Abstract
Recent studies have shown that genes involved in oxidative phosphorylation (OXPHOS) exhibit reduced expression in skeletal muscle of diabetic and prediabetic humans. Moreover, these changes may be mediated by the transcriptional coactivator peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α). By combining PGC-1α-induced genome-wide transcriptional profiles with a computational strategy to detect cis-regulatory motifs, we identified estrogen-related receptor α (Errα) and GA repeat-binding protein α as key transcription factors regulating the OXPHOS pathway. Interestingly, the genes encoding these two transcription factors are themselves PGC-1α-inducible and contain variants of both motifs near their promoters. Cellular assays confirmed that Errα and GA-binding protein a partner with PGC-1α in muscle to form a double-positive-feedback loop that drives the expression of many OXPHOS genes. By using a synthetic inhibitor of Errα, we demonstrated its key role in PGC-1α-mediated effects on gene regulation and cellular respiration. These results illustrate the dissection of gene regulatory networks in a complex mammalian system, elucidate the mechanism of PGC-1α action in the OXPHOS pathway, and suggest that Errα agonists may ameliorate insulin-resistance in individuals with type 2 diabetes mellitus.
Peroxisome proliferator-activated receptor γ coactivator 1α and 1β (PGC-1α and PGC-1β) are transcriptional coactivators involved in the regulation of several key metabolic processes, including mitochondrial biogenesis, adaptive thermogenesis, insulin secretion from the pancreas, and gluconeogenesis (1). PGC-1α is capable of coactivating nearly all known nuclear receptors, as well as many other transcription factors (1). In skeletal muscle, PGC-1α dramatically increases respiration and elevates transcript levels of the insulin-sensitive glucose transporter 4 (2, 3). Moreover, transgenic expression of PGC-1α driven by a muscle-specific promoter results in a drastic switch from glycolytic to oxidative muscle fibers (4). Interestingly, PGC-1α- and PGC-1β-responsive genes involved in oxidative phosphorylation (OXPHOS) show reduced expression in the muscle of patients with type 2 diabetes mellitus (5, 6). These changes are also observed in individuals with impaired glucose tolerance (5), as well as in healthy first degree relatives of diabetics (6), suggesting that these defects are causally linked to the development of insulin resistance. Combined with recent morphologic and functional studies (7–9), these reports indicate that a PGC-1-mediated dysregulation in mitochondrial biogenesis is important in the development of human diabetes.
A pressing question now is to identify the transcription factors that specify the changes observed in muscle: these transcription factors may be sites of genetic variation or misregulation, and may represent important therapeutic targets.
Here, we present an analytical strategy for identifying mammalian cis-regulatory elements by using gene expression profiles. We report the successful application of the technique in discovering the key transcription factors recruited by PGC-1α in mediating OX-PHOS gene expression in muscle. In vitro and cellular assays confirm a model of gene regulation in which the transcription factors estrogen-related receptor α (Errα) and GA repeat-binding protein α (Gabpa) form a double-positive-feedback loop with PGC-1α, driving the robust expression of mitochondrial genes. This circuit appears to be the central network mediating mitochondrial biogenesis through PGC-1α and may have implications in the treatment of type 2 diabetes.
Materials and Methods
Cell Culture and Adenoviral Infection. Mouse myoblasts (C2C12 cells) were cultured and differentiated into myotubes as described (2). After 3 days of differentiation, they were transduced with an adenovirus expressing either GFP or PGC-1α. All experiments were performed in duplicate, and flow cytometry was performed to ensure nearly 100% and equivalent infection titers.
RNA Isolation, Target Preparation, and Hybridization. Cells were harvested before (day 0) as well as 1, 2, and 3 days after viral transduction (all performed in duplicate, yielding 14 samples). RNA isolation, cRNA target preparation, and hybridization to the Affymetrix MG-U74Av2 chip were performed as described (5).
Data Scaling, Visualization, and Annotation Enrichment. Microarray data were subjected to linear scaling by using the median scan as a reference. Scaled time course data are available at www-genome.wi.mit.edu/mpg/PGC_motifs. Data were visualized by using the dchip software package (10), and enrichment by ontology terms was determined with the gosurfer tool, using a P value of 0.01 (11). Mitochondrial genes were defined, based on a recent proteomic survey of organelle in mouse (12).
Promoter Databases. We used the Reference Sequence annotations of mm3 build of the mouse genome (http://genome.ucsc.edu) and the annotation tables for the Affymetrix MG-U74Av2 chip (www.affymetrix.com) to compile a list of 5,034 mouse genes for which there is a 1:1 mapping between the Affymetrix probe set and Reference Sequence. The mouse promoter database consists of the 2,000 base pairs centered on the annotated transcriptional start site (TSS) of these genes. We also performed analyses on a masked promoter database, consisting of the regions within these 2,000 base pairs that are aligned and conserved between mouse and human. We used the mouse/human blastz alignments (mouse mm3 versus human hg15; ref. 13) and only considered the 5,008 promoters for which the mouse:human alignment contained at least 100 bp. We masked the aligned promoters to retain mouse sequence exhibiting at least 70% identity to human across windows of size 10. The median promoter length in the masked database is ≈1,200 bp.
