Fanca contributes to both somatic hypermutation and class switch recombination events in splenic B cells.
Abstract
Fanconi anemia is a rare genetic disorder that can lead to bone marrow failure, congenital abnormalities, and increased risk for leukemia and cancer. Cells with loss-of-function mutations in the FANC pathway are characterized by chromosome fragility, altered mutability, and abnormal regulation of the nonhomologous end-joining (NHEJ) pathway. Somatic hypermutation (SHM) and immunoglobulin (Ig) class switch recombination (CSR) enable B cells to produce high-affinity antibodies of various isotypes. Both processes are initiated after the generation of dG:dU mismatches by activation-induced cytidine deaminase. Whereas SHM involves an error-prone repair process that introduces novel point mutations into the Ig gene, the mismatches generated during CSR are processed to create double-stranded breaks (DSBs) in DNA, which are then repaired by the NHEJ pathway. As several lines of evidence suggest a possible role for the FANC pathway in SHM and CSR, we analyzed both processes in B cells derived from Fanca−/− mice. Here we show that Fanca is required for the induction of transition mutations at A/T residues during SHM and that despite globally normal CSR function in splenic B cells, Fanca is required during CSR to stabilize duplexes between pairs of short microhomology regions, thereby impeding short-range recombination downstream of DSB formation.
To respond to the enormous variety of pathogens they might encounter, B lymphocytes have evolved processes to alter their genetic material to assemble novel functional Ig genes through site-specific V(D)J recombination. Later, contact with antigens can induce two different activation-induced cytidine deaminase (AID)–dependent processes, which are known as somatic hypermutation (SHM) and class switch recombination (CSR). SHM is responsible for the targeted introduction of point mutations into the variable (V) region of the Ig gene, creating Ig variants with enhanced affinity for a particular antigen. CSR allows for exchange of the initial IgM constant region (Cμ) with a downstream constant region (Cδ, Cγ, Cε, or Cα) through deletional recombination to generate different classes of effector antibodies (Alt et al., 2013).
AID, which converts cytosines into uracils, initiates both SHM and CSR by creating dU:dG mismatches (Alt et al., 2013). During SHM, mutations arise from these mismatches through several different mechanisms: (a) DNA replication across the uracil leads to transition mutations; (b) abasic sites generated by the base excision repair glycosylase UNG may be replicated in an error-prone manner by the translesional synthesis (TLS) polymerase REV1 to yield either transition or transversion mutations at the site of the C/G base pair; and (c) mismatch repair (MMR) proteins (MSH2/MSH6 and EXO1) can trigger excision and error-prone resynthesis of short stretches of DNA by the TLS polymerase Polη, thus spreading mutations to surrounding U/G base pairs (Liu and Schatz, 2009). In contrast, during CSR, both base excision repair and MMR proteins induce double-stranded breaks (DSBs) within the switch (S) regions situated upstream of each Ig CH gene. These breaks are then repaired by either the classical nonhomologous end-joining (NHEJ) pathway, which directly rejoins DNA ends with minimal modification of the broken ends, or by the alternative NHEJ pathway, which makes use of sequence microhomologies (MHs) to associate and rejoin distal DSBs (Alt et al., 2013).
Fanconi anemia (FA) is a rare, inherited chromosomal breakage disorder presenting bone marrow failure and cancer predisposition. FA is genetically heterogeneous, with 16 FANC genes (named A through Q) identified to date. After DNA damage or replicative stress, eight FANC proteins (FANCA, -B, -C, -E, -F, -G, -L, and -M) assemble into the FANCcore complex, which is necessary for the monoubiquitination and nuclear foci formation of both FANCD2 and FANCI. Monoubiquitinated FANCD2/FANCI heterodimer functionally or biochemically interacts with FANCD1/BRCA2, FANCN/PALB2, FANCJ/BRIP1, FANCO/RAD51C, FANCP/SXL4, and FANCQ/XPF to eliminate DNA lesions and rescue replication (Kottemann and Smogorzewska, 2013). FANC proteins promote homologous recombination and suppress NHEJ repair (Adamo et al., 2010; Pace et al., 2010). Moreover, FANC pathway disruption has been associated with abnormalities of several proteins involved in SHM or CSR, including the TLS polymerases REV1 and Polη (Kim et al., 2012; Fu et al., 2013; Renaud and Rosselli, 2013), the MMR protein MSH2 (Williams et al., 2011), the helicase BLM, and the DSB repair protein MRE11 (Pichierri et al., 2002, 2004; Naim and Rosselli, 2009). High levels of FANCD2 have been detected in germinal center (GC) cells in the spleen, tonsil, and reactive lymph nodes (Hölzel et al., 2003). Finally, expression of the Fanca mRNA, but not the Fancg mRNA, is specifically increased in GC B cells, which show high levels of SHM and CSR (Heng and Painter, 2008). In light of the above, we decided to analyze SHM and CSR in B cells derived from Fanca−/− mice to determine a possible involvement of FANCA in the process of secondary Ig diversification.
