Abstract
Phosphatase and tensin homolog deleted on chromosome ten (PTEN) is a phosphatase that is frequently altered in cancer. PTEN has phosphatase-dependent and - independent roles; and genetic alterations in PTEN lead to deregulation of protein synthesis, cell cycle, migration, growth, DNA repair, and survival signaling. PTEN localization, stability, conformation, and phosphatase activity are controlled by an array of protein-protein interactions and post-translational modifications. Thus, PTEN-interacting and modifying proteins have profound effects on PTEN’s tumor suppressive functions. Moreover, recent studies identified mechanisms by which PTEN can exit cells, either via exosomal export or secretion, and act on neighboring cells. This review focuses on modes of PTEN protein regulation and ways in which perturbations in this regulation may lead to disease.
Keywords: PTEN, PTEN-Long, PI3K signaling
Importance of PTEN function and regulation
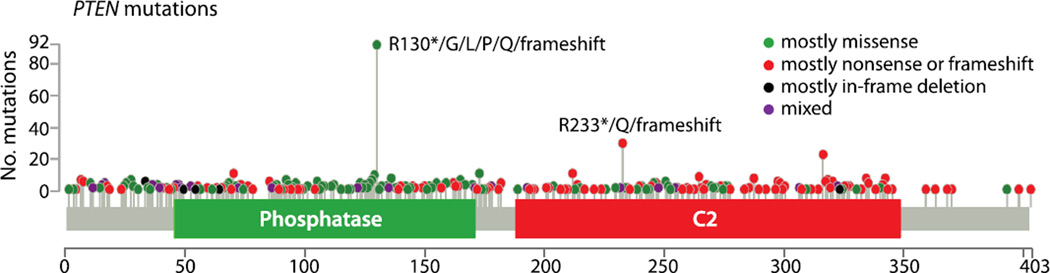
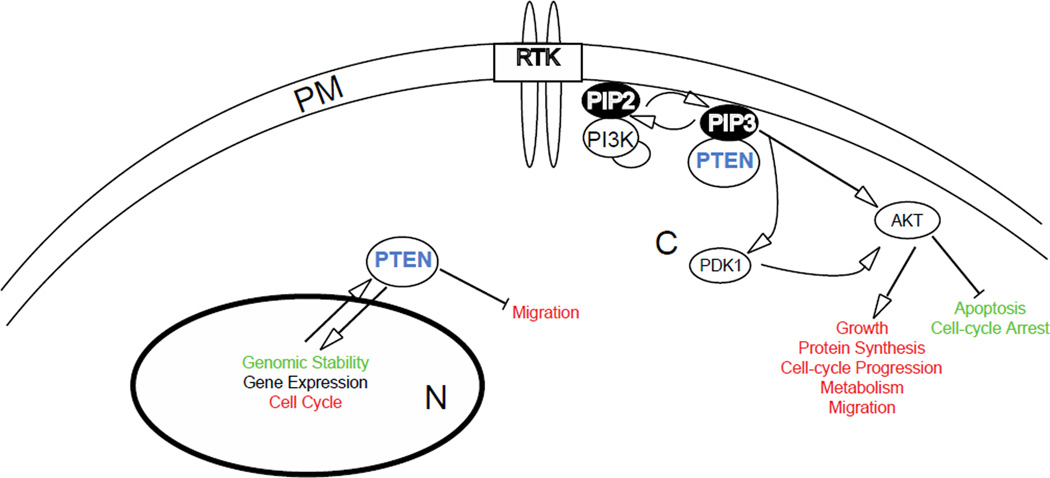
Phosphatase and tensin homologue deleted on chromosome ten (PTEN) was originally identified as a tumor suppressor frequently lost from a region of chromosome 10q23 in a variety of human tumors including those of the brain, breast, and prostate [1, 2]. To date, the COSMIC cancer database currently lists >2700 mutations in PTEN from 28 different tumor types, and the cBio portal of The Cancer Genome Atlas (TCGA) lists 1120 mutations in 27 tumor types (Figure 1) [3, 4] (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic). PTEN is a dual specificity protein and lipid phosphatase, and its primary cellular substrate is the second messenger phosphatidylinositol (3,4,5)-trisphosphate (PIP3), which it hydrolyzes to phosphatidylinositol (4,5)-bisphosphate (PIP2) (Figure 2) [5–8]. PTEN blocks phosphatidylinositol 3 kinase (PI3K) signaling by inhibiting PIP3 dependent processes such as the membrane recruitment and activation of AKT, therefore inhibiting cell survival, growth, and proliferation. PTEN thus occupies a critical node for the inhibition of oncogenic transformation (Figure 2). Mounting evidence indicates that PTEN also has significant PIP3-independent functions. Specifically, PTEN protein phosphatase activity is critical for PTEN mediated inhibition of cellular migration[9]. In glioma cells, it has been shown that the protein phosphatase activity of PTEN is required to induce PTEN phosphorylation and inhibit cellular migration [10]. Furthermore, there is evidence that PTEN phosphatase activity may regulate glioma cell migration by suppressing Src family kinases [11]. It has also been reported that PTEN dephosphorylates focal adhesion kinase, leading to reduced cell migration and spreading in fibroblasts [12]. PTEN also has nuclear functions, which are likely independent of its ability to antagonize PI3K signaling. A variety of proteins have been shown to affect PTEN nuclear localization, thereby impacting PTEN’s ability to act in the nucleus and promote genomic stability [13–15]. In addition to protein regulation, a variety of studies have shown that PTEN is down regulated by promoter methylation in thyroid, breast, lung, endometrial, ovarian, gastric, and brain tumors [13]. PTEN has also been shown to be silenced by the expression of a number of micro-RNAs and non-coding RNAs [13]. These events are critical as it has been demonstrated that subtle changes in the dose of PTEN can have profound effects on tumor susceptibility [16, 17].
Figure 1.
PTEN mutations in cancer. PTEN mutations sites and frequencies were obtained from the cBio Portal of The Cancer Genome Atlas (TCGA). Green dots represent sites for which the majority of alterations are missense mutations. Red dots represent sites for which the majority of alterations are nonsense mutations or frameshifts. Black dots represent sites for which the majoritiy of alterations are in-frame deletions. Purple dots denote sites for which alteration types are mixed [3, 4, 66].
Figure 2.
The functions of PTEN. Phosphatase and Tensin Homologue deleted on chromosome ten (PTEN) is a lipid phosphatase that antagonizes phosphatidlynositol 3 kinase (PI3K) signaling by converting phosphatidylinositol (3,4,5) trisphosphate (PIP3) to phosphatidylinositol (4,5) bisphosphate (PIP2). This negatively regulates Protein Kinase B (AKT), Phosphatidylinositol Dependent Kinase-1 (PDK1) and other PIP3-dependent moieties to inhibit growth, protein synthesis, cell cycle progression, metabolism, and migration and allows for apoptosis and cell cycle arrest down stream of receptor tyrosine kinase (RTK) activation at the cell surface. PTEN also acts in a PI3K-independent manner, inhibiting migration and affecting genomic stability, gene expression, and the cell cycle. Functions that are increased by PTEN are in green whereas those inhibited by PTEN are in red. The nucleus is denoted with an ‘N,’ the plasma membrane with a ‘PM,’ and the cytoplasm with a ‘C.’
