Abstract
New Delhi metallo-β-lactamase (NDM)-producing bacteria are considered potential global health threats. It is necessary to monitor NDM-1 and its variants in clinical isolates in order to understand the NDM-1 epidemic and the impact of its variants on β-lactam resistance. To reduce the lengthy time needed for cloning and expression of NDM-1 variants, a novel PCR-based in vitro protein expression (PCR-P) method was used to detect blaNDM-1 and its variants coding for carbapenemases with different activities (functional variants). The PCR-P method combined a long-fragment real-time quantitative PCR (LF-qPCR) with in vitro cell-free expression to convert the blaNDM-1 amplicons into NDM for carbapenemase assay. The method could screen for blaNDM-1 within 3 h with a detection limit of 5 copies and identify functional variants within 1 day. Using the PCR-P to analyze 5 recent blaNDM-1 variants, 2 functional variants, blaNDM-4 and blaNDM-5, were revealed. In the initial testing of 23 clinical isolates, the PCR-P assay correctly found 8 isolates containing blaNDM-1. This novel method provides the first integrated approach for rapidly detecting the full-length blaNDM-1 and revealing its functional variants in clinical isolates.
INTRODUCTION
Limited antibiotic choices are available for treating patients infected with Enterobacteriaceae, Acinetobacter spp., and other species producing New Delhi metallo-β-lactamase 1 (NDM-1; encoded by the blaNDM-1 gene, nucleotide sequence accession no. KF016990.1) because they are usually resistant to all classes of antibiotics, including carbapenems (1). Much worse, blaNDM-1 is commonly found on plasmids, which could easily transfer between species (2, 3). To date, various kinds of species containing this gene have been isolated and found worldwide (4–6). Therefore, the infections caused by NDM-1-producing bacteria are considered potential global health problems (1). At the same time, along with its spread, variants of NDM-1 are emerging. Until now, 10 NDM variants have been found (7–13; also NDM-9 [nucleotide sequence no. KC999080.1] and NDM-10 [nucleotide sequence accession no. KF361506.1]). Some of the variants, such as NDM-4 and NDM-5, confer higher β-lactam resistance to bacteria than NDM-1 because they pose increased carbapenemase activities compared to that of wild-type NDM-1 (9, 10). In order to better understand the NDM-1 epidemic and the impact of the NDM variants on β-lactam resistance, it is necessary to monitor NDM-1 and its variants in clinical isolates.
Phenotypic techniques for detecting carbapenemase activity, such as the modified Hodge Test (MHT) recommended by the Clinical and Laboratory Standards Institute (CLSI) (14, 15) and the newly developed Carba NP (16), can be used to detect carbapenemase producers, including those producing NDM-1. However, because all of the phenotypic assays detect NDM-1 together with other carbapenemases, specific molecular methods to detect the blaNDM-1 gene have been widely used to indicate the presence of NDM-1-producing bacteria (17–21). These methods share the feature that only a small fragment of the blaNDM-1 gene is PCR amplified, and as a consequence, they can only report the presence of blaNDM-1. To obtain mutation information, PCR amplification of the full-length blaNDM-1 plus DNA sequencing are normally used (11). DNA sequencing can reveal every possible mutation of blaNDM-1 but cannot give function information as to whether the mutations will promote or reduce the ability of the coded NDM to hydrolyze β-lactam (9). For example, the amino acid substitution in NDM-4 is not located in the known active sites of NDM-1, indicating that remote amino acid substitutions might also play a role in the extended activity of NDM-4 (9). Expression and purification of the recombinant NDM are normally used to study the effects of the new mutations, but the process is time consuming (from a few days to weeks).
In this study, a new strategy called PCR-based in vitro protein expression (PCR-P) is explored for rapid detection of the full-length blaNDM-1 and its variants coding for carbapenemases with different activities (functional variants). The PCR-P method consists of a novel long-fragment quantitative PCR (LF-qPCR) to detect the full-length blaNDM-1 in clinical isolates and a function assay of in vitro-synthesized protein to identify whether the LF-qPCR amplicons contain blaNDM-1 or its functional variants through measuring the imipenem degradation rate of the in vitro-synthesized NDM. As far as we know, this is the first assay to integrate the detection of the full-length blaNDM-1 with the identification of blaNDM-1 functional variants.
MATERIALS AND METHODS
Bacterial strains.
