Abstract
Corazonin is a highly conserved neuropeptide hormone of wide-spread occurrence in insects yet is associated with no universally recognized function. After discovery of the corazonin receptor in Drosophila, we identified its ortholog in the moth, Manduca sexta, as a prelude to physiological studies. The corazonin receptor cDNA in M. sexta encodes a protein of 436 amino acids with seven putative transmembrane domains and shares common ancestry with its Drosophila counterpart. The receptor exhibits high sensitivity and selectivity for corazonin when expressed in Xenopus oocytes (EC50 ≈ 200 pM) or Chinese hamster ovary cells (EC50 ≈ 75 pM). Northern blot analysis locates the receptor in peripheral endocrine Inka cells, the source of preecdysis- and ecdysis-triggering hormones. Injection of corazonin into pharate larvae elicits release of these peptides from Inka cells, which induce precocious preecdysis and ecdysis behaviors. In vitro exposure of isolated Inka cells to corazonin (25-100 pM) induces preecdysis- and ecdysis-triggering hormone secretion. Using corazonin receptor as a biosensor, we show that corazonin concentrations in the hemolymph 20 min before natural preecdysis onset range from 20 to 80 pM and then decline over the next 30-40 min. These findings support the role of corazonin signaling in initiation of the ecdysis behavioral sequence. We propose a model for peptide-mediated interactions between Inka cells and the CNS underlying this process in insect development.
Corazonin, a blocked undecapeptide hormone, originally was identified as a cardioaccelerator in the cockroach, Periplaneta americana (1). The identical molecule subsequently was found in the cockroach Nauphoeta cinerea, cricket Gryllus bimaculatus, moths Manduca sexta, Bombyx mori, Galleria mellonella, and fly Drosophila melanogaster (2-5). A closely related peptide [(His)7-corazonin] was identified in locusts Schistocerca americana, Schistocerca gregaria, Locusta migratoria, and stick insect Carausius morosus (1, 6, 7). Corazonin is produced by lateral brain neurosecretory cells projecting to the corpora cardiaca-corpora allata complex and in neurons of the ventral nerve cord (8, 9). These findings demonstrate a widespread distribution and conservation of corazonin structure in diverse insect groups.
Because corazonin is highly conserved with respect to structure and spatial expression pattern, it seems likely that physiological functions would be conserved as well, but no clear pattern of function has emerged. Its cardioacceleratory role in P. americana has not been demonstrated in other insects. Subsequent studies showed that corazonin induces dark color and morphometric phase changes in locusts (6, 8, 10, 11). However, this effect has not been observed in other insects, including G. bimaculatus, B. mori, and G. mellonella (4, 5). A recent report that corazonin reduces the spinning rate of silk during the larva-pupa transition in B. mori indicates potential functions associated with molting and behavior (12). In addition, corazonin is colocalized with the PER protein in brain neurons (13), suggesting a possible role in circadian pacemaking.
The identification of putative hormone G protein-coupled receptors in the Drosophila genome has provided a stimulus for associating orphan receptors with their peptide ligands (14, 15). Of obvious utility in discerning function is the location of receptors in identified target cells and tissues. Identification of the Drosophila gene CG10698 encoding the corazonin receptor (DrmCRZR) was the first step in assigning as-yet-unrecognized functions for this peptide (14, 16). To investigate corazonin action in a physiologically amenable organism, we cloned the DrmCRZR ortholog from M. sexta (MasCRZR) and demonstrate here its sensitivity and selectivity for corazonin. High levels of MasCRZR transcript are detected in Inka cells, which release preecdysis-triggering hormone (PETH) and ecdysis-triggering hormone (ETH) in response to corazonin exposure. These data, combined with evidence that corazonin circulates in the hemolymph just before preecdysis onset, reveal its likely function in initiation of the ecdysis sequence. We propose a model for sequential phases of PETH and ETH release under the control of circulating corazonin and eclosion hormone (EH): Corazonin controls initial release of PETH and ETH, whereas subsequent release of EH induces a massive release of these peptides from Inka cells.
Materials and Methods
Cloning of MasCRZR. MasCRZR was cloned by degenerate PCR, 3′-RACE, and 5′-RACE. For degenerate PCR, primers were selected according to the DrmCRZR protein sequence of the highly conserved regions among DrmCRZR (GenBank accession no. AF522192), D. melanogaster adipokinetic hormone receptor (AKHR) (GenBank accession no. AAF52426), Anopheles gambiae CRZR (GenBank accession no. XP-321555), and B. mori (Bom)AKHR (GenBank accession no. AAL95712) and gonadotropin-releasing hormone receptors (GnRHRs) from eel (GenBank accession no. BAB11961), chicken (GenBank accession no. CAC18674), and rat (GenBank accession no. U92471). Total RNA was isolated from whole bodies of second-instar larvae and used to synthesize cDNA. PCRs with nested sets of degenerate primers were performed with Taq (Invitrogen). PCR products of the expected size were cloned and sequenced at the University of California Riverside Genomics Institute. Full-length cDNA sequences were retrieved by 3′-RACE and 5′-RACE. For 5′-RACE, the protocol developed by Shi et al. (17) was used with minor modifications. The composite clone (-37 to +1,342) containing the entire ORF (1,308 bp) was amplified directly from cDNA by a single round of PCR (35 cycles) by using a Taq (Invitrogen) and Pfu (Stratagene) mixture (0.5 units/0.5 units). The composite clone was subcloned in pcDNA3.1(+) (Invitrogen) for Chinese hamster ovary (CHO) cell expression and pGH19A for Xenopus oocyte expression.