The importance of PTEN in tumor suppression is confirmed by the existence of germline mutations of PTEN in the PTEN Hereditary Tumor Syndromes (PHTS) including Cowden disease, Bannayan-Riley-Ruvalcaba Syndrome, Proteus syndrome, and Proteus like syndrome [18–20]. Patients with PHTS develop benign hamartomas in a variety of organs, and are more likely to develop thyroid or breast cancer [21]. It has been demonstrated that mutations of PTEN in PHTS patients cause deregulation of PI3K signaling and activation of AKT [22]. Mutations in PTEN are also associated with macrocephaly and autism spectrum disorder [23].
Many of the phenotypes seen as a result of PTEN loss in humans have been recapitulated in mice. Pten−/− mice die during embryonic development; however, Pten+/− mice are viable and develop neoplasms of the breast, prostate, adrenal medulla, endometrium, and intestine, as well as lymphomas. Tissue-specific deletion of Pten in mouse models results in cancers of the breast, prostate, lung, bladder, and pancreas [24]. Pten loss has also been shown to cooperate with a variety of mutations to promote tumor development. Deletion of Pten from the tumor microenvironment can also have pro-tumor effects on tumor cells that are wildtype for Pten [25], indicating that PTEN protein produced in one cell can act in neighboring cells.
Genetic alterations in PTEN have profound effects on development and disease; however, modes of PTEN protein regulation such as protein-protein interactions, post translational modifications, and secretion out of the cell, are proving to be equally important for maintaining proper cellular signaling. This review provides a summary of the ways in which PTEN protein function is regulated.
PTEN domains and structure
PTEN encodes a 403-amino acid peptide (Figure 3). The amino acid sequence contains the signature motif of the protein-tyrosine and dual specificity phosphatase catalytic domain, HCXXGXXRS/T, as well as homology to tensin and auxilin [1]. Analysis of the PTEN crystal structure, which includes amino acids 14–351, revealed that amino acids 7–179 make up the phosphatase domain. It contains a deep and wide catalytic pocket with a positive charge that is capable of accommodating phospholipid substrates. Amino acids 186–351 form a C2 domain that lacks the canonical Ca2+ chelating residues and binds to phospholipid membranes independent of calcium. The phosphatase and C2 domains together make up the minimal catalytic region of PTEN. Not included in the crystal structure are the N-terminal phosphatidylinositol-4,5-bisphosphate-binding domain (PBD), a loop in the C2 domain (286–309), and the C-terminal tail (amino acids 353–403) [27]. The PBD, which resides at the extreme N-terminus of PTEN, has been shown to be important for membrane localization and PTEN catalytic activity [28, 29], while the C-terminal tail, which contains a PDZ (PSD-95, Discs-large, ZO-1) binding domain, and is important for PTEN regulation and stability [30].
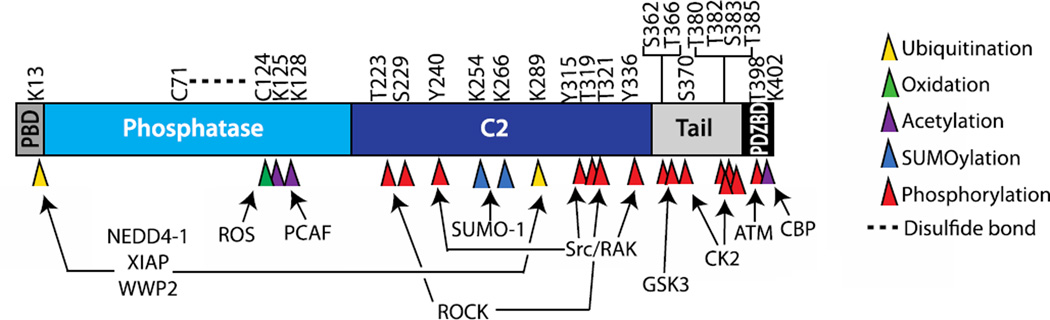
Figure 3.
Post translational modifications of PTEN. PTEN consists of a PIP2-binding domain (PBD), phosphatase domain, C2 domain, tail domain and a PDZ binding domain (PDZbd). Oxidation of PTEN at Cys124 leads to the formation of a disulfide bond with Cys71 (indicated by a broken line) resulting in decreased PTEN activity. PTEN is also acetylated at Lys125 and Lys128 by p300/CREB-binding protein (CBP)-associated factor (PCAF) and at Lys402 by CBP. Ubiquitination of PTEN at Lys13 and Lys 289 by NEDD4-1, X-linked inhibitor of apoptosis (XIAP) and WW domain-containing protein 2 (WWP2) regulates PTEN stability and cellular localization. PTEN SUMOylation at K254 and K266 is critical for PTEN tumor suppressive functions. Dynamic phosphorylation of multiple sites on the C-terminal region of PTEN affects protein stability, phosphatase activity, protein-protein interactions, and cleavage by caspase-3.
PTEN regulation by post-translational modifications
PTEN protein expression and phosphatase activity can be regulated through multiple post-translational modifications (Figure 3). Reversible oxidation of the catalytic cysteine (Cys) 124, such as by hydrogen peroxide treatment, causes the formation of a disulfide bond with Cys71 and results in the inactivation of PTEN phosphatase activity. Conversely, PTEN activity is increased following treatment with ROS scavengers in T-cell acute lymphoblastic leukemia cells [31, 32].
Acetylation has also been shown to regulate PTEN activity. p300/CREB-binding protein (CBP)-associated factor (PCAF, also KAT2B) acetylates PTEN in the catalytic cleft at lysine (Lys) 125 and Lys128. Acetylation of PTEN by PCAF results in decreased catalytic activity, increased AKT phosphorylation, and decreased PTEN-dependent G1 cell cycle arrest [33]. It has also been reported that PTEN is acetylated by CBP on Lys402, which is located in the PDZ binding domain. Acetylation at this site leads to increased binding of human discs large protein (hDLG) and membrane-associated guanylate kinase inverted-2 (MAGI-2) to PTEN, suggesting that PTEN acetylation by CBP regulates protein interactions involving the PDZ-binding domain of PTEN [34].
Ubiquitylation of PTEN also regulates PTEN by promoting protein degradation, nuclear localization, and inhibition of phosphatase activity. Polyubiquitylation by neural precursor cell expressed developmentally down-regulated protein 4 (NEDD4-1), the first identified E3 ubiquitin ligase for PTEN, results in PTEN protein degradation, whereas monoubiquitylation is important for PTEN nuclear transport [35, 36]. Despite this, there is limited evidence of increased Pten protein levels or changes in localization in NEDD4-1 knockout cells, suggesting that other E3 ligases are involved in PTEN ubiquitylation [37, 38]. This is further supported by studies showing that the E3 ligases X-linked inhibitor of apoptosis (XIAP) and WW domain-containing protein 2 (WWP2) can ubiquitylate PTEN [39, 40]. PTEN can also be ubiquitylated by the E3 ligase Ret finger protein (RFP, also TRIM27) at many lysine residues. Interestingly, ubiquitylation can also inhibit PTEN phosphatase activity without altering its stability or localization [41, 42]. Addition of Small Ubiquitin-like Modifiers (SUMO), or SUMOylation, of PTEN has also been implicated in regulating PTEN cellular localization. Specifically, covalent modifications of PTEN at Lys266, located in the C2 domain, enhances PTEN plasma membrane binding, reduces AKT activation and inhibits cellular transformation and tumor progression [43]. Studies have also shown that SUMOlyation at Lys254 is important for PTEN nuclear localization and homologous recombination based repair of DNA double strand breaks [43, 44].