One NDM- producing Acinetobacter baumannii strain (sample no. A. baumannii 65) isolated previously by us (22, 23), along with a non-NDM-1 producer (Escherichia coli 53) as the negative control, were used to optimize the novel PCR-P. As shown in Table 1 and Table S1 in the supplemental material, a collection of 23 carbapenem-resistant strains isolated from different patients in different years by the microbiology laboratory of Guangzhou First Municipal People's Hospital were chosen for testing. The MICs were determined using the Vitek 2 system (BioMerieux, France) and interpreted according to CLSI 2012 guidelines (24). Because the first NDM-1 producer reported in China was A. baumannii (5), A. baumannii isolates were mainly chosen for testing. The results of multilocus sequence typing (MLST) show that the 15 A. baumannii isolates belonged to 5 different known sequence types (STs) and 3 new STs (Table 1) (25). Details are given in Table S2 in the supplemental material.
TABLE 1.
Sample information and results of phenotypic assay, conventional PCR, MLST, LF-qPCR, in vitro-synthesized carbapenemase activity assay, and sequencinga
| Strain or variant | Vitek 2 MIC (mg/liter) of: |
PCRc | STd | Result of indicated step of PCR-P assay |
Sequencing result(s) | ||
|---|---|---|---|---|---|---|---|
| IPM | ETPb | LF-qPCR (CT value) | Antibiotic degradation assay (rate [−%A0/min])e | ||||
| Strains | |||||||
| E. coli 53 (negative control) | ≤1 | 2 | Neg | — | Neg | — | — |
| A. baumannii 65 (positive control) | ≥16 | — | + | New | 13.27 | 1.338 ± 0.135 | WT |
| E. coli 23 | ≤1 | 2 | Neg | — | Neg | — | — |
| E. coli 59 | 2 | ≤0.5 | Neg | — | Neg | — | — |
| E. coli 75 | ≤1 | 2 | Pos | — | 12.89 | 1.393 ± 0.083 | WT |
| K. pneumoniae 17 | ≥16 | ≥8 | Pos | — | 28.24 | 1.344 ± 0.092 | WT |
| K. pneumoniae 19 | 2 | 1 | Neg | — | Neg | — | — |
| K. pneumoniae 76 | 2 | 1 | Neg | — | Neg | — | — |
| E. cloacae 60 | ≤1 | 4 | Neg | — | Neg | — | — |
| E. cloacae 63 | 2 | ≤0.5 | Neg | — | Neg | — | — |
| K. ozaenae 16 | 12 | ≥8 | Pos | — | 14.4 | 1.264 ± 0.134 | WT |
| A. baumannii 26 | ≥16 | — | Neg | 254 | Neg | — | — |
| A. baumannii 30 | ≥16 | — | Neg | 254 | Neg | — | — |
| A. baumannii 31 | ≥16 | — | Pos | 254 | 30.96 | 1.284 ± 0.093 | WT |
| A. baumannii 33 | ≥16 | — | Neg | 254 | Neg | — | — |
| A. baumannii 39 | ≥16 | — | Neg | 254 | Neg | — | — |
| A. baumannii 40 | ≥16 | — | Neg | 254 | Neg | — | — |
| A. baumannii 41 | ≥16 | — | Pos | 254 | 28 | 1.301 ± 0.089 | WT |
| A. baumannii 42 | ≥16 | — | Neg | New | Neg | — | — |
| A. baumannii 59 | 8 | — | Neg | 208 | Neg | — | — |
| A. baumannii 62 | ≥16 | — | Pos | 208 | 24.94 | 1.181 ± 0.245 | WT |
| A. baumannii 63 | ≥16 | — | Pos | 381 | 26.58 | 1.391 ± 0.053 | WT |
| A. baumannii 66 | ≥16 | — | Neg | 368 | Neg | — | — |
| A. baumannii 69 | ≥16 | — | Neg | New | Neg | — | — |
| A. baumannii 70 | ≥16 | — | Pos | 229 | 14.23 | 1.511 ± 0.087 | WT |
| Variantsf | |||||||
| NDM-K211E | — | — | Pos | — | 10.63 | 0.132 ± 0.004 | A611G |
| NDM-2(P28A) | — | — | Pos | — | 15.73 | 1.140 ± 0.069 | C82G |
| NDM-3(D95N) | — | — | Pos | — | 11.52 | 1.120 ± 0.047 | G283A |
| NDM-4(M154L) | — | — | Pos | — | 11.30 | 2.236 ± 0.060 | A460C |
| NDM-5(V88L, M154L) | — | — | Pos | — | 12.63 | 1.773 ± 0.017 | G262T, A460C |
| NDM-6(A233V) | — | — | Pos | — | 13.27 | 1.187 ± 0.105 | C698T |
IPM, imipenem; ETP, ertapenem; ST, sequence type; Pos, positive; Neg, negative; —, not tested; WT, wild type.
Ertapenem has limited activity against A. baumannii.
Conventional PCR to detect blaNDM-1.
New, new ST.
