Abstract
IMPORTANCE
To date, relatively few genes responsible for a fraction of heritability have been identified by means of large genetic association studies of refractive error.
OBJECTIVE
To explore the genetic mechanisms that lead to refractive error in the general population.
DESIGN, SETTING, AND PARTICIPANTS
Genome-wide association studies were carried out in 2 British population-based independent cohorts (N = 5928 participants) to identify genes moderately associated with refractive error.
MAIN OUTCOMES AND MEASURES
Enrichment analyses were used to identify sets of genes overrepresented in both cohorts. Enriched groups of genes were compared between both participating cohorts as a further measure against random noise.
RESULTS
Groups of genes enriched at highly significant statistical levels were remarkably consistent in both cohorts. In particular, these results indicated that plasma membrane (P = 7.64 × 10−30), cell-cell adhesion (P = 2.42 × 10−18), synaptic transmission (P = 2.70 × 10−14), calcium ion binding (P = 3.55 × 10−15), and cation channel activity (P = 2.77 × 10−14) were significantly overrepresented in relation to refractive error.
CONCLUSIONS AND RELEVANCE
These findings provide evidence that development of refractive error in the general population is related to the intensity of photosignal transduced from the retina, which may have implications for future interventions to minimize this disorder. Pathways connected to the procession of the nerve impulse are major mechanisms involved in the development of refractive error in populations of European origin.
Refractive error is the most common ocular disorder.1 It affects about 25% of the adult population in Europe and the United States2 but its prevalence approaches three-quarters among the younger age groups of Southeast Asia.3 Refractive error and in particular one of its manifestations, myopia, is an important risk factor for serious ocular complications and blindness.1,4 Through direct financial costs and indirect costs of loss of productivity, myopia costs societies billions of dollars each year.5
Myopia is by far the most prevalent form of refractive error6 and usually occurs as a result of elongation of the axial length of the eye beyond its focal plane.7 Refraction at birth is almost normally distributed, centered in the hyperopic ranges, but within 5 years of age, the center of the distribution moves away from hyperopia as the standard deviations of that distribution decrease,8 primarily as a result of axial length changes.9,10
The prevalence of refractive error and myopia varies both across geographic regions11 and among different ethnicities living together within the same geographic locations.12,13 This is indicative of both strong environmental influences and genetic predispositions. Higher socioeconomic status,14 education,15 outdoor activity, and especially near work16 are recognized risk factors for myopia and life-course research has recently shown that key prenatal and early childhood factors associated with general growth and development (eg, maternal age, intrauterine growth retardation, smoking in pregnancy, and changing socioeconomic status) are also implicated.17
Yet parental myopia is the strongest predictor for myopia in school-aged children.18 Heritability studies have found that a large portion of the refractive error variation is due to inherited factors.19 Variants at 2 genetic loci, seemingly involved in synaptic transmission and transduction of visual signal,20,21 have been identified through genome-wide association studies (GWAS).22,23
Current knowledge of both environmental and genetic factors predisposing to refractive error is patchy and does not allow us to have an integrated view of the mechanisms that might influence the process of emmetropization. Most complex diseases are polygenic24,25 and because of the relatively low power of real-life GWAS cohorts, true signals can be drowned out by the random noise of false-positive results. However, some of the truly positive associations, albeit at less than genome-wide statistical significance, will generally rank higher in GWAS results because of their inherent role in disease etiology and pathophysiology. Also, by virtue of the biological interactions leading to disease, these genes are likely to share commonalities, such as participation in the same functional classes or known biological classes. Genome-wide association study results of sufficient power would be expected to be enriched for functional gene sets or biological pathways that are of relevance to the phenotype studied.
To further elucidate the potential mechanisms that cause refractive error and myopia in the general population, we carried out a meta-analysis of gene list enrichment analyses of results from 2 separate GWAS of refractive error. To minimize heterogeneity introduced from subtle racial or ethnic differences or exposure to the social environment, we focused on 2 cohorts drawn from the white British general population: the TwinsUK cohort26 and the 1958 British Birth Cohort.27
Methods
The TwinsUK data set comprised 4270 individuals (8% male) with both genotypic and phenotypic information. The phenotypic information consisted of spherical equivalent derived from noncyclopleged autorefraction using an ARM-10 autorefractor (Takagi Ltd). The spherical equivalent was defined as the sphere value (in diopters) plus half of the cylinder value of the same eye. The average of measurements from both eyes was used for the analysis. The mean (SD) spherical equivalent was -0.4 (2.73) diopter, with a range of -25.12 to +9.4 diopters, and the mean age was 53 years. Individuals with previous ocular surgery including cataract and refractive surgery were excluded, as were those with other systemic or ocular conditions that might influence refraction. All participants were unaware of the eye studies at the time of enrollment; they gave informed written consent to participate in genetic association studies. The study and biomedical examination protocols were approved by the St Thomas Hospital Local Research Ethics Committee (TwinsUK) and the South East MultiCentre Research Ethics Committee and the Oversight Committee for the 1958 British Birth Cohort. The research adhered to the tenets of the Declaration of Helsinki.