Phosphorylation events at multiple sites in the C-terminal tail also regulate PTEN activity. Phosphorylation occurs mainly at Ser-366, Ser-370, and at a cluster containing Ser-380, Thr-382, Thr-383, and Ser-385 [30, 45]. Casein kinase 2 (CK2) can phosphorylate PTEN at Ser-370 and at the Ser-380-Ser-385 cluster, while GSK3 has been shown to phosphorylate Ser-362 and Thr-366 [46, 47]. There is also evidence of RhoA-associated kinase (ROCK) phosphorylation of Thr-223, Ser-229, Thr-319 and Thr-321, and Src family kinase mediated tyrosine phosphorylation at Tyr-240, Tyr-315 and Tyr-336 [48–51]. Interestingly, it appears that PTEN phosphorylation at specific sites can enhance phosphorylation at distant sites. For instance, phosphorylation at Ser-370 by CK2 primes PTEN for phosphorylation of Thr-366 by GSK3, while dephosphorylation of Ser-385 leads to reduced phosphorylation of Ser-380, Thr-382, and Thr-383 [47, 52, 53]. There is also evidence that PTEN can autodephosphorylate multiple C-terminal residues through its protein phosphatase activity [26, 54]. Mutational analysis of the Ser-380-Ser-385 cluster revealed that phosphorylation at these sites causes an increase of PTEN half-life but significantly decreases enzymatic activity[30]. Phosphorylation status of this cluster of residues is also important for PTEN’s ability to regulate neuronal spine density [54]. Conversely, Thr-366 phosphorylation leads to decreased PTEN stability and negatively regulates the ability of PTEN to block cellular invasion [26, 52]. PTEN phosphorylation has also been shown to regulate cellular localization, as the introduction of phospho-mimicking mutations at the Ser-380 – Ser-385 cluster results in decreased membrane binding [55]. Phosphorylation of Thr-383 also blocks the ability of PTEN to inhibit cell migration, suggesting that phosphorylation of the Ser-380-Ser385 cluster generally decreases PTEN function [10].
Further studies have revealed that PTEN C-terminal tail phosphorylation is critical for the formation of protein-protein interactions. It has been reported that an intramolecular interaction occurs between the C-terminal tail and the catalytic region of PTEN. Phosphorylation of the Ser-380-Ser-385 cluster is required for this interaction, resulting in a “closed” PTEN conformation that restricts PTEN recruitment to the plasma membrane and PIP3 access [53, 55]. Furthermore, this “closed” conformation appears to mask the PDZ binding domain, thereby blocking PTEN binding to PDZ domain containing proteins such as MAGI-2 [56, 57]. Phosphorylation of other sites outside of the Ser-380-Ser-385 cluster can also regulate protein binding. For instance, phosphorylation at Thr-366 is required for PTEN to bind to and inhibit the oncogene MSP58 [58]. Phosphorylation of the PTEN C-terminal tail also regulates cleavage of PTEN. It has been reported that PTEN is cleaved at multiple sites on the C-terminal tail by caspase-3, and this cleavage is inhibited by CK2-mediated PTEN phosphorylation at Ser-370 and Ser-385. The resulting PTEN cleavage products display decreased stability as well as decreased binding to interacting proteins[59].
Phosphorylation of PTEN can be dynamically regulated by various upstream signaling events. Studies of hypothalamic cells, pancreatic β cells, and hippocampal neurons have shown that the hormone leptin, which plays a key role in the regulation of energy expenditure, can stimulate the phosphorylation of PTEN at multiple sites on the C-terminal tail leading to a reduction in PTEN activity [60, 61]. Phosphorylation of the C-terminal tail is also increased following receptor activation of T-cells. The cell surface protein Programmed cell death-1 (PD-1), which inhibits the T-cell response, reduces PTEN phosphorylation and increases PTEN activity through inhibition of CK2 [62]. Genotoxic stress has also been implicated in the phosphorylation of PTEN at Thr-398 by ataxia telangiectasia mutated (ATM) [44]. Alterations in PTEN phosphorylation status have also been linked to cancer development. For example, PTEN phosphorylation appears to be upregulated in leukemia cancer cells compared to normal thymocytes through decreased CK2 expression and activity [32]. Increased PTEN phosphorylation at Ser-380 has also been observed in gastric tumors, and Notch signaling appears to regulate PTEN phosphorylation in pancreatic cancer cells [63, 64]. This provides a mechanism of PTEN inactivation in cancers where no loss of PTEN expression is observed.
Regulation of PTEN by protein-protein interactions
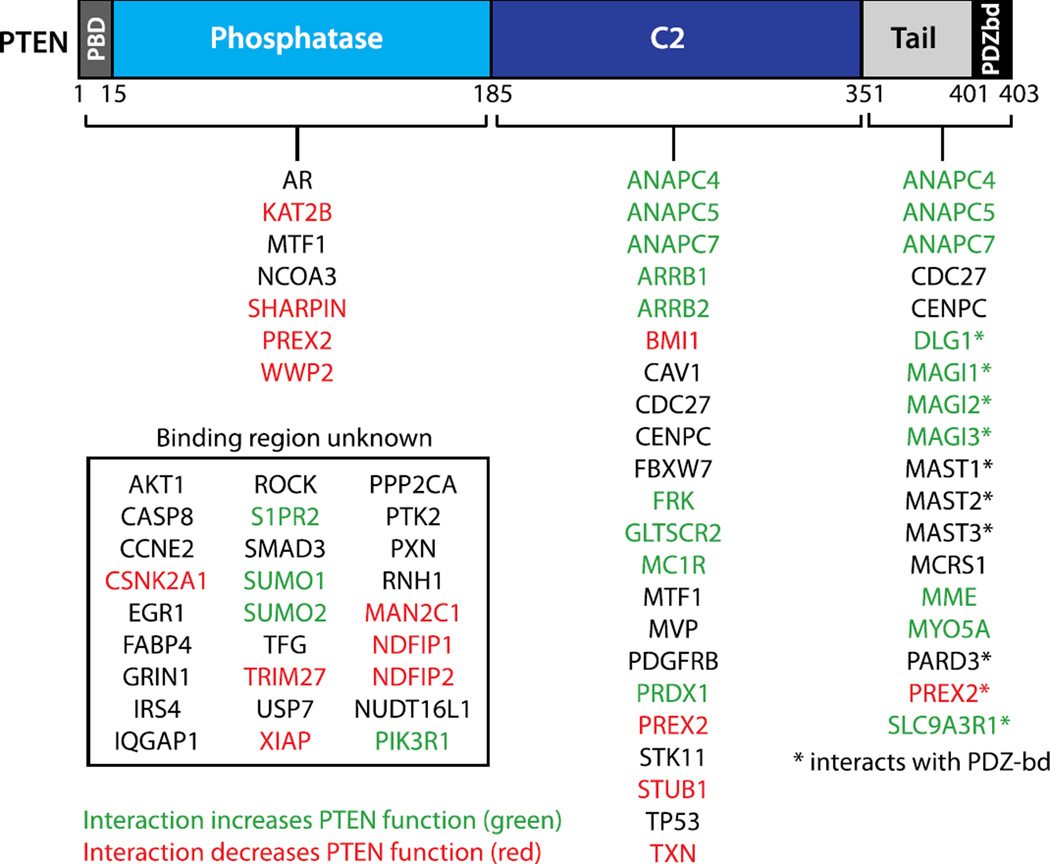
Numerous PTEN-interacting proteins have been discovered (Supplemental Table 1 and Figure 4), and it is becoming increasingly clear that the tumor suppressive functions of PTEN can be regulated by its binding partners. Protein-protein interactions likely help to control PTEN activity during normal development, and such regulatory interactions may go awry in cancer and other pathologic states. Interacting proteins can influence PTEN function through various mechanisms, including mislocalization, altered protein stability or conformation, and regulation of phosphatase activity (Supplemental Table 1).