−%A0/min is the percentage of absorbance decrease per minute. %A0 is the percentage of absorbance relative to the absorbance at time zero (just after imipenem is added).
blaNDM-1 variants were constructed by using a site-directed gene mutagenesis kit and overlap PCR. The mutations in these variants were confirmed by sequencing as shown in the last column.
A site-directed gene mutagenesis kit (Beyotime Institute of Biotechnology, Shanghai) and overlap PCR were used to create 6 blaNDM-1 variants, blaNDM-2 (GenBank accession no. JN112341), blaNDM-3 (JQ734687), blaNDM-4 (JQ348841), blaNDM-5 (JN104597), blaNDM-6 (JQ235754), and blaNDM-K211E (encoding a K-to-E change at position 211), using the primers shown in Table S3 in the supplemental material.
Preparation of DNA templates of clinical isolates.
Both plasmid extracts and genomic DNA were used as the templates for PCR-P. Plasmids were extracted from fresh overnight bacterial cultures with an E.Z.N.A. plasmid minikit (Omega Bio-Tek, United States) according to the protocol suggested by the manufacturer. DNA templates were also obtained by heat lysis (26, 27). Briefly, 1.4 ml bacterial culture (with a McFarland standard of between 3 to 4) was centrifuged, and then the precipitates were resuspended in 0.1 ml sterile MilliQ water and inactivated at 100°C for 10 min.
PCR-P assay procedure.
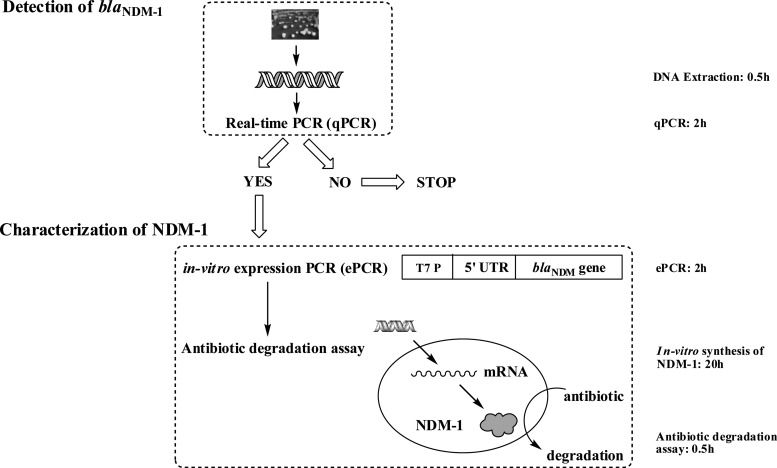
As shown in Fig. 1, the first step of the method was LF-qPCR detection of blaNDM-1 from samples. The major difference of the LF-qPCR from other qPCR methods was that the full-length blaNDM-1 (822 bp) was amplified with the specially designed primers F-rt and R-rt (see Table S3 in the supplemental material). When a positive amplification was detected, the second step was initiated to further confirm the LF-qPCR products and reveal possible functional mutated blaNDM-1 variants. During this step, a second in vitro expression PCR (ePCR) was performed to introduce the expression elements T7 promoter and 5′ untranslated region (5′ UTR), using the primers F-1-UTR and R-1, shown in Table S3. Then, in vitro synthesis of NDM-1 was carried out at 24°C for 20 h using an RTS 100 wheat germ CECF kit (rapid translation system continuous-exchange cell-free kit; 5 Prime, Inc., United States). An equal amount of the lysate without the ePCR amplicon was also incubated simultaneously, as a negative control. Measurements of NDM-1 activities were performed on a Synergy H1 hybrid reader (BioTek, United States) by observing the decrease in optical density at 300 nm (OD300) at 30°C, as described previously (1). The antibiotic degradation rates were compared with that of NDM-1 to identify blaNDM-1 functional variants that possessed either increased or decreased carbapenemase activities. The testing could be finished within 3 h for screening for blaNDM-1 and within 24 h for identifying blaNDM-1 variants. Please see the materials and methods in the supplemental material for the detailed conditions.
FIG 1.
Scheme of the gene-to-protein function assay for rapid identification of blaNDM-1 and functional variants with mutations related to carbapenemase activity.
DNA sequencing and conventional PCR method.
The positive amplicons from the LF-qPCR (containing the full-length blaNDM-1) were sequenced by BGI Tech, Inc. (Wuhan, China). Conventional PCR methods, described previously (5, 7), were used to test for the presence of blaNDM-1 in the isolates, using the primer pairs F-38 and R-344, as well as Pre-A and Pre-B, shown in Table S3 in the supplemental material.
RESULTS
LF-qPCR assay of blaNDM-1.