Genotyping was carried out using 3 genotyping platforms from Illumina: the HumanHap 300k Duo for part of the TwinsUK Cohort and the HumanHap610 quad array for the rest of the cohort, as described elsewhere.22 Stringent quality control measures were applied. Individuals were included if their genotyping success rate exceeded 95% and did not show excess or low heterozygosity (defined by the interval of 0.2-0.4). Single-nucleotide polymorphisms (SNPs) were included in the imputation if they had a genotype success rate of at least 0.95 if their minor allele frequency was superior to 0.005 and at least 0.99 if their minor allele frequency was 0.01 to 0.05. Only SNPs that were within Hardy-Weinberg equilibrium (P > 10−4) and had a minor allele frequency of 0.04 or more were regressed. Imputation of nongenotyped loci was done with IMPUTE28 based on HapMap2 CEU haplotypes. Association between genotypes and phenotypes was computed using the score test as implemented in the program MERLIN,29 as published elsewhere.22
The second data set was a random subset of 1658 individuals from the 1958 British Birth Cohort, with exclusion of individuals with previous ocular surgery or those with other systemic or ocular conditions that might influence refraction.17 The phenotypic information was obtained through noncycloplegic autorefraction (Nikon Retinomax 2) of both eyes of each participant. Spherical equivalent was calculated as described earlier. Genotyping was done in 7 overlapping batches using the Illumina Human1M-Duo chip (2 batches of 1000 and 313 participants), Illumina Infinium HumanHap550 (2 different batches of 770 and 260 participants), Affymetrix 6.0 (317 participants), Affymetrix 5.0 Human SNP Array (299 participants), and the Metabochip30 (1323 participants). Genotypes were aligned in the same strand, the consistency of which was checked using local patterns of linkage disequilibrium between SNPs as implemented in the routine “flip scan” in PLINK.31 Individuals were checked for genotyping success rate (all exceeded 99%) and for excess or low heterozygosity (all participants were checked and found within the predefined interval of 0.2-0.4). Single-nucleotide polymorphisms were included in the analysis if they had a genotype success rate of at least 0.95, were within Hardy-Weinberg equilibrium (P > 10-4), and had a minor allele frequency of 0.04 or more. Imputation was done using MaCH132 and genotypes meeting conventional quality control criteria were analyzed genome-wide using PLINK.31
Results from both the GWAS described earlier were annotated using the ENSEMBL database (www.ensembl.org). We considered that the effect of a SNP may be conferred by its direct or indirect impacts over the whole gene unit, including the transcript and its regulatory regions. The latter are not fully understood and several studies have shown that regulatory elements are often at long distances (or in trans) with the transcript.33 Yet expression quantitative trait loci located within ±100 kilobases (kb) from the transcript are both more frequent and have stronger effects than others.34 Under the assumption that this interval would link the putative effect of a SNP variant with a gene's function, SNPs within 100 kb of a gene's transcript were annotated as part of that gene. Single-nucleotide polymorphisms that were within 100 kb of more than 1 gene were annotated with the gene nearest their genomic location or both genes in cases where the 2 transcripts overlapped. For each of the 2 separate gene list enrichment analyses, genes were assigned a significance according to the strongest associated SNP annotated to them in the respective GWAS. Because the main rationale was to enrich for genes with true association with refractive error, the participating studies needed to have at least a minimum power to detect true effects between 0.1 and 0.5 diopter, consistent with the range of effects observed in previous successful myopia association studies.22,23 In the smaller cohort (1958 British Birth Co-hort), 80% power for that range of alleles and variant frequencies was achieved at α = .001, and for the sake of consistency, this threshold was adopted for both participating cohorts. Only genes whose best-associated SNP passed a preset significance threshold of 0.001 were included in the subsequent enrichment analyses.
Gene list enrichment analyses were used to assess if the genes that were identified as statistically significant in the GWAS participated in specific annotation categories more than by chance. These analyses were carried out using DAVID (the Database for Annotation, Visualization, and Integrated Discovery).35,36 We focused our choice on functional categories for the enrichment of the 3 classes of the Gene Ontology (GO) database (http://www.geneontology.org/) and BioCarta (http://www.biocarta.com/) and KEGG (Kyoto Encyclopedia of Genes and Genomes) (http://www.kegg.jp/) pathways.