Phylogenetic Analysis. clustalx was used to align amino acid sequences of MasCRZR, DrmCRZR, BomAKHR, DrmAKHR, eel GnRHR, chicken GnRHR, and GnRHR (18). CG6111 (putative crustacean cardioactive peptide receptor, GenBank accession no. AAN10041) and AVPR (vasopressin V1a receptor, GenBank accession no. P37288) were included as an outgroup. Rooted trees were produced from the aligned sequences of transmembrane domains 1 to 7 by PAUP (V4.0b8a) by using bootstrap with heuristic search and 1,000 replicates (the optimality criterion: distance).
Xenopus Oocyte and CHO Cell Expression for Functional Analysis. MasCRZR was expressed and assayed in Xenopus oocytes according to described protocols (14). CHO-K1 cells were transiently transfected with an ORF of MasCRZR and codon-optimized aequorin (19). Procedures used for CHO cell transfection and receptor assay were as described (20). Sources of all peptides used in this study were as described (14, 21) except for GnRH, which was purchased from Sigma.
Northern Blot Analysis. Total RNA was extracted from the entire CNS, epidermis, epitracheal glands (EGs) containing Inka cells, trachea, or muscle of pharate pupal M. sexta (dorsal-bar stage) by using the RNeasy minikit (Qiagen, Valencia, CA). For Northern analysis, 10 μg of total RNA was separated on a 1.2% formaldehyde agarose gel and transferred to a nylon membrane (Amersham Pharmacia). The filter was hybridized to an α-32PdCTP-labeled cDNA probe and washed and analyzed by using a Typhoon 9410 PhosphorImager (Amersham Pharmacia).
Corazonin Injection and Observation of Behavioral Responses. Corazonin was injected into pharate fifth-instar larvae just after they absorbed molting fluid (≈6 h before ecdysis). The onset and duration of the ecdysis behavioral sequence were observed under a stereomicroscope.
Effects of corazonin, EH, and ETH on Inka cells were compared in pharate fourth-instar larvae with brown mandibles (≈4 h before expected ecdysis). Larvae were ligated between the first two abdominal segments, and the head with thorax was cut off to remove brain neurosecretory cells producing corazonin and EH. Isolated abdomens were injected with corazonin (1 pmol), EH (1 pmol), or ETH (10 pmol), and initiation of preecdysis was observed under a stereomicroscope. Exactly 60 min after preecdysis onset, entire abdomens were fixed in Bouin's fixative overnight and processed for immunohistochemical staining with PETH antiserum to detect peptide release from Inka cells as described (22).
In Vitro Incubation of EGs and Enzyme Immunoassays. EGs were dissected from pharate pupae ≈3.5 h before natural ecdysis (anterior shrinkage) (21). Glands were incubated individually in 100 μl of Weever's solution containing 0.3% BSA. Different concentrations of corazonin or EH (10 nM to 1 pM) were added to each drop of the saline. Control glands were incubated in saline lacking corazonin or EH. After incubation of glands for 40-45 min, each saline drop was collected, and the amounts of released PETH and ETH were measured by enzyme immunoassays as described (21).
Hemolymph Levels of Corazonin During Ecdysis. By using the time of molting-fluid resorption as a marker (-5 h), hemolymph samples were taken at the following time points: 3 h before preecdysis onset (-180 min), at the onset of intermittent preecdysis (-20 min), and 5, 10, and 30 min after preecdysis onset (time 0). Hemolymph samples (300 μl) were fractionated by reversed-phase high-pressure liquid chromatography (HPLC) [ProStar (Varian) equipped with a Microsorb C-18 column (Rainin Instruments)]. Fractions containing corazonin were assayed for MasCRZR activation by using the CHO-cell-expression system described above. Standard curves were generated in every assay (for example, see Fig. 2C). Each batch of HPLC runs was flanked or preceded by a standard synthetic corazonin run (10 fmol of synthetic corazonin) to ensure accurate elution time of corazonin and to calibrate the efficiency of peptide recovery. To calculate overall recovery from the entire procedure, hemolymph samples were taken at -5 h, spiked with 10 fmol of corazonin before heat treatment, and compared with nonspiked samples. The recovery rate estimated by this method was ≈90%. Details regarding this procedure are available in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Fig. 2.
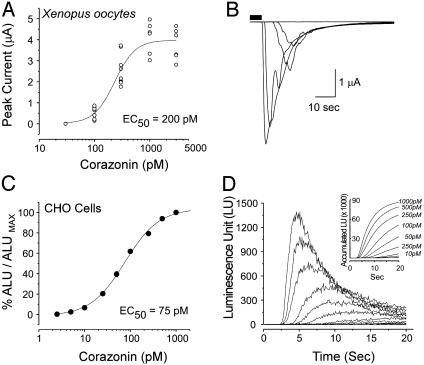
Concentration-response relationships for MasCRZR expressed in Xenopus oocytes (A and B) and CHO cells transiently transfected with aequorin (C and D). (A and C) Concentration-dependent response of MasCRZR to corazonin and EC50 values. (B and D) Corazonin-evoked inward currents measured in Xenopus oocytes and luminescence response [luminescence units (LU)] from CHO cells, respectively. (D Inset) Accumulated luminescence units (ALU) reach a plateau at 1 nM corazonin.
