002) J. Biol. Chem. 277, 13918-13925. [DOI] [PubMed] [Google Scholar]
- 25.Vanacker, J. M., Bonnelye, E., Delmarre, C. & Laudet, V. (1998) Oncogene 17, 2429-2435. [DOI] [PubMed] [Google Scholar]
- 26.Podvinec, M., Kaufmann, M. R., Handschin, C. & Meyer, U. A. (2002) Mol. Endocrinol. 16, 1269-1279. [DOI] [PubMed] [Google Scholar]
- 27.Mootha, V. K., Bunkenborg, J., Olsen, J. V., Hjerrild, M., Wisniewski, J. R., Stahl, E., Bolouri, M. S., Ray, H. N., Sihag, S., Kamal, M., et al. (2003) Cell 115, 629-640. [DOI] [PubMed] [Google Scholar]
- 28.Ichida, M., Nemoto, S. & Finkel, T. (2002) J. Biol. Chem. 277, 50991-50995. [DOI] [PubMed] [Google Scholar]
- 29.Wu, Y., Chin, W. W., Wang, Y. & Burris, T. P. (2003) J. Biol. Chem. 278, 8637-8644. [DOI] [PubMed] [Google Scholar]
- 30.Wang, Y. X., Lee, C. H., Tiep, S., Yu, R. T., Ham, J., Kang, H. & Evans, R. M. (2003) Cell 113, 159-170. [DOI] [PubMed] [Google Scholar]
- 31.Haraguchi, Y., Chung, A. B., Neill, S. & Wallace, D. C. (1994) J. Biol. Chem. 269, 9330-9334. [PubMed] [Google Scholar]
- 32.Villena, J. A., Martin, I., Vinas, O., Cormand, B., Iglesias, R., Mampel, T., Giralt, M. & Villarroya, F. (1994) J. Biol. Chem. 269, 32649-32654. [PubMed] [Google Scholar]
- 33.Evans, M. J. & Scarpulla, R. C. (1989) J. Biol. Chem. 264, 14361-14368. [PubMed] [Google Scholar]
- 34.Diascro, D. D., Jr., Vogel, R. L., Johnson, T. E., Witherup, K. M., Pitzenberger, S. M., Rutledge, S. J., Prescott, D. J., Rodan, G. A. & Schmidt, A. (1998) J. Bone Miner. Res. 13, 96-106. [DOI] [PubMed] [Google Scholar]
- 35.Moyes, C. D. & Hood, D. A. (2003) Annu. Rev. Physiol. 65, 177-201. [DOI] [PubMed] [Google Scholar]
- 36.Harper, M. & Himms-Hagen, J. (2001) Biochim. Biophys. Acta 1504, 159-172. [DOI] [PubMed] [Google Scholar]
- 37.Scarpulla, R. C. (2002) Biochim. Biophys. Acta 1576, 1-14. [DOI] [PubMed] [Google Scholar]
- 38.Lenka, N., Vijayasarathy, C., Mullick, J. & Avadhani, N. G. (1998) Prog. Nucleic Acid Res. Mol. Biol. 61, 309-344. [DOI] [PubMed] [Google Scholar]
- 39.Baar, K., Wende, A. R., Jones, T. E., Marison, M., Nolte, L. A., Chen, M., Kelly, D. P. & Holloszy, J. O. (2002) FASEB J. 16, 1879-1886. [DOI] [PubMed] [Google Scholar]
- 40.Villena, J. A., Carmona, M. C., Rodriguez de la Concepcion, M., Rossmeisl, M., Vinas, O., Mampel, T., Iglesias, R., Giralt, M. & Villarroya, F. (2002) Cell. Mol. Life Sci. 59, 1934-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo, J., Sladek, R., Carrier, J., Bader, J. A., Richard, D. & Giguere, V. (2003) Mol. Cell. Biol. 23, 7947-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bjorntorp, P., Schersten, T. & Fagerberg, S. E. (1967) Diabetologia 3, 346-352. [DOI] [PubMed] [Google Scholar]
- 43.Petersen, K. F., Befroy, D., Dufour, S., Dziura, J., Ariyan, C., Rothman, D. L., DiPietro, L., Cline, G. W. & Shulman, G. I. (2003) Science 300, 1140-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patti, M. E., Butte, A. J., Crunkhorn, S., Cusi, K., Berria, R., Kashyap, S., Miyazaki, Y., Kohane, I., Costello, M., Saccone, R., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 8466-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ek, J., Andersen, G., Urhammer, S. A., Gaede, P. H., Drivsholm, T., Borch-Johnsen, K., Hansen, T. & Pedersen, O. (2001) Diabetologia 44, 2220-2226. [DOI] [PubMed] [Google Scholar]
- 46.Hara, K., Tobe, K., Okada, T., Kadowaki, H., Akanuma, Y., Ito, C., Kimura, S. & Kadowaki, T. (2002) Diabetologia 45, 740-743. [DOI] [PubMed] [Google Scholar]
- 47.Kamei, Y., Ohizumi, H., Fujitani, Y., Nemoto, T., Tanaka, T., Takahashi, N., Kawada, T., Miyoshi, M., Ezaki, O. & Kakizuka, A. (2003) Proc. Natl. Acad. Sci. USA 100, 12378-12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.