The specificity of the primers F-rt and R-rt for blaNDM-1 detection was first evaluated by a BLAST search of the NCBI database (www.ncbi.nlm.nih.gov). No matches to the primers were found except for the blaNDM-1 gene. The specificity of LF-qPCR was further checked using the genomic DNA obtained from the non-NDM-1 producer E. coli 53 (negative control).
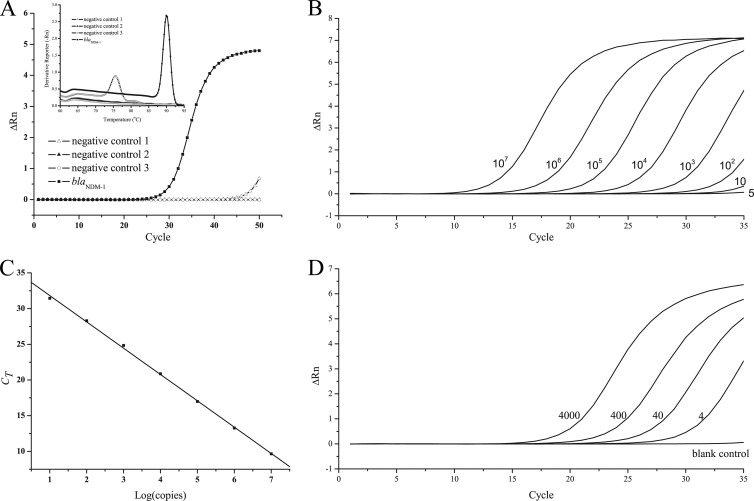
The LF-qPCR amplification generated a threshold cycle (CT) value of more than 40 for the negative control and a CT value of around 26 for 1 × 103 copies of the plasmid DNA extracted from A. baumannii 65, with a single peak in the melting curve (Fig. 2A). The signal obtained for the negative control after many cycles might be due to the formation of primer dimers (with a lower melting temperature, as shown in the inset in Fig. 2A), which is common for qPCR systems based on SYBR green I. Since the LF-qPCR generated a CT value of about 33 for 5 copies (Fig. 2B), which was comparable with the results of other PCR methods (17–19) and low enough for most clinical samples, 35 cycles was chosen as the cutoff. The standard curve, with a linear coefficient (r2) of 0.9995 and a slope of −3.6886 (Fig. 2C), was found by analyzing serial 10-fold dilutions of the plasmid DNA ranging from 10 copies/reaction mixture volume to 1.00 × 107 copies/reaction mixture volume (Fig. 2B). The detection limit was found to be as low as 4 CFU/reaction mixture volume (or 103 CFU/ml), as shown in Fig. 2D, by spiking different amounts of A. baumannii 65 into water.
FIG 2.
LF-qPCR assay of blaNDM-1. (A) Amplification curves and melting curves (inset) of the negative controls and blaNDM-1 (103 copies/reaction mixture volume). (B) Amplification curves of dilutions of the plasmid (copies/reaction mixture volume) extracted from a blaNDM-1-containing A. baumannii strain (A. baumannii 65). (C) Corresponding standard curve of the LF-qPCR assay. (D) Amplification curves of water samples spiked with different concentrations (CFU/reaction mixture volume) of A. baumannii 65. All experiments were repeated three times to determine the sensitivity and linearity of LF-qPCR. ΔRn, baseline-corrected normalized reporter.
ePCR for adding the in vitro expression elements.
In order for the wheat germ system to express protein directly from PCR products, the T7 promoter sequence must be attached to the forward primers, and some 5′ UTR sequences can be used to enhance protein expression (28, 29). Two primer pairs (F-rt-UTR/R1 and F-1-UTR/R-1) with a 5′ UTR sequence (see Table S3 in the supplemental material) were used to amplify the blaNDM-1 amplicon obtained from A. baumannii 65 in the LF-qPCR step. The positions of the primers relative to those of the LF-qPCR primer pair are shown in Fig. S1 in the supplemental material. Both primer pairs could effectively amplify the blaNDM-1 gene, but there were fewer primer dimers and higher amplicon purity when using the primer pair F-1-UTR/R-1 (data not shown). Therefore, primers F-1-UTR and R-1 were used in the experiments described below. After purification, the ePCR amplicons were added into the wheat germ lysate to synthesize NDM.
Characterization of in vitro-synthesized NDM-1 and variants.
