Abstract
Signal recognition particle (SRP) is a ubiquitous ribonucleoprotein complex that targets proteins to endoplasmic reticulum (ER) in eukaryotes. Here we report that Plasmodium falciparum SRP is composed of six polypeptides; SRP9, SRP14, SRP19, SRP54, SRP68 and SRP72 and a 303nt long SRP RNA. We generated four transgenic parasite lines expressing SRP-GFP chimeric proteins and co-localization studies showed the nucleo-cytoplasmic localization for these proteins. The evaluation of the effect of known SRP and nuclear import/export inhibitors on P. falciparum revealed that ivermectin, an inhibitor of importin α/β mediated nuclear import inhibited the nuclear import of PfSRP polypeptides at submicromolar concentration, thereby killing the parasites. These findings provide insights into dynamic structure of P. falciparum SRP and also raise the possibility that ivermectin could be used in combination with other antimalarial agents to control the disease.
Keywords: signal recognition particle, nucleo-cytoplasmic shuttling, ER transport, ivermectin
The protein targeting in Plasmodium falciparum is a complex process that involves an extensive network of rough endoplasmic reticulum (ER), an unstacked Golgi situated anterior to the nucleus along with characteristic apicocomplexan organelles—rhoptries, micronemes and dense granules.1, 2, 3, 4 The biogenesis of the secretory organelles, their relationship to the organelles of higher eukaryotes and various sorting events that allow the proteins to get targeted to the right place are poorly understood in P. falciparum. The signal recognition particle (SRP), a cytoplasmic ribonucleoprotein complex, coordinates the targeting of nascent secretory as well as membrane proteins to the translocation machinery of the cells.5, 6 In addition to the targeting function, SRP also does the elongation arrest or pausing function.7 SRPs have been identified from all the three kingdoms based on their phylogenetically conserved sequences and their structures.8 The eukaryotic SRP is composed of a ∼300 nucleotide 7S RNA to which six distinct polypeptides; SRP9, 14, 19, 54, 68 and 72 are attached.8, 9 SRPs of bacteria are far simpler than its eukaryotic counterpart. In Eschericha coli, SRP consists of a 4.5S RNA to which a single polypeptide, the SRP54 homologue Ffh is attached. SRPs of Leishmania major, Giardia lambia and Trypanosoma brucei contain SRP19, 54, 68 and 72 homologues but do not possess SRP9/14 homologues.10, 11 The molecular and structural studies in mammalian cells have shown that SRP polypeptides; SRP-9, -14, -19, -68 and -72 are imported into the nucleus where they bind SRP RNA.12 The partially assembled SRP is exported out of nucleus in the cytoplasm and is joined by SRP54. The assembled SRP thereafter recognizes the under synthesized polypeptide resulting in elongation arrest.7 It has been shown that the binding of SRP19 to 7S RNA introduces some conformational changes in the RNA molecule, which enables SRP54 to bind it.13, 14 SRP54 acts as a front runner protein in recognizing the signal sequence flaunted by newly synthesized polypeptide chain of translating ribosome. The phenomenon of elongation arrest is considered to be essential, as the cell has to keep pace with the limited number of SRP receptors available on the membrane. The elongation resumes only when the whole conglomeration is transferred to the Sec62 translocon. The importance of components of this protein translocation machinery has been well documented in humans, where mutations in the components of transport machinery have been shown to cause various human diseases, thereby suggesting that this is an important and vital pathway.15
In P. falciparum, although a number of studies have reported the identification of few major components of protein translocation pathway by in silico analysis using Plasmodb data base,16, 17, 18 however, till now Plasmodium protein translocation machinery has not been characterized. In the present study, we identified seven P. falciparum SRP constituents and characterized them biochemically as well as for their sub-cellular distribution at asexual blood stages by generating SRP-GFP transgenic parasite lines. We further investigated the effects of exportin/importin and SRP assembly inhibitors on parasite growth in vitro and studied in detail the mode of action of ivermectin. Our results provide new insights into the molecular organization of P. falciparum SRP and set the stage to further analyze the antimalarial effect of ivermectin clinically.
Results
Identification of P. falciparum SRP components, characterization of PfSRP RNA and PfSRP 9, 14, 19, and 54
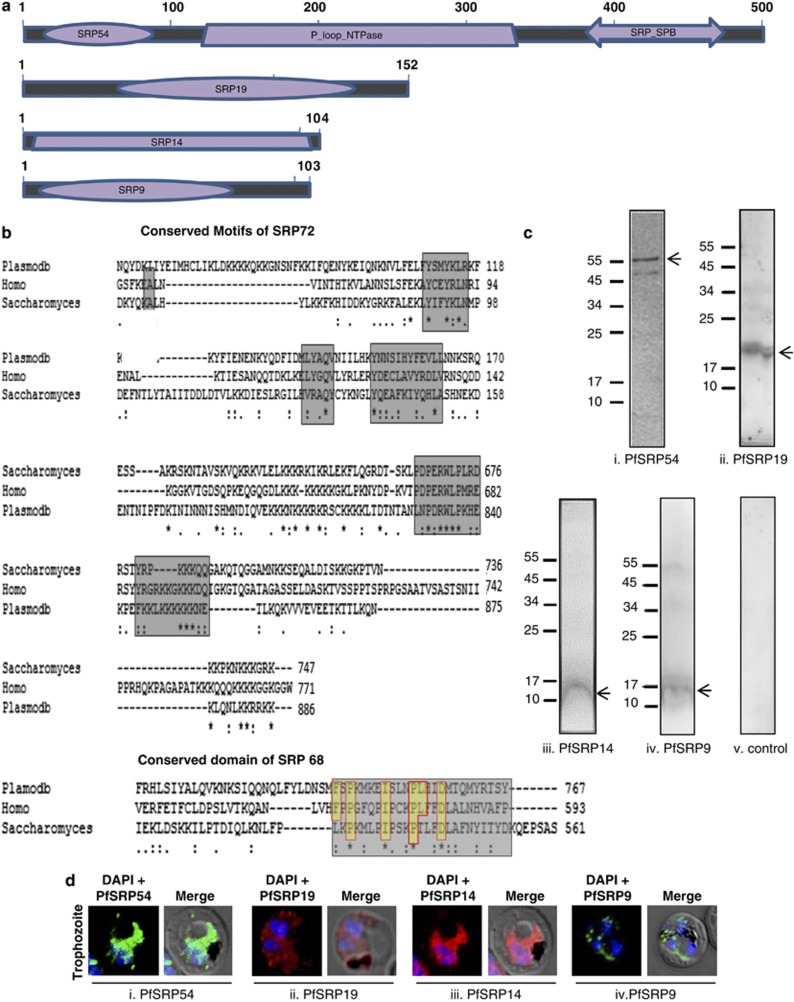
The domain specific search of the recent P. falciparum data base using the amino acid sequences of the human SRP proteins and SRP RNA was performed. The query identified all the predicted human SRP homologues in P. falciparum genome with PlasmoDB numbers PF3D7_0729000 (PfSRP9), PF3D7_1203200 (PfSRP14), PF3D7_1216300 (PfSRP19), PF3D7_1450100 (PfSRP54), PF3D7_0621900 (PfSRP68), PF3D7_1136400 (PfSRP72) and (PfSRP RNA) (Figures 1a and b). PfSRP72 as well as PfSRP68 homologs were identified based on protein–protein and protein–RNA interaction motifs. The annotated PfSRP68 sequence possesses a conserved SRP72 interacting domain, FSPKMKEISLNPLHIDMTQMYRSTSY of the form FSPKMKEISLNPLHIDMTQMYR[TSYL] located at amino acid positions 728–753 in P. falciparum, which harbours the conserved aspartic acid shown to be important for binding of human SRP68 protein.19 PfSRP72 possesses a conserved 7SL RNA binding domain, PDPERWLPLRD of the form PDRWLPKHEK located at amino acid positions 832–840 in P. falciparum. PfSRP72 also possesses the conserved tetrapeptide repeat motif (TPR) consisting of Y110, Q145 and Y153 which has been suggested to mediate interaction with SRP68 (Figure 1b). Together these results suggest that P. falciparum SRP is composed of a∼300 nt RNA and six polypeptides with masses 12.1, 11.8, 18, 55.9, 92.5 and 107.1 kDa. The coding sequences of predicted PfSRP54, PfSRP19, PfSRP14, PfSRP9 and PfSRP RNA were PCR amplified, cloned and sequenced. The sequence analysis of the PCR products showed no differences from the sequences in PlasmoDB data base.