From our analyses, we obtained the Fisher exact test for enrichment, a fold change compared with expectations, and a hypergeometric Bonferroni-corrected value controlling for multiple testing. Fisher exact test results from both analyses were meta-analyzed using the Fisher combined probability method.37 Only results whose fold-change enrichments were going in the same direction in both cohorts (ie, both greater than 1 or both smaller than 1) were considered. To avoid spurious associations, only results that passed Bonferroni multiple-testing correction (P < .05) in each of the 2 data sets were considered for meta-analysis. Finally, because GO entries are not necessarily independent classes but are organized hierarchically with partial or complete overlaps between the different entries, we used tree visualizations generated by the AmiGO tools (http://amigo.geneontology.org) to ascertain intradependencies of the significantly associated GO entries.
Results
We investigated 14 721 entries from the GO database: 9818 biological processes, 1208 cellular component entries, and 3695 different molecular functions. The GO entries are organized hierarchically and often partially overlap with other entries. Each of these categories had between 18 546 (biological processes) and 19 042 (cellular component) unique annotated human genes. We also investigated 202 KEGG and 217 BioCarta pathways.
A total of 3312 unique genes or transcripts were identified for TwinsUK and 2483 in the 1958 British Birth Cohort. Of these, only 2422 genes or transcripts (73.1%) from the TwinsUK data set and 1878 (75.6%) from the 1958 British Birth Cohort had any functional annotation in any of the relevant databases. The number of genes entering the analyses corresponded to roughly 10% of the total number of annotated genes in each of the GO major categories.
With the exception of the BioCarta pathways, in which no significant enrichment was observed in any of the data sets, high levels of entry enrichment were found in all other databases. Despite stringent predefined criteria, there was remarkable overlap between results in both data sets, both in terms of strong probabilities for significant enrichment and, with few exceptions, highly correlated values of fold enrichment for the respective terms (Tables 1, 2, and 3).
Table 1.
Gene List Enrichment Analyses Results From the TwinsUK and the 1958 British Birth Cohort (1958BC) Data sets and Meta-analysis: Gene Ontology (GO) Biological Processesa
| P Value | ||||
|---|---|---|---|---|
| GO Term | GO ID No. | TwinsUK | 1958BC | Meta-analysis |
| Cell-cell adhesion | GO:0007155 | 7.40 × 10–13 | 7.20 × 10–8 | 2.42 × 10–18 |
| Biological adhesion | GO:0022610 | 8.40 × 10–13 | 7.80 × 10–8 | 2.96 × 10–18 |
| Cell morphogenesis involved in differentiation | GO:0000904 | 1.60 × 10–7 | 1.90 × 10–9 | 1.12 × 10–14 |
| Cell morphogenesis | GO:0000902 | 2.80 × 10–8 | 1.60 × 10–8 | 1.63 × 10–14 |
| Synaptic transmission | GO:0007268 | 2.90 × 10–9 | 2.60 × 10–7 | 2.70 × 10–14 |
| Cellular component morphogenesis | GO:0032989 | 1.80 × 10–8 | 8.20 × 10–8 | 5.19 × 10–14 |
| Neuron differentiation | GO:0030182 | 2.20 × 10–6 | 7.00 × 10–10 | 5.41 × 10–14 |
| Ion transport | GO:0006811 | 8.60 × 10–9 | 2.30 × 10–7 | 6.89 × 10–14 |
| Transmission of nerve impulse | GO:0019226 | 5.40 × 10–9 | 4.40 × 10–7 | 8.24 × 10–14 |
| Metal ion transport | GO:0030001 | 3.80 × 10–8 | 6.40 × 10–8 | 8.43 × 10–14 |
| Neuron development | GO:0048666 | 2.00 × 10–6 | 1.80 × 10–9 | 1.23 × 10–13 |
| Cell-cell signaling | GO:0007267 | 3.70 × 10–9 | 1.20 × 10–6 | 1.51 × 10–13 |
| Cation transport | GO:0006812 | 1.50 × 10–7 | 7.70 × 10–8 | 3.82 × 10–13 |
| Cell morphogenesis involved in neuron differentiation | GO:0048667 | 1.20 × 10–6 | 2.00 × 10–8 | 7.77 × 10–13 |
| Regulation of cell death | GO:0010941 | 3.30 × 10–6 | 2.60 × 10–8 | 2.67 × 10–12 |
| Regulation of programmed cell death | GO:0043067 | 4.10 × 10–6 | 6.40 × 10–8 | 7.86 × 10–12 |
| Regulation of system process | GO:0044057 | 1.70 × 10–7 | 2.10 × 10–6 | 1.06 × 10–11 |
| Regulation of apoptosis | GO:0042981 | 5.90 × 10–6 | 1.90 × 10–7 | 3.20 × 10–11 |
| Calcium ion transport | GO:0006816 | 2.90 × 10–6 | 4.20 × 10–7 | 3.46 × 10–11 |
| Axonogenesis | GO:0007409 | 2.70 × 10–6 | 1.50 × 10–6 | 1.10 × 10–10 |
Fisher exact test results from both analyses were meta-analyzed using the Fisher combined probability method.
