Abstract
Dispersal affects the distribution, dynamics and genetic structure of natural populations, and can be significantly different between sexes. However, literature records dealing with the dispersal of migratory birds are scarce, as migratory behaviour can notably complicate the study of dispersal. We used the barn swallow Hirundo rustica as model taxon to investigate patterns of genetic variability in males and in females of a migratory species showing sex-biased dispersal. We collected blood samples (n = 186) over the period 2006 to 2011 from adults (H. r. rustica subspecies) nesting in the same breeding site at either high (Ireland, Germany and Russia) or low (Spain, Italy and Cyprus) latitude across Europe. We amplified the Chromo Helicase DNA gene in all birds in order to warrant a sex-balanced sample size (92 males, 94 females). We investigated both uniparental (mitochondrial ND2 gene) and biparental (microsatellite DNA: 10 loci) genetic systems. The mtDNA provided evidence for demographic expansion yet no significant partition of the genetic variability was disclosed. Nevertheless, a comparatively distant Russian population investigated in another study, whose sequences were included in the present dataset, significantly diverged from all other ones. Different to previous studies, microsatellites highlighted remarkable genetic structure among the studied populations, and pointed to the occurrence of differences between male and female barn swallows. We produced evidence for non-random patterns of gene flow among barn swallow populations probably mediated by female natal dispersal, and we found significant variability in the philopatry of males of different populations. Our data emphasize the importance of taking into account the sex of sampled individuals in order to obtain reliable inferences on species characterized by different patterns of dispersal between males and females.
Introduction
Distribution, dynamics and genetic structure of natural populations can be severely affected by dispersal [1]–[3], i.e. the movement of an organism from its birthplace to its first breeding site (natal dispersal) or from one breeding site to another (breeding dispersal) [4]–[5]. Dispersal may be significantly different between sexes, as has been well documented in birds and mammals [3], [4], [6]. The costs/benefits of such asymmetry usually depend on the life history and the mating system of a given species [7]. In particular, dispersal tends to be female-biased in birds and male-biased in mammals [4], [8], [9]. Given that dispersal can significantly affect gene flow among populations [10], the dispersing sex may appear as genetically less structured. Hence, accounting for sex is fundamental not only to find out potential differences in dispersal [10]–[11] but also to reliably infer the genetic structure of populations, as the latter could be driven mainly by the philopatric sex [12].
The estimate of the dispersal rate in natural populations is often incomplete because it usually requires direct methods with intensive, large-scale and long-term demographic studies [10], [11], [13]. However, recent advances in the genetic techniques allowed researchers to integrate field investigation with molecular DNA analysis. The combined use of markers with different way of inheritance (mitochondrial versus nuclear DNA) represented the most suitable approach to infer discrepancy between the dispersal pattern of males and females. Indeed, differences in the genetic picture drawn by mitochondrial and nuclear markers are expected when sex-biased dispersal occurs [11]. Studies focusing on the application of population genetic tools to infer sex-biased dispersal are well known for vertebrates, more frequently in birds and mammals (e.g.: eiders [14], rodents [15]) than in amphibians and reptiles (e.g.: frogs [16], turtles [17]). Nevertheless, as exhaustively discussed by Møller et al. [18], dispersal of migratory birds is poorly studied because routes can greatly complicate the interpretation of the genetic scenario [19].
The barn swallow Hirundo rustica is a polytypic passerine bird widely distributed throughout most of the northern hemisphere [20]. This species is extensively studied with reference to its morphology and behaviour. For instance, recent studies disclosed significant patterns of morphological differentiation among European populations in a few characters (ventral coloration, tail streamers) known to be under sexual selection [21]–[24]. As far as the migratory behaviour is concerned, European barn swallows can be divided in two main groups: one breeds in south-western Europe and winters in central and western Africa, the other breeds in northern Europe and winters in southern Africa [25]. Differences in morphological and behavioural traits notwithstanding, the occurrence of some degree of genetic differentiation among European H. rustica populations has never been proved by using either mitochondrial or microsatellite DNA markers [23], [24], [26]. However, although the barn swallow is a female-biased dispersal species (males are more philopatric than females, e.g. [27]), the sex of the investigated individuals has never been taken into account in any genetic study focusing on this taxon.
In this work we aim at: (i) analysing the genetic variability of European barn swallow populations over a wide sampling area by means of markers from both uniparental (mitochondrial DNA: mtDNA) and biparental (microsatellite DNA) genetic systems; (ii) testing the consequences of sex-biased dispersal on the population genetic structure by comparing patterns of variation in a well balanced sample of males and females. Overall, the barn swallow represents an excellent model among migratory species to investigate patterns of genetic variation in males and females. While lack of genetic structure is expected for the whole sample size, we predict the occurrence of different genetic pattern between male and female barn swallows [12], [16].
Materials and Methods
Ethics statement
The barn swallow is not an endangered species in all trapping areas of this study. Samples were obtained in the same place (six localities) in different years. Adults were trapped with mist-nets. Samples (one blood droplet) were collected by means of wing venipuncture (brachial/radial/ulnar vein). Birds were not sedated and did not suffer any injury: all of them were released 10 min after blood collection.
We report here below the coordinates of the sampling localities (see also Table S1) together with the information about the permits issued for each specific area: (1) Tullynisk, Offaly (Ireland: 53°07′N, 07°54′W). The licensing authority in Ireland is the National Parks and Wildlife Service (NPWS), which provided an annual (renewable) license for the present study (NPWS references 57/2009 and C41/2010). Separately, the capture of birds is also controlled through the British Trust for Ornithology Ringing Scheme. All licenses and permits were obtained by, and in the name of, A.S. Copland (BTO permit number A5115). All samples were taken from birds at a privately-owned site. Access to this site was arranged through the regional staff of the NPWS, who also have contact details for the owner/manager; (2) Itzehoe (Germany: 53°56′N, 09°31′E). Samples were collected by S. Martens, who is ringer at the Institute for Avian Research “Vogelwarte Helgoland”. Samples were collected at a private farm and future permissions should be requested to the owner of the same; (3) Lake Ladoga (Russian Federation: 60°40′N, 32°56′E). Samples were collected by O. Babushkina at the Ladoga Ornithological Station of the Biological Research Institute of the Saint Petersburg State University. The Ladoga Ornithological Station is comprised within the Nizhne-Svirsky State Reserve. All work at the station adhered to the current legislation of the Russian Federation and to the institutional guidelines of the State University of Saint Petersburg. No specific permits were needed to O. Babushkina. Future permissions should be requested to the Biological Research Institute of the Saint Petersburg State University; (4) Gorliz (Spain: 43°24′N, 02°57′W). Samples were collected by I. Zuberogoitia. He obtained a license to trap barn swallows and collect blood samples by the Department of Medio Ambiente (Diputación Foral de Bizkaia). The same administration manages the experimental farm for the selection of cow races where the sampling was carried out, and all permissions were obtained in the same site. Future permissions should be requested to the same Department; (5) Orti-Bottagone Nature Reserve (Italy: 42°57′N, 10°35′E). Samples were collected by R. Ceccherelli (veterinary, CRUMA, Leghorn). Land is a private property (oasis) of World Wildlife Fund (WWF) and future permissions should be requested to WWF Italy; (6) Polis (Cyprus: 35°02′N, 32°25′E). Samples were collected in a government land by A. Crabtree (Vice Chairman and Ringing Officer of BirdLife Cyprus), who obtained a specific permit from the competent authority of Cyprus (Game & Fauna Service, Ministry of Interior, Nicosia). Future permissions should be requested to the same Ministry.
Biological sampling
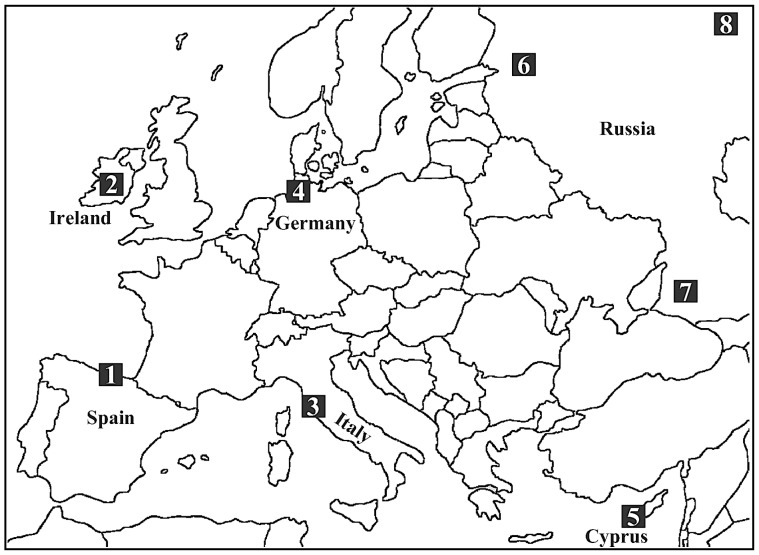
Blood samples (n = 186) were collected in subsequent years (2006–2011) from nesting adults (H. r. rustica subspecies) at the same breeding site in six areas across Europe (Figure 1, Table S1): Gorliz, Spain (SPA, n = 32); Tullynisk, Ireland (IRE, n = 26); Orti-Bottagone Nature Reserve, Italy (ITA, n = 33); Itzehoe, Germany (GER, n = 33); Polis, Cyprus (CYP, n = 32); Lake Ladoga, Russia (RUS, n = 30). One blood droplet was collected on filter paper (Whatman, UK) from each bird by wing venipuncture. Each swallow was ringed and sexed according to a PCR-based assay (see below).