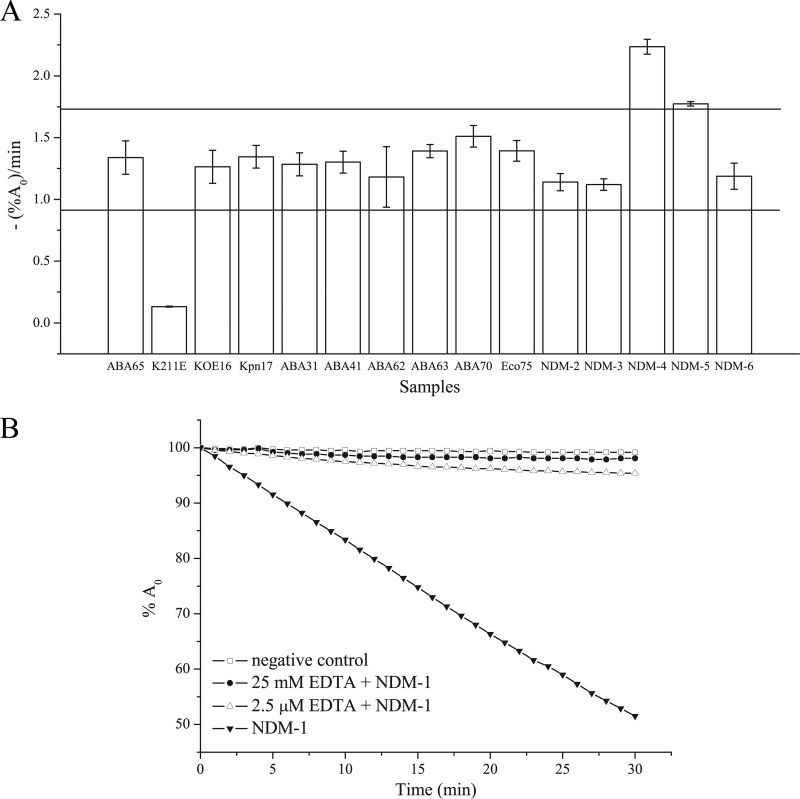
After in vitro synthesis of NDM-1, the wheat germ lysates were used directly for measurement of their reactions with imipenem. As shown by the results in Fig. 3B, the OD300 decreased over time after imipenem was mixed with the lysate containing the blaNDM-1 ePCR amplicon, while no change was found for the negative-control lysate. Inhibition assays also showed that the in vitro-synthesized NDM-1 was strongly inhibited by EDTA. These results demonstrated that NDM-1 was successfully synthesized. Repeated experiments under the same conditions showed that the in vitro-synthesized-NDM assay was reproducible in terms of the imipenem degradation rate, with an intra-assay relative standard deviation (RSD) of less than 10% and an interassay RSD of less than 16%.
FIG 3.
(A) The imipenem degradation rates of the in vitro-synthesized NDM variants (blaNDM-2, -3, -4, -5, and -6 and blaNDM-K211E) and the NDM-1 proteins from the positive clinical isolates in relation to the imipenem degradation rate of the wild-type positive control (A. baumannii strain 65). Data are the averages of three replicate experiments. Error bars show the ±SD. ABA, A. baumannii; KOE, Klebsiella ozaenae; Kpn, Klebsiella pneumoniae; Eco, E. coli. -(%A0)/min, percentage of absorbance decrease per minute. The horizontal lines represent the cutoff values for judging functional variants, which equal the -(%A0)/min value of the wild-type NDM-1 control plus 3 × SD (upper line) and minus 3 × SD (lower line). (B) EDTA inhibition assay for the activity of the in vitro-synthesized NDM-1. %A0, percentage of absorbance relative to the absorbance at time zero (just after imipenem is added).
Using the same procedure, the degradation rates of the lysates containing different NDM variants were tested. As shown by the results in Fig. 3A, NDM-K211E degraded imipenem significantly more slowly than NDM-1, while NDM-4 and NDM-5 were found to have higher carbapenemase activities than NDM-1. Another 3 variants, NDM-2, NDM-3, and NDM-6, showed activities similar to that of NDM-1. These results were consistent with the previously reported low carbapenemase activity of NDM-K211E (30) and the higher activities of NDM-4 and NDM-5 (9, 10).
All the results described above demonstrated that the in vitro-synthesized NDM showed enzymatic properties similar to those of the recombinant NDM-1 and current NDM-1 variants. Therefore, it is possible to use the degradation rate of imipenem to identify the blaNDM-1 variants with different carbapenemase activities.
PCR-P assay of clinical isolates.
As summarized in Table 1, the LF-qPCR found that 8 of the 23 clinical samples were positive for blaNDM-1, which was fully consistent with the results of the conventional PCR test. The typical amplification curves for 4 positive isolates (see Fig. S2A in the supplemental material) showed that similar amplifications were obtained using either the plasmids or the templates prepared using heat lysis. All of the 8 positive isolates showed high-level resistance to imipenem by the Vitek system, except for E. coli sample 75. However, E. coli 75 was found to be resistant to ertapenem, with a MIC of 2 mg/liter (Table 1).