Figure 1.
Schematic representation of domain architecture and expression of PfSRP polypeptides. (a) conserved domains of PfSRP54, PfSRP19, PfSRP14 and PfSRP9 predicted by CDART (http://www.ncbi.nlm.nih.gov/cdd). (b) Aligned representation of conserved domains of PfSRP72 and PfSRP68. Regions marked in yellow show conserved sequences. (c, i-iv) Western blot of expression of PfSRP54, PfSRP19, PfSRP14 and PfSRP9 in P. falciparum as detected by specific anti-PfSRP54, anti-PfSRP19 and anti-PfSRP14 and PfSRP9 sera in the parasite lysate. (c v) Western blot with mice pre-immune sera (d) Immuno-fluorescence staining of PfSRP54, PfSRP19, PfSRP14 and PfSRP9 in P. falciparum detected by specific anti-PfSRP54, anti-PfSRP19 and anti-PfSRP14 and PfSRP9 sera
Expression and purification of recombinant PfSRP 9, 14, 19 and 54
To functionally characterize PfSRP polypeptide homologues, these genes were sub-cloned in E. coli expression vectors pET28a or pET28b. A moderate level of expression was seen for the four recombinant PfSRP proteins. The recombinant proteins were subsequently purified on a Ni-NTA+ column under non-denaturing conditions. The apparent molecular masses of the recombinant proteins PfSRP19, PfSRP14 and PfSRP9 are ∼19, ∼14 and ∼12 kDa respectively (Supplementary Figures S1b–d). In the case of PfSRP54, two additional bands of lower molecular weight were always seen along with the intact protein after purification and all three bands were recognized on a western blot by anti-His antibody, thereby suggesting that these two additional bands were the degradation product of the intact protein (Supplementary Figure S1a, iii). The purified recombinant PfSRP54 was unstable as no intact protein was detected after storage. Therefore, to characterize PfSRP54, two distinct domains covering the entire sequence of PfSRP54; a GTP binding domain (NG domain) and a methionine rich domain (M domain) were cloned in pET28a and expressed. Both the PfSRP54 domains; PfSRP-54NG and PfSRP-54M were purified on a Ni-NTA+ column up to near homogeneity (Supplementary Figures S1a, i and ii). The purified recombinant PfSRP proteins were used to raise antibodies in mice and rats.
Immunoblot analysis of P. falciparum 3D7 parasite lysate using anti-PfSRP54, -PfSRP19, -PfSRP14 and -PfSRP9 antibodies recognized their respective native proteins of ∼54, ∼19, ∼12, and ∼12 kDa corresponding to the actual size of the native proteins. Pre-immune sera failed to detect any band in P. falciparum lysate (Figures 1c, i-v). The specificity of the antibodies was further confirmed by immunolocalization study at asexual blood stages of the parasite. P. falciparum SRP polypeptides are expressed in all three blood stages; ring, trophozoite and schizont (Figures 1d (i-iv) and Supplementary Figures S1e, i-iv). The staining was seen in the parasite cytoplasm and to some extent in the nucleus as well. No staining was observed in parasites with preimmune sera.
PfSRP polypeptides show bi-compartmental (nucleo-cytoplasmic) localization
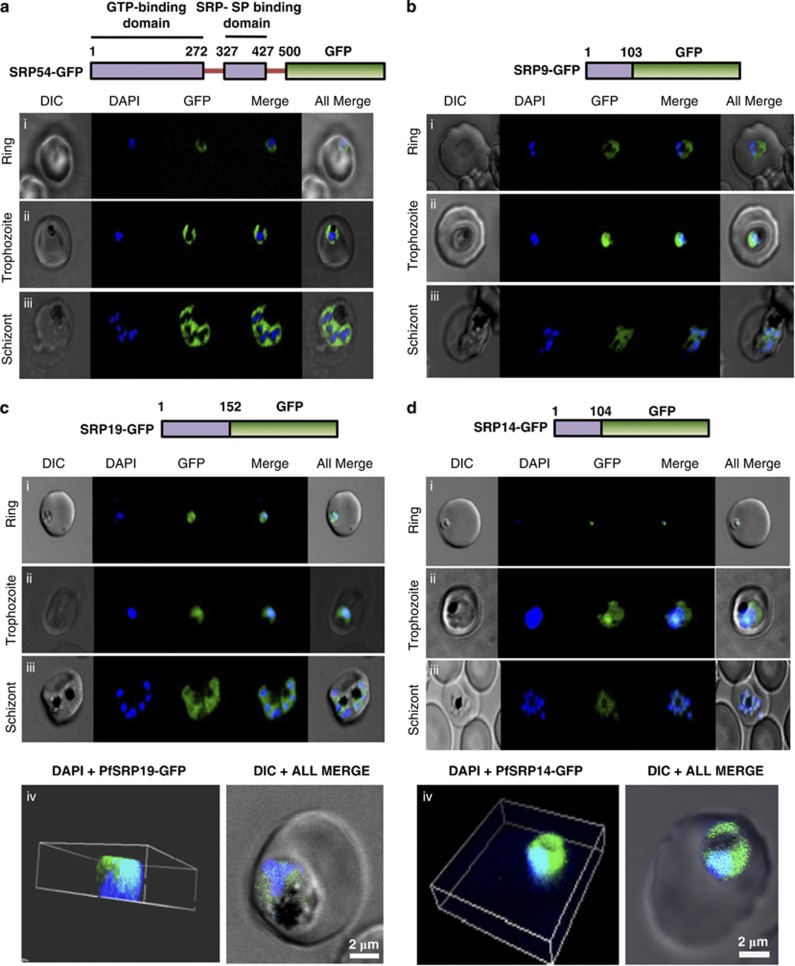
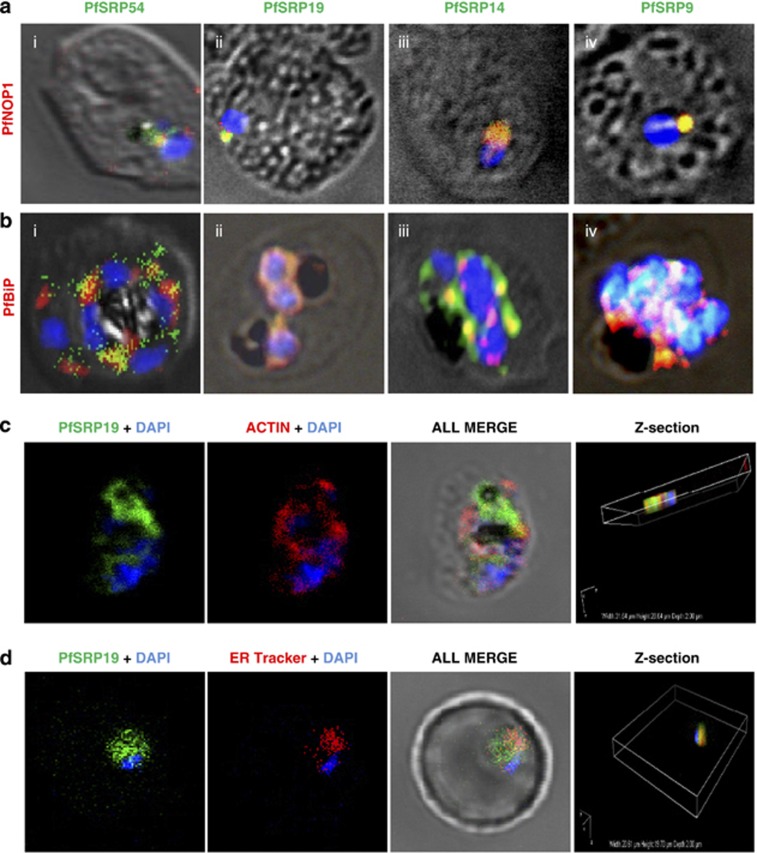