Figure 1. Barn swallow breeding populations sampled in this study.
(1): SPA, Spain; (2): IRE, Ireland; (3): ITA, Italy; (4): GER, Germany; (5): CYP, Cyprus; (6): RUS, Russia. Mitochondrial DNA sequences available in the GenBank were obtained from Russian populations of Krasnodar (7, KRD) and Medvedevo (8, MED) (Table S1).
DNA extraction
Genomic DNA was extracted using the Puregene Core Kit-A (Qiagen, Germany) following the manufacturer's instructions. DNA content and purity were determined with an Eppendorf BioPhotometer (AG Eppendorf, Germany).
Sexing
Chromo Helicase DNA (CHD) gene of ZZ (males) and ZW (females) sexual chromosomes was amplified with primers L1237 (5'-GAGAAACTGTGCAAAACA-3') and H1272 (5'-TCCAGAATATCTTCTGCTCC-3') previously tested in other bird species [28]. PCRs were prepared in 25 µL as in [29] and performed in a MyCycler thermal cycler (Biorad, USA) with the following profile: 3 min at 94°C, 30 cycles of 30 s at 94°C, 1 min at 48°C and 45 s at 72°C, finally 7 min at 72°C. PCR products were run in a 3.5% agarose gel for 60 min together with positive controls for male and female individuals. We set-up the PCR-based procedure by testing 20 barn swallows whose sex (10 males, 10 females) was determined through the inspection of standard morphological traits [30] by one of us (P.M. Politi).
Mitochondrial DNA
Laboratory procedure
The entire mtDNA gene codifying for the second sub-unit of the NADH dehydrogenase (ND2, 1041 bp) was amplified using primers L5216 and H6313 [31]. PCRs (50 µL) were run in a MyCycler thermal cycler (Biorad) as in [29]. PCR products were purified using GenElute PCR Clean-up Kit (Sigma Aldrich, Italy) and directly sequenced on both DNA strands using the BigDye Terminator v. 3.1 Cycle Sequencing Kit on an ABI 3730 DNA automated sequencer (Applied Biosystems, USA) at Genechron (Rome, Italy). We amplified the ND2 gene in a subset of samples (10 for each population, n = 60), and we included in the alignment (clustalx v. 1.81: [32]) 16 sequences from the GenBank (Russia: Krasnodar Kray, KRD, n = 7 and Arkhangel'sk Oblast', MED, n = 9: [33]) (Figure 1, Table S1).
Population genetic inferences
We used dnasp v. 5.1 [34] to infer the mtDNA haplotypes. A network was constructed using dna alignment v. 1.3.3.1 (2003–2013 Fluxus Technology, UK) and the Median Joining method of [35] with network v. 4.5.1.0 (2004–2009 Fluxus Technology). We used arlequin v. 3.5.1 [36] to: (i) calculate the haplotype diversity (h), the nucleotide diversity (π) and the mean number of pairwise differences (k); (ii) investigate the partition of the mtDNA diversity (Analysis of the Molecular Variance, amova) among and within the populations using the PhiST analogous to Wright's F-statistics (1000 permutations) [37]; (iii) compute the average genetic distance among populations (1000 replicates with the TN93 algorithm) [38].
Historical demography
Inferences of historical demography were obtained using dnasp and different statistics as described in [39]. The analysis included (i) males and females plus the GenBank sequences (n = 60+16 = 76), (ii) only the males (n = 27, no GenBank entries), and (iii) only the females (n = 33, no GenBank entries). Ramirez-Soriano et al. [40] investigated the statistical power of a wide range of statistics computed on DNA polymorphism data in detecting a sudden population expansion, a sudden contraction or a bottleneck. They found that the most powerful tests were those based on haplotype frequencies, including the F S of Fu [41] and the R 2 statistic [42]. In this study, the significance of the F S and R 2 statistics was investigated by examining the null distribution of 5000 coalescence simulations using dnasp. Only significant negative F S and positive R 2 values were retained as evidence of population expansion [39]. We also computed the Tajima's D [43]. Nevertheless, [42] reported that R 2 statistic has a greater power than the Tajima's D or F S to detect population expansion when the sample size is small (∼10). Furthermore, the McDonald-Kreitman test [44] as implemented in dnasp was conducted for the entire dataset to investigate the deviation from an equal ratio of non-synonymous (Ka) to synonymous (Ks) fixed substitutions. Specifically for this test we used two US H. r. erythrogaster samples as outgroup (UWBM 78832 and UWBM 80547 from the University of Washington Burke Museum of Natural History, Seattle, USA; GenBank accession codes: HF548593-94).
The Mismatch Distribution (MD) of mtDNA pairwise differences was also examined using arlequin (males + females, males only, females only). The more ragged the shape of the distribution the closer was the population to a stationary model of constant size over a long period (raggedness index, r) [45]. The MD test uses the observed parameters of the expansion to perform coalescent simulations and to create new estimates of the same parameters. Departure from a model of sudden expansion was tested for each population by summing the squared differences (SSD) between observed and estimated MD [46], [47].
Microsatellite DNA
Laboratory procedure
All samples (n = 186) were investigated at 10 loci of the microsatellite DNA (Short Tandem Repeats, STR) reported in [48], [49]. PCRs (12.5 µL) were performed as in [50] (Table 1). Gene sizing was carried out at the Research Centre of Clinical and Molecular Genetics (Pisa, Italy) on an ABI Prism 3730 DNA automated sequencer using genescan (Applied Biosystems). For the statistical analyses we used either the whole sample size (n = 186) or males (n = 92) and females (n = 94) separately.
Table 1. The characteristics of the investigated STR loci are shown.
| Locus | T M (°C) | Size-range (bp) | Repeat motif | A | HO | HE | P ID | P IDsib |
| Hir7 | TD 52-50 | 215–273 | (CT)2 | 25 | 0.69 | 0.94 | 7.07×10−3 | 2.82×10−1 |
| Hir24 | TD 56-54 | 194–238 | (AGTG)4 | 12 | 0.46 | 0.88 | 2.06×10−4 | 9.06×10−2 |
| Hir10 | TD 52-50 | 147–201 | (GTTT)4 | 13 | 0.75 | 0.85 | 8.79×10−6 | 3.06×10−2 |
| Hru5 | TD 52-50 | 110–140 | (GT)9 | 15 | 0.66 | 0.84 | 4.01×10−7 | 1.05×10−2 |
| Hir20 | TD 50-48 | 231–275 | (ATAG)9 | 15 | 0.76 | 0.84 | 1.90×10−8 | 3.61×10−3 |
| Hir6 | TD 56-52 | 178–218 | (TCTA)11 | 11 | 0.80 | 0.84 | 9.19×10−10 | 1.24×10−3 |
| Hir11 | TD 50-48 | 166–214 | (GATA)7 | 12 | 0.78 | 0.83 | 4.71×10−11 | 4.34×10−4 |
| Hir4 | TD 56-54 | 259–297 | (GTTT)5 | 16 | 0.39 | 0.79 | 3.22×10−12 | 1.63×10−4 |
| Hir5 | TD 50-48 | 213–233 | (GTTT)4 | 6 | 0.45 | 0.71 | 4.48×10−13 | 6.98×10−5 |
| Hir15 | TD 50-48 | 209–249 | (ATGT)2 | 9 | 0.55 | 0.47 | 8.16×10−14 | 3.29×10−5 |
T M (°C), annealing temperature; TD, touch-down PCR; A, number of alleles per locus; H O, mean observed heterozygosity; H E, mean expected heterozygosity; P ID, probability that two individuals drawn at random share identical genotypes; P IDsib, probability of identity among siblings. STR loci are sorted according to the increasing order of their P ID (P IDsib) single-locus values (i.e., the locus at the top is the most informative one), and a sequentially multi-loci P ID (P IDsib) is reported for each locus.
Genetic variability and relatedness
The discriminatory power of the whole set of STR loci was evaluated with gimlet v. 1.3.3 [51] by estimating the probability that two individuals drawn at random from the populations showed identical multilocus genotypes by chance (P ID and P ID sib: for the latter, we assumed sibling relationships) [52], [53]. Moreover, all loci were investigated using micro-checker v. 2.2.3 [54] to check for null alleles, allele dropout and scoring errors due to stuttering. Arlequin, fstat v. 2.9.3 [55] and genepop v. 3.4 [56] were used in order to: (i) compute the number of alleles per locus, the number of unique alleles and the allelic richness; (ii) calculate expected (H E) and observed (H O) heterozygosity; (iii) infer deviations from both Hardy-Weinberg Equilibrium (HWE) and Linkage Disequilibrium (LE) (10 000 dememorisations, 100 batches, 5000 iterations per batch); (iv) estimate gene flow (N em, effective number of migrants per generation) via the private allele method of Slatkin [57]; (v) investigate the partition of the STR diversity within and among populations by amova; (vi) infer the degree of genetic differentiation among populations by estimating the average F ST distance values. Bonferroni correction [58] was adopted to adjust the significance level of each test. The average F ST distance values were plotted on the first two axes of a Principal Component Analysis (PCA) using statistica 5.0/W (Statsoft Inc., USA).
Population genetic structure
Bayesian clustering analysis was performed with structure v. 2.3.4 [59] to investigate the spatial structure of the genetic diversity. We focused on identifying the K (unknown) clusters of origin of the sampled individuals and to simultaneously assign them to each cluster. We assumed correlated allele frequencies and we used a prior population information option to take the sampling locality into account [60]. All simulations were run with 106 Markov Chain of Monte-Carlo iterations, following a burn-in period of 105 iterations, and were replicated five times per each K-value (1 to 12). The number of clusters that best fitted to the data was chosen using the formula of Evanno et al. [61]. An identification threshold to each cluster was selected (Qi = 0.80) as in [62].