These LF-qPCR-positive samples were further tested with the in vitro-synthesized-carbapenemase assay. As shown by the results in Fig. 3A, the enzyme activities were found to be within ±3 times the standard deviation (SD) of the results for NDM-1 for all the samples. After 25 mM EDTA was added to the lysates, the imipenem degradation was inhibited (data not shown). Therefore, the PCR-P results showed that all of the 8 isolates contained blaNDM-1 but none contained functional variants. Fully consistent with the PCR-P results, the DNA sequencing results confirmed no mutations in the full length of the blaNDM-1 gene of the 8 clinical isolates.
DISCUSSION
As a mobile genetic resistance gene, blaNDM-1 could transfer between species and mutate under either the selection pressure of β-lactam or natural evolution during the spread. Clearly, we should pay more attention to the functional blaNDM-1 variants, i.e., the variants whose mutations could affect the carbapenemase activities they encode, since they can confer different levels of resistance to β-lactams. Therefore, an ideal monitoring assay should not only identify the presence of blaNDM-1 but also reveal functional blaNDM-1 variants. In view of these requirements, the current widely used PCR sequencing method is not sufficient, because phenotypic testing (expression of recombinant protein) is needed to confirm whether a new variant identified by sequencing has an alteration in the function of its coded protein (14, 31).
Since new variants are expected to emerge due to the worldwide spread of blaNDM-1, the advantage of direct identification of functional variants would be especially useful for practical applications, which is the rationale behind our work to design the PCR-P approach. As shown above, the LF-qPCR detection of the full length of blaNDM-1 was sensitive and fast in screening for the presence of blaNDM-1 in clinical isolates. The amplified long fragment not only facilitates the sensitivity of detection using SYBR green, which can reduce the cost compared to other qPCRs based on probes (17, 19), but it can also be used as the template for the in vitro-synthesized-carbapenemase assay for finding functional variants. Furthermore, the in vitro-synthesized-carbapenemase assay provides a few advantages not available by other methods. First, the positive degradation to imipenem can be used for confirmation of the LF-qPCR result. Because false amplified fragments will not result in active carbapenemase, this step could help to reduce the false-positive rate of the LF-qPCR due to nonspecific amplification. Second, functional blaNDM-1 variants can be revealed directly by comparing the imipenem degradation rates of the synthesized NDMs to that of NDM-1, as shown in Fig. 3A. Finally, the PCR-P assay avoids the need with the recombinant methods to study the properties of blaNDM-1 variants. The in vitro expression can be finished within 1 day, while the conventional recombinant protein procedures would take several days.
Besides blaNDM-1, many other resistance-coding genes, such as the plasmid-mediated blaAMP-C gene and blaFIM-1 gene, are monitored by PCR approaches (32, 33). Based on a similar strategy, a PCR-P assay could be developed for detecting these genes and functional variants through measuring the activity of the in vitro-synthesized lactamases.
However, because the PCR-P assay is basically a molecular-testing method, clinical laboratories will need molecular expertise to perform the assay. Another obstacle might be the cost, which is higher than the costs of the phenotypic methods. Therefore, we foresee that the PCR-P assay would be best utilized as a complement to a phenotypic method, such as Carba NP, for identifying blaNDM-1 or its functional variants.
In conclusion, with its capabilities of rapidly detecting the presence of the full-length blaNDM-1 gene and revealing blaNDM-1 functional variants, the PCR-P assay provides a unique approach for monitoring blaNDM-1 epidemics and functional variants in clinical isolates.
Supplementary Material
ACKNOWLEDGMENTS
We thank Xiaomian Zhou from the Guangzhou First Municipal People's Hospital for his help on this project. We also thank Rongzhang Hao and Xuejun Ma (Center for Disease Control, China) for useful discussions.
This work was supported by the Infectious Disease Control Research Program of the Ministry of Health of China (2009ZX10003-019) and the Key Laboratory on Emerging Infectious Diseases and Biosafety in Wuhan.
Footnotes
Published ahead of print 26 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03363-13.