Table 2.
Gene List Enrichment Analyses Results From the TwinsUK and the 1958 British Birth Cohort (1958BC) Data sets and Meta-analysis: Gene Ontology (GO) Molecular Functionsa
| P Value | ||||
|---|---|---|---|---|
| GO Term | GO ID No. | TwinsUK | 1958BC | Meta-analysis |
| Calcium ion binding | GO:0005509 | 3.90 × 10–6 | 2.40 × 10–11 | 3.55 × 10–15 |
| Gated channel activity | GO:0022836 | 1.10 × 10–10 | 1.00 × 10–6 | 4.15 × 10–15 |
| Cation channel activity | GO:0005261 | 7.90 × 10–10 | 9.80 × 10–7 | 2.77 × 10–14 |
| Substrate-specific channel activity | GO:0022838 | 9.90 × 10–10 | 5.00 × 10–6 | 1.68 × 10–13 |
| Ion channel activity | GO:0005216 | 3.70 × 10–10 | 2.30 × 10–5 | 2.84 × 10–13 |
| Metal ion transmembrane transporter activity | GO:0046873 | 1.10 × 10–8 | 1.50 × 10–6 | 5.40 × 10–13 |
| Passive transmembrane transporter activity | GO:0022803 | 3.70 × 10–9 | 9.50 × 10–6 | 1.12 × 10–12 |
| Channel activity | GO:0015267 | 3.30 × 10–9 | 1.70 × 10–5 | 1.77 × 10–12 |
| Calcium channel activity | GO:0005262 | 1.70 × 10–7 | 7.10 × 10–7 | 3.71 × 10–12 |
| Voltage-gated cation channel activity | GO:0022843 | 5.00 × 10–8 | 3.20 × 10–5 | 4.51 × 10–11 |
| Calmodulin binding | GO:0005516 | 2.00 × 10–6 | 1.10 × 10–5 | 5.62 × 10–10 |
| Voltage-gated ion channel activity | GO:0005244 | 1.60 × 10–6 | 1.60 × 10–5 | 6.50 × 10–10 |
| Voltage-gated channel activity | GO:0022832 | 1.60 × 10–6 | 1.60 × 10–5 | 6.50 × 10–10 |
Fisher exact test results from both analyses were meta-analyzed using the Fisher combined probability method.
Table 3.
Gene List Enrichment Analyses Results From the TwinsUK and the 1958 British Birth Cohort (1958BC) Data sets and Meta-Analysis: Gene Ontology (GO) Cellular Components and KEGG Pathways (Bottom 2 Rows)a
| P Value | ||||
|---|---|---|---|---|
| GO Term | GO ID No. | TwinsUK | 1958BC | Meta-analysis |
| Plasma membrane part | GO:0044459 | 2.40 × 10–15 | 4.40 × 10–17 | 7.64 × 10–30 |
| Plasma membrane | GO:0005886 | 1.40 × 10–14 | 2.10 × 10–11 | 1.69 × 10–23 |
| Intrinsic to plasma membrane | GO:0031226 | 3.30 × 10–15 | 5.20 × 10–9 | 9.17 × 10–22 |
| Integral to plasma membrane | GO:0005887 | 1.00 × 10–14 | 2.20 × 10–8 | 1.12 × 10–20 |
| Neuron projection | GO:0043005 | 1.70 × 10–6 | 1.70 × 10–11 | 1.13 × 10–15 |
| Synapse | GO:0045202 | 4.60 × 10–7 | 1.70 × 10–9 | 2.80 × 10–14 |
| Cell projection | GO:0042995 | 2.00 × 10–5 | 4.70 × 10–11 | 3.35 × 10–14 |
| Synapse part | GO:0044456 | 1.30 × 10–6 | 1.50 × 10–7 | 5.90 × 10–12 |
| Cell junction | GO:0030054 | 3.80 × 10–6 | 1.80 × 10–6 | 1.83 × 10–10 |
| Extracellular matrix | GO:0031012 | 1.70 × 10–5 | 4.20 × 10–7 | 1.90 × 10–10 |
| Cation channel complex | GO:0034703 | 1.60 × 10–6 | 1.50 × 10–5 | 6.11 × 10–10 |
| Axon guidance | map04360 | 1.70 × 10–6 | 5.10 × 10–4 | 1.90 × 10–8 |
| Arrhythmogenic right ventricular cardiomyopathy | map05412 | 6.40 × 10–5 | 5.10 × 10–5 | 6.70 × 10–8 |
Abbreviation: KEGG, Kyoto Encyclopedia of Genes and Genomes.