To decipher the sub-cellular localization of P. falciparum SRP polypeptides, four transgenic parasite lines expressing PfSRP54, PfSRP19, PfSRP14 and PfSRP9 as chimeric proteins with GFP were generated. Figures 2a–d, upper panels show the schematic of the PfSRP54-GFP, PfSRP19-GFP, PfSRP14-GFP and PfSRP9-GFP fusion constructs used for transfecting the parasite. The localizations of PfSRPs-GFP by fluorescence microscopy of live parasites were also investigated. At the early stage of development >20 h (ring stage), a crescent shaped staining was observed that changed into a ring of fluorescence around the nuclear envelop at the trophozoite stage (Figures 2a–d, lower panels i and ii). In later stages, staining was quite intense and spread through-out the cytoplasm (Figures 2a–d, lower panel iii). At schizont stage, extensive ER branching creating a mesh-like network in some sections was observed (Figures 2a–d, lower panel iii). The PfSRP54 staining was mainly observed in cytosol (Figure 2a, lower panels). In comparison to PfSRP54 distribution in the nucleus, staining for PfSRP19, PfSRP14 and PfSRP9 in subnuclear areas was quite intense at the asexual blood stages particularly at the trophozoite stage (Figures 2b–d, lower panel ii, Supplementary Figure S2). The inspection of transgenic parasites by confocal microscopy (mid-depth Z slice) also revealed the nucleo-cytoplasmic distribution of PfSRP-chimeric proteins (Figures 2c and d, lower panel iv). To further confirm the bi-compartmental localization of PfSRP polypeptides, co-localization experiments for the parasite SRP polypeptides with PfNOP1, a known nucleolar marker20 or with PfBiP, an ER marker21 were performed with their respective antibodies. A considerable overlap in staining was observed between three PfSRP polypeptides; PfSRP19, -14, and -9 with PfNOP1 and with PfBiP protein (Figures 3a and b, i-iv). Importantly, considerable co-localization of PfSRP54 with PfNOP1 in early stages of parasite development i.e. merozoite and ring stages (Figure 3a, i and Supplementary Figure S3) was observed. In addition considerable co-localization between the GFP protein and the cytoplasmic marker Pfactin, along with ER tracker (invitrogen) was also observed (Figures 3c and d). These results collectively suggest that the parasite's SRPs reside in nucleus and ER membrane.
Figure 2.
Localisation of GFP fused PfSRP54, PfSRP19, PfSRP14 and PfSRP9 polypeptides. Upper panels of a, b, c and d show schematic representation of respective GFP fused PfSRP polypeptides. Lower panels show the expression of PfSRP polypeptides in three asexual blood stages of P. falciparum i.e. ring, trophozoite and schizont stage. Panel c(iv) and d(iv) show three dimensional reconstruction of confocal Z-stack merged images of GFP fused PfSRP19 and PfSRP14 (green), respectively, along with nuclear stain DAPI (blue)
Figure 3.
Co-staining of PfSRP polypeptides with nucleolar marker PfNOP1, ER marker PfBiP and cytoplasmic marker Pfactin. (a, b and c) P. falciparum infected erythrocytes stained with anti-PfSRP antibodies (green), anti-PfNOP1 (red), anti-PfBiP (red) and anti-Pfactin (red) respectively. (d) localization of GFP fused SRP19 (green) with ER tracker (red). DAPI is used for nuclear staining (blue)
PfSRP assembly involves protein–RNA and protein–protein interactions
In vitro binding studies in mammalian cells have revealed that SRP RNA is at the centre of structure of SRP complex.9 Protein–RNA interaction by an RNA binding assay and protein–protein interactions by bacterial two hybrid analysis were performed with PfSRP RNA and between PfSRP proteins to understand the assembly of P. falciparum signal recognition particle.