Results
Identification of the Manduca CRZR. Nested PCR using degenerate primers based on highly conserved regions among DrmCRZR, DrmAKHR, BomAKHR, and GnRHRs amplified a ≈300-bp fragment from cDNA prepared from whole second-instar larvae. Sequencing and blast analysis of the PCR fragment using the Drosophila local database (http://flybase.net/blast) showed that it shares high sequence similarity with DrmCRZR. Subsequent 3′-RACE and 5′-RACE experiments yielded a cDNA of 2,309 bp containing a 1,308-bp ORF flanked by in-frame stop codons at positions -9 (5′ end) and +1,308 (3′ end) (GenBank accession no. AY369029). The predicted 49.7-kDa protein contains seven putative transmembrane domains and shows 56% identity (71% similarity) and 30% identity (52% similarity) to DrmCRZR and BomAKHR, respectively (Fig. 8, which is published as supporting information on the PNAS web site). A blastp search of the NCBI local database revealed that DrmCRZR is the most closely related protein; the Manduca protein is henceforth referred to as MasCRZR.
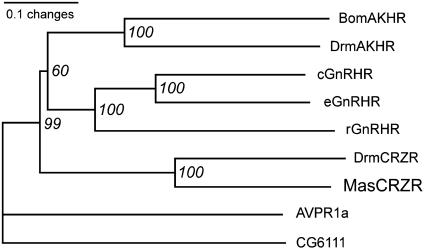
Phylogenetic analysis of MasCRZR and other G protein-coupled receptors shows that MasCRZR is grouped with DrmCRZR with a 100% bootstrap value (Fig. 1). CRZRs occur within a monophyletic clade including GnRHRs and AKHRs with 99% bootstrap value.
Fig. 1.
Phylogram of MasCRZR and its related G protein-coupled receptors. The length of each branch represents the number of changes between each sequence. The percentage of 1,000 bootstrap replicates supporting each node was shown. For details of the molecules analyzed, see Materials and Methods. cGnRHR, chicken GnRHR; eGnRHR, eel GnRHR; rGnRHR, rat GnRHR.
Functional Characterization of MasCRZR. MasCRZR was expressed and assayed for sensitivity to peptides by using two heterologous expression systems, Xenopus oocytes and CHO cells. Inward currents recorded in voltage-clamped oocytes injected with cRNA containing the MasCRZR ORF (-37 to +1,342) were highly specific for corazonin and proportional to the amount of peptide in the bathing medium (Fig. 2B). MasCRZR was insensitive to 11 additional peptides tested at concentrations up to 10 μM (neuromedin-U8, neurotensin, GnRH, MasCap2b, SCPB, MasETH, Mas-PETH, Mas myoinhibiting peptide, crustacean cardioactive peptide, Tabanus atratus AKH, and FMRFamide). Because oocytes expressing MasCRZR were desensitized by even trace amounts of corazonin, peak currents in response to increasing concentrations of corazonin were averaged from individual oocyte applications. The lowest effective concentration of the peptide was 100 pM, and the estimated EC50 is ≈200 pM (Fig. 2A).
CHO cells expressing both codon-optimized aequorin (19) and MasCRZR provided a more discriminating pharmacological analysis of MasCRZR. Ligand-mediated activation of the receptor produced elevation of intracellular Ca2+ and luminescence responses via activation of an aequorin-coelentrazine complex (20, 23). In CHO cells, MasCRZR had the same ligand selectivity as in Xenopus oocytes; no other peptides including 100 nM EH evoked responses. The EC50 for corazonin determined in CHO cells was ≈75 pM (Fig. 2C).
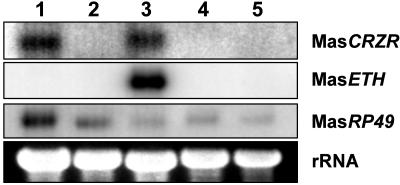
The MasCRZR Is Expressed in Inka Cells. Expression of MasCRZR in Inka cells was confirmed by Northern blot analysis. Among five different tissue samples isolated from pharate pupae (CNS, epidermis, EGs, muscle, and trachea), only EGs containing Inka cells and the CNS had detectable levels of MasCRZR mRNA (Fig. 3). The size of the MasCRZR transcript was estimated at ≈2.6 kilonucleotides (knt) according to Northern analysis, corresponding to the entire cDNA (2.3 kbp) isolated by PCR.
Fig. 3.
Expression of the CRZR (MasCRZR) in pharate pupae of M. sexta. Northern blot analysis was carried out by using 10 μg of total RNA from the CNS (lane 1), epidermis (lane 2), EGs (lane 3), muscle (lane 4), and trachea (lane 5). The membrane was hybridized with different molecular probes labeled with [α-32P]dCTP. MasRP49 and rRNA serve as RNA loading controls. MasETH is an Inka-cell-specific gene. Note that the MasCRZR transcript was detected only in EGs containing Inka cells and the CNS.