Fisher exact test results from both analyses were meta-analyzed using the Fisher combined probability method.
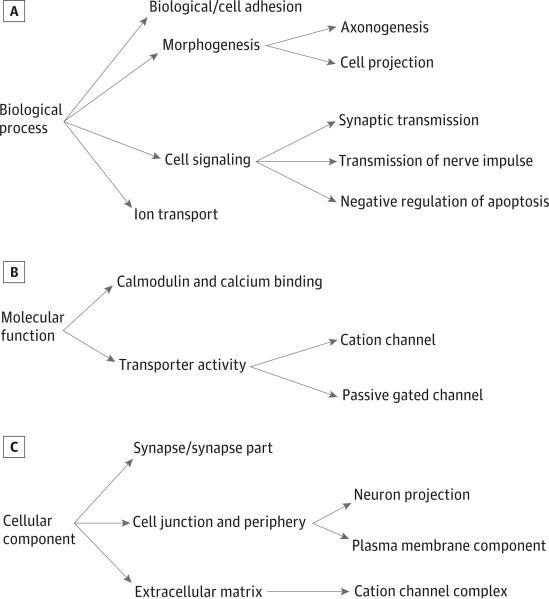
Across the different GO categories, strong suggestions of association between sensory pathways and refractive error were found. Among the most associated GO entries were adhesion of cells (P = 2.42 × 10-18), synapse (P = 2.80 × 10-14) and synaptic transmission (P = 2.70 × 10-14), and ion channel (P = 2.84 × 10−13) and passive voltage-gated channel activity (P = 6.50 × 10−10). Most of these suggest sensory participation in refractive error pathophysiology. Interestingly, however, there was a strong morphogenesis presence of highly enriched genes in our analysis: cell (P = 1.63 × 10-14) and axon (P = 1.10 × 10-10) morphogenesis and the axon guidance KEGG pathway (P = 1.90 × 10-8). A simplified scheme of the associated GO entries taking into account their dependencies is shown in the Figure.
Figure.
Scheme of the Main Gene Ontology Entries Significantly Enriched in TwinsUK and 1958 British Birth Cohort Data sets
Discussion
Functional inferences from GWAS data can provide some insight into the molecular mechanisms that influence emmetropization, the highly active process of self-regulation of axial growth in response to changes in the focal plane.7 Visually guided regulation appears to act at a local level and persists even if communication between the retina and the central nervous system is eliminated.38 Local defocus involving only parts of the retina causes differential local elongation.39 Cell adhesion and junction functional categories showed some of the highest statistical enrichment in both cohorts, consistent with highly significant associations previously reported for the GJD2 gene with refractive error.23 The visual signal starts when photons are absorbed by the photopigment present in rod or cone photoreceptors, eventually leading to hyperpolarization of these cells and a reduction in glutamate release.40,41 Photoreceptors synapse with bipolar cells and horizontal cells at the outer plexiform layer. The connexin 36 protein, coded by the GJD2 gene, appears to play an important stabilizing role here.42-44 Also, RASGRF1 expression localizes to these layers, with knockout mice showing reduced retinal responses.21 RASGRF1 has also been associated with refractive error.22 Synaptic transmission GO entries and channel activity as well as calcium and calmodulin (calcium is important in both synaptic transmission and retinal adaptation) showed consistent patterns of highly significant enrichment in both data sets. Thus, the results of this study, taken together with the previously reported genetic associations, lend strong support to the notion that alterations in transmission of the visual signal, even at the first synapse, play a significant role in the development of refractive error.
Gene Ontology entries are often imprecise annotation categories describing known gene functionalities but lacking the precise mechanisms by which these genes act coherently together. KEGG pathways, on the other hand, are less extensive but better characterized entries from a molecular point of view. Interestingly, the 2 KEGG pathways that met our conservative significance criteria were both related to electrophysiologically active cells involved in signal transmission. Axon guidance is the most strongly enriched category and may suggest a role for axonal/presynaptic alterations affecting synaptic transmission. The arrhythmogenic right ventricular cardiomyopathy enrichment in our data sets seems to suggest that at least some of the genes and their protein products instrumental in visual signaling pathways have multiple roles in the organism, affecting cellular electrical activity in other contexts, organs, or systems, which adds strength to life-course epidemiological observations, for example, relating general growth to ocular growth.17 Clearly, future drug development to prevent myopia progression will need to target pathways specific to the failure of emmetropization, akin to the actions of the HMGCoA reductase enzyme targeted by statins, rather than more general pathways. The hope is that identification of pathways through this gene enrichment process will identify potential targets, which might not have been found by GWAS or other genetic studies.