RESULTS AND DISCUSSION
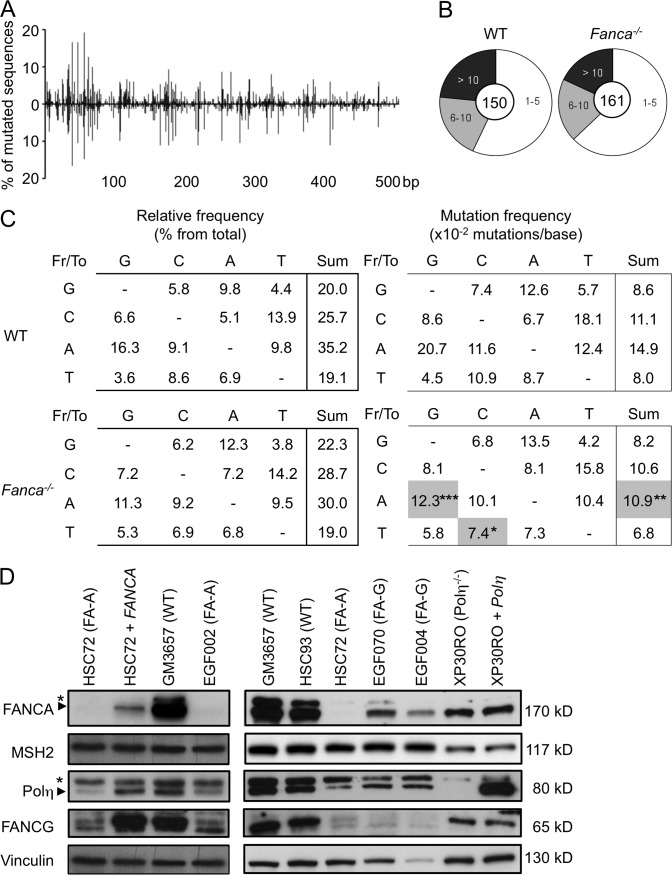
Reduced A/T transitions during SHM in Fanca−/− mice
To determine the impact of Fanca loss-of-function on SHM, we compared the levels and patterns of somatic mutation in the nonselected intronic region flanking the rearranged VHDJH4 genes (JH4 intronic region) in Peyer’s patch peanut agglutininhigh (PNAhigh) B cells isolated from WT and Fanca−/− mice. To calculate mutation frequency, the accumulated number of unique mutations was divided by the theoretical maximum number of the corresponding type of mutation to correct for base composition (MacCarthy et al., 2009). We observed that the distributions of mutations along the analyzed region were similar in Fanca−/− and WT mice (Fig. 1 A) but that the overall mutation frequency was significantly lower (P < 10−3) in B cells from Fanca−/− mice (Table 1). However, Fanca−/− mice showed a consistent but nonsignificant decrease in the proportion of highly mutated sequences (>10 mutations) and an increase in the proportion of sequences with <5 mutations (Fig. 1 B). Consequently, the reduced mutation frequency we observed could be accounted for solely by the reduction in the number of highly mutated sequences. Alternatively, the number of DNA lesions generated by AID or the number of mutations introduced during the course of SHM could prevent Fanca−/− cells from proliferating normally within the GC, leading to an apparent reduction in mutation frequency; i.e., the accumulation, not the generation, of mutations may be compromised in Fanca−/− mice.
Figure 1.
Reduced A/T transitions during SHM in Fanca−/− mice. (A) Distribution of mutations in the JH4 intronic region (506 bp) that was amplified from Peyer’s patch PNAhigh B cells isolated from WT and Fanca−/− mice. (B) Proportion of sequences with numbers of mutations per clone (the central circle shows the total number of analyzed sequences) from WT (n = 5) and Fanca−/− mice (n = 5). (C) The spectrum of base substitutions is expressed as a percentage of the total number of mutations (left), and the frequency of mutation (right) was corrected for base composition. Gray boxes denote a significant decrease in mutation frequency compared with WT (the χ2 test was applied according to the SHMTool algorithm; *, P < 0.05; **, P < 10−3; ***, P < 10−4). Data for the SHM are from five independent experiments. (D) Differential expression of Polη in FANCA- and FANCG-deficient cells. Whole cell extracts were prepared from the indicated human lymphoblast cell lines and analyzed by immunoblotting for the expression of Polη, MSH2, FANCA, FANCG, and Vinculin (asterisks indicate nonspecific bands). Representative data from two independent experiments are shown.
Table 1.
Global analysis of unique mutation frequencies of the JH4 intronic region
| JH4 intronic region | WT | Fanca−/− | P-value |
| Number of mice | 5 | 5 | |
| Mutated sequences analyzed | 150 | 161 | |
| Unique mutations (total) | 787 | 665 | |
| Overall mutation frequency | 10.5 x 10−2 | 8.8 x 10−2 | 0.0008 |
| A/T mutations | 11.2 x 10−2 | 8.7 x 10−2 | 0.0001 |
| Transversions | 9.1 x 10−2 | 8.2 x 10−2 | 0.2637 |
| Transitions | 15.5 x 10−2 | 9.7 x 10−2 | <10−5 |
| Motif | |||
| WA | 33.7 x 10−2 | 19.7 x 10−2 | <10−4 |
| TW | 14.7 x 10−2 | 9.5 x 10−2 | 0.1637 |
| G/C mutations | 9.4 x 10−2 | 9.0 x 10−2 | 0.6013 |
| Transversions | 6.9 x 10−2 | 6.4 x 10−2 | 0.5355 |
| Transitions | 14.5 x 10−2 | 14.3 x 10−2 | 0.9504 |
| Motif | |||
| WRC | 53.3 x 10−2 | 44.0 x 10−2 | 0.327 |
| GYW | 36.8 x 10−2 | 36.8 x 10−2 | 1.0 |
All mutation frequencies were calculated according to the SHMTool algorithm. Bold denotes a significant decrease in the mutation frequency compared with WT. Underlining indicates the mutated residues in the motifs.
However, supporting a direct, specific role for Fanca during SHM, we observed that the frequency of mutations at A/T residues was significantly reduced in Fanca−/− mice (P = 10−4; Table 1), and this reduction was associated with a highly significant (P < 10−5) decrease in the frequency of transitions at these sites (Table 1). In contrast, transversions at A/T sites were similar in both Fanca−/− and WT mice (Table 1 and Fig. 1 C). Analysis of the patterns of nucleotide substitution showed a strong reduction in A to G transitions (P < 10−4) and a less robust, but still significant, decrease in T to C transitions (P < 0.05) in Fanca−/− mice (Fig. 1 C). Moreover, mutations in residues within WA motifs (W = A or T; underlining indicates the mutated residues) were significantly decreased in Fanca−/− mice (P < 10−4; Table 1), whereas no significant difference was detected in the mutation frequency of TW motifs, indicating a targeting of WA motifs on the transcribed strand. In contrast, the mutation frequency at G/C residues as well as the frequencies of transversions or transitions at these sites was unchanged in the absence of Fanca (Table 1). Furthermore, the types of base substitutions and the mutation frequencies at hotspots for AID (WRC/GYW) were also similar between the two groups of mice (Table 1 and Fig. 1 C), indicating that AID activity and targeting were normal in the absence of Fanca.
TLS polymerases have been shown to be involved in SHM (Weill and Reynaud, 2008), with Polη being the sole A/T mutator during the SHM process (Delbos et al., 2005). Additionally, WA motifs and the complementary TW motif have been shown to be hotspots for Polη activity at the Ig locus (Rogozin et al., 2001), with a preference for A to G substitutions within WA motifs on the nontranscribed strand (Mayorov et al., 2005). Therefore, the altered mutation pattern we observed in Fanca−/− B cells may be partially caused by previously described effects of FANC pathway loss-of-function on Polη (Fu et al., 2013; Renaud and Rosselli, 2013).