Corazonin Induces Ecdysis Behaviors. Specific expression of the CRZR in Inka cells suggested that it may mediate release of PETH and ETH (24-26). We tested this by injecting corazonin into pharate fifth-instar larvae just after resorption of molting fluid or ≈6 h before ecdysis. After injection of corazonin (0.5, 2, or 10 pmol), preecdysis behavior occurred in ≈40-90 min followed by ecdysis behavior (Table 1). The delay from neuropeptide injection to onset of preecdysis behavior suggests that corazonin does not act directly on the nervous system but possibly through release of PETH and ETH.
Table 1. Injection of corazonin accelerated the onset of Preecdysis and ecdysis behaviors in pharate fifth-instar larvae of M. sexta.
| Corazonin injection, pmol | n | Preecdysis latency, h* | Ecdysis latency, h* |
|---|---|---|---|
| 10 | 7 | 0.7 ± 0.2 | 3.1 ± 0.8 |
| 2 | 5 | 0.7 ± 0.3 | 2.8 ± 1.0 |
| 0.5 | 4 | 1.5 ± 0.3 | 3.5 ± 0.9 |
| Controls | 8 | 3.7 ± 0.3 | 4.7 ± 0.4 |
| Natural | 6 | 3.6 ± 0.1 | 4.3 ± 0.2 |
All fifth instar larvae were injected immediately after they absorbed molting fluid from their old head capsule (air-filled head capsule).
Mean ± SD.
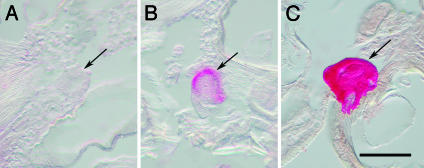
Blood-borne EH induces the ecdysis sequence through release of ETH from Inka cells (21, 27, 28), whereas ETH acts directly on the CNS to activate preecdysis and ecdysis behaviors (24, 25). Therefore, we compared the effects of corazonin injection into isolated abdomens of pharate fourth-instar larvae with those elicited by EH and ETH injection. The effects of these peptides were quantified in two ways. First, we compared the latency from injection to onset of preecdysis. Second, we examined PETH immunoreactivity (IR) of Inka cells 60 min after peptide-induced preecdysis. To detect release of peptide hormones from Inka cells, we used an antiserum to PETH, which reacts specifically with amidated C termini of PETH and ETH (25). Injection of either corazonin (1 pmol) or EH (1 pmol) induced preecdysis in ≈30 min, which was accompanied by reduction or complete abolition of PETH IR in ≈60% of Inka cells examined (Figs. 4 A and B and Table 2). These results indicate that corazonin and EH elicit behaviors by causing PETH and ETH release from Inka cells. In contrast, ETH injection evoked preecdysis behavior within 3-6 min but had little or no effect on PETH IR in Inka cells (Fig. 4C and Table 2), consistent with its direct actions on the CNS (25).
Fig. 4.
Corazonin (1 pmol) injection into isolated abdomens of pharate fourth-instar larvae (4 h before ecdysis) induced the release of PETH and ETH from Inka cells, followed by the onset of preecdysis in ≈30 min. PETH IR was depleted (A) or reduced considerably (B) in Inka cells 60 min later. Injection of ETH (10 pmol) into control isolated abdomens induced preecdysis in 4-10 min through direct action on the CNS; thus, endogenous PETH and ETH were not released from Inka cells even 60 min after the initiation of behavior (C). (Scale bar, 150 μm.)
Table 2. Behavioral and immunohistochemical responses of Inka cells to injection of corazonin, EH, or ETH into isolated abdomens of pharate fourth-instar larvae.
| Preecdysis latency, min*
|
PETH-IR in Inka cells
|
|||
|---|---|---|---|---|
| Peptides injected, pmol | n | +++ | − | |
| Corazonin, 1 | 12 | 27 ± 3 | 60 | 80 |
| EH, 1 | 10 | 28 ± 3 | 54 | 64 |
| ETH, 10 | 12 | 6 ± 2 | 124 | 20 |
Injected corazonin and EH causes Inka cells to release PETH and ETH, which act on the CNS to induce preecdysis with a latency of ≈30 min. PETH IR is visualized 1 h later. Direct action of injected ETH on the CNS elicits preecdysis in 4-10 min, and thus a majority of Inka cells do not release their content. +++, strong IR; −, no or very weak IR.
Mean ± SD.
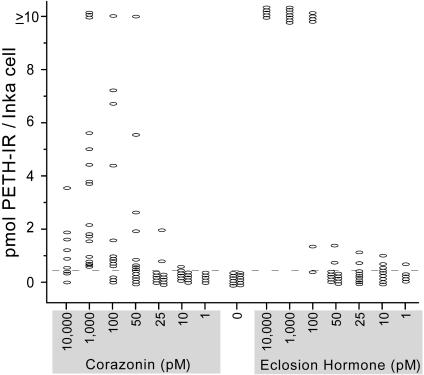
Corazonin Induces Graded Release of PETH and ETH from Isolated Inka Cells. Induction of precocious preecdysis and ecdysis behaviors by corazonin injection is accompanied by depletion of PETH and ETH IR in Inka cells. We therefore assayed for direct actions of corazonin on Inka cells in vitro by measuring the appearance of PETH/ETH IR in the bathing medium by using an enzyme immunoassay. Our results show that corazonin elicits secretion of Inka cell peptides at concentrations as low as 25 pM (Fig. 5). The percentage of Inka cells responding to 50 pM corazonin was ≈50%, and at 100 pM, 80% of cells responded.