Enrichment analysis based on GWAS data cannot irrefutably prove involvement or lack of involvement of specific pathways or molecular mechanisms. First, both enrichment and genetic association studies are methods of a probabilistic nature, where the right balance between high-sensitivity thresholds and high specificity is complicated by the relatively high level of multiple testing. In this work, we used some very conservative methods to control for multiple testing by requiring Bonferroni significant probabilities and consistent replication in both data sets. This option may have favored specificity at the expense of sensitivity. For example, the glutamate receptor ontology, which could be related to glutamate transmission of the signal to bipolar cells, narrowly missed inclusion because it did not reach Bonferroni-corrected statistical significance in the smaller 1958 British Birth Cohort data set. Second, we deliberately opted to minimize heterogeneity, both genetic and environmental, by studying enrichment in 2 populations with similar demographic characteristics. The high complexity of ethnic and environmental factors leading to refractive error45 makes it unlikely that the mechanisms we identified are either undisputedly universal or exclusive across the world. Finally, enrichment for annotated functional classes always has the inherent publication bias risks because existing knowledge at any given moment may cover different areas unevenly within the same scientific discipline.
Notwithstanding these potential limitations, our work provides some clues as to the most likely mechanisms involved in refractive error and emmetropization. These results were surprising in 2 different ways. First, they were remarkably in agreement with what we know so far about photosignal transduction and transmission. Second, the results between the larger TwinsUK data set and the much smaller 1958 British Birth Cohort were very consistent in the direction of changes and magnitude. This reflects the robustness of the association and enrichment signals, lending additional support to the notion that the results of this study represent a credible advance in our endeavor to identify the pathophysiology of refractive error at a molecular level. The consistency of enrichment analyses is encouraging and may be used more widely in the future, both to improve knowledge on multigenic mechanisms of complex disease and to analyze GWAS in a way that limits their focus to smaller prioritized groups of genes. This would reduce the high dimensionality of SNP data sets and increase the power to detect more heritable risk factors associated with clinical phenotypes.
Acknowledgments
Funding/Support: Phenotyping for the 1958 British Birth Cohort was funded by the Medical Research Council Health of the Public grant (principal investigators, C. Power, PhD, and D. P. Strachan, MD) and the genetic studies, by the Wellcome Trust (grant 083478 to Dr Rahi), with additional personal funding (Dr Hysi) by the Ulverscroft Vision Research Group. The Centre for Paediatric Epidemiology and Biostatistics was supported in part by the Medical Research Council in its capacity as the MRC Centre of Epidemiology for Child Health. Research at the UCL Institute of Child Health, Great Ormond Street Hospital for Children, Moorfields Eye Hospital NHS Foundation Trust, and UCL Institute of Ophthalmology receives a proportion of their funding from the National Institute for Health Research Biomedical Research Centres funding scheme. TwinsUK received funding from the Wellcome Trust; the European Union MyEuropia Marie Curie Research Training Network; Guide Dogs for the Blind Association; the European Community's FP7 (grant HEALTHF22008201865GEFOS); ENGAGE (grant HEALTHF42007201413); the FP-5 GenomEUtwin Project (grant QLG2CT200201254); US National Institutes of Health/National Eye Institute (grant 1RO1EY018246, principal investigator, T. L. Young, MD); NIH Center for Inherited Disease Research; and the National Institute for Health Research comprehensive Biomedical Research Centre award to Guy's and St Thomas' National Health Service Foundation Trust partnering with King's College London. Dr Hysi is the recipient of a Fight for Sight ECI award. Dr Williams acknowledges financial support from the TFC Frost Charitable Trust. Dr Mahroo is a recipient of the Fight for Sight New Lecturers' Grant Award.
Role of the Sponsor: The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Author Contributions: Dr Hysi had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Hysi, Young, Mackey, Hammond.
Acquisition of data: Hysi, Cumberland, Young, Mackey, Rahi, Hammond.
Analysis and interpretation of data: Hysi, Mahroo, Cumberland, Wojciechowski, Williams, Hammond. Drafting of the manuscript: Hysi, Hammond. Critical revision of the manuscript for important intellectual content: All authors.
Statistical analysis: Hysi, Cumberland, Wojciechowski.
Obtained funding: Young, Mackey, Rahi, Hammond. Administrative, technical, or material support: Hysi, Mahroo, Williams, Mackey, Hammond.
Study supervision: Rahi, Hammond.
Conflict of Interest Disclosures: None reported.
Disclaimer: The views expressed are those of the authors and not necessarily those of the National Health Service, the National Institute for Health Research, or the Department of Health.