Figure 4.
PTEN interacting proteins. The binding regions for PTEN-interacting proteins are shown. PTEN-interactors for which the sites of PTEN binding are unknown are listed in the inset box. PTEN-interactors that increase PTEN function are shown in green, and interactors that decrease PTEN function are shown in red.
Many interacting partners can activate PTEN, and this occurs through a variety of mechanisms. One such mechanism is to increase PTEN stability. Two examples that have potential cancer relevance are the interactions with melanocortin-1 receptor (MC1R) and the Fyn-Related Kinase (FRK), both of which interfere with PTEN ubiquitylation [51, 65]. MC1R is primarily expressed in melanocytes and is thought to play a significant role in melanoma development. It is able to bind to PTEN and prevent ubiquitylation by the E3 ligase WWP2, thereby preventing PTEN degradation. A series of mutations have been identified in MC1R that confer susceptibility to melanoma. Interestingly, these MC1R variants are unable to bind PTEN, resulting in higher activation of the PI3K pathway upon UV exposure, and cooperation with BRAF-V600E to induce melanomagenesis in melanocytes [65]. Similarly, FRK is a tyrosine kinase that has been shown to phosphorylate PTEN and antagonize NEDD4-1 ubiquitylation of PTEN, most likely by interfering with NEDD4-1 binding. Overexpression of FRK in human breast cancer cells reduces invasion and anchorage independent growth, while knockdown increases anchorage independent growth and promotes tumor formation in a mouse xenograft model [51]. Consistent with their potential tumor suppressive functions as PTEN activators, analysis of the TCGA cBio portal revealed that these two genes are deleted in various tumor types. The highest rate of deletion for both proteins is in prostate cancer, where it was reported that MC1R and FRK were deleted in 8.5% and 7.3% of cases, respectively [3, 4, 66].
Protein-protein interactions can also modulate PTEN activation through regulation of PTEN localization. For example, the scaffolding proteins Membrane Associated Guanylate Kinase, WW and PDZ Domain Containing 1b (MAG1b), MAGI2, and MAGI3 facilitate the lipid phosphatase activity of PTEN by binding to the C-terminal PDZ binding domain of PTEN and recruiting it to signaling complexes at the membrane [56, 67–69]. RhoA-dependent activation of PTEN at the posterior edge of a chemotaxing cell also requires PTEN recruitment to the membrane. This regulation is likely occurring through the RhoA-dependent kinase, ROCK1, which is able to bind and phosphorylate PTEN [50]. RhoA activation also leads to the interaction of PTEN with β-arrestins, which are scaffolding proteins that activate PTEN phosphatase activity. Interestingly, β-arrestins were also found to block the inhibitory effect of PTEN on cell migration, pointing to differential regulation of multiple PTEN functions [66]. Additionally, PTEN is recruited to platelet derived growth factor receptor (PDGFR) at the membrane by the adaptor protein Na+/H+ exchanger regulatory factor (NHERF, also SLC9A3R1), dampening the activation of the pathway [70]. PTEN is also activated downstream of epidermal growth factor (EGF), which stimulates the binding of p85 to PTEN, activating the lipid phosphatase activity [71]. Ovarian cancer-associated mutations in PIK3R1/2, the gene encoding p85, disrupt p85 binding and destablize PTEN, resulting in an increase in AKT phosphorylation [72]. Both of these studies suggest that the p85-PTEN interaction is important for the ability of PTEN to antagonize PI3K signaling. Lastly, neutral endopeptidase (NEP) regulates both PTEN membrane recruitment and stability to increase PTEN activity in prostate cancer cells [73].
Several proteins that bind PTEN and inhibit its lipid phosphatase activity have been discovered. It has been shown that one such PTEN-interactor is PtdIns(3,4,5)P3-dependent RAC exchanger factor 2a (PREX2a), a widely-expressed guanine nucleotide exchange factor for RAC GTPase [74]. PREX2a binds PTEN via two interfaces: the PREX2a IP4P domain binds tightly to the PTEN PDZ-binding domain, while the PREX2a DHPH domain interacts with the catalytic phosphatase and C2 domain of PTEN and inhibits PTEN lipid phosphatase activity [75]. Thus, PREX2a amplifies PI3K signaling and may be co-opted in cancer as a means to inhibit PTEN function. PREX2a is upregulated in various cancers and cooperates with mutant PI3K to transform human breast cells [74]. Furthermore, sequencing analyses reveal that PREX2a is mutated or amplified in more than 25% of melanomas [4], and PREX2 mutants have been shown to accelerate tumor formation of immortalized melanocytes [76].
Another negative regulator of PTEN is shank-interacting protein-like 1 (SHARPIN), a member of the NF-ĸB-activating linear ubiquitin chain assembly complex [77]. SHARPIN binds strongly to the PTEN catalytic domain but also interacts weakly with the PTEN C-terminus [78]. The ubiquitin-like domain of SHARPIN is sufficient to attenuate PTEN lipid phosphatase activity and enhance PI3K signaling, and SHARPIN expression enhances xenograft tumor formation through inhibition of PTEN function [78]. PTEN is co-localized with SHARPIN in PTEN-positive human primary cervical cancer tissues, and SHARPIN expression is associated with increased AKT activation in cervical cancer [78]. Thus, SHARPIN may contribute to oncogenesis via suppression of PTEN activity.
Cytosolic α-mannosidase 2C1 (MAN2C1) is another example of a PTEN-interacting protein that interferes with PTEN action. MAN2C1 functions as a catabolic enzyme for the breakdown of free oligosaccharides, and new evidence suggests that it has a separate role as a regulator of apoptosis [79]. In cancer, overexpression of MAN2C1 promotes tumor growth and metastasis, while down-regulation of MAN2C1 delays growth and induces apoptosis [79–82]. MAN2C1 binds PTEN and inhibits its lipid phosphatase activity, however MAN2C1 may also impair PTEN function by preventing its recruitment to the cell membrane [83]. Furthermore, MAN2C1 is up-regulated in PTEN-positive prostate cancers and increased MAN2C1 expression is associated with AKT activation and elevated risk of prostate cancer recurrence [83]. Thus, in the absence of PTEN mutation, up-regulation of MAN2C1 may be one mechanism by which tumors down-regulate PTEN function.