Motif Discovery. For a given day, genes from the microarray are ordered on the basis of expression difference between GFP and PGC-1α (we use the signal-to-noise ratio as our difference metric). Each gene is annotated for the presence of a motif in the promoter by searching for exact k-mers (where k = 6, 7, 8, or 9) or for selected motifs of interest. We use the Mann–Whitney rank sum statistic U to determine whether the distribution of differential expression for those genes with a given motif differs from those genes lacking the motif. When working with promoters of unequal length (e.g., the masked promoter database), a more appropriate null hypothesis for the Mann–Whitney statistic is that the probability of detecting a motif in a promoter is proportional to its length. To assess the significance of a motif with rank sum U that appears in C promoters, we use Monte Carlo simulation (with 1,000 samples) to estimate the null distribution of U for a sample of C ranks drawn randomly, without replacement, given relative weights proportional to the promoter lengths. For large C (C > 10) and a reasonable distribution of promoter lengths, U is approximately normally distributed. Promoter databases and the motifs associated with differential expression (motifADE) source code. Complete results are available at www-genome.wi.mit.edu/mpg/PGC_motifs.
Cotransfection Assays. The Errα and the Gabpa promoters and the Gabpa intron 1 were amplified from genomic mouse DNA by PCR, and were subsequently cloned into the pGL3 reporter gene vectors (Promega). Transfections, reporter gene assays, and site-directed mutagenesis were performed as described (14). Simultaneously with the transfection, cells were treated with vehicle (0.1% DMSO) or 1 μM of the inhibitor, 3-[4-(2,4-bis-trifluoromethyl-benzyloxy)-3-methoxy-phenyl]-2-cyano-N-(5-trifluoromethyl-[1,3,4]thiadiazol-2-yl)-acrylamide (XCT790), for 48 h. Motifs for Errα and Gabpa were found at the following positions (core motif is in bold): Errα site at -585 CCGCAGTGACCTTGAGTTTTG -565 and Gabpa site at -41 GGAGGAAGCGGAGTAGGAAGCA -20 in the Errα promoter, Errα site at -614 GCGCCGTGACCTTGGGCTGCC -594 in the Gabpa promoter, and Gabpa sites at +3 GAGTTGCGGACG +14 and +51 TGGTTCCGGGGC +62 in intron 1 of the Gabpa gene. These motifs were either directly identified by motifADE, or, in the case of the Gabpa-binding site in the Errα promoter, were found by looking for other Gabpa motifs that have been described (15).
Semiquantitative Real-Time PCR. Primers for target genes were designed by using the primerexpress software (Applied Biosystems). Relative gene expression was calculated with the ΔΔCt method by using SYBR green on an iCycler (Bio-Rad).
Respiration Assays. For these assays, C2C12 myotubes were infected with adenovirus encoding for GFP or PGC-1α, and were treated with vehicle or XCT790. Two days after infection, total and uncoupled respiration were measured as published (16).
Results
Discovering motifADE. Systematic identification of transcription factors involved in biological processes in mammals remains a largely unsolved problem (17). A promising approach relates genome-wide expression profiles to promoter sequences to discover influential cis motifs (18–21). Such methods have yielded impressive results in simple organisms such as yeast, but it has been challenging to extend these algorithms to mammalian genomes, where intergenic regions are large, annotation of gene structure is imperfect, and DNA sequence can be highly repetitive. Most of these methods seek motifs by comparison with a fixed background model of nucleotide composition (which fails to represent the fluctuations seen in large genomes) or by comparison between two sets of genes (which is likely to capture only very sharp differences). Furthermore, many of these methods assume that the expression data are normally distributed, which may not always be true.
To overcome some of these obstacles, we devised a simple, nonparametric strategy for identifying motifADE (Fig. 1A). The algorithm involves three steps: (i) ranking genes based on differential expression between two conditions; (ii) given a candidate motif, identifying the subset of genes whose promoter regions contains the motif; and (iii) testing by means of a nonparametric, rank sum statistic whether these genes tend to appear toward the top or bottom of the ranked list (indicating association) or are randomly distributed on the list. motifADE may be applied to a specific candidate motif of interest or to the list of all possible motifs of a given size (in which case the significance level should be adjusted to reflect multiple hypothesis testing). By using a nonparametric scoring procedure, we do not make assumptions about the distribution of the expression data. Furthermore, by considering the entire rank-ordered list, the promoters without the motif implicitly provide a background of DNA composition for comparison, and there is no need to demarcate or cluster a group of genes as being “responsive.” The method can operate on a traditional promoter database or even a database of promoters that have been masked based on evolutionary conservation.
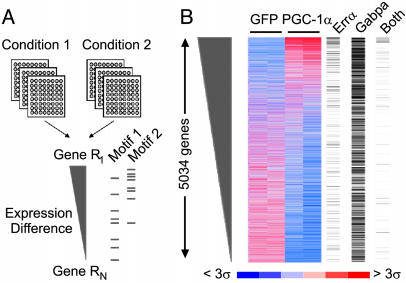
Fig. 1.