REFERENCES
- 1.Pizzarello L, Abiose A, Ffytche T, et al. VISION 2020: The Right to Sight. a global initiative to eliminate avoidable blindness. Arch Ophthalmol. 2004;122(4):615–620. doi: 10.1001/archopht.122.4.615. [DOI] [PubMed] [Google Scholar]
- 2.Kempen JH, Mitchell P, Lee KE, et al. Eye Diseases Prevalence Research Group. The prevalence of refractive errors among adults in the United States, Western Europe, and Australia. Arch Ophthalmol. 2004;122(4):495–505. doi: 10.1001/archopht.122.4.495. [DOI] [PubMed] [Google Scholar]
- 3.He M, Zeng J, Liu Y, Xu J, Pokharel GP, Ellwein LB. Refractive error and visual impairment in urban children in southern china. Invest Ophthalmol Vis Sci. 2004;45(3):793–799. doi: 10.1167/iovs.03-1051. [DOI] [PubMed] [Google Scholar]
- 4.Saw SM, Gazzard G, Shih-Yen EC, Chua WH. Myopia and associated pathological complications. Ophthalmic Physiol Opt. 2005;25(5):381–391. doi: 10.1111/j.1475-1313.2005.00298.x. [DOI] [PubMed] [Google Scholar]
- 5.Smith TS, Frick KD, Holden BA, Fricke TR, Naidoo KS. Potential lost productivity resulting from the global burden of uncorrected refractive error. Bull World Health Organ. 2009;87(6):431–437. doi: 10.2471/BLT.08.055673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vitale S, Ellwein L, Cotch MF, Ferris FL III, Sperduto R. Prevalence of refractive error in the United States, 1999-2004. Arch Ophthalmol. 2008;126(8):1111–1119. doi: 10.1001/archopht.126.8.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegwart JT, Jr, Norton TT. Perspective: how might emmetropization and genetic factors produce myopia in normal eyes? Optom Vis Sci. 2011;88(3):365–372. doi: 10.1097/OPX.0b013e31820b053d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mutti DO, Mitchell GL, Jones LA, et al. Axial growth and changes in lenticular and corneal power during emmetropization in infants. Invest Ophthalmol Vis Sci. 2005;46(9):3074–3080. doi: 10.1167/iovs.04-1040. [DOI] [PubMed] [Google Scholar]
- 9.Wallman J, Winawer J. Homeostasis of eye growth and the question of myopia. Neuron. 2004;43(4):447–468. doi: 10.1016/j.neuron.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Zadnik K, Mutti DO, Mitchell GL, Jones LA, Burr D, Moeschberger ML. Normal eye growth in emmetropic schoolchildren. Optom Vis Sci. 2004;81(11):819–828. doi: 10.1097/01.opx.0000145028.53923.67. [DOI] [PubMed] [Google Scholar]
- 11.Negrel AD, Maul E, Pokharel GP, Zhao J, Ellwein LB. Refractive Error Study in Children: sampling and measurement methods for a multi-country survey. Am J Ophthalmol. 2000;129(4):421–426. doi: 10.1016/s0002-9394(99)00455-9. [DOI] [PubMed] [Google Scholar]
- 12.Saw SM, Katz J, Schein OD, Chew SJ, Chan TK. Epidemiology of myopia. Epidemiol Rev. 1996;18(2):175–187. doi: 10.1093/oxfordjournals.epirev.a017924. [DOI] [PubMed] [Google Scholar]
- 13.Baldwin WR. A review of statistical studies of relations between myopia and ethnic, behavioral, and physiological characteristics. Am J Optom Physiol Opt. 1981;58(7):516–527. doi: 10.1097/00006324-198107000-00002. [DOI] [PubMed] [Google Scholar]
- 14.Wong TY, Foster PJ, Johnson GJ, Seah SK. Education, socioeconomic status, and ocular dimensions in Chinese adults: the Tanjong Pagar Survey. Br J Ophthalmol. 2002;86(9):963–968. doi: 10.1136/bjo.86.9.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Au Eong KG, Tay TH, Lim MK. Education and myopia in 110,236 young Singaporean males. Singapore Med J. 1993;34(6):489–492. [PubMed] [Google Scholar]
- 16.Saw SM, Zhang MZ, Hong RZ, Fu ZF, Pang MH, Tan DT. Near-work activity, night-lights, and myopia in the Singapore-China study. Arch Ophthalmol. 2002;120(5):620–627. doi: 10.1001/archopht.120.5.620. [DOI] [PubMed] [Google Scholar]
- 17.Rahi JS, Cumberland PM, Peckham CS. Myopia over the lifecourse: prevalence and early life influences in the 1958 British birth cohort. Ophthalmology. 2011;118(5):797–804. doi: 10.1016/j.ophtha.2010.09.025. [DOI] [PubMed] [Google Scholar]
- 18.Jones LA, Sinnott LT, Mutti DO, Mitchell GL, Moeschberger ML, Zadnik K. Parental history of myopia, sports and outdoor activities, and future myopia. Invest Ophthalmol Vis Sci. 2007;48(8):3524–3532. doi: 10.1167/iovs.06-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopes MC, Andrew T, Carbonaro F, Spector TD, Hammond CJ. Estimating heritability and shared environmental effects for refractive error in twin and family studies. Invest Ophthalmol Vis Sci. 2009;50(1):126–131. doi: 10.1167/iovs.08-2385. [DOI] [PubMed] [Google Scholar]