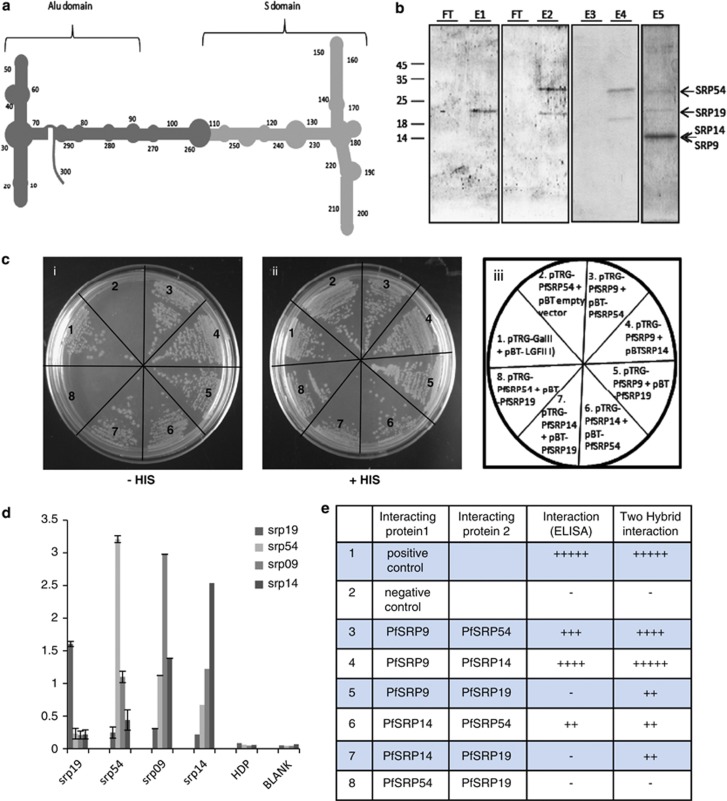
The results of protein/RNA interaction studies showed that PfSRP19 bound to the S domain of PfSRP RNA (Figure 4a) as well as to the full length SRP RNA (Figure 4b, lane E1). Similarly, PfSRP54 also bound the S domain and intact PfSRP RNA similar to PfSRP19 protein (Figure 4b, lanes E2 and E4). Moreover, PfSRP-9, -14, -19 and -54 together bound strongly to the intact full length PfSRP RNA (Figure 4b, lane E5). However, PfSRP RNA was unable to bind to a non-SRP protein, PfHDP (25 kDa) (Figure 4b, lane E3). These results support identification of PfSRP genes in the present study as the real constituents of the P. falciparum SRP machinery and further suggest that PfSRP RNA like its other homologs provides a backbone for the coordinated assembly of PfSRP proteins.
Figure 4.
Interaction between PfSRP polypeptides and SRPRNA. (a) schematic diagram of secondary structure of PfSRP RNA showing the Alu domain and S domain (b). Binding of SRP RNA to SRP 19 and M domain of SRP 54. FT represents the lanes with flow through. Elutes of S domain of SRP RNA binding (by DEAE sepharose method) showing the presence of SRP19 (E1), SRP19 and M-domain of SRP54( E2). Elutes of full SRP RNA binding (by DEAE sepharose method) with non-SRP protein PfHDP (E3) and PfSRP19 and SRP54 (E4), and all four PfSRP54 (M-domain), PfSRP19, PfSRP14, PfSRP9 (E5). (c) Interaction between protein components of PfSRP polypeptides using Bacterial Two Hybrid system. Growth of cotransformed reporter strain XL1-blue in dual selective (i) and non-selective (ii) medium, (iii) shows the plasmid constructs used to cotransform the reporter strain. (d) Interaction between protein components of PfSRP polypeptides using ELISA. X-axis shows the coated protein and Y-axis is absorbance read at 490 nm. (e) Table showing summary of interactions among all PfSRP polypeptides using both ELISA and Bacterial Two Hybrid system
To know whether PfSRP assembly also involves protein–protein interactions besides protein–RNA interactions, direct interaction studies between PfSRP54, -19, -14 and -9 using an ELISA based approach or by a well established bacterial two-hybrid interaction approach were performed. Varying degrees of interactions among the four SRP polypeptides were observed but no interaction between PfSRP19 and PfSRP54 was seen (Figures 4c–e and Supplementary Figure S4). The associations between these PfSRP proteins in vivo were also illustrated by the co-localization studies performed at asexual blood stages of the parasite using their respective antibodies (Supplementary Figure S2).
Effect of aflatoxin B1, leptomycin B and ivermectin on PfSRP distribution and parasite growth
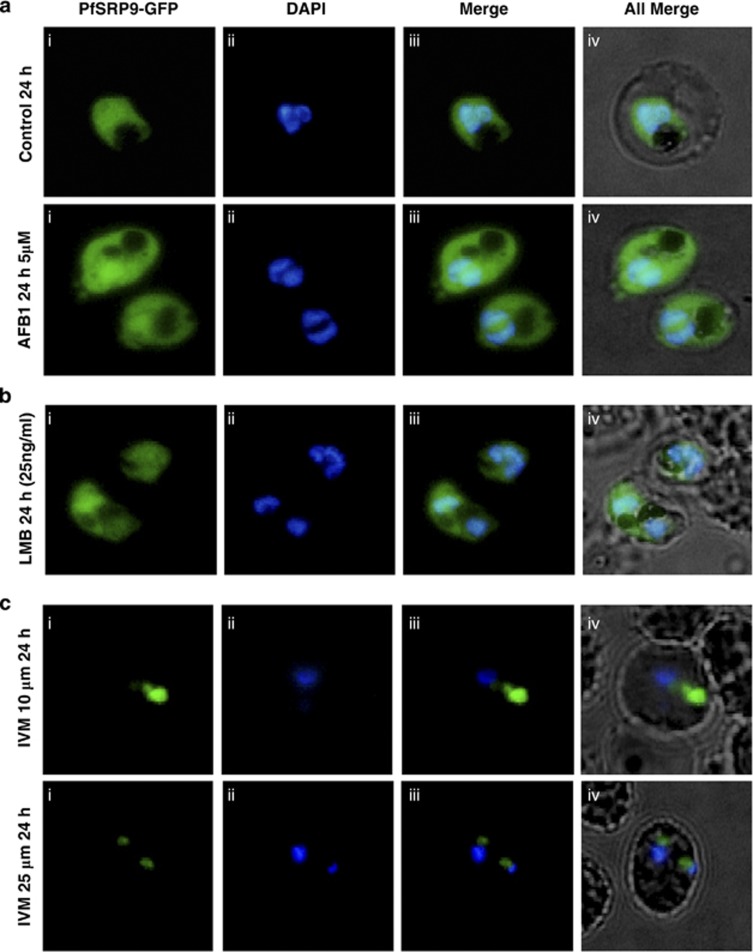
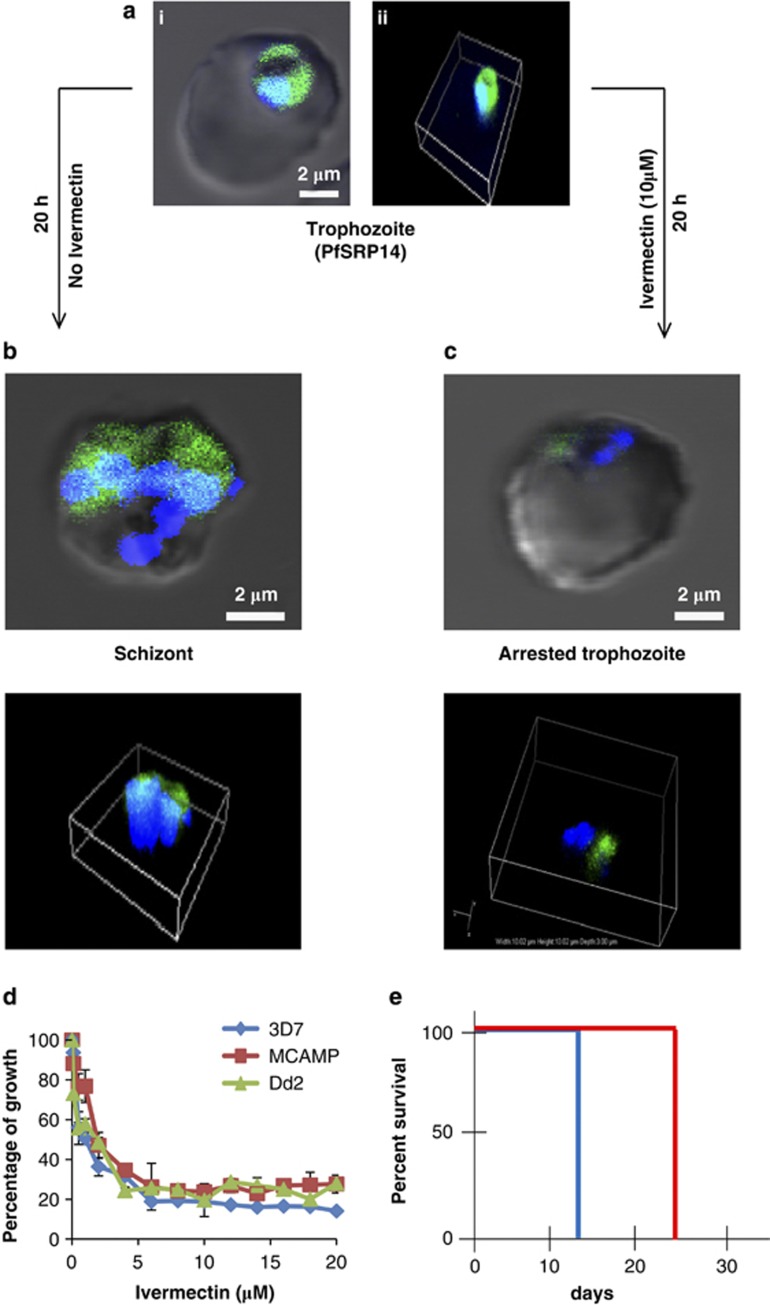