Sex-biased dispersal
In order to test for possible differences in the dispersal rate between males and females, fstat was used to calculate five different parameters: F IS, F ST, relatedness (R), mean (mAI C) and variance (vAI C) of the assignment index (AI C) within each sex [10]. The latter estimates the probability that a given genotype originates from the population where it was sampled, and the statistical significance is determined by a two-tailed test using 10 000 randomizations. Low mAI C and high vAI C values are interpreted for the dispersing sex. While F ST and R are expected to be larger in the philopatric than in the dispersing sex, the opposite occurs for F IS. Nevertheless, the power of these statistics depends on dispersal rates, bias intensity, sampling design and the number of loci [10].
Results
Sexing
The CHD gene was amplified in all barn swallows: 92 birds were identified as males (single PCR product, ca. 200 bp) while 94 as females (two PCR products, ca. 200 and 240 bp).
Mitochondrial DNA
Population genetics
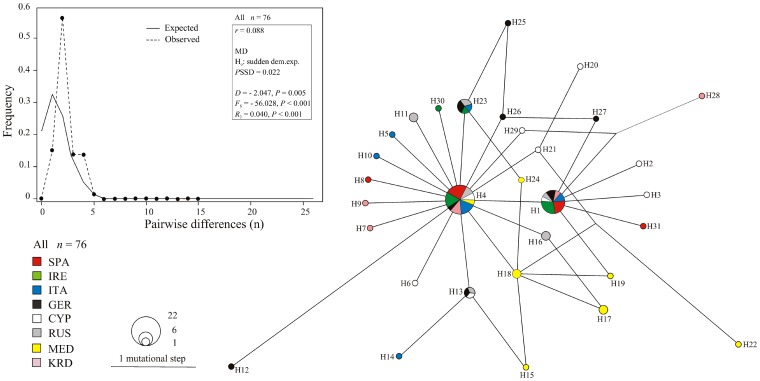
The alignment of 76 (60+16) ND2 sequences produced 31 haplotypes (H1-H31: GenBank accession codes HF548562-HF548592) including 27 polymorphic sites. Estimates of all parameters are summarized in Table S2. SPA and IRL populations showed the lowest number of haplotypes as well as the lowest values of haplotype diversity (h), average number of pairwise differences (k) and nucleotide diversity (π). The highest number of haplotypes was found in CYP and MED. The Median Joining network showed that all barn swallow populations were genetically admixed with no noticeable divergence among haplotypes (Figure 2). In particular, two haplotypes (H1 and H4) were common to all populations. The 88.4% of the mtDNA variability was partitioned within populations while the 11.6% among them (PhiST = 0.131, P<0.001: data not shown). Only MED significantly diverged from all other populations (PhiST range: 0.28–0.38, all P<0.001, Table S3). When amova was performed either without KRD and MED populations or excluding MED only, we found that the partition of the mtDNA variability among populations decreased to 1.86% and 1.33%, respectively (PhiST = 0.02 and 0.01, respectively, P>0.05: data not shown). Finally, when all the estimated parameters were computed by including in the analysis males and females separately, all results matched those produced using the entire dataset (data not shown).
Figure 2. Median-Joining network of barn swallow populations computed on mtDNA haplotypes with network.
Size of circles is proportional to the haplotype frequency. The colour of each population is indicated as well as the number of each haplotype. A length bar to compute the number of mutational changes was provided (see also Table S1). The inset showed Mismatch Distribution (MD) of the mtDNA pairwise differences (observed: dotted; expected: line) computed on the whole dataset (n = 76). The expected curve was obtained from simulated values computed from the data under the model of demographic expansion (H0). The Harpending's raggedness index (r) was given with the P value of the SSD test as well as Tajima's D, Fu's F S and R 2 statistics.
Historical demography
With the sole exception of the SSD test (P = 0.022), the statistics we used (D = −2.047, F S = −56.028 and R 2 = 0.040: all P<0.001) could not exclude the occurrence of a demographic expansion for the entire sample size (n = 76) (Figure 2). Indeed, the bell-shaped (r = 0.088) curve obtained for the Mismatch Distribution (MD) of the pairwise differences suggested a rapid population growth. When the demographic parameters were computed using males (n = 27) and females (n = 33) (GenBank entries excluded) separately, males showed exactly the same trend as the whole dataset (D = −1.970, F S = −13.178 and R 2 = 0.056: all P<0.001; SSD test: r = 0.158, P = 0.023), while for the females a demographic expansion was suggested by all statistics (D = −1.571, F S = −18.449 and R 2 = 0.064: all P<0.001; SSD test: r = 0.123, P = 0.063). The McDonald-Kreitman test was not significant for the whole sample size (Fisher exact test, P = 0.16).
Microsatellite DNA: all individuals
Genetic variability
All STR loci employed were highly polymorphic. In the entire sample size (n = 186) the STR panel was powerful in discriminating individuals (P ID = 8.16×10−14 and P IDsib = 3.29×10−5, Table 1), as values lower than 0.001 can be considered as satisfactory [53]. Micro-checker did not provide evidence for allele dropout or scoring errors due to stuttering, although three loci (Hir4, Hir7 and Hir24) showed an excess of homozygotes for most of the allele-size classes, thus pointing to the possible presence of null alleles (data not shown). The total number of alleles at each locus ranged between 6 and 25 (Hir5 and Hir7, respectively), with a mean of 13.4 alleles per locus (Table 1).
The average values of H O were smaller than H E for each locus (Fisher exact test, P<0.001 all loci, Table 1) and ranged between 0.39 and 0.80 (Hir4 and Hir6, respectively: Table 1). There was no evidence of LE at any pair of loci after sequential Bonferroni correction (P>0.05, all comparisons: data not shown). Both the number of alleles and gene diversity of each locus pointed to a very high degree of genetic variability. Allelic richness ranged between 7.9 and 9.9 (SPA and CYP, respectively: Table 2); CYP showed the highest number of private alleles (n = 8, Table 2). Average levels of H O and H E across all loci and populations were relatively homogeneous and showed very restricted ranges (H O: 0.60–0.67, H E: 0.79–0.82, Table 2). H O values pointed to a deficiency of heterozygotes in all populations, which was confirmed by departure from HWE (P<0.001 all loci, Table 2). One locus in IRE, two loci in SPA, three loci in GER and RUS, and four loci in ITA and CYP were not in HWE (P<0.001, data not shown). However, we found that no locus deviated from HWE in all populations, and when the Bonferroni's correction was taken into account only three loci (Hir4, Hir7 and Hir24) deviated from HWE in some populations yet not in all (P<0.001, data not shown).
Table 2. The genetic variability of the STR loci for each population.
| Population | n | n a | A r | A u | H O | H E | P HWE | χ2 (df) | F ST | |
| All | SPA | 32 | 8.3 | 7.9 | 3 | 0.63 | 0.80 | <0.001 | ∞ (20) | 0.014 |
| IRE | 26 | 9.4 | 9.3 | 4 | 0.67 | 0.80 | <0.001 | ∞ (20) | 0.014 | |
| ITA | 33 | 10.0 | 9.4 | 4 | 0.60 | 0.81 | <0.001 | ∞ (20) | 0.014 | |
| GER | 33 | 9.5 | 9.0 | 2 | 0.65 | 0.81 | <0.001 | ∞ (20) | 0.014 | |
| CYP | 32 | 10.4 | 9.9 | 8 | 0.62 | 0.82 | <0.001 | ∞ (20) | 0.013 | |
| RUS | 30 | 9.3 | 8.9 | 2 | 0.61 | 0.79 | <0.001 | ∞ (20) | 0.014 | |
| Males | SPA | 16 | 7.3 | 6.8 | 5 | 0.61 | 0.79 | <0.001 | ∞ (20) | 0.018 |
| IRE | 14 | 8.0 | 7.8 | 6 | 0.70 | 0.82 | 0.001 | 45.2 (20) | 0.017 | |
| ITA | 16 | 7.9 | 7.5 | 2 | 0.62 | 0.80 | <0.001 | ∞ (20) | 0.014 | |
| GER | 16 | 8.3 | 7.9 | 4 | 0.68 | 0.83 | <0.001 | ∞ (20) | 0.017 | |
| CYP | 16 | 8.4 | 7.9 | 5 | 0.60 | 0.81 | <0.001 | ∞ (20) | 0.017 | |
| RUS | 14 | 6.6 | 6.5 | 1 | 0.59 | 0.77 | <0.001 | 54.5 (20) | 0.019 | |
| Females | SPA | 16 | 7.1 | 6.6 | 1 | 0.66 | 0.80 | 0.001 | 53.2 (20) | 0.022 |
| IRE | 12 | 6.8 | 6.6 | 3 | 0.63 | 0.77 | <0.001 | ∞ (20) | 0.024 | |
| ITA | 17 | 8.1 | 7.1 | 3 | 0.58 | 0.81 | <0.001 | ∞ (20) | 0.022 | |
| GER | 17 | 7.6 | 6.8 | 1 | 0.62 | 0.79 | <0.001 | ∞ (20) | 0.022 | |
| CYP | 16 | 9.0 | 7.9 | 8 | 0.64 | 0.82 | <0.001 | ∞ (20) | 0.021 | |
| RUS | 16 | 8.4 | 7.3 | 4 | 0.62 | 0.79 | <0.001 | 68.7 (20) | 0.022 |
The genetic variability of the STR loci for each population was computed considering all individuals and male and female genotypes separately: n, sample size; n a, average number of alleles/locus; A r, allelic richness; A u, number of unique alleles; H O, observed heterozygosity; H E, expected heterozygosity; P HWE, probability value for the Hardy-Weinberg Equilibrium test; χ2 test with relative degrees of freedom (df) (Fisher exact test, all loci). Departure from HWE was significant in all populations after Bonferroni correction (α = 0.05, α' = 0.05/60 = 0.0008).