- 20.Deans MR, Volgyi B, Goodenough DA, Bloomfield SA, Paul DL. Connexin36 is essential for transmission of rod-mediated visual signals in the mammalian retina. Neuron. 2002;36(4):703–712. doi: 10.1016/s0896-6273(02)01046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernández-Medarde A, Barhoum R, Riquelme R, et al. RasGRF1 disruption causes retinal photoreception defects and associated transcriptomic alterations. J Neurochem. 2009;110(2):641–652. doi: 10.1111/j.1471-4159.2009.06162.x. [DOI] [PubMed] [Google Scholar]
- 22.Hysi PG, Young TL, Mackey DA, et al. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat Genet. 2010;42(10):902–905. doi: 10.1038/ng.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solouki AM, Verhoeven VJ, van Duijn CM, et al. A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat Genet. 2010;42(10):897–901. doi: 10.1038/ng.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park JH, Wacholder S, Gail MH, et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 2010;42(7):570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Benyamin B, McEvoy BP, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moayyeri A, Hammond CJ, Valdes AM, Spector TD. Cohort profile: TwinsUK and Healthy Ageing Twin Study. Int J Epidemiol. 2013;42(1):76–85. doi: 10.1093/ije/dyr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Power C, Elliott J. Cohort profile: 1958 British birth cohort (National Child Development Study). Int J Epidemiol. 2006;35(1):34–41. doi: 10.1093/ije/dyi183. [DOI] [PubMed] [Google Scholar]
- 28.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 29.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. MERLIN: rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 30.Voight BF, Kang HM, Ding J, et al. The Metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8(8):e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34(8):816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang L, Morar N, Dixon AL, et al. A cross-platform analysis of 14,177 expression quantitative trait loci derived from lymphoblastoid cell lines. Genome Res. 2013;23(4):716–726. doi: 10.1101/gr.142521.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dixon AL, Liang L, Moffatt MF, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39(10):1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 35.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):3. [PubMed] [Google Scholar]
- 37.Fisher RA. Statistical Methods for Research Workers. Oliver & Boyd; Edinburgh, Scotland: 1925. [Google Scholar]
- 38.Schaeffel F, Troilo D, Wallman J, Howland HC. Developing eyes that lack accommodation grow to compensate for imposed defocus. Vis Neurosci. 1990;4(2):177–183. doi: 10.1017/s0952523800002327. [DOI] [PubMed] [Google Scholar]
- 39.Diether S, Schaeffel F. Local changes in eye growth induced by imposed local refractive error despite active accommodation. Vision Res. 1997;37(6):659–668. doi: 10.1016/s0042-6989(96)00224-6. [DOI] [PubMed] [Google Scholar]
- 40.Tomita T. Electrical activity of vertebrate photoreceptors. Q Rev Biophys. 1970;3(2):179–222. doi: 10.1017/s0033583500004571. [DOI] [PubMed] [Google Scholar]
- 41.Yau KW. Phototransduction mechanism in retinal rods and cones: the Friedenwald Lecture. Invest Ophthalmol Vis Sci. 1994;35(1):9–32. [PubMed] [Google Scholar]
- 42.Dedek K, Schultz K, Pieper M, et al. Localization of heterotypic gap junctions composed of connexin45 and connexin36 in the rod pathway of the mouse retina. Eur J Neurosci. 2006;24(6):1675–1686. doi: 10.1111/j.1460-9568.2006.05052.x. [DOI] [PubMed] [Google Scholar]
- 43.Feigenspan A, Janssen-Bienhold U, Hormuzdi S, et al. Expression of connexin36 in cone pedicles and OFF-cone bipolar cells of the mouse retina. J Neurosci. 2004;24(13):3325–3334. doi: 10.1523/JNEUROSCI.5598-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee EJ, Han JW, Kim HJ, et al. The immunocytochemical localization of connexin 36 at rod and cone gap junctions in the guinea pig retina. Eur J Neurosci. 2003;18(11):2925–2934. doi: 10.1046/j.1460-9568.2003.03049.x. [DOI] [PubMed] [Google Scholar]
- 45.Wojciechowski R. Nature and nurture: the complex genetics of myopia and refractive error. Clin Genet. 2011;79(4):301–320. doi: 10.1111/j.1399-0004.2010.01592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]