A number of inhibitors/drugs such as aflatoxin B1 (AFB1), leptomycin B (LMB) and ivermectin have been shown to either interact with SRP components or inhibit protein nuclear export/import machinery.22, 23, 24 We tested the effect of these inhibitors on transgenic parasite lines. AFB1 up to 5 μM and LMB up to 100 ng/ml had no significant effect on PfSRP9-GFP protein distribution and on parasite growth (Figures 5a and b and Supplementary Figures S5a and b). Ivermectin, an inhibitor of importin α/β, at 10 μM to 25 μM showed pronounced effects 24 h after the treatment on PfSRP9-GFP transgenic parasites (Figure 5c). The distribution of GFP-fused PfSRP9 chimeric protein was concentrated in parasite cytoplasm in the treated parasites, in comparison to the untreated parasite culture where the chimeric protein showed the nucleo-cytoplasmic distribution. The effect was more pronounced at 25 μM concentration of ivermectin (Figure 5c). The removal of ivermectin from the culture media after 24 h did not alter the growth of the parasite i.e. effect of the drug was irreversible. A similar block in nucleo-cytoplasmic shuttling was observed for GFP fused PfSRP14 transgenic parasites (Figures 6b and c and supplementary videos 1 and 2). Subsequently, the effect of ivermectin on parasite growth was assessed by the SYBR green based assay on different P. falciparum drug resistant strains; chloroquine (CQ)/mefloquine resistant (DD2 and MCamp) along with a chloroquine sensitive strain P. falciparum 3D7. Ivermectin exhibited anti-plasmodial activity against all the three strains Dd2, MCamp and 3D7 in a dose dependent manner with IC50 values of 2.85, 1.92 and 1.56 μM respectively (Figure 6d). We further tested the anti-malarial activity of ivermectin in vivo with P. berghei infected mice by administering a 0.5 mg/kg dose of ivermectin using a modified Thompson test (3 consecutive days of dosing). As seen in Figure 6e, ivermectin alone suppressed day 4 parasitemias by ∼40% compared to placebo-treated controls. All four mice administered with ivermectin survived up to 23 days, while the control mice died on day 13 (Supplementary Figure S5c). Together these results suggest that ivermectin can be considered for the development as an antimalarial, which can be used in combination with other drugs.
Figure 5.
Effect of aflatoxin B1, leptomycin B and ivermectin, on nucleo-cytoplasmic shuttling of GFP fused PfSRP9. Synchronised ring stage infected erythrocytes at 2% initial parasitemia were treated with these inhibitors and imaged at different time intervals. (a) upper panel, DMSO treated control parasite after 24 h of treatment, lower panel, AflatoxinB1 (AFB1, 10 μM ) treated parasite at 24 h after treatment. (b) LeptomycinB (LMB, 25 ng/ml ) treated parasite at 24 h after treatment and (c) parasite after 24 h of ivermectin (IVM) treatment at10 μM and 25 μM, respectively
Figure 6.