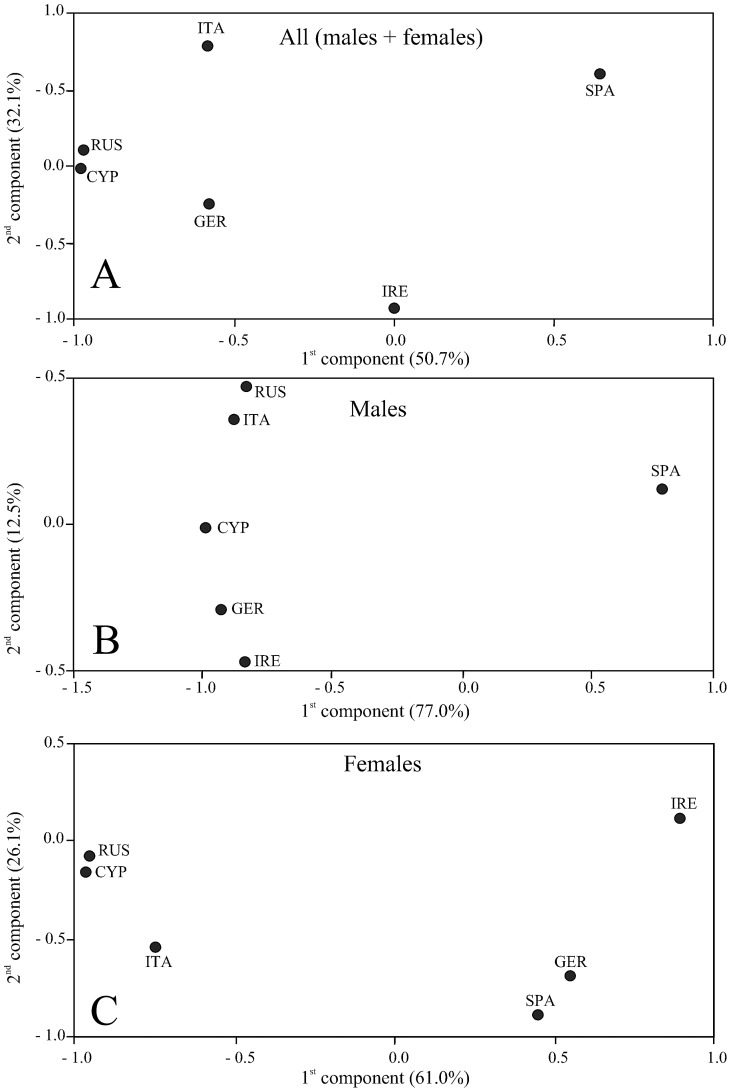
We found that 98.6% of the total STR variability was partitioned within populations and 1.38% among them (F ST = 0.014, P<0.001). In the PCA plot reported in Figure 3A, the first two components explained the 82.8% of the total variability, SPA being the most diverging population (e.g., versus CYP and RUS, F st = 0.026 and 0.027, respectively: P<0.001). Non significant F st distance values were found among ITA, GER, CYP and RUS (gene flow range: 3.87–6.07, Table 3).
Figure 3. The Principal Component Analysis performed using the average pairwise F ST distance values among STR genotypes.
The percentage of total variance explained by each of the first two components is given. (A) All individuals. (B) Only males. (C) Only females.
Table 3. Estimates of population pairwise genetic distance values (F ST) and number of migrants (N em).
| SPA | IRE | ITA | GER | CYP | RUS | ||
| All | SPA | - | 3.24 | 4.05 | 3.01 | 4.09 | 2.96 |
| IRE | 0.024** | - | 4.23 | 3.43 | 3.31 | 2.80 | |
| ITA | 0.009 | 0.019* | - | 5.43 | 4.37 | 6.07 | |
| GER | 0.017* | 0.012* | 0.008 | - | 4.58 | 3.87 | |
| CYP | 0.026** | 0.016* | 0.006 | 0.010 | - | 4.53 | |
| RUS | 0.027** | 0.019* | 0.005 | 0.010 | 0.002 | - | |
| Males | SPA | - | 2.89 | 2.22 | 2.57 | 1.69 | 2.59 |
| IRE | 0.037* | - | 2.07 | 2.51 | 1.96 | 1.69 | |
| ITA | 0.022* | 0.012 | - | 2.08 | 2.36 | 2.91 | |
| GER | 0.029* | 0.001 | 0.001 | - | 3.03 | 2.09 | |
| CYP | 0.050** | 0.012 | 0.008 | 0.004 | - | 4.09 | |
| RUS | 0.039* | 0.020 | 0.004 | 0.012 | 0.012 | - | |
| Females | SPA | - | 1.67 | 3.44 | 2.58 | 3.22 | 2.37 |
| IRE | 0.021 | - | 1.79 | 2.18 | 3.05 | 2.75 | |
| ITA | 0.012 | 0.034* | - | 3.13 | 3.38 | 2.76 | |
| GER | 0.009 | 0.027* | 0.025* | - | 3.71 | 3.77 | |
| CYP | 0.019 | 0.036* | 0.008 | 0.029* | - | 3.95 | |
| RUS | 0.031* | 0.048** | 0.015 | 0.028* | 0.005 | - |
Estimates of population pairwise genetic distance (F ST) and number of migrants (N em) were computed for all individuals, only males and only females. Below diagonal: F ST pairwise distance values. Above diagonal: the effective number of migrants per generation (N em); *, P<0.05; **, P<0.001; others, P>0.05.
Population genetic structure
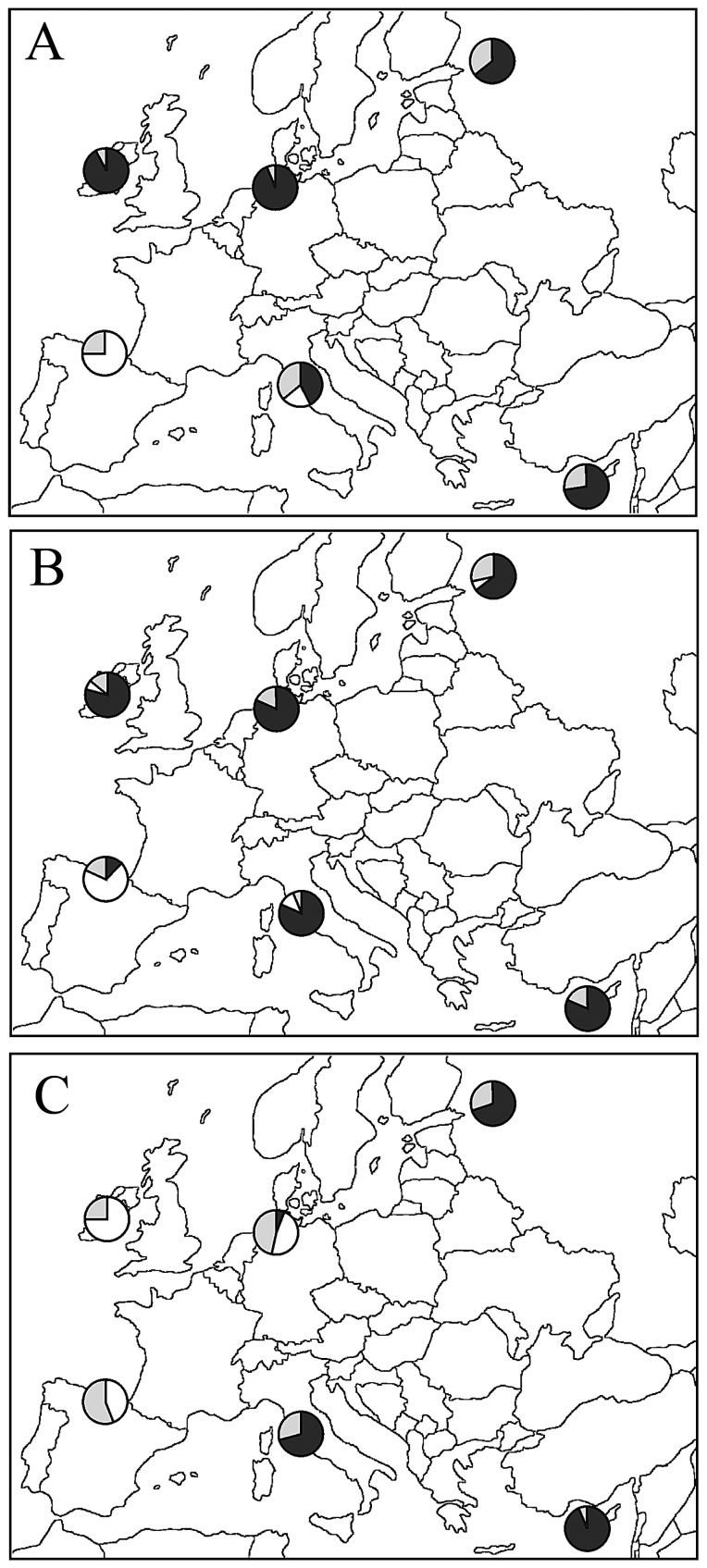
In Figure 4A the whole sample size was taken into account. Bayesian clustering analysis indicated that barn swallows could be divided into two genetic groups (K = 2, see also Table S4). Genetic differentiation was strong between SPA and all other populations, and Spanish individuals were mostly assigned to the cluster II (QI = 0.16; QII = 0.84). While ITA showed the highest number of barn swallows with admixed genotype (n = 12: QI = 0.59 and QII = 0.41, see Table S4), a slight differentiation was found between (IRE + GER) and (CYP + RUS) population pairs, yet their individuals were all assigned to the cluster I (QI range: 0.82–0.94). When we excluded from the Bayesian clustering analysis the STR loci showing null alleles and deviating from HWE (Hir4, Hir7, and Hir24: see above), barn swallows were assigned to two genetic groups and results matched those obtained when the entire set of loci was taken into account (data not shown).