Schematic overview of motifADE and application to the PGC-1α time course. (A) The motifADE strategy. It begins with a list of genes ordered on the basis of differential expression across two conditions. Each gene is then annotated for the presence of a given motif in the promoter region. A nonparametric statistic is used to assess whether genes with the motif tend to rank high on this list. In this example, genes with Motif 1 are randomly distributed on the list, whereas genes with Motif 2 tend to rank high, suggesting an association between Motif 2 and the differential expression. (B) C2C12 cells were infected with an adenovirus expressing either GFP (control) or with PGC-1α and were profiled over a 3-day period. Experiments were performed in duplicate and relative gene expression measures are shown. Genes are ranked according to the difference in expression between PGC-1α and GFP on day 3. Mouse genes having a perfect Errα motif (5′-TGACCTTG-3′), a perfect Gabpa/b motif (5′-CTTCCG-3′), or both motifs are labeled with a black bar on the right side of the correlogram.
Binding Sites for Errα and Gabpa Are the Top-Scoring Motifs Associated with the PGC-1α Transcriptional Program. To identify motifs related to PGC-1α action, we infected mouse C2C12 muscle cells with an adenovirus expressing PGC-1α and obtained gene expression profiles for 12,488 genes at 0, 1, 2, and 3 days after infection. We found 649 genes that were induced at least 1.5-fold (nominal P < 0.05) at day 3. As expected, these genes were enriched for genes involved in carbohydrate metabolism and the mitochondrion (1). Interestingly, many genes involved with protein synthesis (gene ontology terms: protein biosynthesis, mitochondrial ribosome, and ribosome) are also induced.
We then applied motifADE to study the 5,034 mouse genes for which we have measures of gene expression, as well as reliable annotations of the TSS. For each gene, the target region was defined to be a 2-kb region centered on the TSS. We then tested all possible k-mers ranging in size from k = 6 to k = 9 nucleotides for association with differential expression on each of the 3 days of the time course. A total of 20 motifs achieved high statistical significance (P < 0.001, after Bonferroni correction for multiple hypothesis testing) and these were almost exclusively related to two distinct motifs (Table 1, and Table 2, which is published as supporting information on the PNAS web site). The first motif, 5′-TGACCTTG-3′ was significant on days 1, 2, and 3 (adjusted P = 2.1 × 10-6,2.9 × 10-9, and 7.7 × 10-7, respectively). It corresponds to the published binding site for the orphan nuclear receptor Errα (22), which is known to be capable of being coactivated by PGC-1α and -1β (23–25). The Errα gene is known to be involved in metabolic processes, based on studies showing that knockout mice have reduced body weight and peripheral fat tissue, as well as altered expression of genes involved in metabolic pathways (26). The second motif is 5′-CTTCCG-3′ (adjusted P = 8.9 × 10-9), which is the top-scoring motif on day 3. It corresponds to the published binding site for Gabpa (27), which complexes with Gabpb (15) to form the heterodimer, nuclear respiratory factor-2 (NRF-2), a factor known to regulate the expression of some OXPHOS genes (28). Interestingly, the reverse complements of these motifs did not score as well, suggesting a preference for the orientation of these motifs, and some occurrences of the motifs were found downstream of the TSS. Whereas each of these motifs is individually associated with PGC-1α, our analyses suggest that a gene having both motifs typically ranks higher on the list of differentially expressed genes than genes with only one of the motifs (Fig. 6, which is published as supporting information on the PNAS web site), suggesting that the two motifs might have an additive or synergistic effect.
Table 1. Motifs associated with differential expression on days 1, 2, and 3.
| Day | Motif | Frequency | Nominal P value | Adjusted P value | Annotation | Ref. |
|---|---|---|---|---|---|---|
| 1 | TGACCTTG | 0.07 | 3.15 × 10-11 | 2.06 × 10-6 | Errα | 22 |
| TGACCTTGA | 0.02 | 4.59 × 10-10 | 1.20 × 10-4 | Errα | ||
| 2 | TGACCTTG | 0.07 | 4.44 × 10-14 | 2.91 × 10-9 | Errα | 22 |
| TGACCTT | 0.16 | 3.62 × 10-12 | 5.93 × 10-8 | Errα | ||
| TGACCT | 0.45 | 1.46 × 10-11 | 5.97 × 10-8 | NR half-site | 34 | |
| GACCTTG | 0.16 | 7.92 × 10-11 | 1.30 × 10-6 | Errα | ||
| GACCTT | 0.41 | 1.42 × 10-9 | 5.81 × 10-6 | Errα | ||
| TTGACC | 0.27 | 2.42 × 10-7 | 9.92 × 10-4 | Errα | ||
| 3 | CTTCCG | 0.33 | 2.19 × 10-12 | 8.97 × 10-9 | Gabpa | 35 |
| TGACCTTG | 0.07 | 1.17 × 10-11 | 7.66 × 10-7 | Errα | 22 | |
| TGACCTT | 0.16 | 1.23 × 10-10 | 2.02 × 10-6 | Errα | ||
| CCCGCC | 0.54 | 2.04 × 10-8 | 8.36 × 10-5 | |||
| GCGGCG | 0.43 | 3.78 × 10-8 | 1.55 × 10-4 | |||
| AGGTCA | 0.42 | 3.90 × 10-8 | 1.60 × 10-4 | NR half-site | 34 | |
| CTTCCGG | 0.16 | 1.95 × 10-8 | 3.19 × 10-4 | Gabpa | ||
| TTCCGG | 0.31 | 1.09 × 10-7 | 4.46 × 10-4 | Gabpa | ||
| GGGGCG | 0.54 | 1.24 × 10-7 | 5.08 × 10-4 | |||
| TTCCGCT | 0.07 | 3.30 × 10-6 | 5.41 × 10-4 | Gabpa | ||
| GCCGGC | 0.42 | 1.57 × 10-7 | 6.44 × 10-4 | |||
| ACTTCCG | 0.09 | 5.11 × 10-8 | 8.38 × 10-4 | Gabpa |
motiFADE was performed by using the mouse promoter database on each of days 1, 2, and 3. All motifs achieving a Bonferroni-corrected P value < 1 × 10-3 are shown. Annotations of the motif and the literature references, when available, are indicated. NR, nuclear receptor.