As the Fancg−/− mice did not show defects in SHM (Krijger et al., 2010), we asked whether Fanca differs from Fancg in Polη expression/stabilization. To address this question, B220+/Gl7+ cells were sorted from Peyer’s patches isolated from Fanca−/−, Fancg−/−, and WT mice, and total RNA was prepared and subjected to semiquantitative RT-PCR. We failed to observe differences in the abundance of Polη transcripts in either Fanca−/− or Fancg−/− mice compared with WT mice (not depicted). In the absence of reliable antibodies against mouse Polη, we examined Polη levels in human FANCA- or FANCG-deficient lymphoblasts by immunoblot. Polη is highly reduced in FANCA-deficient lymphoblasts, and its expression was rescued by the ectopic expression of the WT FANCA cDNA (Fig. 1 D, compare HSC-72 with HSC-72 + FANCA). On the contrary, we observed only a moderate Polη reduction in FANCG-deficient cells (Fig. 1 D). Consistent with a previous study (Garcia-Higuera et al., 2000), we observed that FA-A cells failed to express significant levels of FANCG, whereas FANCA remained detectable in FA-G cells (Fig. 1 D). This result suggests that the reduced Polη levels in FANCG-deficient cells were likely the result of FANCA destabilization in the absence of FANCG. Although Polη protein is reduced in FANCG-deficient cells, Fancg−/− mice are fully able to induce A/T mutations (Krijger et al., 2010). This result is consistent with a previous study showing that a strong reduction in cellular Polη levels is mandatory to affect the rate of A/T mutations (Faili et al., 2004). The expression of MSH2, another crucial factor for generating A/T mutations during SHM, is not altered in the absence of either FANCA or FANCG (Fig. 1 D).
It is interesting to note that in the absence of Fanca, the observed percentage of transitions and transversions within A/T bases cannot be explained exclusively by Polη reduction, suggesting that others factors, yet unknown, involved in A/T mutation pathway are also affected. In conclusion, our observations demonstrate that loss of Fanca specifically affects the induction of A/T mutations during SHM.
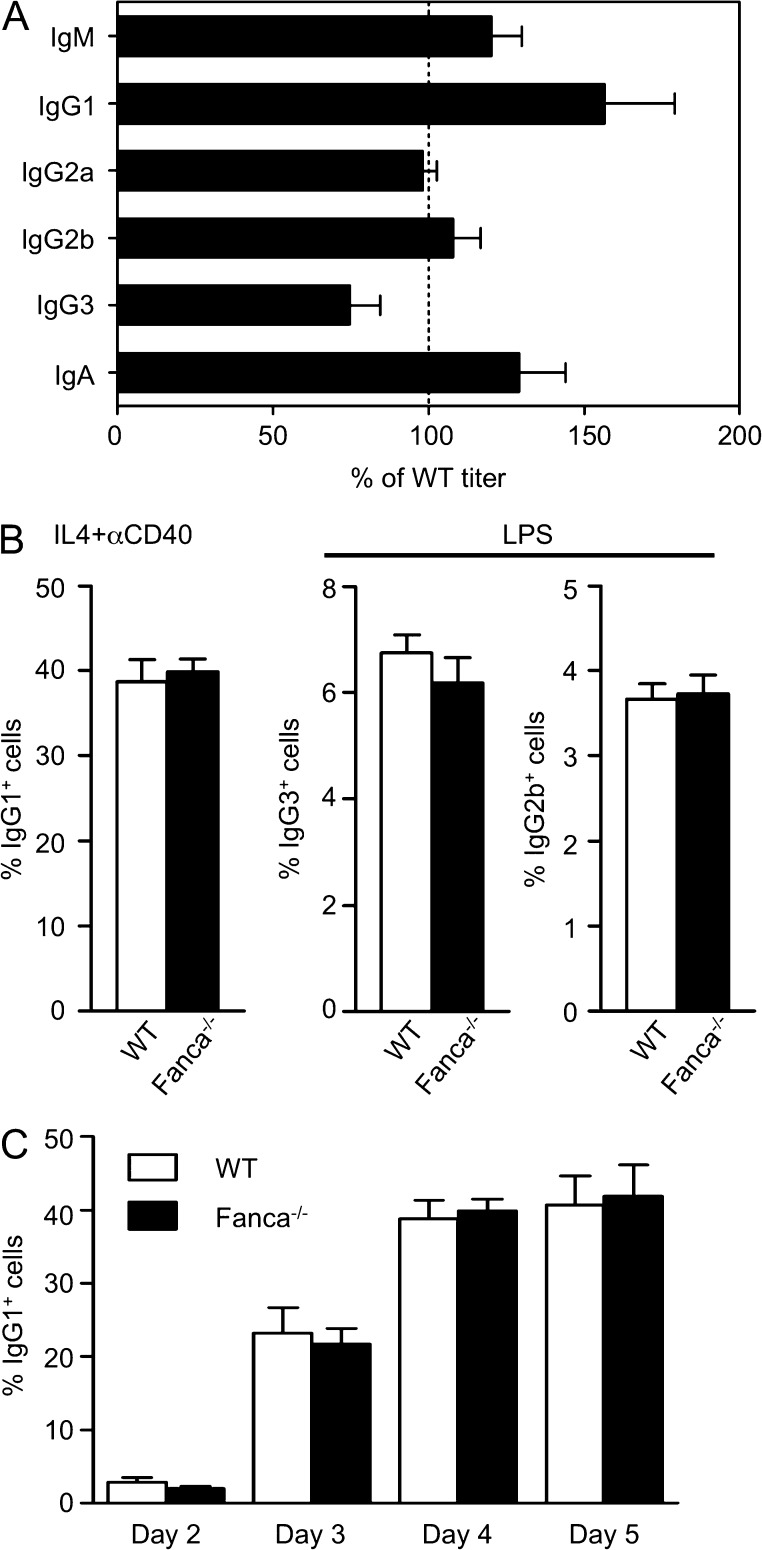
Robust CSR in Fanca−/− mice
To determine whether loss of Fanca affects Ig class switching, we determined the levels of IgM, IgG subclasses, and IgA in the serum from Fanca−/− and WT mice at 8–12 wk of age. All Ig isotype titers were similar between WT and Fanca−/− mice, indicating that Fanca−/− B cells are proficient for Ig secretion (Fig. 2 A). We next examined the intrinsic ability of Fanca−/− B cells to undergo CSR in vitro. Splenic B cells from Fanca−/− and WT mice were stimulated with either IL-4 plus αCD40 to induce switching from IgM to IgG1 or with LPS to induce switching to IgG3 and IgG2b. Stimulated B cells were cultured for 4 d and analyzed by flow cytometry for surface expression of IgG1, IgG3, and IgG2b. Fanca−/− B cells were able to switch to all isotypes examined (Fig. 2 B). We also examined the kinetics of in vitro switching to IgG1 in WT and Fanca−/− purified splenic B cells stimulated with αCD40 plus IL-4 for 2–5 d, and we observed that Fanca−/− and WT B cells switched to IgG1 at similar frequencies (Fig. 2 C). Building on previous observations made in Fancg−/− mice (Krijger et al., 2010), these data indicate that loss of the FANC pathway does not impair secondary Ig diversification and secretion.