Figure 4. Bayesian admixture analysis as inferred using structure.
The ΔK calculated according to Evanno et al. [61] was optimal for K = 2 in all computations. Each population was represented by a pie chart whose segments were proportional to the number of specimens assigned to cluster I (black), to cluster II (white) or which showed admixed genotypes (grey). Threshold value for assignment to each cluster was Qi = 0.80 (Table S4). (A) All individuals. (B) Only males. (C) Only females.
Microsatellite DNA: males versus females
Genetic variability
Average levels of H O and H E across all loci and populations were relatively homogeneous and showed deficiency of heterozygotes in both male and female barn swallows (Fisher exact test, P<0.001 all loci, Table 2). IRE and CYP showed the highest number of private alleles in males and females, respectively (Table 2). Females showed global F ST values higher than males in all populations (on average, females: F ST = 0.022, P<0.001; males: F ST = 0.017, P = 0.003; Table 2). The majority of the total STR variability was partitioned within populations in both males and females (98.2% and 97.8%, respectively: data not shown). Differences between sexes were found in the PCA plot of average F ST distance values. Males were highly differentiated along the 1st component (77.0% of the STR diversity: Figure 3B), while in the females the 2nd component explained a significant portion of the total variability (26.1%: Figure 3C). In males, SPA was the only divergent population with F ST values ranging from 0.022 to 0.050 (versus ITA and CYP, respectively, P<0.05: Table 3). Significant F ST distance values were disclosed among other pairs of populations in females but not in males (Table 3). In females, the largest genetic distance values were found between IRE and ITA (0.034), IRE and CYP (0.036) and IRE and RUS (0.048) (all P<0.05, Table 3), whereas no genetic differentiation was disclosed among ITA, CYP and RUS as well as among IRE, SPA and GER. Gene flow was, on average, higher in females than in males (average N em: females = 2.92, males = 2.45, Table 3).
Population genetic structure
Bayesian clustering analysis performed in males and females separately suggested the occurrence of two groups (K = 2, Figure 4B, 4C; see also Table S4). For each population, males and females showed a largely different genetic make-up that could be inferred only partially when the entire sample size was included in the analysis (see above). When we used only the genotypic data inferred from the males, SPA diverged from all other populations and showed the highest membership value to the cluster II (QI = 0.21; QII = 0.79, Figure 4B). No similar evidence was observed in the other populations: in these latter, most of the individuals were assigned to the cluster I (QI range: 0.75–0.92) and only a very few were assigned to the cluster II or showed admixed genotype (Figure 4B, Table S4). When we used only the genotypic data inferred from the females, two different groups were disclosed. One included SPA, IRE and GER, which showed the highest assignment values to the cluster I (QI = 0.67, 0.84 and 0.70, respectively: Figure 4C). The other comprised ITA, CYP and RUS, which showed the highest assignment values to the cluster II (QII = 0.83, 0.89 and 0.78, respectively: Figure 4C, Table S4). When SPA was excluded from the Bayesian clustering analysis, the pattern showed by the females was very similar to that inferred using all populations (Figure S1 versus Figure 4C): in ITA, CYP and RUS the majority of the individuals were included in the black cluster, while IRE and GER included many birds assigned to the white cluster. On the contrary, when the SPA birds were excluded, a not negligible level of genetic diversity came to the fore among the males (Figure S1 versus Figure 4B), and pointed to the occurrence of three genetic groups (IRE + GER, ITA + CYP, and RUS), although the F ST distance values were not significantly different among each other (not shown).
Dispersal
Males and females were tested for sex-biased dispersal using either all populations separately or keeping SPA on its own and grouping all of the other ones. In the first case, females showed higher inter-population F IS and vAI C values than males (females: F IS = 0.225, vAI C = 10.982; males: F IS = 0.217, vAI C = 8.829, Table 4). However, these differences were not statistically significant (all parameters). In the second case, F ST (females: F ST = 0.004; males: F ST = 0.028, P = 0.015) and R values (females: R = 0.007; males: R = 0.045, P = 0.017) were significantly different and suggested female-biased dispersal. This finding was supported further by higher values of F IS in the females than in the males as well as by the negative value of mAI C in the females (F IS = 0.234, mAI C = −0.040); however, these differences were not statistically significant (all parameters, Table 4).
Table 4. Sex-biased dispersal tests.
| All populations | SPA versus all others | |||||
| Males | Females | P -value | Males | Females | P -value | |
| F IS | 0.217 | 0.225 | 0.389 | 0.217 | 0.234 | 0.243 |
| F ST | 0.011 | 0.015 | 0.763 | 0.028 | 0.004 | 0.015* |
| R | 0.017 | 0.025 | 0.752 | 0.045 | 0.007 | 0.017* |
| mAI C | −0.052 | 0.051 | 0.594 | 0.041 | −0.040 | 0.444 |
| vAI C | 8.829 | 10.982 | 0.147 | 15.580 | 11.110 | 0.898 |
F IS, F ST, relatedness (R), mean assignment index (mAI C) and variance of mean assignment index (vAI C) were estimated on males and females considering either single populations or by separating Spanish barn swallows from all other ones. Significance (P) (two-tailed test) was assessed using the randomization method of [10]; *, P<0.05.
Discussion
There are a few studies focusing on migratory bird species and relying on the use of molecular DNA markers (e.g., sandhill crane [63]; black-throated blue warbler [64]; reed warbler [65]). This likely occurs because species with elevated mobility are expected to show a higher level of gene flow and a weaker genetic structure than sedentary ones [66]–[68]). However, none of these studies accounted for sex in the genetic analysis. Only Ortego et al. [12] investigated the correspondence between population genetic structure and natal dispersal by analyzing males and females separately but in a non-migratory passerine (Cyanistes caeruleus, blue tit). On the contrary, studies focusing on other vertebrate species with sex-biased dispersal and taking into account the sex of individuals in the genetic analysis are known [16], [17], [69]. In the barn swallow, the use of either mitochondrial or microsatellite DNA pointed to the admixed genetic structure of the European populations investigated so far [23], [24], [26], and significant genetic (mtDNA) distances were found only among barn swallows of different continents [33]. Nevertheless, in this species, the genetic pattern of males and females has never been compared.
Mitochondrial DNA
The mtDNA analysis did not suggest any significant structuring of the genetic variability (Figure 2), as it might be expected in a species with female-biased dispersal and male philopatry [69]. The star-like shape of the network, which included two ancestral haplotypes (H1 and H4), as well as the bell-shaped curve of the MD were consistent with a recent demographic expansion of the studied barn swallow populations (Figure 2, Table S2) [26], [33]. Such a scenario was statistically well supported (Fig. 2: D = −2.047, F S = −56.028 and R 2 = 0.040, all P<0.001), with the SSD statistic (P = 0.022) being an exception. However, although all tests we employed may be sensitive to unknown structure within populations, [70] stressed that the SSD statistic was actually the less powerful. Furthermore, when only the females were analyzed, a model of expansion was supported also by the SSD statistic (P = 0.063). Overall, we felt confident in considering that such a deviation from neutrality was very likely due to demographic changes rather than selective processes, as the McDonald-Kreitman test was not significant in the entire dataset [71]). This result is in agreement with the relatively recent barn swallow range and demographic expansion due to the proliferation of human settlements providing widespread availability of suitable nest sites [33].
The 11.6% of total mtDNA variability was partitioned among populations, a value much higher than that (<2%) found by Dor et al. [24], who analyzed populations of H. r. rustica and H. r. transitiva (Middle East). However, we feel confident that this result was due to the divergence of MED, the easternmost and northernmost population in our sampling scheme. Indeed, when MED was excluded from the analysis the mtDNA genetic structure promptly disappeared (Figure 1, Table S3).
Microsatellite DNA: general overview
The loci of the microsatellite DNA investigated in this study showed a pattern of variability similar to that reported by [47], [48], with very high degree of polymorphism and low level of relatedness among the genotyped barn swallows (Table 1). Microsatellites did not show any evidence of Linkage Disequilibrium. However, a significant departure from HWE was found in all populations due to a deficiency of heterozygotes (Table 2). This result was possibly caused by either the occurrence of null alleles at a few STR loci or the Wahlund effect [72], which, in turn, pointed to sub-structuring due to sourcing from different populations with different allele frequencies [73]. As discussed by [63], who studied migratory sandhill cranes (Grus canadensis), a deficiency of heterozygotes may be common in migratory species when a given population consists of local and immigrant individuals with different origin. Overall, departures from HWE would seem also a deficit intrinsic to the barn swallow, which, however, do not necessarily affect the result of the genetic analysis. Indeed, Bayesian procedure does not require perfect equilibria to cluster individuals, yet it attempts to minimize such departures within groups [73]. Rodríguez-Ramilo et al. [74] evaluated the accuracy of some Bayesian clustering methods when both Hardy-Weinberg and Linkage Equilibria were not fully respected. They found that structure could reliably determine the correct number of clusters also for F ST values as low as 0.01; hence, we did not exclude any STR locus from our analysis. Nevertheless, we also showed that the output of structure did not change when Hir4, Hir7 and Hir24 loci were ruled out (see Results).