Errα and Gabpa Motifs Are Evolutionarily Conserved and Enriched Upstream of OXPHOS Genes. We next repeated motifADE analysis by using a “masked” promoter database (Table 3, which is published as supporting information on the PNAS web site). We still considered the 2,000 base pairs centered on the TSS, but we only considered those nucleotides aligned and conserved between mouse and human. Still, the top ranking motifs on days 1 and 3 were related to Errα (adjusted P = 4.8 × 10-6, P = 1.2 × 10-11, respectively) and to Gabpa (day 3 P = 3.1 × 10-11), providing additional support that these motifs are biologically relevant.
The Errα and Gabpa motifs are particularly enriched upstream of a coregulated subset of OXPHOS genes, which exhibit reduced expression in human diabetes (5, 6). Whereas the top-scoring Errα motif (5′-TGACCTTG-3′ or its reverse complement) only occurs in 12% of the promoters in the database, in 29% of the PGC-responsive genes (i.e., those genes induced at least 1.5-fold on day 3), and in 27% of the mitochondrial genes, they are found in 52% of the coregulated subset of OXPHOS genes (significance of enrichment, P = 1 × 10-4). Approximately one-half of these sites are perfectly conserved in the syntenic region in human (data not shown). The top-scoring Gabpa-binding sites (5′-CTTCCG-3′ or its reverse complement) are much more common (62% of all promoters of the database and in 79% of the PGC-responsive genes), however, they too show significant enrichment in the coregulated subset of OXPHOS genes (89%, P = 0.02).
Errα and Gabpa Are Themselves Induced by PGC-1α. The above results suggest that Errα and Gabpa may be the key transcriptional factors mediating PGC-1α action in muscle. In this connection, it is notable that based on the microarray data, both Errα and Gabpa are themselves induced ≈2-fold (P < 0.01) on day 1 after expression of PGC-1α, which is consistent with previous studies (2, 23). Moreover, careful analysis of the Errα and Gabpa genes suggest that each contain potential binding sites for both transcription factors within the vicinity of their promoters. The Errα gene has the Errα motif, as well as a conserved variant of the Gabpa-binding site (27) upstream of the TSS, whereas the Gabpa gene has an Errα site upstream of the TSS and a conserved variant of the Gabpa-binding site in its first intron. These results raise the possibility that Errα and Gabpa may regulate their own expression, and that of each other.
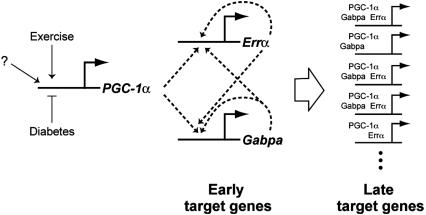
A Transcriptional Switch Driving Mitochondrial Biogenesis. Taken together, the systematic analysis of the transcriptional program driven by PGC-1α in skeletal muscle suggests a model (Fig. 2) in which increases in PGC-1α protein levels results in increased transcriptional activity of Gabpa and Errα on their own promoters, leading to a stable increase in the expression of these two factors by means of a double-positive-feedback loop. These two factors, perhaps in combination with PGC-1α, are then crucial in the induction of downstream target genes, many of which have binding sites for these motifs (Fig. 2). Such a circuit may serve as a regulatory switch, analogous to a feed-forward loop that plays a key role in the early stages of endomesodermal development in sea urchin (29).
Fig. 2.
Proposed model of mechanism of action of PGC-1α. PGC-1α is a highly regulated gene that responds to external stimuli, e.g., reduced in diabetes and increased after exercise. When PGC-1α levels rise, the expression of Errα and Gabpa are immediately induced by means of a double-positive-feedback loop. These levels rise, and over the course of 2–3 days, these factors couple with PGC-1α to induce the expression of NRF-1 and hundreds of downstream targets, such as OXPHOS and other mitochondrial genes that are enriched for these transcription factor-binding sites.