Figure 2.
Robust CSR in Fanca−/− mice. (A) Serum from Fanca−/− (n = 5) and WT mice (n = 7) was collected and analyzed by ELISA for the indicated IgM, IgG subclasses, and IgA. The results are displayed as mean ± SEM values for the Fanca−/− titer as a percentage of WT. Data are representative of two independent experiments with at least five mice per group. (B) Splenic B cells were isolated from Fanca−/− and WT mice and stimulated in vitro with IL-4 and anti-CD40 or LPS. The expression of IgG1, IgG3, and IgG2b was determined 4 d later by flow cytometry. Bar graphs show the mean percentages of cells expressing the indicated IgG ± SEM. Data are representative of three independent experiments with at least three mice per group. (C) Splenic B cells were isolated from Fanca−/− and WT mice and stimulated in vitro with IL-4 and anti-CD40. Surface IgG1 expression was determined by flow cytometry on days 2, 3, 4, and 5 after stimulation. Bar graphs show the mean percentages of IgG1+ cells ± SEM. Data are representative of five independent experiments with at least five mice per group.
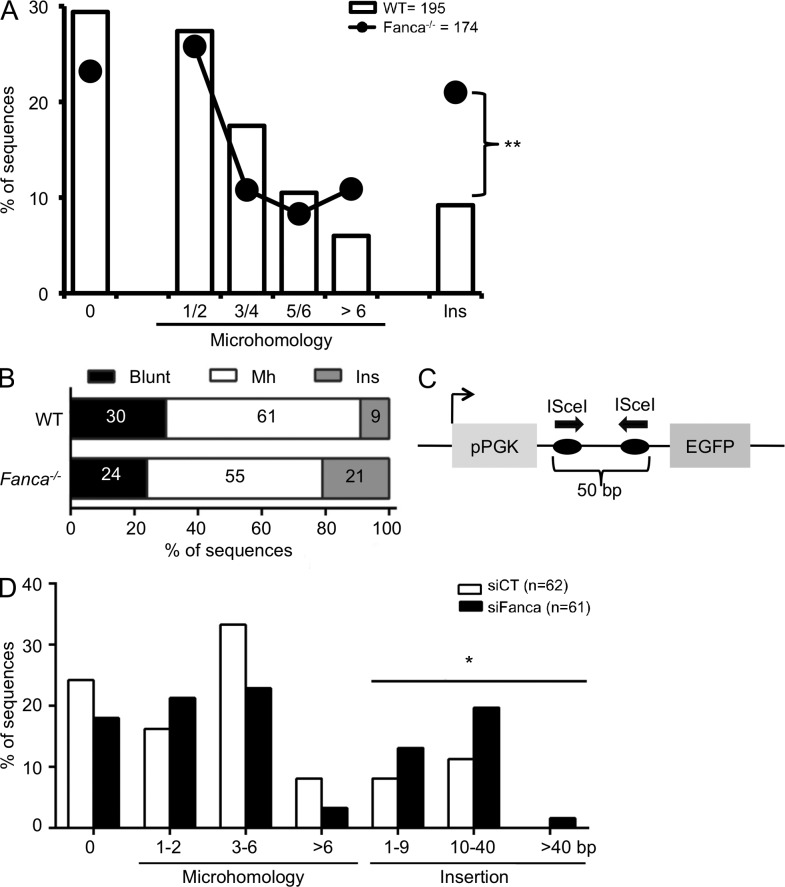
Altered CSR junctions in Fanca−/− mice
To address the role of Fanca in the repair of the DSBs created during CSR, we analyzed Sμ-Sγ1 junctions from Fanca−/− and WT splenic B cells after activation with αCD40 plus IL-4 for 4 d. Fanca−/− B cells displayed a slight decrease in the frequency of direct and short MH (up to 6 bp) junctions (Fig. 3 A) and an increase in the frequency of long MH (>7 bp) junctions, although the mean MH length was similar in the two groups of mice (2.45 ± 0.23 bp in Fanca−/− vs. 2.29 ± 0.19 bp in WT mice). Moreover, sequence analysis revealed that the probability of nucleotide insertions at junctions was significantly higher in Fanca−/− than in WT mice (Fig. 3, A and B). Finally, analysis of breakpoint distribution within the S regions showed a similar pattern between WT and Fanca−/− mice (not depicted), supporting a specific role for Fanca in broken DNA end processing during CSR.
Figure 3.
Altered CSR junctions in Fanca−/− mice. (A) Splenic B cells from WT (n = 6) and Fanca−/− (n = 6) mice were stimulated with anti-CD40 and IL-4, and 4 d later, Sμ–Sγ1 junctions were amplified by PCR from genomic DNA and then sequenced. Percentage of sequences with blunt joins (0), indicated MH, and junctional insertions (Ins) are shown. Data from six independent experiments were pooled and analyzed by the χ2 test (**, P < 10−3). (B) A plot representing the relative frequency of sequences with blunt joins, MH, or insertions for WT and Fanca−/− B cells from A. (C) A schematic representation of the I-SceI chromosomal reporter. (D) The GCV6 cells containing the I-SceI substrate described in C were transfected with siCT or siFANCA for 48 h and subsequently with I-SceI–expressing plasmid. 3 d later, I-SceI junctions were amplified from genomic DNA and then sequenced. Percentage of sequences with blunt joins (0), indicated MH, or insertions at the I-SceI–induced NHEJ junction are reported. Analyzed sequences were obtained from two independent experiments (*, P < 10−2).