Microsatellite DNA: males + females, males only, females only
Santure et al. [23] and Dor et al. [24] investigated H. rustica including European populations with different morphology and migratory behaviour. Nevertheless, both mitochondrial and microsatellite DNA markers did not disclose any significant genetic structure. By contrasting an equal number of males and females from six localities across Europe, we show significantly more population structure in males than females. This difference in structure can be explained by dependence of dispersal on sex (Figure 3, 4; Tables 3, S4). Different from [23], in our study the pairwise F ST computations as well as the PCA and the Bayesian clustering analysis revealed a genetic picture never before reported. While we cannot exclude that the discrepancy between the two studies could be due to the different distribution of the sampling sites, it should be noted that Santure et al. [23] used a set of STR markers (6 loci) smaller than that (10 loci) employed in the present study, and did not provide sex ratio of the investigated sample. Hence, it seems likely that our analysis has more power to detect any genetic structuring of populations than that performed by [23]. Overall, the PCA carried out by using the average pairwise F ST distance values computed among all populations as well as the Bayesian clustering analysis (males + females), pointed to the strong divergence between SPA and all other populations (Figure 3A, 4A, Table 3). This result was even more evident when we used only the male genotypes (Figure 3B, 4B, Table 3). For instance, in the Bayesian analysis, most of the SPA individuals grouped together in the cluster II, whereas the males from all of the other populations were mainly assigned to the cluster I (Figure 4B). Balbontín et al. [27] obtained similar results in a field study. They investigated long-term trends in natal dispersal of northern (Denmark) and southern (Spain) Europe barn swallow populations. They found female-biased natal dispersal in both populations and male philopatry six times higher in the Spanish than in the Danish population. In our study, the genetic differentiation showed by the SPA population could be due to a particularly high rate of natal philopatry of males. We would like to stress that in the barn swallow the choice of the first breeding site is crucial, as it can determine where an individual will reproduce for the rest of its life. Balbontín et al. [27] suggested that the probability of philopatry, which is related to the fitness in terms of longevity, may depend on ecological factors related to the breeding site, although both the livestock farming and the architecture of rural buildings could also influence the choice between philopatry and dispersal [75]. Compared to the Spanish population, the lack of differentiation among all of the other ones could be due to the lower natal philopatry of their males. These populations, a mix of resident (philopatric) and immigrant (dispersing) individuals, showed reciprocal small genetic distances (Figure 3B, Table 3), and suggested that the barn swallow's dispersal behaviour should be regarded as a rather plastic trait [27]. However, when the SPA population was excluded from the Bayesian clustering analysis (only males), a not negligible degree of genetic differentiation was disclosed across Europe (Figure S1 versus Figure 4B). Although this result could seem related to the occurrence of differences in the migratory routes as we have suggested for the females (see next paragraph), we feel more confident in stating that male philopatry in each population is the primary cause for the population genetic structure we have found.
Genetic structure and migratory behaviour (males versus females)
When only the females were taken into account the genetic scenario was strikingly different compared to that inferred using the males or the entire sample size. The average F ST distance values (Table 3; with related PCA of Figure 3C) as well as the Bayesian clustering (Figure 4C) marked out two groups of females: one (central to western Europe) included SPA, IRE and GER, the other (central to eastern Europe) comprised ITA, CYP and RUS. The genetic distance between the two groups was significant (F ST = 0.020, P<0.001, data not shown). Admixed genotypes were more frequent and gene flow level higher in the females than in the males (Figure 4, Tables 3, S4), this pointing to a higher dispersal rate and a lower philopatry of the first group compared to the second (female-biased dispersal) [4], [8], [9], [27].
When the clustering of the females was considered the occurrence of a significant latitudinal component could not be ruled out (e.g., compare SPA versus IRE and CYP versus RUS). This, in turn, suggested that gene flow among populations could be partially influenced by the axis of migration. However, the divergence between ITA and GER also suggests that dispersal does not strictly occur along a North-South axis. On the other hand, the genetic differentiation between eastern and western populations resembles what has been observed for other passerine species showing a clear migratory divide in central Europe [19], [76]. While clear evidences for the existence of a migratory divide do lack in the barn swallow, ringing data indicate that the autumn migratory routes of eastern and western populations of this species are different. Barn swallows from western Europe head for the Iberian Peninsula, while those of eastern populations travel down throughout the eastern Mediterranean and the Middle East. Again, barn swallows from central Europe may travel South straight across the Mediterranean or south-west to Spain [20]. This pattern of migration roughly parallels the results obtained by Ambrosini et al. [25] regarding the migratory connectivity in H. rustica. This analysis, which was carried out on a large dataset of ringing recoveries, produced weight for the existence of two main clusters: one includes birds breeding in south-west Europe and wintering in central Africa, the other comprises birds breeding in northern and eastern Europe and wintering in southern Africa. The genetic relationships disclosed in the present study, on one side, between RUS and CYP and, on the other, between SPA, GER and IRE, fit to the groups described by [25] and to the main migratory movements summarized in [20]. Furthermore, mark-recapture data provided by BirdLife Cyprus (A.C. Author pers. comm. 2012) also point to a single migration route between RUS and CYP. Whereas according to Ambrosini et al. [25] ITA should be separated from the RUS and CYP populations, our results point to the occurrence of some genetic kinship among the females of these countries (Figure 4A, Table S4). Overall, this may either indicate that the pattern of gene flow is only partially related to the migratory movement or suggest a poorly documented migratory route that connects central Italy, north-west Russia and Cyprus. Actually, the geographic area of birds recovered in Italy or ringed in Italy and recovered abroad is huge and include several eastern and south-eastern countries (e.g., Turkey and Greece, [77]). These wide connections, probably due to the position of the Italian Peninsula in the Mediterranean, should be further investigated by extending the number of the Italian sampling locations in order to attempt to disentangle a very complex pattern of gene flow.
When only the males were considered, the very high philopatry of the Spanish population forced the whole genetic scenario by isolating the latter against all of the other ones (see previous paragraph). On the contrary, when the SPA males were excluded, we disclosed slight differences among the studied populations that pointed to the occurrence of three groups: (IRE + GER), (ITA + CYP) and RUS (Figure S1). This pattern showed some degree of correspondence with the female clustering, thus suggesting that the mechanisms driving the direction of dispersal movements are probably the same in both sexes. Nevertheless, being aware that further investigations are needed, we suggest do not over-interpreting these results.
Sex-biased dispersal
Cases of inconsistency between population structures inferred from mtDNA or from STR variability are well known. A greater population differentiation can be detected using nuclear rather than mitochondrial markers (e.g.: [78], [79], this study) but the opposite may occur as well (e.g.: [71], [80]). Although both natal and breeding dispersal may enable high connectivity among populations [1], [2], [5], [6], breeding dispersal is virtually absent in the barn swallow [27]. Hence, sex-biased natal dispersal represents the best explanation fitting the observed discordance, although tests for the whole dataset failed to obtain significance for all parameters (Table 4). However, as stressed by [10], their overall outcome must be discussed in light of the amount of combined factors that may affect the statistical power, namely the dispersal rate and the bias intensity, the sampling design and the number of investigated STR loci [16], [81]. For instance, when the dispersal rate and the sex-bias are not strong vAI C seems the most powerful test, followed by mAI C and F ST, which work better with less than 20 loci [10]. In this study, vAI C was higher in the females than in the males, thus pointing to the occurrence of female-biased dispersal. When the tests were performed and SPA was separated from all other populations, R and F ST values were significantly lower in the females than in the males, mAI C value was negative (Table 4) and F IS value was lower in males than in females, thus suggesting high philopatry for the Spanish males.
In conclusion, we partially confirmed published data reporting the lack of a significant partition of diversity using mtDNA markers, while, in contrast to previous studies, we detected significant genetic structure by using nuclear DNA markers. The different scenarios observed between sexes could be explained with the non-random patterns of gene flow likely mediated by female natal dispersal and significant variability in male philopatry among barn swallow populations. Our results emphasize the importance of taking into account the sex of sampled individuals to obtain unbiased results on species showing a different pattern of dispersal between males and females.
Supporting Information
Bayesian admixture analysis as inferred using structure performed excluding the SPA population in male and female barn swallows. ΔK was optimal for K = 2, all computations. Each population was represented by a pie chart whose segments were proportional to the number of individuals assigned to cluster I (black), cluster II (white) or which showed admixed genotypes (grey). Threshold value for assignment to each cluster was Qi = 0.80.
(DOC)
The sample size ( n = 186) of this study and the mtDNA sequences downloaded from the GenBank ( n = 16). Population (Pop), country, region, locality (latitude/longitude, Lat/Long), type of tissue, sample size for STR/mtDNA analysis, and the number of ND2 mtDNA haplotypes are given. The number of male (M) and female (F) individuals genotyped with mitochondrial and STR markers was given (unavailable on line for MED population). GenBank accession codes for KRD and MED populations are reported in [33].