While protein interactions often regulate the activity of PTEN, it is also true that PTEN can regulate the function of its binding partners via these interactions. For example, in the nucleus PTEN has been shown to interact with four different components of APC/C (Anaphase Promoting Complex/Cyclosome), which is an E3 ubiquitin ligase that regulates the degradation of many important proteins in the cell cycle pathway [84]. PTEN was found to promote the binding of APC/C to CDH1, which is critical for the ubiquitinase activity of the complex. In addition, the PTEN-APC/C interactions appear to play a role in the ability of PTEN to suppress cell growth and in the induction of cellular senescence that occurs as a result of PTEN loss. Interestingly, the effect of PTEN on APC/C occurs independently of its phosphatase activity [84].
In sum, it is apparent that protein-protein interactions play important roles in controlling PTEN function. Many PTEN binding partners are mutated in cancer, which may disrupt PTEN function and promote cancer development. For example, PIK3R1 and PREX2, two PTEN interactors that affect PTEN lipid phosphatase activity, are mutated in approximately 33% of uterine tumors and 24% of melanomas, respectively [3, 4, 71, 74]. Thus, these interacting proteins may exert oncogenic or tumor suppressive effects, depending on whether they enhance or restrain PTEN function.
PTEN-LONG: a secreted PTEN
Regulation of the intracellular localization and activity of PTEN has been well described. However, several studies have now demonstrated that PTEN is able to exit and exist outside the cell [85, 86]. One study shows that the canonical PTEN protein is packaged into exosomes and is transferred from one cell to another by way of vesicles [85]. Another study identified a translational variant of PTEN, named PTEN-Long, which is translated from an alternate start site within the 5’ region of PTEN mRNA. The alternatively translated region (ATR) of PTEN-Long adds an additional 173 N-terminal amino acids to the canonical protein, and the PTEN-Long ATR is evolutionarily conserved [86]. PTEN-Long is secreted and can be detected in human serum and plasma. Furthermore, the PTEN-Long ATR contains a polyarginine stretch with homology to known cell permeable peptides, which allows it to enter cells and inhibit PI3K signaling both in vitro and in vivo (Figure 5). Thus, PTEN-Long can act like a therapeutic agent and cause tumor regression in several mouse tumor models. PTEN-Long activity may share the same mechanisms of regulation as canonical PTEN, including the post-translational modifications and protein-protein interactions mentioned above. However, alternative mechanisms of regulation may exist. Cellular control of PTEN-Long translation and trafficking are likely to be tightly regulated and complex processes. Furthermore, interacting proteins may exist that bind to the PTEN-Long ATR and therefore specifically regulate PTEN-Long function. This is supported by Malaney et al, who performed computational analysis of the PTEN-Long ATR and predicted that it is largely unstructured, and may have the potential to interact with a wide array of other proteins [87].
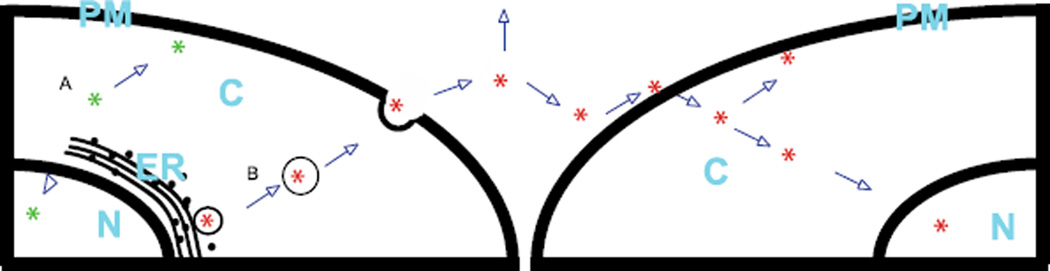
Figure 5.
Trafficking of PTEN-LONG. (A) PTEN-LONG (Green *) is translated and can act in its cell of origin in a manner similar to canonical PTEN. PTEN-Long can act in the nucleus ‘N’, cytoplasm ‘C,’ or at the plasma membrane ‘PM.’ (B) Translation of PTEN-Long (Red *) occurs at the endoplasmic reticulum ‘ER’ and the synthesized protein is transported in the lumen of secretory vesicle. These vesicles then travel to the membrane where they fuse and release PTEN-LONG into the extracellular space. Once outside the cell, PTEN-LONG can interact with extracellular proteins and lipids, as well as with heparinated glycoproteins on the cell surface in order to enter cells. Once inside the cell, PTEN-LONG can act much like canonical PTEN and traffic within the cytoplasm or migrate to the nucleus, therefore increasing the intracellular dose of PTEN in the recipient cell.
Concluding Remarks
Given the importance of PTEN in normal development and disease, regulating the location and level of active PTEN in a cell is critical for maintaining proper homeostasis. Many of the mechanisms of PTEN regulation involve post-translational modifications and protein-protein interactions. However, new evidence showing that PTEN is present in different cellular and extracellular compartments suggests that novel modes of regulation may exist that have not yet been identified. Understanding the myriad of ways in which PTEN activity can be regulated remains a critical avenue for future studies, as it is through these mechanisms that we may come to understand how to modulate PTEN activity for therapeutic benefit.
Supplementary Material
PTEN interacting proteins and the PTEN domain to which they bind. References to the papers that describe these interactions are listed.
Highlights.
PTEN is an antagonist of PI3K signaling.
PTEN is regulated by PTMs that affect its localization, stability and activity.
Many proteins interact with PTEN, both positively and negatively regulating its activity.