Fig. 5.
Distinctive responses of Inka cells to corazonin and EH in vitro. Each oval indicates the response of an individual Inka cell, and maximal responses are ≥10 pmol. Increasing concentrations of corazonin (25-1,000 pM) cause increasing rates of PETH and ETH secretion from most Inka cells, although only in a few cases do Inka cells produce a maximal secretory response (≥10 pmol). High concentrations of corazonin (e.g., 10 nM) suppress Inka cell release, presumably because of rapid receptor desensitization. In contrast, EH induces only minor secretory responses at concentrations ≤50 pM but predominantly maximal responses at levels ≥100 pM. The dashed line indicates maximum amounts of PETH and ETH released from untreated control Inka cells.
Comparison of Inka cell secretory responses to corazonin with those elicited by EH revealed interesting differences. Low concentrations of EH (1-50 pM) caused minimal secretion, with most cells showing no response; slightly higher concentrations of EH (≥100 nM) caused virtually all responders to generate maximal rates of secretion leading to depletion (≥10 pmol per Inka cell). In contrast, corazonin exposure even at high concentrations (100-1,000 pM) caused only a few cells to reach maximum levels of release. At 10 nM, corazonin actually caused lower levels of release, an effect that may relate to desensitization of the receptor, as observed in oocytes.
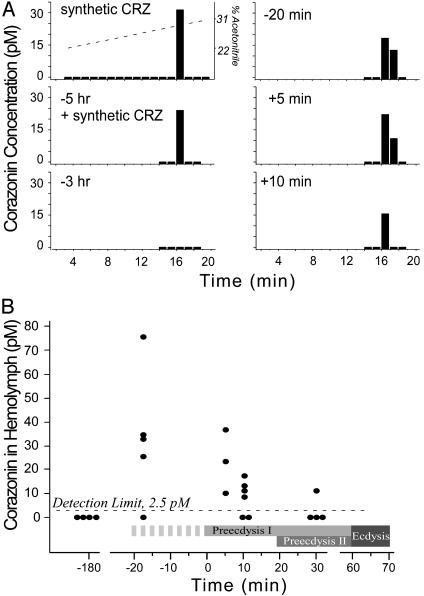
Corazonin Is Released Before Preecdysis Onset. The foregoing evidence indicates that corazonin is capable of initiating the ecdysis sequence through a direct action on Inka cells. If such an action is physiologically relevant, the peptide should circulate in the hemolymph at the appropriate time. We developed a sensitive assay using MasCRZR expressed in CHO cells as a biosensor for detection of corazonin levels in the hemolymph. Preliminary experiments indicated that crude hemolymph extracts contained components causing either false-positives or interference with the positive corazonin response. Consequently, we purified each hemolymph sample by reversed-phase HPLC before the receptor assay. Using synthetic corazonin as a standard, we found the detection limit for a 300-μl hemolymph sample from one animal to be ≈2.5 pM (≈700 amol). Under the HPLC conditions developed for these studies, native circulating corazonin and synthetic corazonin (10 fmol) added to control hemolymph samples coeluted at 16-17 min (Fig. 6A). Using this assay, we examined hemolymph levels of corazonin at -180, -20, +5, +10, and +30 min, with time 0 defined as onset of preecdysis (Fig. 6B). Corazonin was absent in hemolymph samples collected at -180 min, but levels were markedly elevated in samples collected at -20 min, corresponding to the time larvae begin to show intermittent, weak preecdysis-I like contractions in the last two abdominal segments. After this time point, corazonin levels declined gradually over the next 40-50 min. Specifically, hemolymph samples collected at -20 min (four of five) and +5 min (two of three) contained physiologically effective levels of corazonin (25-80 pM), whereas those taken at +10 and +30 min had <25 pM corazonin (Fig. 6B).
Fig. 6.
Quantification of circulating hemolymph corazonin levels before and during the ecdysis sequence of pharate fifth-instar Manduca larvae using the CHO receptor assay as a biosensor. (A) Representative HPLC chromatograms of synthetic corazonin (10 fmol) alone (Top Left) and hemolymph samples. A hemolymph sample taken at -5 h was spiked with 10 fmol of synthetic corazonin (Middle Left). Hemolymph samples taken at -3h (Bottom Left) showed no trace of the peptide, but samples taken at -20 min (Top Right), +5 min (Middle Right), +10 min (Bottom Right) showed clearly elevated levels. The acetonitrile gradient is indicated by the dotted line (Top Left). (B) Hemolymph levels of corazonin are plotted against the behavioral schedule of the natural ecdysis sequence, including preecdysis I, preecdysis II, and ecdysis. The onset of preecdysis I was designated as time 0 (0 min), which was preceded by an ≈20-min-long period of intermittent preecdysis-like contractions (hatched pattern). The detection limit of the receptor assay (2.5 pM) is indicated with a dashed line; samples below the detection limit are considered negative for corazonin content.
Discussion
MasCRZR: Phylogenetics, Pharmacology, and Tissue-Specific Expression. The MasCRZR shows high sequence similarity to its Drosophila counterpart, and phylogenetic analysis indicates that both receptors share common ancestry. These receptors also share a common pharmacological profile. Assays using the Xenopus oocyte-expression system showed that DrmCRZR and MasCRZR have EC50 values of ≈1 and ≈0.2 nM corazonin, respectively, and both were insensitive to other peptides assayed up to 1 μM concentration (see also refs. 14 and 16). The strong conservation of primary structure and pharmacological profile between fly and moth CRZRs supports their roles as authentic receptors for corazonin. Such conservation of both ligand and receptor structure is remarkable, considering that flies and moths diverged ≈58 million years ago (29). Conservation of function therefore is anticipated also.