(DOC)
Estimates of mtDNA genetic diversity (averagez± SD) as computed for each population and for the whole dataset. Sites: number of segregating sites.
(DOC)
Average mtDNA pairwise distance values (PhiST) as computed among all populations; *, P <0.05; **, P <0.001; others, P >0.05.
(DOC)
Proportion of estimated membership of all populations to each cluster (QI and QII) as inferred by structure with K = 2 ( Figure 4 ). Computations were performed using: (i) all individuals (n = 186), (ii) males (n = 92) and (iii) females (n = 94). The number of specimens assigned to the cluster I (n I) and II (n II) was reported as well as the number of individuals showing admixed genotype (n mix). The threshold value for admixed assignment was Qi = 0.80.
(DOC)
Acknowledgments
We are grateful to R. Ceccherelli (CRUMA, Livorno) for the blood sampling in Orti-Bottagone Nature Reserve (Italy). The authors wish to thank S. Birks and the University of Washington Burke Museum of Natural History (Seattle, USA) for the loan of barn swallow tissues (H. r. erythrogaster: UWBM 78832, UWBM 80547). The authors also wish to thank: I. Byrkjedal and the Museum of Zoology of the University of Bergen (Norway); G. Frisk and the Swedish Museum of Natural History of Stockholm (Sweden); J. Fjeldsa, J.B. Kristensen and the Zoological Museum of the University of Copenhagen (Denmark). However, samples obtained from these three museums were investigated at the Cyt-b gene (partial sequence) and were not included in the present study. The authors are grateful to R. Ambrosini (Università degli Studi di Milano Bicocca, Milan, Italy) for his comments on an early draft of the manuscript, and to N. Saino (Università di Milano, Milan, Italy) for suggestions about the molecular sexing. Comments of two anonymous reviewers as well as of the Associate Editor improved an earlier draft of the manuscript.
Funding Statement
The Anastasios G. Leventis Foundation (Nicosia, Cyprus) funded this research with two grants (2008-2010, 2010-2012). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bohonak AJ (1999) Dispersal, gene flow and population structure. Q Rev Biol 74: 21–45. [DOI] [PubMed] [Google Scholar]
- 2. Belliure J, Sorci G, Møller AP, Clobert J (2000) Dispersal distances predict subspecies richness in birds. J Evolution Biol 13: 480–487. [Google Scholar]
- 3. Clobert J, Danchin E, Dhondt AA, Nichols JD (2009) Informed dispersal, heterogeneity in animal dispersal syndromes and the dynamics of spatially structured populations. Ecol Lett 12: 197–209. [DOI] [PubMed] [Google Scholar]
- 4. Greenwood PJ (1980) Mating systems, philopatry and dispersal in birds and mammals. Anim Behav 28: 1140–1162. [Google Scholar]
- 5. Paradis E, Baillie SR, Sutherland WJ, Gregory RD (1998) Patterns of natal and breeding dispersal in birds. J Anim Ecol 67: 518–536. [Google Scholar]
- 6.Clobert J, Danchin E, Dhondt AA, Nichols JD (2001) Dispersal. Oxford University Press. [Google Scholar]
- 7. Perrin N, Mazalov V (2000) Local competition, inbreeding, and the evolution of sex-biased dispersal. Am Nat 155: 116–127. [DOI] [PubMed] [Google Scholar]
- 8. Greenwood PJ, Harvey PH (1982) The natal and breeding dispersal of birds. Ann Rev Ecol Syst 13: 1–21. [Google Scholar]
- 9.Newton I (2003) The speciation and biogeography of birds. Amsterdam Academic Press. [Google Scholar]
- 10. Goudet J, Perrin N, Waser P (2002) Tests for sex-biased dispersal using bi-parentally inherited genetic markers. Mol Ecol 11: 1103–1114. [DOI] [PubMed] [Google Scholar]
- 11. Prugnolle F, de Meeus T (2002) Inferring sex-biased dispersal from population genetic tools: a review. Heredity 88: 161–165. [DOI] [PubMed] [Google Scholar]
- 12. Ortego J, García-Navas V, Ferrer ES, Sanz JJ (2011) Genetic structure reflects natal dispersal movements at different spatial scales in the blue tit, Cyanistes caeruleus . Anim Behav 82: 131–137. [Google Scholar]
- 13. Koenig WD, van Vuren D, Hooge PN (1996) Detectability, philopatry and the evolution of dispersal distances in vertebrates. Trends Ecol Evol 11: 514–518. [DOI] [PubMed] [Google Scholar]
- 14. Scribner KT, Petersen MR, Fields RL, Talbot SL, Pearce JM, et al. (2001) Sex-biased gene flow in spectacled eiders (Anatidae): inferences from molecular markers with contrasting modes of inheritance. Evolution 55: 2105–2115. [DOI] [PubMed] [Google Scholar]
- 15. Le Galliard JF, Rémy A, Ims RA, Lambin X (2012) Patterns and processes of dispersal behaviour in arvicoline rodents. Mol Ecol 21: 505–523. [DOI] [PubMed] [Google Scholar]
- 16. Palo JU, Lesbarrères D, Schmeller DS, Primmer CR, Merilä J (2004) Microsatellite marker data suggest sex-biased dispersal in the common frog Rana temporaria . Mol Ecol 13: 2865–2869. [DOI] [PubMed] [Google Scholar]
- 17. Paquette SR, Louis EE Jr, Lapointe FJ (2010) Microsatellite analyses provide evidence of male-biased dispersal in the radiated tortoise Astrochelys radiate (Chelonia: Testudinidae). J Hered 101: 403–412. [DOI] [PubMed] [Google Scholar]
- 18. Itonaga N, Köppen U, Plath M, Wallschläger D (2010) Breeding dispersal directions in the white stork (Ciconia ciconia) are affected by spring migration routes. J Ethol 28: 393–397. [Google Scholar]
- 19. Møller AP, Garamszegi LZ, Peralta-Sánchez J, Soler JJ (2011) Migratory divides and their consequences for dispersal, population size and parasite-host interactions. J Evolution Biol 24: 1744–1755. [DOI] [PubMed] [Google Scholar]
- 20.Turner A (2006) The barn swallow. Ed T. and A.D. Poyser, London, UK.
- 21.Møller AP (1994) Sexual selection and the barn swallow. Oxford University Press. [Google Scholar]
- 22. Roff DA, Mousseau T, Møller AP, de Lope F, Saino N (2004) Geographic variation in the G matrices of wild populations of the barn swallow. Heredity 93: 8–14. [DOI] [PubMed] [Google Scholar]
- 23. Santure AW, Ewen JG, Sicard D, Roff DA, Møller AP (2010) Population structure in the barn swallow, Hirundo rustica: a comparison between neutral DNA markers and quantitative traits. Biol J Linn Soc 99: 306–314. [Google Scholar]
- 24. Dor R, Safran RJ, Vortman Y, Lotem A, McGowan A, et al. (2012) Population genetics and morphological comparisons of migratory European (Hirundo rustica rustica) and sedentary East-Mediterranean (Hirundo rustica transitiva) barn swallows. J Hered 103: 55–63. [DOI] [PubMed] [Google Scholar]
- 25. Ambrosini R, Møller AP, Saino N (2009) A quantitative measure of migratory connectivity. J Theor Biol 257: 203–211. [DOI] [PubMed] [Google Scholar]
- 26. Dor R, Safran RJ, Sheldon FH, Winkler DW, Lovette IJ (2010) Phylogeny of the genus Hirundo and the barn swallow subspecies complex. Mol Phylogenet Evol 56: 409–418. [DOI] [PubMed] [Google Scholar]
- 27. Balbontín J, Møller AP, Hermosell IG, Marzal A, Reviriego M, et al. (2009) Geographic patterns of natal dispersal in barn swallows Hirundo rustica from Denmark and Spain. Behav Ecol Sociobiol 63: 1197–1205. [Google Scholar]
- 28. Kahn NW, St John J, Quinn TW (1998) Chromosome-specific intron size differences in the avian CHD gene provide a simple and efficient method for sexing birds. Auk 115: 1074–1078. [Google Scholar]
- 29. Barbanera F, Negro JJ, Di Giuseppe G, Bertoncini F, Cappelli F, et al. (2005) Analysis of the genetic structure of red-legged partridge (Alectoris rufa, Galliformes) populations by means of mitochondrial DNA and RAPD markers: a study from central Italy. Biol Conserv 122: 275–287. [Google Scholar]
- 30.Svensson L (1975) Identification guide to European passerines, 2nd edn. Naturhistoriska Riksmuseet, Stockholm, Sweden.