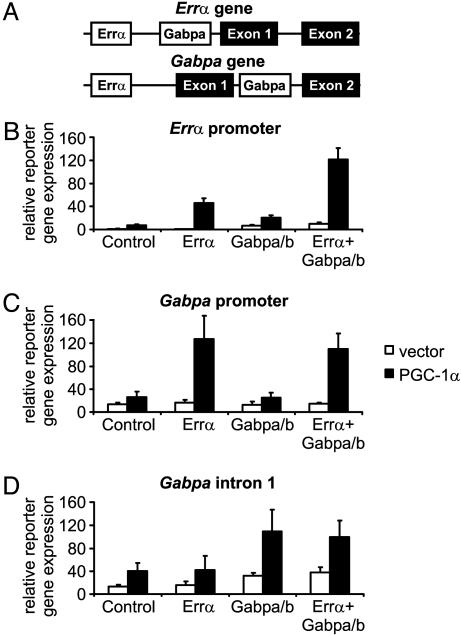
Experimental Validation of the Model. We experimentally tested this model of gene regulation. To determine whether Errα and Gabpa are indeed key transcriptional partners of PGC-1α, we tested whether they are coactivated by PGC-1α on the Errα and Gabpa promoters (Fig. 3). We cloned appropriate portions of these promoters into expression vectors and tested these factors' ability to drive expression of a reporter gene, in the presence and absence of PGC-1α. The results confirm the predicted coactivation between PGC-1α and the two transcription factors in muscle cells. Moreover, the results for the Errα promoter fragment containing both sites show that the transcription factors act additively or even synergistically (Fig. 3).
Fig. 3.
Errα and Gabpa cooperate with PGC-1α to induce their own expression. (A) Putative Errα and Gabpa motifs 1 kb upstream and downstream of the Errα and the Gabpa TSSs. (B–D) C2C12 cells were transfected with a reporter gene plasmid containing 2 kb of the Errα promoter (B), Gabpa promoter (C), or Gabpa intron 1 (D), together with expression plasmids for Errα, Gabpa, Gabpb1, and PGC-1α. Forty-eight hours after transfection, reporter gene levels were determined and were normalized to β-galactosidase levels.
We next sought to explore the functional role of Errα in executing the PGC-1α-mediated response, by using a recently developed, selective Errα inverse agonist, called XCT790 (referred to below as the inhibitor; Fig. 7, which is published as supporting information on the PNAS web site). This compound does not significantly inhibit or activate other nuclear receptors at doses near its IC50 (583 nM) for Errα, although higher concentrations (>3.3 μM) have been found to weakly activate PPARγ (R.A.H., personal communication). In an affinity chromatography assay, XCT790 specifically reduced the interaction between PGC-1α and Errα, whereas PGC-1α binding to another nuclear receptor, the hepatic nuclear factor 4α was not affected (Fig. 8, which is published as supporting information on the PNAS web site).
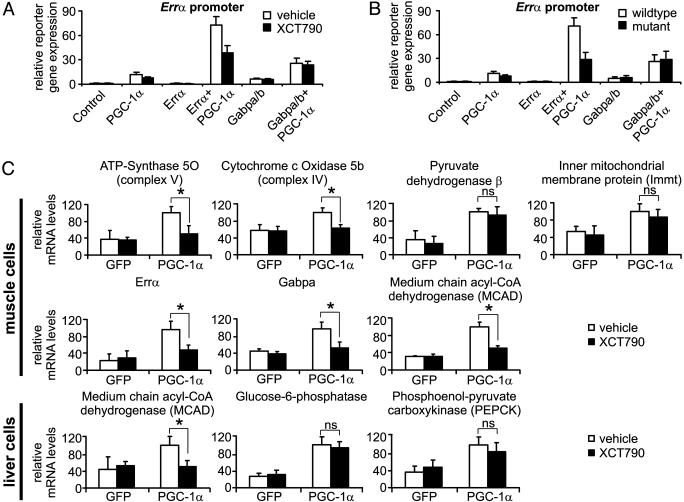
The inhibitor was first used to probe the role of Errα in activation of Errα and Gabpa, by repeating the above described experiments in the presence or absence of the inhibitor at a dose slightly above its IC50. The inhibitor markedly reduced activation of the Errα promoter by Errα and PGC-1α, but had no effect on Gabpa/b-driven reporter gene expression (Fig. 4A). The effect of the Errα inhibitor resembles the effect of site-directed mutagenesis of the Errα-binding site on the Errα promoter (Fig. 4B).
Fig. 4.
A small-molecule inhibitor of Errα inhibits the response to PGC-1α. (A) C2C12 myoblasts transfected with wild-type Errα promoter and the respective expression plasmids were treated with vehicle (0.1% DMSO) or 1 μM XCT790 for 48 h before reporter gene levels were measured. (B) C2C12 cells were transfected with wild-type or Errα-promoter harboring a mutated Errα motif (mutant), together with expression plasmids for Errα, Gabpa, Gabpb1, and PGC-1α. After 48 h, reporter gene levels were determined and normalized to β-galactosidase levels. (C) C2C12 myotubes were infected with GFP- or PGC-1α adenovirus and were treated with vehicle (0.1% DMSO) or 1 μM XCT790 for 1 day. Relative expression levels of several genes were then determined by semiquantitative real-time PCR and were normalized against 18S rRNA levels. (D) Fao rat hepatoma cells were infected with adenoviral GFP or PGC-1α and were treated with vehicle (0.1% DMSO) or 1 μM XCT790 for 1 day before relative gene expression levels were measured by real-time PCR, and were normalized against 18S rRNA levels. *, P < 0.05. NS, not significant.