Northern analysis detected the transcript for MasCRZR in Inka cells and in the CNS. These spatial expression patterns of MasCRZR reflect peripheral and possible central roles for corazonin, similar to those described for EH (21, 30). Occurrence of MasCRZR in Inka cells is of considerable functional significance and supports a role for circulating corazonin in regulation of the ecdysis sequence. The abundant presence of MasCRZR mRNA in the CNS is consistent with corazonin localization in neurons of the ventral nerve cord (8, 13, 31). It seems possible if not likely that centrally released corazonin modulates the activity of central neurons during ecdysis sequence.
Distinctive Inka Cell Responses to Corazonin and EH. Both corazonin and EH act directly on Inka cells to cause secretion of PETH and ETH. However, whereas EH causes an “all-or-nothing” secretory response and peptide depletion, corazonin-induced secretion is graded and rarely maximal even within the most efficacious concentration range (100-1,000 pM). Significantly, when exposed to 10 nM corazonin, Inka cells exhibited reduced rates of release. It thus seems that corazonin-induced peptide release is self-limiting, whereas EH-induced secretion usually results in depletion of Inka cell peptide content (21, 25, 27). On the other hand, low corazonin levels (25-50 pM) were more effective than those of EH (Fig. 5).
Differences in the way corazonin and EH cause peptide release from Inka cells likely is associated with different types of signal transduction. It is well known that EH causes elevation of cGMP and intracellular calcium in Inka cells, leading to depletion of their peptide stores (21, 27, 28). Once activated by EH, it seems that signal transduction in Inka cells operates in a regenerative or self-sustainable manner, leading to massive release. In contrast, Inka cell responses to corazonin do not involve elevation of cGMP and are subject to desensitization, which would tend to hold the secretory response in check during the early phases of preecdysis.
The high variability in Inka cell responses to corazonin and EH is remarkable. We obtained similar results in a previous study of EH action on Inka cells (21). It is likely that such variability arises from the fact that Inka cells chosen for bioassays are taken ≈3.5 h before preecdysis onset so as to minimize false-positives, which arise as the time of preecdysis approaches. Acquisition of competence for secretion in Inka cells occurs at approximately -5 h, but synchronization of this process is imprecise (32). Therefore, it seems plausible that Inka cells used in our in vitro bioassays have variable sensitivity to corazonin and EH and hence release different amounts of PETH and ETH. However, because each Inka cell contains ≈20 pmol of PETH and ETH (25), secretion from only a few competent cells would be enough to establish a physiological effective concentration of PETH and ETH for initiation of the ecdysis sequence.
Timing of Corazonin Release and Hemolymph Levels. Natural preecdysis is preceded by intermittent, weak preecdysis behavior. This behavior begins ≈20-30 min before full-blown preecdysis I behavior. Because no defined morphological or behavioral markers are known between molting-fluid resorption (-5 h) and onset of preecdysis (time 0) in pharate larvae, we estimated that intermittent preecdysis-like contractions begin at -20 min in our study. Nevertheless, the length of intermittent preecdysis-like behavior is variable between individual larvae. Therefore, variation in corazonin concentrations recorded in hemolymph samples could be caused by the inevitable variability in staging larvae. In addition, according to our results from in vivo corazonin-injection experiments, the latency between corazonin injection and preecdysis onset was ≈30 min (Table 1). In this context, we propose that the major release of corazonin likely occurs ≈10-20 min before obvious intermittent preecdysis, which is estimated to be at ≈30-40 min before onset of preecdysis I.
A Model for Sequential Phases of PETH and ETH Release Regulated by Corazonin and EH. Several lines of evidence support corazonin as an initiator of the ecdysis sequence. First, the receptor for corazonin (MasCRZR) is expressed in Inka cells. Second, injection of corazonin into Manduca larvae causes PETH and ETH release from Inka cells. Third, incubation of isolated EGs with corazonin in vitro leads to PETH and ETH secretion at peptide concentrations in the range of the EC50 for MasCRZR expressed in CHO cells. Fourth, corazonin concentrations in the hemolymph are elevated at -20 min (intermittent preecdysis) and remain significantly high at +5 min (preecdysis I). Corazonin levels at -20 min were clearly high enough to cause PETH and ETH release from Inka cells in vitro.
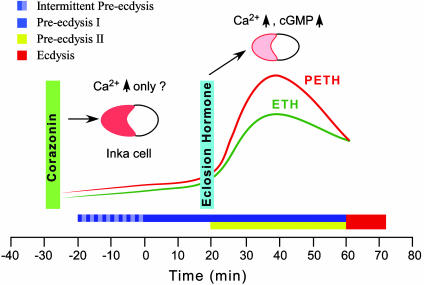
Based on these findings, we propose a model for hormonal regulation of the ecdysis sequence (Fig. 7). At approximately -30 min, the brain releases corazonin through the corpora cardiaca-corpora allata into hemolymph (Fig. 6). CRZR activation in Inka cells leads to an initial phase of low-level PETH and ETH release. Gradual accumulation of PETH and ETH in the hemolymph results in intermittent preecdysis lasting for 10-30 min followed by preecdysis onset. Thereafter, circulating ETH activates ventromedial cells in the brain, causing release of EH into hemolymph (approximately +20 min). Circulating EH, after a delay, causes massive secretory activity in Inka cells and depletion of PETH and ETH through elevation of cGMP and intracellular calcium (21, 25, 27). These events lead to maximum levels of PETH and ETH in the hemolymph and ultimately the transition from preecdysis to ecdysis.