REFERENCES
- 1.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis. 11:355–362. 10.1016/S1473-3099(11)70059-7 [DOI] [PubMed] [Google Scholar]
- 3.Moellering RC., Jr 2010. NDM-1—a cause for worldwide concern. N. Engl. J. Med. 363:2377–2379. 10.1056/NEJMp1011715 [DOI] [PubMed] [Google Scholar]
- 4.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10:597–602. 10.1016/S1473-3099(10)70143-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Zhou Z, Jiang Y, Yu Y. 2011. Emergence of NDM-1-producing Acinetobacter baumannii in China. J. Antimicrob. Chemother. 66:1255–1259. 10.1093/jac/dkr082 [DOI] [PubMed] [Google Scholar]
- 6.Shibl AM, Memish ZA, Al-Agamy MH, Senok AC, Begum S, Assiri A. 2013. Emergence of OXA-48 and NDM-1 positive Klebsiella pneumoniae in Riyadh, Saudi Arabia. Int. J. Antimicrob. Agents 42:S83–S84. 10.1016/S0924-8579(13)70375-3 [DOI] [PubMed] [Google Scholar]
- 7.Kaase M, Nordmann P, Wichelhaus TA, Gatermann SG, Bonnin RA, Poirel L. 2011. NDM-2 carbapenemase in Acinetobacter baumannii from Egypt. J. Antimicrob. Chemother. 66:1260–1262. 10.1093/jac/dkr135 [DOI] [PubMed] [Google Scholar]
- 8.Rogers BA, Sidjabat HE, Silvey A, Anderson TL, Perera S, Li J, Paterson DL. 2013. Treatment options for New Delhi metallo-beta-lactamase-harboring enterobacteriaceae. Microb. Drug Resist. 19:100–103. 10.1089/mdr.2012.0063 [DOI] [PubMed] [Google Scholar]
- 9.Nordmann P, Boulanger AE, Poirel L. 2012. NDM-4 metallo-β-lactamase with increased carbapenemase activity from Escherichia coli. Antimicrob. Agents Chemother. 56:2184–2186. 10.1128/AAC.05961-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hornsey M, Phee L, Wareham DW. 2011. A novel variant, NDM-5, of the New Delhi metallo-beta-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother. 55:5952–5954. 10.1128/AAC.05108-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williamson DA, Sidjabat HE, Freeman JT, Roberts SA, Silvey A, Woodhouse R, Mowat E, Dyet K, Paterson DL, Blackmore T, Burns A, Heffernan H. 2012. Identification and molecular characterisation of New Delhi metallo-beta-lactamase-1 (NDM-1)- and NDM-6-producing Enterobacteriaceae from New Zealand hospitals. Int. J. Antimicrob. Agents 39:529–533. 10.1016/j.ijantimicag.2012.02.017 [DOI] [PubMed] [Google Scholar]
- 12.Gottig S, Hamprecht AG, Christ S, Kempf VA, Wichelhaus TA. 2013. Detection of NDM-7 in Germany, a new variant of the New Delhi metallo-beta-lactamase with increased carbapenemase activity. J. Antimicrob. Chemother. 68:1737–1740. 10.1093/jac/dkt088 [DOI] [PubMed] [Google Scholar]
- 13.Tada T, Miyoshi-Akiyama T, Dahal RK, Sah MK, Ohara H, Kirikae T, Pokhrel BM. 2013. NDM-8 metallo-beta-lactamase in a multidrug-resistant Escherichia coli strain isolated in Nepal. Antimicrob. Agents Chemother. 57:2394–2396. 10.1128/AAC.02553-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mochon AB, Garner OB, Hindler JA, Krogstad P, Ward KW, Lewinski MA, Rasheed JK, Anderson KF, Limbago BM, Humphries RM. 2011. New Delhi metallo-β-lactamase (NDM-1)-producing Klebsiella pneumoniae: case report and laboratory detection strategies. J. Clin. Microbiol. 49:1667–1670. 10.1128/JCM.00183-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girlich D, Poirel L, Nordmann P. 2012. Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J. Clin. Microbiol. 50:477–479. 10.1128/JCM.05247-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nordmann P, Poirel L, Dortet L. 2012. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 18:1503–1507. 10.3201/eid1809.120355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diene SM, Bruder N, Raoult D, Rolain JM. 2011. Real-time PCR assay allows detection of the New Delhi metallo-beta-lactamase (NDM-1)-encoding gene in France. Int. J. Antimicrob. Agents 37:544–546. 10.1016/j.ijantimicag.2011.02.006 [DOI] [PubMed] [Google Scholar]