Effect of Ivermectin on localisation of GFP fused PfSRP14 and growth of parasite. (a) i and ii show three dimensional reconstruction of confocal Z-stack merged images of GFP fused control trophozoite with DAPI (blue) as nuclear marker merged with DIC. (b) and (c), show the three dimensional reconstruction of confocal Z-stack merged images of GFP fused PfSRP14 with DAPI as nuclear marker merged with DIC in control and Ivermectin (10 μM) treated parasite at 24 h after treatment. (d) dose dependent effect of ivermectin on growth of P. falciparum 3D7, MCamp and Dd2 by SYBR Green assay (e) In-vivo effect of ivermectin against P. berghei in mice
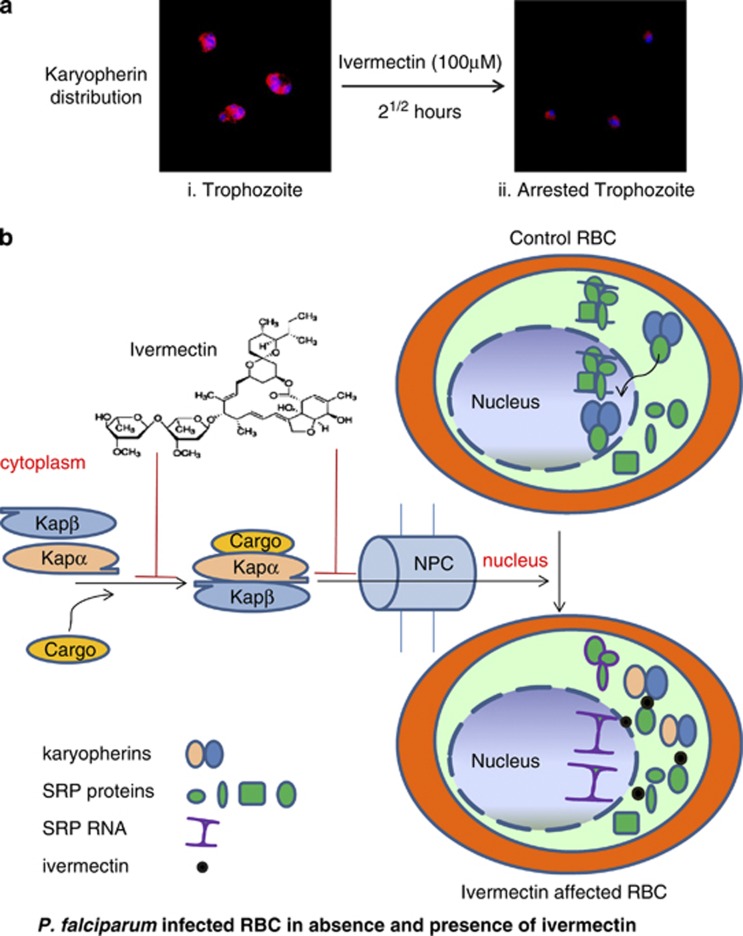
Ivermectin is a known inhibitor of importin α/β mediated nuclear transport.24 P. falciparum has only two important nuclear import factors; karyopherins α and β in its genome, while the human genome encodes numerous importins. We next analysed the sub-cellular distribution of karyopherin β in ivermectin treated asexual blood stages of P. falciparum using anti-Pfkaryopherin β antibody.25 Anti-Pfkaryopherin β recognized the parasite specific karyopherin β in untreated parasites and staining was mainly seen in and around the nucleus (Figure 7a, i). Intriguingly, parasites treated with ivermectin for 21/2 hours at 100 μM concentration or with 5 μM concentration for 20 h showed restricted staining in the cytoplasm (Figure 7a, ii and supplementary Figures S6a and b). Faint or no staining for PfSRP9 protein was observed at 20 h post exposure to 25 μM of ivermectin, mainly because almost all the parasites were dead after this treatment (Supplementary Figure S6c). Together these results suggest that ivermectin inhibits malaria parasite development by blocking nucleo-cytoplasmic shuttling of PfSRP components.
Figure 7.
Effect of Ivermectin on localisation of karyopherin β, proposed mechanism of cargo import by karyopherin α- karyopherin β pathway and possible mode of action of ivermectin. (a, i), control parasite with karyopherin β (red) and DAPI (blue) and ii, ivermectin (100 μM) treated parasites after 21/2 h. (b) Karyopherin α and β form a dimer and attach with the cargo and pass through the nuclear pore complex into nucleus where the cargo detaches itself from the karyopherins which are recycled into cytoplasm via RanGTP. SRP subunits join SRPRNA in the nucleus and the whole conglomerate is transported back to the cytoplasm where it becomes functional. Ivermectin blocks the parasite growth by inhibiting nucleo-cytoplasmic shuttling of SRPs essential for parasite survival either by affecting at the stage of karyopherin dimerisation or at the level of SRP attachment with Karyopherin dimer which inturn hampers the entry of SRP to its site of RNA assembly in the nucleus. Cell death occurs because of piling up of proteins in the cytoplasm
Discussion
Recognition of the signal sequence in the ribosome nascent-chain (RNC) complex by SRP is the first step in protein secretion.26 SRP couples the synthesis of membrane/secretory proteins and their transport across the ER membrane in eukaryotes27, 28 or across the bacterial plasma membrane8 and chloroplast thylakoid membranes.29 Considerable variations have been observed in the constituents of SRPs in different organisms.10 Among the protozoan parasites, T. brucei and Leptomonas collosoma possess an unusual SRP that comprises two SRP RNAs; 7SL RNA and a tRNA like molecule and three SRP proteins; SRP19, SRP72 and SRP68, while the bacterial SRPs are of reduced complexity consisting of a small SRP RNA and a single protein SRP54, also referred as Ffh.10 In the present study, in silico analysis revealed that the P. falciparum SRP is more similar to the mammalian SRP as it consists of six polypeptides; PfSRP72, PfSRP68, PfSRP54, PfSRP19, PfSRP14 and PfSRP9 and a single 303 nucleotide long PfSRP RNA. In comparison to the previous in silico studies, in the present study two additional PfSRP polypeptides; PfSRP68 and PfSRP72 were identified in the P. falciparum genome. It is important to mention here that the other members of the Alveolata to which P. falciparum belongs such as Theileria, Eimeria as well as members of the Euglenozoa such as Trypanosoma and Leishmania, lack one or two of the six SRP polypeptides. For example SRP9/14 has not been identified in L. major, T. cruzi, Thelileria annulata, and Giardia lamblia.10, 11 A number of structural and protein–RNA interactions studies for E. coli and mammalian SRPs have provided insights into the assembly of SRPs and also delineated the interaction motifs (residues) between SRP54 and SRP RNA.30, 31, 32 However, our understanding of SRPs in lower eukaryotes has been limited. Here we identified and report some of the essential features of P. falciparum SRP. The results demonstrate that all the components of mammalian SRP are conserved in the P. falciparum genome. We further show that Plasmodium SRP assembly process appears to be similar to that of human SRP assembly. The binding of PfSRP19 to PfSRP RNA most likely induces conformational changes in PfSRP RNA that subsequently binds to other components; such as PfSRP54, PfSRP14 and PfSRP9. Additionally, we demonstrate that during PfSRP assembly various PfSRP proteins also interact with each other, presumably to stabilize the PfSRP.
The sub-cellular localization studies performed for PfSRP proteins showed a clear nucleolar as well as cytoplasmic distribution for PfSRP9, PfSRP14 and PfSRP19 polypeptides at asexual blood stages by anti-PfSRPs antibodies and by GFP fluorescence analysis in transgenic lines. This was further confirmed by co-localization experiments with the nucleolar and ER markers. Importantly, PfSRP54 also showed nucleoplasmic distribution, although its level in the nucleus was low in comparison to other PfSRP polypeptides. Significantly, we could also observe PfSRP54 co-localization with PfNOP1, a nucleolar marker especially in earlier stages of development. It has been proposed that nucleolus is the site of assembly and/or interaction between the families of mammalian ribonucleoproteins involved in protein synthesis, in addition to ribosomes themselves.33, 34, 35 However, in mammalian cells, SRP54 does not seem to have a nucleolar phase.12 Based on these findings, it has been suggested that in mammalian cells, SRP is partially assembled in the nucleus and it is piggybacked with ribosomal units for export from the nucleus.36 In the cytoplasm, the partially assembled SRP is joined by SRP54 that recognizes the nascent signal sequence.33 Based on the data for the localization of PfSRP proteins and mammalian SRP proteins, it seems more likely that in the case of PfSRP, complete assembly may occur in nucleus and this assembled complex is then transported to cytoplasm to perform ER targeting.