The role of Errα was next investigated in the regulation of endogenous PGC-1α target genes. PGC-1α was expressed in C2C12 myotubes, treated with inhibitor or vehicle, and we measured the expression level of specific genes on day 1 by using RT-PCR. We studied five genes that contain variants of the Errα motif, including two of the coregulated subset of OXPHOS genes, Gabpa, and Errα, as well as medium-chain acyl-CoA dehydrogenase (MCAD), a published target of Errα (30). The expression of all of these genes is induced by PGC-1α, and this induction is diminished by the inhibitor (Fig. 4C). The specificity of the response was confirmed by using two genes that are induced by PGC-1α that lack obvious Errα-binding sites (Fig. 4C). Together, these results suggest that a subset of PGC-1α genes are regulated by Errα, primarily the target genes containing an Errα motif. The fact that the inhibition of these genes by the ERRα inverse agonist was not complete suggests that other factors might also be involved in this regulation. To ensure that the effects of the Errα inhibitor were not due to activation of PPARγ, control experiments were performed by using the synthetic PPARγ agonist rosiglitazone, and confirmed that it had no effect on the expression of these genes (Fig. 9, which is published as supporting information on the PNAS web site).
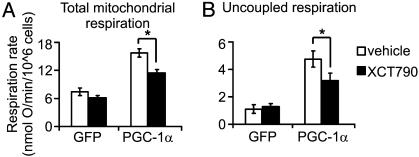
Errα Inhibition Reduces the PGC-1α Program in Cellular Assays. The role of Errα in the PGC-1α-mediated physiologic response was characterized by using functional assays of mitochondrial respiration. The Errα inhibitor potently diminishes PGC-1α induction of total cellular respiration and also significantly reduces PGC-1α-triggered increases in uncoupled respiration (Fig. 5). The data suggest that inhibition of Errα elicits certain aspects of a diabetic phenotype in cultured muscle cells.
Fig. 5.
Total and uncoupled mitochondrial respiration are inhibited by the synthetic Errα-inverse agonist XCT790. (A and B) C2C12 myotubes were infected with GFP or PGC-1α adenovirus and were treated with vehicle (0.1% DMSO) or 1 μM XCT790 for 2 days before total mitochondrial respiration (A) and uncoupled respiration (B) was determined. *, P < 0.05 in paired t test.
Inhibition of Errα Does Not Affect Gluconeogenesis in Liver. We extended the analysis to cells derived from liver, where PGC-1α has been shown to induce genes of the fasting response, including those involved in gluconeogenesis and β-oxidation of fatty acids (1). These responses are generally elevated in poorly controlled diabetics. As expected, MCAD and two key genes related to gluconeogenesis (glucose-6-phosphatase and phosphoenol-pyruvate carboxykinase) were all induced by PGC-1α. However, the Errα antagonist repressed PGC-1α induction of MCAD but not the gluconeogenic genes (Fig. 4D).
NRF-1 Is Downstream of Errα and Gabpa. Finally, we investigated the relationship of NRF-1 to Errα function in muscle cells. NRF-1 has been suggested to be a key transcription factor involved in mito-chondrial biogenesis (31), and is induced and coactivated by PGC-1α in this process (2). However, what has been puzzling is that transgenic overexpression of NRF-1 in muscle is not sufficient to induce mitochondrial biogenesis (32). By using the synthetic Errα inverse agonist, we found that induction of NRF-1 by PGC-1α is actually downstream of the initial Errα-mediated events (Fig. 10, which is published as supporting information on the PNAS web site). Interestingly, motifADE analysis of the NRF-1 consensus motif (5′-GCGCAYGCGC-3′ or reverse complement) yields a score that is significant if considered as a test of single hypothesis (nominal P = 0.001 on day 3). Overall, these results suggest that NRF-1 is an important but relatively late mediator of the mito-chondrial transcriptional program induced by PGC-1α.
Discussion
In this study, we have used a computational strategy to systemat-ically dissect a mammalian transcriptional circuit central to cellular energetics. The results above have computational, biological, and medical implications.
First, the motifADE algorithm provides a simple, nonparametric approach for discovering cis elements by considering differential gene expression. The method makes very few assumptions about the statistical properties of DNA composition or about the distribution of gene expression. The method is flexible, and as we have shown, can easily incorporate “masked” or “phylogenetically foot-printed” promoters from cross-species comparisons. With additional cross-species comparisons, it should be possible to interrogate larger promoter regions (33). Moreover, the method may be useful for discovering motifs associated with human disease data sets, such as healthy versus sick, or treated versus control, etc. Clearly, the method has some limitations. For example, generic, experimental strategies will be needed to systematically determine the occupancy of newly identified motifs. Moreover, a motif may be missed if it lies outside the target promoter region, or if a functional binding site is too degenerate for our motif search strategy.