To validate the previous observation, we used a genetically modified cell line bearing a single copy of an NHEJ reporter construct inserted into the genome (Fig. 3 C). The reporter contained two opposite-facing I-SceI target sites separated by 50 bp, which allowed us to determine rejoining efficiency of DSBs induced at these two sites. siRNA-mediated knockdown of FANCA did not significantly affect the frequency of GFP-positive cells retrieved after I-SceI–mediated induction of DSBs (not depicted), demonstrating that I-SceI–mediated cleavage and rejoining events did not require FANCA. However, we noticed that in the absence of FANCA, as during CSR, the frequency of MH junctions was mildly decreased and the frequency of nucleotide insertions at junctions was significantly increased (Fig. 3 D). Again, the mean length of the MHs was not affected by FANCA depletion (2.25 ± 0.33 bp in siFANCA- vs. 2.62 ± 0.35 bp in siCT-treated cells). In summary, loss of Fanca function decreased the use of short junctional MHs and increased junctional insertions during repair.
Insertional joining can occur when the overhangs at each end of a DSB are unstable, resulting in the dissociation and reannealing of DNA ends and the addition of a short stretch of newly synthesized nucleotides closer to the break site (Chan et al., 2010). Different scenarios can explain our findings. First, considering only the DNA end modifications that occur during CSR, the phenotype associated with Fanca loss at S junctions is reminiscent of that observed in both MSH2- and MSH3-deficient mice, which display increases in junctional insertions (Schrader et al., 2002; Li et al., 2004). In addition, it has been proposed that during single-strand annealing in yeast, the MSH2-MSH3 heterodimer binds to transient MH regions to stabilize the annealed intermediate or to signal to the Rad1-Rad10 endonuclease (homologues of mammalian XPF and ERCC1) to cleave the overhangs (Sugawara et al., 1997). Thus, in the case of weak DNA end annealing, FANCA could play a similar stabilizing role during the processing of DSBs created downstream of AID. Second, as a requirement for FANCA in XPF-ERCC1 endonuclease recruitment to DNA damage has been reported (Kumaresan and Lambert, 2000), it is possible that Fanca may be required during CSR to eliminate single-stranded DNA overhangs at regions of matched MHs by recruiting/loading the necessary endonuclease. Finally, the insertion of multiple nucleotides at junctions may reflect a nontemplated addition of nucleotides during NHEJ by Polμ, a TdT-like polymerase, which Fanca may oppose by promoting annealing.
Therefore, Fanca may enhance the stability of relatively short matched MH sequences, permitting the optimal assembly/action of MSH2-MSH3. In turn, this would facilitate the recruitment of endonucleases to trim overhangs and avoid the inappropriate access of polymerases to DNA ends released from unstable aligned MH regions.
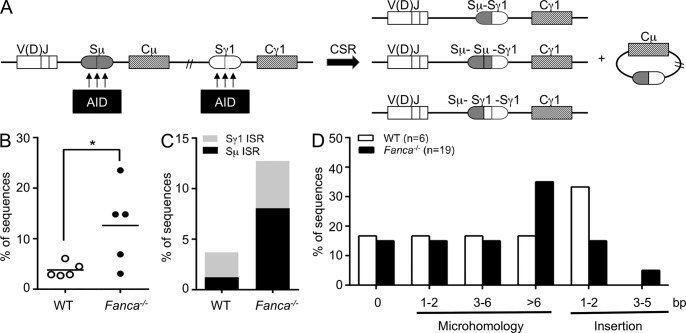
Increased intra-switch recombination (ISR) in Fanca−/− mice
AID activity generates multiple DSBs within a given S region, and CSR joins the most upstream DSB with the most downstream DSB from two different S regions, which can be separated by as much as 200 kb of DNA (Fig. 4 A). The intervening double-stranded DNA fragments are then degraded or joined to form excision circles. However, some rejoined sequences could be involved in unusual insertions of DNA fragments internal to an S region. This can result from local rejoining or ISR and can lead to the presence of Sμ-Sμ-Sγ1 or Sμ-Sγ1-Sγ1 sequence junctions, as opposed to the canonical Sμ-Sγ1 sequence (Reina-San-Martin et al., 2007). Remarkably, we found that the frequency of ISR was significantly higher in Fanca−/− than in WT B cells (12.6 vs. 3.8%; Fig. 4 B). The levels of Sμ ISR in Fanca−/− B cells (8.1%) were much higher than in WT B cells (1.2%), whereas the levels of Sγ1 ISR in Fanca−/− B cells were twofold greater than in WT B cells (4.7 vs. 2.5%; Fig. 4 C). These findings suggest that ISR at the Sμ region may occur more frequently than at the Sγ1 region, consistent with previously published data showing that Sμ appears to be intrinsically preferred by AID compared with other downstream S regions during CSR (Alt et al., 2013). Considering the small number of ISR junctions we obtained as well as the fact that the levels of Sμ ISR appeared to be higher than those for Sγ1 in WT mice, we cannot rule out the possibility that Fanca specifically inhibits ISR at Sμ to a greater extent than at downstream S regions. Interestingly, we also observed increased insertions of long sequences (>10 bp) at junctions from siFANCA-transfected cells bearing an intrachromosomal NHEJ reporter system (Fig. 3 D). Strikingly, with the exception of one junction that showed a large insertion from another region of the construct, these long inserted sequences originated from the 50-bp I-SceI–excised sequence. Again, this finding is consistent with our observations of ISR during the CSR process in Fanca−/− mice.
Figure 4.