PTEN can be secreted and can enter cells from the outside thereby changing the effective dose of PTEN.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li J, et al. PTEN, a Putative Protein Tyrosine Phosphatase Gene Mutated in Human Brain, Breast, and Prostate Cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 2.Steck PA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nature genetics. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 3.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li DM, Sun H. TEP1, encoded by a candidate tumor suppressor locus, is a novel protein tyrosine phosphatase regulated by transforming growth factor beta. Cancer research. 1997;57:2124–2129. [PubMed] [Google Scholar]
- 6.Myers MP, et al. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. The Journal of biological chemistry. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 8.Stambolic V, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 9.Leslie NR, et al. PtdIns(3,4,5)P(3)-dependent and -independent roles for PTEN in the control of cell migration. Current biology : CB. 2007;17:115–125. doi: 10.1016/j.cub.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raftopoulou M, et al. Regulation of cell migration by the C2 domain of the tumor suppressor PTEN. Science. 2004;303:1179–1181. doi: 10.1126/science.1092089. [DOI] [PubMed] [Google Scholar]
- 11.Dey N, et al. The protein phosphatase activity of PTEN regulates SRC family kinases and controls glioma migration. Cancer research. 2008;68:1862–1871. doi: 10.1158/0008-5472.CAN-07-1182. [DOI] [PubMed] [Google Scholar]
- 12.Tamura M, et al. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science. 1998;280:1614–1617. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- 13.Song MS, et al. The functions and regulation of the PTEN tumour suppressor. Nature reviews. Molecular cell biology. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 14.Cully M, et al. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nature reviews. Cancer. 2006;6:184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 15.Mayo LD, Donner DB. The PTEN, Mdm2, p53 tumor suppressor-oncoprotein network. Trends in biochemical sciences. 2002;27:462–467. doi: 10.1016/s0968-0004(02)02166-7. [DOI] [PubMed] [Google Scholar]
- 16.Alimonti A, et al. Subtle variations in Pten dose determine cancer susceptibility. Nature genetics. 2010;42:454–458. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen-Li H, et al. Reduction of Pten dose leads to neoplastic development in multiple organs of Pten (shRNA) mice. Cancer biology & therapy. 2010;10:1194–1200. doi: 10.4161/cbt.10.11.13814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liaw D, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nature genetics. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 19.Arch EM, et al. Deletion of PTEN in a patient with Bannayan-Riley-Ruvalcaba syndrome suggests allelism with Cowden disease. American journal of medical genetics. 1997;71:489–493. [PubMed] [Google Scholar]
- 20.Zhou X, et al. Association of germline mutation in the PTEN tumour suppressor gene and Proteus and Proteus-like syndromes. Lancet. 2001;358:210–211. doi: 10.1016/s0140-6736(01)05412-5. [DOI] [PubMed] [Google Scholar]
- 21.Marsh DJ, et al. Allelic imbalance, including deletion of PTEN/MMACI, at the Cowden disease locus on 10q22-23, in hamartomas from patients with Cowden syndrome and germline PTEN mutation. Genes, chromosomes & cancer. 1998;21:61–69. doi: 10.1002/(sici)1098-2264(199801)21:1<61::aid-gcc8>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 22.Zhou XP, et al. Germline inactivation of PTEN and dysregulation of the phosphoinositol-3-kinase/Akt pathway cause human Lhermitte-Duclos disease in adults. American journal of human genetics. 2003;73:1191–1198. doi: 10.1086/379382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butler MG, et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. Journal of medical genetics. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hollander MC, et al. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nature reviews. Cancer. 2011;11:289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trimboli AJ, et al. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature. 2009;461:1084–1091. doi: 10.1038/nature08486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tibarewal P, et al. PTEN protein phosphatase activity correlates with control of gene expression and invasion, a tumor-suppressing phenotype, but not with AKT activity. Science signaling. 2012;5:ra18. doi: 10.1126/scisignal.2002138. [DOI] [PubMed] [Google Scholar]
- 27.Lee JO, et al. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 28.Walker SM, et al. The tumour-suppressor function of PTEN requires an N-terminal lipid-binding motif. The Biochemical journal. 2004;379:301–307. doi: 10.1042/BJ20031839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campbell RB, et al. Allosteric activation of PTEN phosphatase by phosphatidylinositol 4,5-bisphosphate. The Journal of biological chemistry. 2003;278:33617–33620. doi: 10.1074/jbc.C300296200. [DOI] [PubMed] [Google Scholar]
- 30.Vazquez F, et al. Phosphorylation of the PTEN tail regulates protein stability and function. Molecular and cellular biology. 2000;20:5010–5018. doi: 10.1128/mcb.20.14.5010-5018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee SR, et al. Reversible inactivation of the tumor suppressor PTEN by H2O2. The Journal of biological chemistry. 2002;277:20336–20342. doi: 10.1074/jbc.M111899200. [DOI] [PubMed] [Google Scholar]
- 32.Silva A, et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability. The Journal of clinical investigation. 2008;118:3762–3774. doi: 10.1172/JCI34616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okumura K, et al. PCAF modulates PTEN activity. The Journal of biological chemistry. 2006;281:26562–26568. doi: 10.1074/jbc.M605391200. [DOI] [PubMed] [Google Scholar]
- 34.Ikenoue T, et al. PTEN acetylation modulates its interaction with PDZ domain. Cancer research. 2008;68:6908–6912. doi: 10.1158/0008-5472.CAN-08-1107. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trotman LC, et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128:141–156. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fouladkou F, et al. The ubiquitin ligase Nedd4-1 is dispensable for the regulation of PTEN stability and localization. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:8585–8590. doi: 10.1073/pnas.0803233105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drinjakovic J, et al. E3 ligase Nedd4 promotes axon branching by downregulating PTEN. Neuron. 2010;65:341–357. doi: 10.1016/j.neuron.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Themsche C, et al. X-linked inhibitor of apoptosis protein (XIAP) regulates PTEN ubiquitination, content, and compartmentalization. The Journal of biological chemistry. 2009;284:20462–20466. doi: 10.1074/jbc.C109.009522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maddika S, et al. WWP2 is an E3 ubiquitin ligase for PTEN. Nature cell biology. 2011;13:728–733. doi: 10.1038/ncb2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JT, et al. RFP-mediated ubiquitination of PTEN modulates its effect on AKT activation. Cell research. 2013;23:552–564. doi: 10.1038/cr.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maccario H, et al. Ubiquitination of PTEN (phosphatase and tensin homolog) inhibits phosphatase activity and is enhanced by membrane targeting and hyperosmotic stress. The Journal of biological chemistry. 2010;285:12620–12628. doi: 10.1074/jbc.M109.072280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang J, et al. SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane. Nature communications. 2012;3:911. doi: 10.1038/ncomms1919. [DOI] [PubMed] [Google Scholar]
- 44.Bassi C, et al. Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science. 2013;341:395–399. doi: 10.1126/science.1236188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller SJ, et al. Direct identification of PTEN phosphorylation sites. FEBS letters. 2002;528:145–153. doi: 10.1016/s0014-5793(02)03274-x. [DOI] [PubMed] [Google Scholar]