The movement of proteins between the nucleus and cytoplasm is an essential process and has been shown to be critical for the disease states such as parasitic diseases, viral diseases and oncogenesis. Although seven importin αs and >20 importin βs have been described in human along with a wide range of NLS/NES sequences that are required for this transport, functional role(s) of each transporter in specific protein shuttling across nuclear membrane is not still understood.23 Currently, the exportin/CRM1 inhibitor, Leptomycin B and importin α/β inhibitor, ivermectin are the only accepted inhibitors being used to inhibit nuclear transport in mammalian cells. We used these inhibitors along with aflatoxin B1 to study their effect on PfSRP polypeptides and their nucleo-cytoplasmic shuttling. Importantly, neither Leptomycin B, nor aflatoxin B1 affected the localization and distribution of PfSRP-GFP polypeptides and these treated parasites grew normally in in vitro culture. In comparison, ivermectin, a specific inhibitor of importin α/β blocked the nucleo-cytoplasmic shuttling of SRP polypeptides; PfSRP9-GFP and PfSRP14-GFP chimera proteins were mainly seen in the parasite cytoplasm in the ivermectin treated parasites. Ivermectin showed a potent inhibition in the growth of P. falciparum 3D7 as well as other drug resistant strains, Mcamp and DD2. Ivermectin treated P. falciparum parasites also showed a dramatic change in the level as well as the distribution of karyophorin β. Limited in vivo studies in a P. berghei mice model further demonstrated the antimalarial effect of ivermectin.
Ivermectin has been shown to inhibit replication of HIV-1 and Dengue virus by blocking the shuttling of nuclear proteins mediated by importin α/β. In nematodes, ivermectin is known to bind and activate chloride ion channels,37 a phenomenon which has been shown to be responsible for its anti-parasitic effect. Ivermectin also causes cell death in leukemia cells by inducing the chloride dependent membrane hyperpolarization. Based on these studies, it has been proposed that ivermectin affects the shuttling of importin α/β by inducing the chloride dependent polarization of parasite nuclear membrane that in turn inhibits the nuclear import of SRP polypeptides. Since karyopherin α/β are the only known importins present in P. falciparum, it is likely that ivermectin treatment is arresting parasite growth by blocking the shuttling of SRP polypeptides and some other nuclear proteins (Figure 7b). Ivermectin has recently been shown to inhibit the sporogony of P. falciparum in Anopheles gambiae.38 Although a detailed in vivo analysis will be required for the use of ivermectin as an antimalarial agent, however given the prior use of ivermectin and its safety record in humans and animals, it can be considered in combination therapy with other antimalarials. In conclusion, the results presented here provide insights into the malaria parasite PfSRP and show that ivermectin an anti-parasitic has the potential to treat multi-drug resistant malaria.
Materials and Methods
Generation of PfSRP-GFP lines
To generate P. falciparum's SRPs-GFP lines, genes corresponding to PfSRP54, -19, -14, -9 were PCR amplified and cloned into pSSPF2. The plasmids were transfected into P. falciparum 3D7 line and transfected cells were selected as described in detail in SI text.
Analysis of protein–RNA and protein–protein interactions
In-vitro interactions between PfSRPs and PfSRP RNA were performed using well established RNA binding assay as described in SI text. Protein–protein interactions between four PfSRP components were performed using bacterial two hybrid system and ELISA as described in SI text.
Effect of inhibitors on PfSRP-GFP transgenic lines
P. falciparum culture was synchronised by two consecutive sorbitol treatments at an interval of four hours. The parasite culture was incubated with different concentrations of leptomycinB, aflatoxin B1 and ivermectin as described in SI text.
Growth inhibition assay on P. falciparum 3D7, Dd2 and mCAMP strains
Parasite culture with an initial parasitemia of 0.8% was treated with different doses of leptomycin B, aflatoxin B1 and ivermectin in 96 well plate and new ring formation was monitored as described in SI text.
Acknowledgments
We thank Christen M Klinger, University of Alberta, Canada for help with the SRP68 and SRP72 sequence alignment. This work is partially supported by Department of Science and Technology grant. Infra-structural support from the Department of Biotechnology, Government of India is gratefully acknowledged.
Glossary
- AFB1
aflatoxin B1
- CQ
chloroquine
- ER
endoplasmic reticulum
- ELISA
enzyme-linked immunosorbent assay
- GFP
green fluorescent protein
- HDP
heme detoxification protein
- IVM
ivermectin
- LMB
leptomycin B
- SRP
signal recognition particle
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by A Stephanou
Supplementary Material
References
- Lingelbach KR. Plasmodium falciparum: a molecular view of protein transport from the parasite into the host erythrocyte. Exp Parasitol. 1993;76:318–327. doi: 10.1006/expr.1993.1039. [DOI] [PubMed] [Google Scholar]
- Van Dooren GG, Waller RF, Joiner KA, Roos DS, McFadden GI. Traffic jams: protein transport in Plasmodium falciparum. Parasitol Today. 2000;16:421–427. doi: 10.1016/s0169-4758(00)01792-0. [DOI] [PubMed] [Google Scholar]
- Nacer A, Berry L, Slomianny C, Mattei D. Plasmodium falciparum signal sequences: simply sequences or special signals. Int. J. Parasitol. 2001;31:1371–1379. doi: 10.1016/s0020-7519(01)00253-3. [DOI] [PubMed] [Google Scholar]
- Lingelbach K, Przyborski JM. The long and winding road: protein trafficking mechanisms in the Plasmodium falciparum infected erythrocyte. Mol Biochem Parasitol. 2006;147:1–8. doi: 10.1016/j.molbiopara.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum. II. Signal recognition protein (SRP) mediates the selective binding to microsomal membranes of in-vitro-assembled polysomes synthesizing secretory protein. J Cell Biol. 1981;91:551–556. doi: 10.1083/jcb.91.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol. 1981;91:557–561. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakkaraju AK, Mary C, Scherrer A, Johnson AE, Strub K. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell. 2008;133:440–451. doi: 10.1016/j.cell.2008.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- Gundelfinger ED, Di Carlo M, Zopf D, Melli M. Structure and evolution of the 7SL RNA component of the signal recognition particle. EMBO J. 1984;3:2325–2332. doi: 10.1002/j.1460-2075.1984.tb02134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool MR. Signal recognition particles in chloroplasts, bacteria, yeast and mammals. Mol Membr Biol. 2005;22:3–15. doi: 10.1080/09687860400026348. [DOI] [PubMed] [Google Scholar]