Increased ISR in Fanca−/− mice. (A) A model for the joining of AID-generated DSBs between S regions during switch to IgG1. AID generates multiple DSBs within a given S region, and CSR joins two DSBs from two different S regions, producing canonical Sµ-Sγ1 junctions. The intervening double-stranded DNA fragments are then degraded or joined to form excision circles; occasionally some rejoined sequences exhibiting unusual junctions (e.g., Sµ-Sµ-Sγ1 or Sµ-Sγ1-Sγ1) resulting from local rejoining or ISR are observed. (B) Frequency of ISR in WT and Fanca−/− B cells induced to switch to IgG1 by anti-CD40 and IL-4 for 4 d. ISR was examined by analysis of Sμ-Sγ1 junctions. Horizontal bars indicate the means (*, P < 0.05 with the two-tailed Student’s t test). (C) Bar graph shows the percentage of Sμ and Sγ1 ISR in WT and Fanca−/− B cells from B. (D) Percentage of sequences exhibiting blunt joins (0), indicated MH, or insertions at the ISR junction. Data for the ISR analysis are based on five mice per genotype from five independent experiments.
Essentially all of the isolated ISR junctions were joined by the alternative NHEJ, representing 16 of 19 junctions in Fanca−/− B cells and 5 of 6 junctions in WT B cells that were either insertion or MH mediated (Fig. S1). Additionally, there were no significant differences in the sizes of the deletions or recombination products at ISR junctions between Fanca−/− and WT B cells (not depicted). However, an increased usage of long MH regions was observed at ISR junctions from Fanca−/− B cells (Fig. 4 D and Fig. S1), further supporting our hypothesis that Fanca plays a role in alignment stabilization during MH-mediated joining.
Considering the similar distributions of breakpoints within the S regions of WT and Fanca−/− mice (not depicted) as well as our observations regarding SHM, these data clearly support the notion that AID functions properly in the absence of Fanca. Consequently, the increased ISR observed in Fanca−/− B cells was likely caused by defects in DNA end management. Therefore, the unstable alignment of short MH regions in the absence of Fanca could lead to enhanced joining of DNA ends within the same S region with longer MH regions.
Alternatively, as has been reported (Kottemann and Smogorzewska, 2013), after exposure to genotoxic drugs, FANC pathway deficiency may result in the inappropriate recruitment of NHEJ factors to DSB ends leading to (a) “over-stability” of intervening DNA stretches from a given S region, thereby protecting them from degradation; (b) an increase in the joining of small DNA fragments at the expense of large chromosomal fragments; and (c) changes in chromatin structure at DSB ends to enhance short-range recombination. Collectively, these results suggest that FANCA suppresses short-range joining during CSR.
Concluding remarks
Our results show that loss of Fanca function is associated with molecular alterations in both SHM and CSR. The observed alterations had a modest impact on Ig production, likely because of the intricate processes of production and selection that B cells must undergo before becoming fully competent. Although FA patients do not display severe immunological problems, a study has described the presence of immunological abnormalities in subsets of these patients, including reductions in the absolute numbers of B lymphocytes in the blood and decreases in both IgM and IgG serum isotypes (Korthof et al., 2013).
Our data support a specific role for Fanca in the SHM process that contributes to the generation of mutations at A/T residues, potentially through the regulation of Polη expression/stabilization. A molecular analysis of the CSR-mediated junction clearly indicated a role for Fanca in control of the CSR process. In particular, Fanca appears to participate in the stabilization of duplexes between two short MH regions. Additionally, our findings suggest a requirement for Fanca in inhibiting short-range recombination but not CSR-like long-range recombination.
MATERIALS AND METHODS
Mice.
Fanca−/− and Fancg−/− mice and their WT littermates (129Ola/FVB background) were generated as previously described (Cheng et al., 2000). The Ethics Committee for Animal Experimentation, in accordance with French regulations, approved all of the procedures in this study.
SHM analysis.
Peyer’s patches were prepared from 8–12-wk-old Fanca−/− and WT mice. Cells from the Peyer’s patches were stained with a PE-conjugated anti–mouse B220 rat mAb (RA3-6B2; BioLegend) and FITC-conjugated PNA (eBioscience). GC B220+PNAhigh B cells were sorted using a high-output MoFlo cell sorter (Dako). The JH4 intron-flanking rearranged VH sequences were amplified using a mixture of five VH primers (designed to amplify most the majority of the mouse VH families) and a downstream primer, enabling the determination of 506 bp of the noncoding sequence, as reported previously (Delbos et al., 2005). Sequence alignments were performed using CodonCode Aligner 3.7 using NCBI Nucleotide sequence NT_114985.3 (nucleotides 300967–301472) as the reference sequence. Analysis of the mutated sequences was performed using the SHMTool webserver.
ELISA.
Serum levels of IgM, IgG1, IgG2a, IgG2b, IgG3, and IgA from 8–12-wk-old Fanca−/− and WT mice were measured using the Mouse Immunoglobulin Isotyping ELISA kit (BD).
In vitro CSR assay.
Splenic B cells were isolated from individual 8–12-wk-old Fanca−/− and WT mice by negative selection using the Mouse B Cell Isolation kit (Miltenyi Biotec). The cells were cultured in complete RPMI 1640 medium (15% FBS, 20 mM Hepes, 1 mM sodium pyruvate, 2 mM l-glutamine, 1× nonessential amino acids, 100 µM 2-mercaptoethanol, and 1% antibiotics) and stimulated with either 1 µg/ml α-CD40 (eBioscience) plus 20 ng/ml IL-4 (PeproTech) to induce CSR to IgG1 or 25 µg/ml LPS (serotype O111:B4 Escherichia coli; Sigma-Aldrich) to induce CSR to IgG3 and IgG2b. Cultured cells were maintained daily at a density of 5 × 105 cells/ml. Cells were collected on various days for FACS analysis of Ig surface expression using anti–mouse IgG1 (RMG1-1; BioLegend), anti–mouse IgG3 (R40-82; BD), or anti–mouse IgG2b (RMG2b-1; BioLegend) and PE-conjugated anti–mouse CD45R (B220) rat mAbs (RA3-6B2; BioLegend).
Switch junction analysis.
Genomic DNA was extracted from B cells activated with α-CD40 plus IL-4 for 4 d using the DNeasy Blood and Tissue kit (QIAGEN). Sµ-Sγ1 junctions were amplified using nested PCR with iProof High-Fidelity DNA Polymerase (Bio-Rad Laboratories) and the previously described primers and reaction conditions (Begum et al., 2007). PCR products (300–700 bp) were subsequently cloned into the Zero Blunt vector (Invitrogen) and sequenced. The switch junctions were identified by aligning the PCR sequences with the Sµ (MUSIGCD07) and the Sγ1 (MUSIGHANB) reference sequences using BLAST (NCBI).