- 46.Torres J, Pulido R. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. The Journal of biological chemistry. 2001;276:993–998. doi: 10.1074/jbc.M009134200. [DOI] [PubMed] [Google Scholar]
- 47.Al-Khouri AM, et al. Cooperative phosphorylation of the tumor suppressor phosphatase and tensin homologue (PTEN) by casein kinases and glycogen synthase kinase 3beta. The Journal of biological chemistry. 2005;280:35195–35202. doi: 10.1074/jbc.M503045200. [DOI] [PubMed] [Google Scholar]
- 48.Koul D, et al. Motif analysis of the tumor suppressor gene MMAC/PTEN identifies tyrosines critical for tumor suppression and lipid phosphatase activity. Oncogene. 2002;21:2357–2364. doi: 10.1038/sj.onc.1205296. [DOI] [PubMed] [Google Scholar]
- 49.Lu Y, et al. Src family protein-tyrosine kinases alter the function of PTEN to regulate phosphatidylinositol 3-kinase/AKT cascades. The Journal of biological chemistry. 2003;278:40057–40066. doi: 10.1074/jbc.M303621200. [DOI] [PubMed] [Google Scholar]
- 50.Li Z, et al. Regulation of PTEN by Rho small GTPases. Nature cell biology. 2005;7:399–404. doi: 10.1038/ncb1236. [DOI] [PubMed] [Google Scholar]
- 51.Yim EK, et al. Rak functions as a tumor suppressor by regulating PTEN protein stability and function. Cancer cell. 2009;15:304–314. doi: 10.1016/j.ccr.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maccario H, et al. PTEN is destabilized by phosphorylation on Thr366. The Biochemical journal. 2007;405:439–444. doi: 10.1042/BJ20061837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Odriozola L, et al. Regulation of PTEN activity by its carboxyl-terminal autoinhibitory domain. The Journal of biological chemistry. 2007;282:23306–23315. doi: 10.1074/jbc.M611240200. [DOI] [PubMed] [Google Scholar]
- 54.Zhang XC, et al. Functional analysis of the protein phosphatase activity of PTEN. The Biochemical journal. 2012;444:457–464. doi: 10.1042/BJ20120098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rahdar M, et al. A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:480–485. doi: 10.1073/pnas.0811212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tolkacheva T, et al. Regulation of PTEN binding to MAGI-2 by two putative phosphorylation sites at threonine 382 and 383. Cancer research. 2001;61:4985–4989. [PubMed] [Google Scholar]
- 57.Vazquez F, et al. Phosphorylation of the PTEN tail acts as an inhibitory switch by preventing its recruitment into a protein complex. The Journal of biological chemistry. 2001;276:48627–48630. doi: 10.1074/jbc.C100556200. [DOI] [PubMed] [Google Scholar]
- 58.Okumura K, et al. Cellular transformation by the MSP58 oncogene is inhibited by its physical interaction with the PTEN tumor suppressor. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2703–2706. doi: 10.1073/pnas.0409370102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Torres J, et al. Phosphorylation-regulated cleavage of the tumor suppressor PTEN by caspase-3: implications for the control of protein stability and PTEN-protein interactions. The Journal of biological chemistry. 2003;278:30652–30660. doi: 10.1074/jbc.M212610200. [DOI] [PubMed] [Google Scholar]
- 60.Ning K, et al. A novel leptin signalling pathway via PTEN inhibition in hypothalamic cell lines and pancreatic beta-cells. The EMBO journal. 2006;25:2377–2387. doi: 10.1038/sj.emboj.7601118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moult PR, et al. Leptin regulates AMPA receptor trafficking via PTEN inhibition. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:4088–4101. doi: 10.1523/JNEUROSCI.3614-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patsoukis N, et al. PD-1 increases PTEN phosphatase activity while decreasing PTEN protein stability by inhibiting casein kinase 2. Molecular and cellular biology. 2013;33:3091–3098. doi: 10.1128/MCB.00319-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang Z, et al. Reduced expression of PTEN and increased PTEN phosphorylation at residue Ser380 in gastric cancer tissues: a novel mechanism of PTEN inactivation. Clinics and research in hepatology and gastroenterology. 2013;37:72–79. doi: 10.1016/j.clinre.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 64.Vo K, et al. Targeting notch pathway enhances rapamycin antitumor activity in pancreas cancers through PTEN phosphorylation. Molecular cancer. 2011;10:138. doi: 10.1186/1476-4598-10-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cao J, et al. MC1R is a potent regulator of PTEN after UV exposure in melanocytes. Molecular cell. 2013;51:409–422. doi: 10.1016/j.molcel.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lima-Fernandes E, et al. Distinct functional outputs of PTEN signalling are controlled by dynamic association with beta-arrestins. The EMBO journal. 2011;30:2557–2568. doi: 10.1038/emboj.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu X, et al. Evidence for regulation of the PTEN tumor suppressor by a membrane-localized multi-PDZ domain containing scaffold protein MAGI-2. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:4233–4238. doi: 10.1073/pnas.97.8.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu Y, et al. Interaction of the tumor suppressor PTEN/MMAC with a PDZ domain of MAGI3, a novel membrane-associated guanylate kinase. The Journal of biological chemistry. 2000;275:21477–21485. doi: 10.1074/jbc.M909741199. [DOI] [PubMed] [Google Scholar]
- 69.Kotelevets L, et al. Implication of the MAGI-1b/PTEN signalosome in stabilization of adherens junctions and suppression of invasiveness. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2005;19:115–117. doi: 10.1096/fj.04-1942fje. [DOI] [PubMed] [Google Scholar]
- 70.Takahashi Y, et al. PTEN tumor suppressor associates with NHERF proteins to attenuate PDGF receptor signaling. The EMBO journal. 2006;25:910–920. doi: 10.1038/sj.emboj.7600979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chagpar RB, et al. Direct positive regulation of PTEN by the p85 subunit of phosphatidylinositol 3-kinase. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5471–5476. doi: 10.1073/pnas.0908899107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheung LW, et al. High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer discovery. 2011;1:170–185. doi: 10.1158/2159-8290.CD-11-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sumitomo M, et al. Synergy in tumor suppression by direct interaction of neutral endopeptidase with PTEN. Cancer cell. 2004;5:67–78. doi: 10.1016/s1535-6108(03)00331-3. [DOI] [PubMed] [Google Scholar]
- 74.Fine B, et al. Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science. 2009;325:1261–1265. doi: 10.1126/science.1173569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hodakoski C, et al. Regulation of PTEN Inhibition by the pleckstrin homology domain of PREX2 during insulin signaling and glucose homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2014 doi: 10.1073/pnas.1213773111. IN PRESS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Berger MF, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ikeda F, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.He L, et al. Shank-interacting protein-like 1 promotes tumorigenesis via PTEN inhibition in human tumor cells. The Journal of clinical investigation. 2010;120:2094–2108. doi: 10.1172/JCI40778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang L, Suzuki T. Dual functions for cytosolic alpha-mannosidase (Man2C1): its down-regulation causes mitochondria-dependent apoptosis independently of its alpha-mannosidase activity. The Journal of biological chemistry. 2013;288:11887–11896. doi: 10.1074/jbc.M112.425702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yue W, et al. Suppression of 6A8 alpha-mannosidase gene expression reduced the potentiality of growth and metastasis of human nasopharyngeal carcinoma. International journal of cancer. Journal international du cancer. 2004;108:189–195. doi: 10.1002/ijc.11536. [DOI] [PubMed] [Google Scholar]