- Lustig Y, Goldshmidt H, Uliel S, Michaeli S. The Trypanosoma brucei signal recognition particle lacks the Alu-domain-binding proteins: purification and functional analysis of its binding proteins by RNAi. J Cell Sci. 2005;118:4551–4562. doi: 10.1242/jcs.02578. [DOI] [PubMed] [Google Scholar]
- Politz JC, Yarovoi S, Kilroy SM, Gowda K, Zwieb C, Pederson T. Signal recognition particle components in the nucleolus. Proc Natl Acad Sci USA. 2000;97:55–60. doi: 10.1073/pnas.97.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oubridge C, Kuglstatter A, Jovine L, Nagai K. Crystal structure of SRP19 in complex with the S domain of SRP RNA and its implication for the assembly of the signal recognition particle. Mol Cell. 2002;9:1251–1261. doi: 10.1016/s1097-2765(02)00530-0. [DOI] [PubMed] [Google Scholar]
- Hainzl T, Huang S, Sauer-Eriksson AE. Structure of the SRP19 RNA complex and implications for signal recognition particle assembly. Nature. 2002;417:767–771. doi: 10.1038/nature00768. [DOI] [PubMed] [Google Scholar]
- Zimmermann R, Müller L, Wullich B. Protein transport into the endoplasmic reticulum: mechanisms and pathologies. Trends Mol Med. 2006;12:567–573. doi: 10.1016/j.molmed.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Tuteja R. Unraveling the components of protein translocation pathway in human malaria parasite Plasmodium falciparum. Arch Biochem Biophys. 2007;467:249–260. doi: 10.1016/j.abb.2007.08.031. [DOI] [PubMed] [Google Scholar]
- Rosenblad MA, Zwieb C, Samuelsson T. Identification and comparative analysis of components from the signal recognition particle in protozoa and fungi. BMC Genomics. 2004;5:5. doi: 10.1186/1471-2164-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen ES, et al. The tmRDB and SRPDB resources. Nucleic Acids Res. 2006;34:163–168. doi: 10.1093/nar/gkj142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakhiaeva E, Hinck CS, Hinck AP, Zwieb C. Characterization of the SRP68/72 interface of human signal recognition particle by systematic site-directed mutagenesis. Protein Sci. 2009;18:2183–2195. doi: 10.1002/pro.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo LM, Rocha EP, Mancio-Silva L, Prevost C, Hernandez-Verdun D, Scherf A. The unusually large Plasmodium telomerase reverse-transcriptase localizes in a discrete compartment associated with the nucleolus. Nucleic Acids Res. 2005;33:1111–1122. doi: 10.1093/nar/gki260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Koski G, Harada M, Aikawa M, Zheng H. Induction and localization of Plasmodium falciparum stress proteins related to the heat shock protein 70 family. Mol Biochem Parasitol. 1991;48:47–58. doi: 10.1016/0166-6851(91)90163-z. [DOI] [PubMed] [Google Scholar]
- Singh J, Singh S, Dani HM, Sharma R, Steinberg P. Interactions of aflatoxin B1 with SRP components can disrupt protein targeting. Cell Biochem Funct. 2005;23:9–13. doi: 10.1002/cbf.1120. [DOI] [PubMed] [Google Scholar]
- Alavian CN, Politz JC, Lewandowski LB, Powers CM, Pederson T. Nuclear export of signal recognition particle RNA in mammalian cells. Biochem Biophys Res Commun. 2004;313:51–55. doi: 10.1016/j.bbrc.2003.11.126. [DOI] [PubMed] [Google Scholar]
- Wagstaff KM, Sivakumaran H, Heaton SM, Harrich D, Jans DA. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem J. 2012;443:851–856. doi: 10.1042/BJ20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohmmed A, Kishore S, Patra KP, Dasaradhi PV, Malhotra P, Chauhan VS. Identification of karyopherin beta as an immunogenic antigen of the malaria parasite using immune mice and human sera. Parasite Immunol. 2005;27:197–203. doi: 10.1111/j.1365-3024.2005.00759.x. [DOI] [PubMed] [Google Scholar]
- Walter P, Lingappa VR. Mechanism of protein translocation across the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1986;2:499–516. doi: 10.1146/annurev.cb.02.110186.002435. [DOI] [PubMed] [Google Scholar]
- Kurzchalia TV, Wiedmann M, Girshovich AS, Bochkareva ES, Bielka H, Rapoport TA. The signal sequence of nascent preprolactin interacts with the 54K polypeptide of the signal recognition particle. Nature. 1986;320:634–636. doi: 10.1038/320634a0. [DOI] [PubMed] [Google Scholar]
- High S, Flint N, Dobberstein B. Requirements for the membrane insertion of signal-anchor type proteins. J Cell Biol. 1991;113:25–34. doi: 10.1083/jcb.113.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuenemann D, Gupta S, Persello-Cartieaux F, Klimyuk VI, Jones JD, Nussaume L, et al. A novel signal recognition particle targets light-harvesting proteins to the thylakoid membranes. Proc Natl Acad Sci USA. 1998;95:10312–10316. doi: 10.1073/pnas.95.17.10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Kabe Y, Wada T, Asai A, Handa H. A new mechanism of 6-((2-(dimethylamino)ethyl)amino)-3-hydroxy-7H-indeno(2,1-c)quinolin-7-one dihydrochloride (TAS-103) action discovered by target screening with drug-immobilized affinity beads. Mol Pharmacol. 2008;73:987–994. doi: 10.1124/mol.107.043307. [DOI] [PubMed] [Google Scholar]
- Ataide SF, Schmitz N, Shen K, Ke A, Shan SO, Doudna JA, et al. The crystal structure of the signal recognition particle in complex with its receptor. Science. 2011;331:881–886. doi: 10.1126/science.1196473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Becker T, Pool MR, Spahn CM, Grassucci RA, Frank J, et al. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- Ciufo LF, Brown JD. Nuclear export of yeast signal recognition particle lacking Srp54p by the Xpo1p/Crm1p NES-dependent pathway. Curr Biol. 2000;10:1256–1264. doi: 10.1016/s0960-9822(00)00743-0. [DOI] [PubMed] [Google Scholar]
- Siegel V, Walter P. Functional dissection of the signal recognition particle. Trends Biochem Sci. 1988;13:314–316. doi: 10.1016/0968-0004(88)90127-2. [DOI] [PubMed] [Google Scholar]
- Phillips GJ, Silhavy TJ. The E. coli ffh gene is necessary for viability and efficient protein export. Nature. 1992;359:744–746. doi: 10.1038/359744a0. [DOI] [PubMed] [Google Scholar]
- Jacobson MR, Pederson T. Localization of signal recognition particle RNA in the nucleolus of mammalian cells. Proc Natl Acad Sci. 1998;95:7981–7986. doi: 10.1073/pnas.95.14.7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottesen EA, Campbell WC. Ivermectin in human medicine. J Antimicrob Chemother. 1994;34:195–203. doi: 10.1093/jac/34.2.195. [DOI] [PubMed] [Google Scholar]
- Kobylinski KC, Foy BD, Richardson JH. Ivermectin inhibits the sporogony of Plasmodium falciparum in Anopheles gambiae. Malar J. 2012;11:381. doi: 10.1186/1475-2875-11-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.