I-SceI chromosomal NHEJ substrate.
The GCV6 cell line (provided by B. Lopez and J. Guirouilh-Barbat, Centre National de la Recherche Scientifique UMR 8200, Villejuif, France) was derived from SV40-transformed GM639 human fibroblasts containing the NHEJ substrate, as described in Fig. 3 C. The cells were cultured and transfected with specific siRNA and I-SceI expression plasmids (pBASce), according to previously described protocols (Rass et al., 2009). In brief, 48 h after transfection of the siRNA (final concentration 20 nM), cells were transfected with 1 µg pBASce using 4 µl JetPEI (Polyplus). 3 h after I-SceI transfection, the media was replaced. 72 h after I-SceI transfection, GFP expression, from the product of successful joining, was measured using flow cytometry. The FANCA SMARTpool siRNA was obtained from Thermo Fisher Scientific, and a control (non-targeted) siRNA (5′-CGUCGACGGAAUACUUCGATT-3′) was purchased from Eurogentec. The expression levels of FANCA and hemagglutinin-tagged I-SceI were verified using Western blotting analysis with antibodies against FANCA (Bethyl), HA (Covance), and vinculin (Abcam). I-SceI junctions were amplified from genomic DNA using the iProof High-Fidelity DNA Polymerase (Bio-Rad Laboratories) with the PGK1, 5′-TCGCACACATTCCACATCCACCG-3′; and eGFP-int, 5′-TCTTGTAGTTGCCGTCGTCCTTGA-3′. PCR products (400–800 bp) were cloned into the Zero Blunt vector (Invitrogen) and sequenced.
Cell culture.
The following cell lines were used in this study: EBV-immortalized lymphoblasts from FA-A (HSC72 and EGF002) and FA-G (EGF004 and EGF070) patients; HSC72 + FANCA cells are derived from the HSC72 cell line stably transfected with the WT FANCA cDNA; EBV-transformed lymphoblasts from a normal donor (WT; GM3657, HSC93); and the SV40-transformed fibroblasts XP30RO (XP-V) and XP30ROpolh (XP-V), which stably express the POLH cDNA. With the exception of GM3657 (Coriell Cell Repositories), all of the FA cell lines were provided by M. Buchwald (Hospital for Sick Children, Toronto, Ontario, Canada), J. Soulier (Saint-Louis Hospital, Paris, France), and B.A. Cox (Oregon Health and Science University, Portland, OR). The XP-V cell lines were provided by P. Kannouche (Institut Gustave Roussy, Villejuif, France). Lymphoblasts were maintained in RPMI (Gibco) media supplemented with 12% heat-inactivated FCS, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C under 5% CO2. Fibroblasts were grown in Eagle’s minimal essential medium (MEM; Gibco) containing 10% FCS, l-glutamine, and penicillin/streptomycin at 37°C under 5% CO2.
Western blotting analysis.
Whole-cell extracts were prepared in lysis buffer (50 mM Tris, pH 7.5, 20 mM NaCl, 1 mM MgCl2, 0.1% SDS, protease inhibitor cocktail, and 50 U/ml Benzonase [EMD Millipore]). Western blotting analysis was performed using the following antibodies: Polη (Abcam), FANCA (Bethyl Laboratories, Inc.), FANCG (Fanconi Anemia Research Fund), MSH2 (Bethyl Laboratories, Inc.), and vinculin (Abcam).
Online supplemental material.
Fig. S1 shows examples of ISR junctions from Fanca−/− and WT B cells. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20131637/DC1.
Supplementary Material
Acknowledgments
We thank B. Lopez and J. Guirouilh-Barbat for the generous gift of the GCV6 cell line as well as all the members of the F. Rosselli team for their helpful discussions and advice.
This work was supported by grants from La Ligue Contre le Cancer, Agence Nationale de la Recherche (ANR-08-GENO-0013), and INCa-DGOS-Inserm 6043.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AID
- activation-induced cytidine deaminase
- CSR
- class switch recombination
- DSB
- double-stranded break
- FA
- Fanconi anemia
- GC
- germinal center
- ISR
- intra-switch recombination
- MH
- microhomology
- MMR
- mismatch repair
- NHEJ
- nonhomologous end-joining
- PNA
- peanut agglutinin
- SHM
- somatic hypermutation
- TLS
- translesional synthesis
References
- Adamo A., Collis S.J., Adelman C.A., Silva N., Horejsi Z., Ward J.D., Martinez-Perez E., Boulton S.J., La Volpe A. 2010. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol. Cell. 39:25–35 10.1016/j.molcel.2010.06.026 [DOI] [PubMed] [Google Scholar]
- Alt F.W., Zhang Y., Meng F.-L., Guo C., Schwer B. 2013. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 152:417–429 10.1016/j.cell.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum N.A., Izumi N., Nishikori M., Nagaoka H., Shinkura R., Honjo T. 2007. Requirement of non-canonical activity of uracil DNA glycosylase for class switch recombination. J. Biol. Chem. 282:731–742 10.1074/jbc.M607439200 [DOI] [PubMed] [Google Scholar]
- Chan S.H., Yu A.M., McVey M. 2010. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 6:e1001005 10.1371/journal.pgen.1001005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N.C., van de Vrugt H.J., van der Valk M.A., Oostra A.B., Krimpenfort P., de Vries Y., Joenje H., Berns A., Arwert F. 2000. Mice with a targeted disruption of the Fanconi anemia homolog Fanca. Hum. Mol. Genet. 9:1805–1811 10.1093/hmg/9.12.1805 [DOI] [PubMed] [Google Scholar]
- Delbos F., De Smet A., Faili A., Aoufouchi S., Weill J.-C., Reynaud C.-A. 2005. Contribution of DNA polymerase η to immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 201:1191–1196 10.1084/jem.20050292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faili A., Aoufouchi S., Weller S., Vuillier F., Stary A., Sarasin A., Reynaud C.-A., Weill J.-C. 2004. DNA polymerase η is involved in hypermutation occurring during immunoglobulin class switch recombination. J. Exp. Med. 199:265–270 10.1084/jem.20031831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D., Dudimah F.D., Zhang J., Pickering A., Paneerselvam J., Palrasu M., Wang H., Fei P. 2013. Recruitment of DNA polymerase eta by FANCD2 in the early response to DNA damage. Cell Cycle. 12:803–809 10.4161/cc.23755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I., Kuang Y., Denham J., D’Andrea A.D. 2000. The Fanconi anemia proteins FANCA and FANCG stabilize each other and promote the nuclear accumulation of the Fanconi anemia complex. Blood. 96:3224–3230 [PubMed] [Google Scholar]