Fig. 7.
Model for sequential phases of Inka cell regulation by corazonin and EH. Corazonin is released from the brain through the corpora cardiaca-corpora allata into the hemolymph ≈30 min before preecdysis onset (-30 min). Blood-borne corazonin binds to the MasCRZR in Inka cells to elicit initial, low-level secretion of PETH and ETH, resulting in intermittent preecdysis-like contractions (beginning at approximately -20 min) and subsequent preecdysis I behavior (time 0). Accumulating levels of ETH in the hemolymph activate ventromedial cells to release EH (+20 min), which in turn acts back on Inka cells to induce cGMP elevation and depletion of PETH and ETH. The Inka cell secretory response to corazonin is likely mediated by mobilization of Ca2+, whereas EH causes both Ca2+ and cGMP production in these cells (21, 27, 28).
Our results illuminate a potentially general function for corazonin in insects, namely as a hormonal factor regulating initiation of the ecdysis sequence. Because ETH occurs in Inka cells across widely diverging insect taxa (33), it will be very interesting to determine whether corazonin regulates secretion of Inka cells in insect orders other than Lepidoptera.
Another intriguing function for corazonin may be the coupling of ecdysis and/or eclosion to the circadian clock. In some insects including moths, eclosion is gated according to a circadian clock located in the brain (34, 35). Interestingly, neurosecretory cells producing corazonin recently have been shown to express the circadian rhythm gene period in Manduca (13). It therefore is possible that corazonin could be involved in the circadian gating of adult eclosion in some moth species.
Multiple Hormonal Signals Regulate Inka Cell Secretion in Insects. Our findings also show that multiple hormonal signals regulate release of PETH and ETH from Inka cells. It also seems possible that some of these pathways may operate with or without the contribution of EH or corazonin. For example, a relatively high proportion of EH-knockout Drosophila larvae perform all ecdyses (ref. 36 and Y.P., unpublished data), which suggests that, at least in Drosophila, additional signaling pathways inducing ETH release from Inka cells may exist. Other examples include the absence of corazonin in brain neurosecretory cells of some representatives of Coleoptera (beetles) and in albino locusts. Nevertheless, these insects complete their life cycles without developmental abnormalities despite this absence (6, 8, 11). Because insects represent the most diverse group of organisms on Earth, it is not surprising that diversity in regulation of ecdysis could occur. Indeed, we have isolated other peptides from the brain and gut that cause precocious preecdysis and ecdysis behaviors when injected into Manduca larvae (I.S.-V. and D.Z., unpublished data).
In summary, we have characterized the molecular and pharmacological properties of the MasCRZR and demonstrated its likely role in signaling between the brain and Inka cells important for initiation of ecdysis behaviors. It is also possible that this function provides a more generalized, conserved role for a cryptic neuropeptide that occurs in many insect orders. Finally, this study provides an example for how identification and characterization of a peptide receptor can lead to definition of a previously unknown function for its ligand.
Supplementary Material
Acknowledgments
We thank Timothy Kingan and Sarjeet Gill for sage advice and critical reading of the manuscript. This work was supported by National Institutes of Health Grants AI 40555 and GM 67310 and Vedecká Grantová Agentúra Grant 2/7168/20.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CRZR, corazonin receptor; Drm, Drosophila melanogaster; Mas, Manduca sexta; PETH, preecdysis-triggering hormone; ETH, ecdysis-triggering hormone; EH, eclosion hormone; AKH, adipokinetic hormone; AKHR, AKH receptor; Bom, Bombyx mori; GnRH, gonadotropin-releasing hormone; GnRHR, GnRH receptor; CHO, Chinese hamster ovary; EG, epitracheal gland; IR, immunoreactivity.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY369029).