Second, the analyses above indicate that the immediate effects of PGC-1α on OXPHOS genes in muscle are largely mediated through Errα and Gabpa. Recent studies (25) have shown that PGC-1β can also coactivate Errα, and initial application of motifADE to cells expressing PGC-1β suggests that these same factors are also important in regulating this program (data not shown). Together, the data imply a model of gene regulation in which PGC-1α (and likely PGC-1β) initially induces the expression of Errα and Gabpa, by means of a double-positive-feedback mechanism (Fig. 2). These transcription factors are then expressed at higher levels and are themselves coactivated by PGC-1 to induce downstream genes such as NRF-1 and members of OXPHOS. Certainly, other transcription factors and regulators, not identified in the current study, are involved in the mitochondrial biogenesis program. Whereas previous studies have shown that PGC-1 interacts with and/or induces 15–20 transcription factors in various physiological settings [including Errα and Gabpa (2, 23–25)], the present study points to Errα and Gabpa as being especially important early in the time course in muscle.
Finally, the results suggest a potential approach to the treatment of type 2 diabetes. Recent studies (5, 6, 8, 9) in diabetic and prediabetic humans have demonstrated that there is a consistent decrease in the expression of genes of OXPHOS phosphorylation that are responsive to PGC-1α and PGC-1β, and that treatments that induce PGC-1α (such as exercise) lead to increased expression of OXPHOS genes and improved insulin sensitivity. On its face, this result might argue for developing therapeutic approaches that raise the transcriptional activity of PGC-1. However, PGC-1 activates many different pathways in many tissues, and such approaches may suffer from lack of specificity. For example, global transgenic overexpression of PGC-1β in mice results in resistance to obesity induced by a high-fat diet or by a genetic abnormality, although the contribution of PGC-1β expression in muscle has not been explored (25). On the other hand, a global knockout of Errα also causes a leaner phenotype and resistance to high-fat-diet-induced obesity (26). Thus, the identification of the critical roles of Errα and Gabpa in mediating the transcriptional program altered in human diabetic muscle may offer a more specific target. Errα plays a key role in muscle cells but does not appear to affect genes involved in gluconeogenesis in liver cells. This finding implies that synthetic Errα agonists, particularly those that promote docking of PGC-1α on Errα, may serve to ameliorate insulin resistance in human diabetic muscle without exacerbating hyperglycemia in the liver. Because Errα is an orphan nuclear receptor, it may be an attractive, “druggable” target for diabetes and other human metabolic disorders.
Supplementary Material
Acknowledgments
We thank H. Bolouri for valuable discussions. This work was supported in part by National Institutes of Health grants (to B.M.S.). V.K.M. was supported by the Howard Hughes Medical Institute as a Physician Post-doctoral Fellow, and C.H. was supported by a Fellowship of the Schweizerische Stiftung für Medizinisch-Biologische Stipendien, the Swiss Academy of Medical Sciences, and the Swiss National Science Foundation.
Abbreviations: OXPHOS, oxidative phosphorylation; PGC-1, peroxisome proliferator-activated receptor γ coactivator-1; Errα, estrogen-related receptor α; Gabp, GA repeat-binding protein; motifADE, motifs associated with differential expression; TSS, transcriptional start site; NRF, nuclear respiratory factor; MCAD, medium-chain acyl-CoA dehydrogenase; XCT790, 3-[4-(2,4-bis-trifluoromethyl-benzyloxy)-3-methoxy-phenyl]-2-cyano-N-(5-trifluoromethyl-[1,3,4]thiadiazol-2-yl)-acrylamide.
References
- 1.Puigserver, P. & Spiegelman, B. M. (2003) Endocr. Rev. 24, 78-90. [DOI] [PubMed] [Google Scholar]
- 2.Wu, Z., Puigserver, P., Andersson, U., Zhang, C., Adelmant, G., Mootha, V., Troy, A., Cinti, S., Lowell, B., Scarpulla, R. C., et al. (1999) Cell 98, 115-124. [DOI] [PubMed] [Google Scholar]
- 3.Michael, L. F., Wu, Z., Cheatham, R. B., Puigserver, P., Adelmant, G., Lehman, J. J., Kelly, D. P. & Spiegelman, B. M. (2001) Proc. Natl. Acad. Sci. USA 98, 3820-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin, J., Wu, H., Tarr, P. T., Zhang, C. Y., Wu, Z., Boss, O., Michael, L. F., Puigserver, P., Isotani, E., Olson, E. N., et al. (2002) Nature 418, 797-801. [DOI] [PubMed] [Google Scholar]