- Heng T.S.P., Painter M.W.; Immunological Genome Project Consortium. 2008. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9:1091–1094 10.1038/ni1008-1091 [DOI] [PubMed] [Google Scholar]
- Hölzel M., van Diest P.J., Bier P., Wallisch M., Hoatlin M.E., Joenje H., de Winter J.P. 2003. FANCD2 protein is expressed in proliferating cells of human tissues that are cancer-prone in Fanconi anaemia. J. Pathol. 201:198–203 10.1002/path.1450 [DOI] [PubMed] [Google Scholar]
- Kim H., Yang K., Dejsuphong D., D’Andrea A.D. 2012. Regulation of Rev1 by the Fanconi anemia core complex. Nat. Struct. Mol. Biol. 19:164–170 10.1038/nsmb.2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korthof E.T., Svahn J., Peffault de Latour R., Terranova P., Moins-Teisserenc H., Socié G., Soulier J., Kok M., Bredius R.G., van Tol M., et al. 2013. Immunological profile of Fanconi anemia: a multicentric retrospective analysis of 61 patients. Am. J. Hematol. 88:472–476 10.1002/ajh.23435 [DOI] [PubMed] [Google Scholar]
- Kottemann M.C., Smogorzewska A. 2013. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 493:356–363 10.1038/nature11863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijger P.H.L., Wit N., van den Berk P.C.M., Jacobs H. 2010. The Fanconi anemia core complex is dispensable during somatic hypermutation and class switch recombination. PLoS ONE. 5:e15236 10.1371/journal.pone.0015236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaresan K.R., Lambert M.W. 2000. Fanconi anemia, complementation group A, cells are defective in ability to produce incisions at sites of psoralen interstrand cross-links. Carcinogenesis. 21:741–751 10.1093/carcin/21.4.741 [DOI] [PubMed] [Google Scholar]
- Li Z., Scherer S.J., Ronai D., Iglesias-Ussel M.D., Peled J.U., Bardwell P.D., Zhuang M., Lee K., Martin A., Edelmann W., Scharff M.D. 2004. Examination of Msh6- and Msh3-deficient mice in class switching reveals overlapping and distinct roles of MutS homologues in antibody diversification. J. Exp. Med. 200:47–59 10.1084/jem.20040355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Schatz D.G. 2009. Balancing AID and DNA repair during somatic hypermutation. Trends Immunol. 30:173–181 10.1016/j.it.2009.01.007 [DOI] [PubMed] [Google Scholar]
- MacCarthy T., Roa S., Scharff M.D., Bergman A. 2009. SHMTool: a webserver for comparative analysis of somatic hypermutation datasets. DNA Repair (Amst.). 8:137–141 10.1016/j.dnarep.2008.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayorov V.I., Rogozin I.B., Adkison L.R., Gearhart P.J. 2005. DNA polymerase η contributes to strand bias of mutations of A versus T in immunoglobulin genes. J. Immunol. 174:7781–7786 [DOI] [PubMed] [Google Scholar]
- Naim V., Rosselli F. 2009. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat. Cell Biol. 11:761–768 10.1038/ncb1883 [DOI] [PubMed] [Google Scholar]
- Pace P., Mosedale G., Hodskinson M.R., Rosado I.V., Sivasubramaniam M., Patel K.J. 2010. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 329:219–223 10.1126/science.1192277 [DOI] [PubMed] [Google Scholar]
- Pichierri P., Averbeck D., Rosselli F. 2002. DNA cross-link-dependent RAD50/MRE11/NBS1 subnuclear assembly requires the Fanconi anemia C protein. Hum. Mol. Genet. 11:2531–2546 10.1093/hmg/11.21.2531 [DOI] [PubMed] [Google Scholar]
- Pichierri P., Franchitto A., Rosselli F. 2004. BLM and the FANC proteins collaborate in a common pathway in response to stalled replication forks. EMBO J. 23:3154–3163 10.1038/sj.emboj.7600277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass E., Grabarz A., Plo I., Gautier J., Bertrand P., Lopez B.S. 2009. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 16:819–824 10.1038/nsmb.1641 [DOI] [PubMed] [Google Scholar]
- Reina-San-Martin B., Chen J., Nussenzweig A., Nussenzweig M.C. 2007. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1−/− B cells. Eur. J. Immunol. 37:235–239 10.1002/eji.200636789 [DOI] [PubMed] [Google Scholar]
- Renaud E., Rosselli F. 2013. FANC pathway promotes UV-induced stalled replication forks recovery by acting both upstream and downstream Polη and Rev1. PLoS ONE. 8:e53693 10.1371/journal.pone.0053693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin I.B., Pavlov Y.I., Bebenek K., Matsuda T., Kunkel T.A. 2001. Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nat. Immunol. 2:530–536 10.1038/88732 [DOI] [PubMed] [Google Scholar]
- Schrader C.E., Vardo J., Stavnezer J. 2002. Role for mismatch repair proteins Msh2, Mlh1, and Pms2 in immunoglobulin class switching shown by sequence analysis of recombination junctions. J. Exp. Med. 195:367–373 10.1084/jem.20011877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Pâques F., Colaiácovo M., Haber J.E. 1997. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc. Natl. Acad. Sci. USA. 94:9214–9219 10.1073/pnas.94.17.9214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill J.-C., Reynaud C.-A. 2008. DNA polymerases in adaptive immunity. Nat. Rev. Immunol. 8:302–312 10.1038/nri2281 [DOI] [PubMed] [Google Scholar]
- Williams S.A., Wilson J.B., Clark A.P., Mitson-Salazar A., Tomashevski A., Ananth S., Glazer P.M., Semmes O.J., Bale A.E., Jones N.J., Kupfer G.M. 2011. Functional and physical interaction between the mismatch repair and FA-BRCA pathways. Hum. Mol. Genet. 20:4395–4410 10.1093/hmg/ddr366 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.