References
- 1.Veenstra, J. A. (1989) FEBS Lett. 250, 231-234. [DOI] [PubMed] [Google Scholar]
- 2.Veenstra, J. A. (1991) Peptides (Tarrytown, NY) 12, 1285-1289. [DOI] [PubMed] [Google Scholar]
- 3.Veenstra, J. A. (1994) Biochem. Biophys. Res. Commun. 204, 292-296. [DOI] [PubMed] [Google Scholar]
- 4.Hansen, I. A., Sehnal, F., Meyer, S. R. & Scheller, K. (2001) Insect Mol. Biol. 10, 341-346. [DOI] [PubMed] [Google Scholar]
- 5.Hua, Y., Ishibashi, J., Saito, H., Tawfik, A. I., Sakakibara, M., Tanaka, Y., Derua, R., Waelkens, E., Baggerman, G., De Loof, A., et al. (2000) J. Insect Physiol. 46, 853-860. [DOI] [PubMed] [Google Scholar]
- 6.Tawfik, A. I., Tanaka, S., De Loof, A., Schoofs, L., Baggerman, G., Waelkens, E., Derua, R., Milner, Y., Yerushalmi, Y. & Pener, M. P. (1999) Proc. Natl. Acad. Sci. USA 96, 7083-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Predel, R., Kellner, R. & Gade, G. (1999) Eur. J. Entomol. 96, 275-278. [Google Scholar]
- 8.Roller, L., Tanaka, Y. & Tanaka, S. (2003) Cell Tissue Res. 312, 393-406. [DOI] [PubMed] [Google Scholar]
- 9.Veenstra, J. A. & Davis, N. T. (1993) Cell Tissue Res. 274, 57-64. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka, S., Zhu, D. H., Hoste, B. & Breuer, M. (2002) J. Insect Physiol. 48, 1065-1074. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka, S. (2000) Appl. Entomol. Zool. (Jpn.) 35, 509-517. [Google Scholar]
- 12.Tanaka, Y., Hua, Y., Roller, L. & Tanaka, S. (2002) J. Insect Physiol. 48, 707-714. [DOI] [PubMed] [Google Scholar]
- 13.Wise, S., Davis, N. T., Tyndale, E., Noveral, J., Folwell, M. G., Bedian, V., Emery, I. F. & Siwicki, K. K. (2002) J. Comp. Neurol. 447, 366-380. [DOI] [PubMed] [Google Scholar]
- 14.Park, Y., Kim, Y. J. & Adams, M. E. (2002) Proc. Natl. Acad. Sci. USA 99, 11423-11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hewes, R. S. & Taghert, P. H. (2001) Genome Res. 11, 1126-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cazzamali, G., Saxild, N. & Grimmelikhuijzen, C. (2002) Biochem. Biophys. Res. Commun. 298, 31-36. [DOI] [PubMed] [Google Scholar]
- 17.Shi, X., Karkut, T., Chahmanhkah, M., Alting-Mees, M., Hemmingsen, S. M. & Hegedus, D. (2002) BioTechniques 32, 480-482. [DOI] [PubMed] [Google Scholar]
- 18.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vernon, W. I. & Printen, J. A. (2002) BioTechniques 33, 730, 732, 734. [DOI] [PubMed] [Google Scholar]
- 20.Park, Y., Kim, Y. J., Dupriez, V. & Adams, M. E. (2003) J. Biol. Chem. 278, 17710-17715. [DOI] [PubMed] [Google Scholar]
- 21.Kingan, T. G., Gray, W., Žitňan, D. & Adams, M. E. (1997) J. Exp. Biol. 200, 3245-3256. [DOI] [PubMed] [Google Scholar]
- 22.Zitnanova, I., Adams, M. E. & Žitňan, D. (2001) J. Exp. Biol. 204, 3483-3495. [DOI] [PubMed] [Google Scholar]
- 23.Le Poul, E., Hisada, S., Mizuguchi, Y., Dupriez, V. J., Burgeon, E. & Detheux, M. (2002) J. Biomol. Screen. 7, 57-65. [DOI] [PubMed] [Google Scholar]
- 24.Žitňan, D., Kingan, T. G., Hermesman, J. L. & Adams, M. E. (1996) Science 271, 88-91. [DOI] [PubMed] [Google Scholar]
- 25.Žitňan, D., Ross, L. S., Zitnanova, I., Hermesman, J. L., Gill, S. S. & Adams, M. E. (1999) Neuron 23, 523-535. [DOI] [PubMed] [Google Scholar]
- 26.Park, Y., Filippov, V., Gill, S. S. & Adams, M. E. (2002) Development (Cambridge, U.K.) 129, 493-503. [DOI] [PubMed] [Google Scholar]
- 27.Ewer, J., Gammie, S. C. & Truman, J. W. (1997) J. Exp. Biol. 200, 869-881. [DOI] [PubMed] [Google Scholar]
- 28.Kingan, T. G., Cardullo, R. A. & Adams, M. E. (2001) J. Biol. Chem. 276, 25136-25142. [DOI] [PubMed] [Google Scholar]
- 29.Riek, E. F. (1970) in The Insects of Australia; A Textbook for Students and Research Workers, ed. Waterhouse, D. F. (Melbourne Univ. Press, Victoria, Australia), pp. 168-186.
- 30.Gammie, S. C. & Truman, J. W. (1999) J. Exp. Biol. 202, 343-352. [DOI] [PubMed] [Google Scholar]
- 31.Cantera, R., Veenstra, J. A. & Nassel, D. R. (1994) J. Comp. Neurol. 350, 559-572. [DOI] [PubMed] [Google Scholar]
- 32.Kingan, T. G. & Adams, M. E. (2000) J. Exp. Biol. 203, 3011-3018. [DOI] [PubMed] [Google Scholar]
- 33.Žitňan, D., Zitnanova, I., Spalovská, I., Takac, P., Park, Y. & Adams, M. E. (2003) J. Exp. Biol. 206, 1275-1289. [DOI] [PubMed] [Google Scholar]
- 34.Truman, J. W. & Riddiford, L. M. (1970) Science 167, 1624-1626. [DOI] [PubMed] [Google Scholar]
- 35.Truman, J. W. (1972) J. Comp. Physiol. 81, 99-114. [Google Scholar]
- 36.McNabb, S. L., Baker, J. D., Agapite, J., Steller, H., Riddiford, L. M. & Truman, J. W. (1997) Neuron 19, 813-823. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.