- 5.Mootha, V. K., Lindgren, C. M., Eriksson, K. F., Subramanian, A., Sihag, S., Lehar, J., Puigserver, P., Carlsson, E., Ridderstrale, M., Laurila, E., et al. (2003) Nat. Genet. 34, 267-273. [DOI] [PubMed] [Google Scholar]
- 6.Patti, M. E., Butte, A. J., Crunkhorn, S., Cusi, K., Berria, R., Kashyap, S., Miyazaki, Y., Kohane, I., Costello, M., Saccone, R., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 8466-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelley, D. E., He, J., Menshikova, E. V. & Ritov, V. B. (2002) Diabetes 51, 2944-2950. [DOI] [PubMed] [Google Scholar]
- 8.Petersen, K. F., Befroy, D., Dufour, S., Dziura, J., Ariyan, C., Rothman, D. L., DiPietro, L., Cline, G. W. & Shulman, G. I. (2003) Science 300, 1140-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sreekumar, R., Halvatsiotis, P., Schimke, J. C. & Nair, K. S. (2002) Diabetes 51, 1913-1920. [DOI] [PubMed] [Google Scholar]
- 10.Schadt, E. E., Li, C., Ellis, B. & Wong, W. H. (2001) J. Cell. Biochem. Suppl. 37, 120-125. [DOI] [PubMed] [Google Scholar]
- 11.Zhong, S., Li, C. & Wong, W. H. (2003) Nucleic Acids Res. 31, 3483-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mootha, V. K., Bunkenborg, J., Olsen, J. V., Hjerrild, M., Wisniewski, J. R., Stahl, E., Bolouri, M. S., Ray, H. N., Sihag, S., Kamal, M., et al. (2003) Cell 115, 629-640. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz, S., Kent, W. J., Smit, A., Zhang, Z., Baertsch, R., Hardison, R. C., Haussler, D. & Miller, W. (2003) Genome Res. 13, 103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handschin, C., Rhee, J., Lin, J., Tarr, P. T. & Spiegelman, B. M. (2003) Proc. Natl. Acad. Sci. USA 100, 7111-7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batchelor, A. H., Piper, D. E., de la Brousse, F. C., McKnight, S. L. & Wolberger, C. (1998) Science 279, 1037-1041. [DOI] [PubMed] [Google Scholar]
- 16.St-Pierre, J., Lin, J., Krauss, S., Tarr, P. T., Yang, R., Newgard, C. B. & Spiegelman, B. M. (2003) J. Biol. Chem. 278, 26597-26603. [DOI] [PubMed] [Google Scholar]
- 17.Qiu, P. (2003) Biochem. Biophys. Res. Commun. 309, 495-501. [DOI] [PubMed] [Google Scholar]
- 18.Tavazoie, S., Hughes, J. D., Campbell, M. J., Cho, R. J. & Church, G. M. (1999) Nat. Genet. 22, 281-285. [DOI] [PubMed] [Google Scholar]
- 19.Liu, X. S., Brutlag, D. L. & Liu, J. S. (2002) Nat. Biotechnol. 20, 835-839. [DOI] [PubMed] [Google Scholar]
- 20.Conlon, E. M., Liu, X. S., Lieb, J. D. & Liu, J. S. (2003) Proc. Natl. Acad. Sci. USA 100, 3339-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bussemaker, H. J., Li, H. & Siggia, E. D. (2001) Nat. Genet. 27, 167-171. [DOI] [PubMed] [Google Scholar]
- 22.Johnston, S. D., Liu, X., Zuo, F., Eisenbraun, T. L., Wiley, S. R., Kraus, R. J. & Mertz, J. E. (1997) Mol. Endocrinol. 11, 342-352. [DOI] [PubMed] [Google Scholar]
- 23.Schreiber, S. N., Knutti, D., Brogli, K., Uhlmann, T. & Kralli, A. (2003) J. Biol. Chem. 278, 9013-9018. [DOI] [PubMed] [Google Scholar]
- 24.Huss, J. M., Kopp, R. P. & Kelly, D. P. (2002) J. Biol. Chem. 277, 40265-40274. [DOI] [PubMed] [Google Scholar]
- 25.Kamei, Y., Ohizumi, H., Fujitani, Y., Nemoto, T., Tanaka, T., Takahashi, N., Kawada, T., Miyoshi, M., Ezaki, O. & Kakizuka, A. (2003) Proc. Natl. Acad. Sci. USA 100, 12378-12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo, J., Sladek, R., Carrier, J., Bader, J. A., Richard, D. & Giguere, V. (2003) Mol. Cell. Biol. 23, 7947-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chinenov, Y., Coombs, C. & Martin, M. E. (2000) Gene 261, 311-320. [DOI] [PubMed] [Google Scholar]
- 28.Virbasius, J. V. & Scarpulla, R. C. (1994) Proc. Natl. Acad. Sci. USA 91, 1309-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davidson, E. H., Rast, J. P., Oliveri, P., Ransick, A., Calestani, C., Yuh, C. H., Minokawa, T., Amore, G., Hinman, V., Arenas-Mena, C., et al. (2002) Science 295, 1669-1678. [DOI] [PubMed] [Google Scholar]
- 30.Sladek, R., Bader, J. A. & Giguere, V. (1997) Mol. Cell. Biol. 17, 5400-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scarpulla, R. C. (2002) Gene 286, 81-89. [DOI] [PubMed] [Google Scholar]
- 32.Baar, K., Song, Z., Semenkovich, C. F., Jones, T. E., Han, D. H., Nolte, L. A., Ojuka, E. O., Chen, M. & Holloszy, J. O. (2003) FASEB J. 17, 1666-1673. [DOI] [PubMed] [Google Scholar]
- 33.Kellis, M., Patterson, N., Endrizzi, M., Birren, B. & Lander, E. S. (2003) Nature 423, 241-254. [DOI] [PubMed] [Google Scholar]
- 34.Stunnenberg, H. G. (1993) BioEssays 15, 309-315. [DOI] [PubMed] [Google Scholar]
- 35.Chinenov, Y., Henzl, M. & Martin, M. E. (2000) J. Biol. Chem. 275, 7749-7756. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.