- 18.Liu W, Zou D, Li Y, Wang X, He X, Wei X, Shao C, Li X, Shang W, Yu K, Liu D, Guo J, Yin Z, Yuan J. 2012. Sensitive and rapid detection of the new Delhi metallo-beta-lactamase gene by loop-mediated isothermal amplification. J. Clin. Microbiol. 50:1580–1585. 10.1128/JCM.06647-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naas T, Ergani A, Carrer A, Nordmann P. 2011. Real-time PCR for detection of NDM-1 carbapenemase genes from spiked stool samples. Antimicrob. Agents Chemother. 55:4038–4043. 10.1128/AAC.01734-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuzon G, Naas T, Bogaerts P, Glupczynski Y, Nordmann P. 2012. Evaluation of a DNA microarray for the rapid detection of extended-spectrum beta-lactamases (TEM, SHV and CTX-M), plasmid-mediated cephalosporinases (CMY-2-like, DHA, FOX, ACC-1, ACT/MIR and CMY-1-like/MOX) and carbapenemases (KPC, OXA-48, VIM, IMP and NDM). J. Antimicrob. Chemother. 67:1865–1869. 10.1093/jac/dks156 [DOI] [PubMed] [Google Scholar]
- 21.Vasoo S, Cunningham SA, Kohner PC, Mandrekar JN, Lolans K, Hayden MK, Patel R. 2013. Rapid and direct real-time detection of blaKPC and blaNDM from surveillance samples. J. Clin. Microbiol. 51:3609–3615. 10.1128/JCM.01731-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang YM, Ye HF, Zhang WH, Chen HL, Zhou XM. 2011. Detection of New Delhi metallo-β-lactamase 1 gene in Klebsiella ozaenae and Acinetobacter baumannii. Int. J. Lab. Med. 32:1407–1409 (In Chinese.) [Google Scholar]
- 23.Zhou XM, Liu D, Huang HL. 2011. NDM-1(+) bacteria screening using capillary PCR and direct PCR amplification method. Int. J. Lab. Med. 32:1404–1406 (In Chinese.) [Google Scholar]
- 24.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial susceptibility testing: 22nd informational supplement. CLSI document M100–S22. CLSI, Wayne, PA [Google Scholar]
- 25.Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodriguez-Valera F. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43:4382–4390. 10.1128/JCM.43.9.4382-4390.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams-Haduch JM, Paterson DL, Sidjabat HE, Pasculle AW, Potoski BA, Muto CA, Harrison LH, Doi Y. 2008. Genetic basis of multidrug resistance in Acinetobacter baumannii clinical isolates at a tertiary medical center in Pennsylvania. Antimicrob. Agents Chemother. 52:3837-3843. 10.1128/AAC.00570-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang C, Yuan J.-f., Shi Z.-l. 2007. Molecular epidemiological investigation of infectious hypodermal and hematopoietic necrosis virus and taura syndrome virus in Penaeus vannamei cultured in China. Virologica Sinica 22:380–388. 10.1007/s12250-007-0036-x [DOI] [Google Scholar]
- 28.Akbergenov R, Zhanybekova S, Kryldakov RV, Zhigailov A, Polimbetova NS, Hohn T, Iskakov BK. 2004. ARC-1, a sequence element complementary to an internal 18S rRNA segment, enhances translation efficiency in plants when present in the leader or intercistronic region of mRNAs. Nucleic Acids Res. 32:239–247. 10.1093/nar/gkh176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou M, Geng X, Chen J, Wang X, Wang D, Deng J, Zhang Z, Wang W, Zhang XE, Wei H. 2011. Rapid colorimetric testing for pyrazinamide susceptibility of M. tuberculosis by a PCR-based in-vitro synthesized pyrazinamidase method. PLoS One 6:e27654. 10.1371/journal.pone.0027654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang Z, Li L, Wang Y, Chen L, Kong X, Hong Y, Lan L, Zheng M, Guang-Yang C, Liu H, Shen X, Luo C, Li KK, Chen K, Jiang H. 2011. Molecular basis of NDM-1, a new antibiotic resistance determinant. PLoS One 6:e23606. 10.1371/journal.pone.0023606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barczak AK, Gomez JE, Kaufmann BB, Hinson ER, Cosimi L, Borowsky ML, Onderdonk AB, Stanley SA, Kaur D, Bryant KF, Knipe DM, Sloutsky A, Hung DT. 2012. RNA signatures allow rapid identification of pathogens and antibiotic susceptibilities. Proc. Natl. Acad. Sci. U. S. A. 109:6217–6222. 10.1073/pnas.1119540109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geyer CN, Reisbig MD, Hanson ND. 2012. Development of a TaqMan multiplex PCR assay for detection of plasmid-mediated AmpC β-lactamase genes. J. Clin. Microbiol. 50:3722–3725. 10.1128/JCM.02038-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollini S, Maradei S, Pecile P, Olivo G, Luzzaro F, Docquier JD, Rossolini GM. 2013. FIM-1, a new acquired metallo-beta-lactamase from a Pseudomonas aeruginosa clinical isolate from Italy. Antimicrob. Agents Chemother. 57:410–416. 10.1128/AAC.01953-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.