- 31. Sorenson MD, Ast JC, Dimcheff DE, Yuri T, Mindell DP (1999) Primers for a PCR-based approach to mitochondrial genome sequencing in birds and other vertebrates. Mol Phylogenet Evol 12: 105–114. [DOI] [PubMed] [Google Scholar]
- 32. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zink RM, Pavlova A, Rohwer S, Drovetski SV (2006) Barn swallows before barns: population histories and intercontinental colonization. Proc R Soc B 273: 1245–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Librado P, Rozas J (2009) DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- 35. Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16: 37–48. [DOI] [PubMed] [Google Scholar]
- 36. Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10: 564–567. [DOI] [PubMed] [Google Scholar]
- 37. Wright S (1965) The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19: 395–420. [Google Scholar]
- 38. Tamura K, Nei M (1993) Estimation of the number of the nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10: 512–526. [DOI] [PubMed] [Google Scholar]
- 39. Pilkington MM, Wilder JA, Mendez FL, Cox MP, Woerner A, et al. (2008) Contrasting signatures of population growth for mitochondrial DNA and Y chromosomes among human populations in Africa. Mol Biol Evol 25: 517–525. [DOI] [PubMed] [Google Scholar]
- 40. Ramírez-Soriano A, Ramos-Onsins SE, Rozas J, Calafell F, Navarro A (2008) Statistical power analysis of neutrality tests under demographic expansions, contractions and bottlenecks with recombination. Genetics 179: 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147: 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ramos-Onsins SE, Rozas J (2002) Statistical properties of new neutrality tests against population growth. Mol Biol Evol 19: 2092–2100. [DOI] [PubMed] [Google Scholar]
- 43. Tajima F (1989) A statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McDonald JH, Kreitman M (1991) Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. [DOI] [PubMed] [Google Scholar]
- 45. Harpending HC, Sherry ST, Rogers AR, Stoneking M (1993) Genetic structure of ancient human populations. Curr Anthropol 34: 483–496. [Google Scholar]
- 46. Schneider S, Excoffier L (1999) Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics 152: 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Excoffier L (2004) Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model. Mol Ecol 13: 853–864. [DOI] [PubMed] [Google Scholar]
- 48. Primmer CR, Møller AP, Ellegren H (1995) Resolving genetic relationships with microsatellite markers-a parentage testing system for the swallow Hirundo rustica . Mol Ecol 4: 493–498. [DOI] [PubMed] [Google Scholar]
- 49. Tsyusko OV, Peters MB, Hagen C, Tuberville TD, Mousseau TA, et al. (2007) Microsatellite markers isolated from barn swallows (Hirundo rustica). Mol Ecol Notes 7: 833–835. [Google Scholar]
- 50. Barbanera F, Marchi C, Guerrini M, Panayides P, Sokos C, et al. (2009a) Genetic structure of Mediterranean chukar (Alectoris chukar, Galliformes) populations: conservation and management implications. Naturwissenschaften 96: 1203–1212. [DOI] [PubMed] [Google Scholar]
- 51. Valiere N (2002) Gimlet: a computer program for analyzing genetic individual identification data. Mol Ecol Notes 2: 377–379. [Google Scholar]
- 52. Peatkau D, Waits L, Clarkson PL, Craighead L, Vyse E, et al. (1998) Variation in genetic diversity across the range of north American brown bears. Conserv Biol 12: 418–429. [Google Scholar]
- 53. Waits LS, Luikart G, Taberlet P (2001) Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Mol Ecol 10: 249–256. [DOI] [PubMed] [Google Scholar]
- 54. Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4: 535–538. [Google Scholar]
- 55.Goudet J (2001) Fstat, a program to estimate and test gene diversities and fixation indices (Version 2.9.3). http://www.unil.ch/izea/softwares/fstat.html.
- 56. Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86: 248–249. [Google Scholar]
- 57. Slatkin M (1985) Gene flow in natural populations. Ann Rev Ecol Syst 16: 393–430. [Google Scholar]
- 58. Hochberg Y (1988) A sharper Bonferroni procedure for multiple tests of significance. Biometrika 75: 800–802. [Google Scholar]
- 59. Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Res 9: 1322–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software Structure: a simulation study. Mol Ecol 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- 62. Vähä JP, Primmer CR (2006) Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Mol Ecol 15: 63–72. [DOI] [PubMed] [Google Scholar]
- 63. Jones KL, Krapu GL, Brandt DA, Ashley MV (2005) Population genetic structure in migratory sandhill cranes and the role of Pleistocene glaciations. Mol Ecol 14: 2645–2657. [DOI] [PubMed] [Google Scholar]
- 64. Davis LA, Roalson EH, Cornell KL, McClanahan KD, Webster MS (2006) Genetic divergence and migration patterns in a North American passerine bird: implications for evolution and conservation. Mol Ecol 15: 2141–2152. [DOI] [PubMed] [Google Scholar]
- 65. Procházka P, Stokke BG, Jensen H, Fainová D, Bellinvia E, et al. (2011) Low genetic differentiation among reed warbler Acrocephalus scirpaceus populations across Europe. J Avian Biol 42: 103–113. [Google Scholar]
- 66.Rockwell RF, Barrowclough GF (1987) Gene flow and the genetic structure of populations. In: Cooke F, Buckley PA, editors. Avian genetics. London Academic Press, pp. 223–255. [Google Scholar]
- 67. Arguedas N, Parker PG (2000) Seasonal migration and genetic population structure in house wrens. Condor 102: 517–528. [Google Scholar]
- 68. Webster MS, Marra PP, Haig SM, Bensch S, Holmes RT (2002) Links between worlds: unravelling migratory connectivity. Trends Ecol Evol 17: 76–83. [Google Scholar]
- 69. Blair ME, Melnick DJ (2012) Genetic evidence for dispersal by both sexes in the Central American squirrel monkey, Saimiri oerstedii citrinellus . Am J Primatol 74: 37–47. [DOI] [PubMed] [Google Scholar]
- 70. Ptak SE, Przeworski M (2002) Evidence for population growth in humans is confounded by fine-scale population structure. Trends Genet 18: 559–563. [DOI] [PubMed] [Google Scholar]
- 71. Barbanera F, Zuffi MAL, Guerrini M, Gentilli A, Tofanelli S, et al. (2009) Molecular phylogeography of the asp viper Vipera aspis (Linnaeus, 1758) in Italy: evidence for introgressive hybridization and mitochondrial DNA capture. Mol Phylogenet Evol 52: 103–114. [DOI] [PubMed] [Google Scholar]
- 72. Wahlund S (1928) Zusammensetzung von Population und Korrelationserscheinung vom Standpunkt der Vererbungslehre aus betrachtet. Hereditas 11: 65–106. [Google Scholar]
- 73. Randi E, Tabarroni C, Rimondi S, Lucchini V, Sfougaris A (2003) Phylogeography of the rock partridge (Alectoris graeca). Mol Ecol 12: 2201–2214. [DOI] [PubMed] [Google Scholar]
- 74. Rodríguez-Ramilo ST, Toro MA, Fernández J (2009) Assessing population genetic structure via the maximisation of genetic distance. Genet Sel Evol 41: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ambrosini R, Bolzern AM, Canova L, Arieni S, Møller AP, et al. (2002) The distribution and colony size of barn swallows in relation to agricultural land use. J Appl Ecol 39: 524–534. [Google Scholar]
- 76. Pérez-Tris J, Bensch S, Carbonell R, Helbig A, Tellería JL (2004) Historical diversification of migration patterns in a passerine bird. Evolution 58: 1819–1832. [DOI] [PubMed] [Google Scholar]
- 77.Spina F, Volponi S (2008) Atlante della Migrazione degli Uccelli in Italia. Ministero dell'Ambiente e della Tutela del Territorio e del Mare, Istituto Superiore per la Protezione e la Ricerca Ambientale (ISPRA).
- 78. Piertney SB, MacColl ADC, Bacon PJ, Racey PA, Lambin X, et al. (2000) Matrilineal genetic structure and female mediated gene flow in red grouse (Lagopus lagopus scoticus): an analysis using mitochondrial DNA. Evolution 54: 279–289. [DOI] [PubMed] [Google Scholar]
- 79. Johnson JA, Toepfer JE, Dunn PO (2003) Contrasting patterns of mitochondrial and microsatellite population structure in fragmented populations of greater prairie-chickens. Mol Ecol 12: 3335–3347. [DOI] [PubMed] [Google Scholar]
- 80. Borden WC, Stepien CA (2006) Discordant population genetic structuring of smallmouth bass, Micropterus dolomieu Lacepède, in Lake Erie based on mitochondrial DNA sequences and nuclear DNA microsatellites. J Great Lakes Res 32: 242–257. [Google Scholar]
- 81. Lampert KP, Rand AS, Mueller UG, Ryan MJ (2003) Fine-scale genetic pattern and evidence for sex-biased dispersal in the túngara frog, Physalaemus pustulosus . Mol Ecol 12: 3325–3334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bayesian admixture analysis as inferred using structure performed excluding the SPA population in male and female barn swallows. ΔK was optimal for K = 2, all computations. Each population was represented by a pie chart whose segments were proportional to the number of individuals assigned to cluster I (black), cluster II (white) or which showed admixed genotypes (grey). Threshold value for assignment to each cluster was Qi = 0.80.
(DOC)
The sample size ( n = 186) of this study and the mtDNA sequences downloaded from the GenBank ( n = 16). Population (Pop), country, region, locality (latitude/longitude, Lat/Long), type of tissue, sample size for STR/mtDNA analysis, and the number of ND2 mtDNA haplotypes are given. The number of male (M) and female (F) individuals genotyped with mitochondrial and STR markers was given (unavailable on line for MED population). GenBank accession codes for KRD and MED populations are reported in [33].
(DOC)
Estimates of mtDNA genetic diversity (averagez± SD) as computed for each population and for the whole dataset. Sites: number of segregating sites.
(DOC)
Average mtDNA pairwise distance values (PhiST) as computed among all populations; *, P <0.05; **, P <0.001; others, P >0.05.
(DOC)
Proportion of estimated membership of all populations to each cluster (QI and QII) as inferred by structure with K = 2 ( Figure 4 ). Computations were performed using: (i) all individuals (n = 186), (ii) males (n = 92) and (iii) females (n = 94). The number of specimens assigned to the cluster I (n I) and II (n II) was reported as well as the number of individuals showing admixed genotype (n mix). The threshold value for admixed assignment was Qi = 0.80.
(DOC)