- 81.Tian Y, et al. Inhibition of alpha-mannosidase Man2c1 gene expression suppresses growth of esophageal carcinoma cells through mitotic arrest and apoptosis. Cancer science. 2008;99:2428–2434. doi: 10.1111/j.1349-7006.2008.01019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiang ZG, et al. hMan2c1 transgene promotes tumor progress in mice. Transgenic research. 2010;19:67–75. doi: 10.1007/s11248-009-9299-3. [DOI] [PubMed] [Google Scholar]
- 83.He L, et al. alpha-Mannosidase 2C1 attenuates PTEN function in prostate cancer cells. Nature communications. 2011;2:307. doi: 10.1038/ncomms1309. [DOI] [PubMed] [Google Scholar]
- 84.Song MS, et al. Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a phosphatase-independent manner. Cell. 2011;144:187–199. doi: 10.1016/j.cell.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Putz U, et al. The tumor suppressor PTEN is exported in exosomes and has phosphatase activity in recipient cells. Science signaling. 2012;5:ra70. doi: 10.1126/scisignal.2003084. [DOI] [PubMed] [Google Scholar]
- 86.Hopkins BD, et al. A secreted PTEN phosphatase that enters cells to alter signaling and survival. Science. 2013;341:399–402. doi: 10.1126/science.1234907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Malaney P, et al. The PTEN Long N-tail is intrinsically disordered: increased viability for PTEN therapy. Molecular bioSystems. 2013;9:2877–2888. doi: 10.1039/c3mb70267g. [DOI] [PubMed] [Google Scholar]
- 88.Mistafa O, et al. Purinergic receptor-mediated rapid depletion of nuclear phosphorylated Akt depends on pleckstrin homology domain leucine-rich repeat phosphatase, calcineurin, protein phosphatase 2A, and PTEN phosphatases. The Journal of biological chemistry. 2010;285:27900–27910. doi: 10.1074/jbc.M110.117093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lin HK, et al. Regulation of androgen receptor signaling by PTEN (phosphatase and tensin homolog deleted on chromosome 10) tumor suppressor through distinct mechanisms in prostate cancer cells. Mol Endocrinol. 2004;18:2409–2423. doi: 10.1210/me.2004-0117. [DOI] [PubMed] [Google Scholar]
- 90.Fan C, et al. PTEN inhibits BMI1 function independently of its phosphatase activity. Molecular cancer. 2009;8:98. doi: 10.1186/1476-4598-8-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Crockett DK, et al. Analysis of phosphatase and tensin homolog tumor suppressor interacting proteins by in vitro and in silico proteomics. Proteomics. 2005;5:1250–1262. doi: 10.1002/pmic.200401046. [DOI] [PubMed] [Google Scholar]
- 92.Caselli A, et al. Some protein tyrosine phosphatases target in part to lipid rafts and interact with caveolin-1. Biochemical and biophysical research communications. 2002;296:692–697. doi: 10.1016/s0006-291x(02)00928-2. [DOI] [PubMed] [Google Scholar]
- 93.Shen WH, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 94.Sotelo NS, et al. A functional network of the tumor suppressors APC, hDlg, and PTEN, that relies on recognition of specific PDZ-domains. Journal of cellular biochemistry. 2012;113:2661–2670. doi: 10.1002/jcb.24141. [DOI] [PubMed] [Google Scholar]
- 95.Adey NB, et al. Threonine phosphorylation of the MMAC1/PTEN PDZ binding domain both inhibits and stimulates PDZ binding. Cancer research. 2000;60:35–37. [PubMed] [Google Scholar]
- 96.Lu D, et al. Microsomal prostaglandin E synthase-1 inhibits PTEN and promotes experimental cholangiocarcinogenesis and tumor progression. Gastroenterology. 2011;140:2084–2094. doi: 10.1053/j.gastro.2011.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gorbenko O, et al. Identification of novel PTEN-binding partners: PTEN interaction with fatty acid binding protein FABP4. Molecular and cellular biochemistry. 2010;337:299–305. doi: 10.1007/s11010-009-0312-1. [DOI] [PubMed] [Google Scholar]
- 98.Yang C, et al. PTEN suppresses the oncogenic function of AIB1 through decreasing its protein stability via mechanism involving Fbw7 alpha. Molecular cancer. 2013;12:21. doi: 10.1186/1476-4598-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Okahara F, et al. Regulation of PTEN phosphorylation and stability by a tumor suppressor candidate protein. The Journal of biological chemistry. 2004;279:45300–45303. doi: 10.1074/jbc.C400377200. [DOI] [PubMed] [Google Scholar]
- 100.Jurado S, et al. PTEN is recruited to the postsynaptic terminal for NMDA receptor-dependent long-term depression. The EMBO journal. 2010;29:2827–2840. doi: 10.1038/emboj.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gunaratne J, et al. Protein interactions of phosphatase and tensin homologue (PTEN) and its cancer-associated G20E mutant compared by using stable isotope labeling by amino acids in cell culture-based parallel affinity purification. The Journal of biological chemistry. 2011;286:18093–18103. doi: 10.1074/jbc.M111.221184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Valiente M, et al. Binding of PTEN to specific PDZ domains contributes to PTEN protein stability and phosphorylation by microtubule-associated serine/threonine kinases. The Journal of biological chemistry. 2005;280:28936–28943. doi: 10.1074/jbc.M504761200. [DOI] [PubMed] [Google Scholar]
- 103.Lin MC, et al. PTEN interacts with metal-responsive transcription factor 1 and stimulates its transcriptional activity. The Biochemical journal. 2012;441:367–377. doi: 10.1042/BJ20111257. [DOI] [PubMed] [Google Scholar]
- 104.Yu Z, et al. PTEN associates with the vault particles in HeLa cells. The Journal of biological chemistry. 2002;277:40247–40252. doi: 10.1074/jbc.M207608200. [DOI] [PubMed] [Google Scholar]
- 105.van Diepen MT, et al. MyosinV controls PTEN function and neuronal cell size. Nature cell biology. 2009;11:1191–1196. doi: 10.1038/ncb1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Howitt J, et al. Ndfip1 regulates nuclear Pten import in vivo to promote neuronal survival following cerebral ischemia. The Journal of cell biology. 2012;196:29–36. doi: 10.1083/jcb.201105009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mund T, Pelham HR. Regulation of PTEN/Akt and MAP kinase signaling pathways by the ubiquitin ligase activators Ndfip1 and Ndfip2. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11429–11434. doi: 10.1073/pnas.0911714107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.von Stein W, et al. Direct association of Bazooka/PAR-3 with the lipid phosphatase PTEN reveals a link between the PAR/aPKC complex and phosphoinositide signaling. Development. 2005;132:1675–1686. doi: 10.1242/dev.01720. [DOI] [PubMed] [Google Scholar]
- 109.Mahimainathan L, Choudhury GG. Inactivation of platelet-derived growth factor receptor by the tumor suppressor PTEN provides a novel mechanism of action of the phosphatase. The Journal of biological chemistry. 2004;279:15258–15268. doi: 10.1074/jbc.M314328200. [DOI] [PubMed] [Google Scholar]
- 110.Cao J, et al. Prdx1 inhibits tumorigenesis via regulating PTEN/AKT activity. The EMBO journal. 2009;28:1505–1517. doi: 10.1038/emboj.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Haier J, Nicolson GL. PTEN regulates tumor cell adhesion of colon carcinoma cells under dynamic conditions of fluid flow. Oncogene. 2002;21:1450–1460. doi: 10.1038/sj.onc.1205213. [DOI] [PubMed] [Google Scholar]
- 112.Tamura M, et al. PTEN interactions with focal adhesion kinase and suppression of the extracellular matrix-dependent phosphatidylinositol 3-kinase/Akt cell survival pathway. The Journal of biological chemistry. 1999;274:20693–20703. doi: 10.1074/jbc.274.29.20693. [DOI] [PubMed] [Google Scholar]
- 113.Kim YJ, et al. PTEN modulates miR-21 processing via RNA-regulatory protein RNH1. PloS one. 2011;6:e28308. doi: 10.1371/journal.pone.0028308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sanchez T, et al. PTEN as an effector in the signaling of antimigratory G protein-coupled receptor. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4312–4317. doi: 10.1073/pnas.0409784102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yang L, et al. Na(+)/H(+) exchanger regulatory factor 1 (NHERF1) is required for the estradiol-dependent increase of phosphatase and tensin homolog (PTEN) protein expression. Endocrinology. 2011;152:4537–4549. doi: 10.1210/en.2011-1207. [DOI] [PubMed] [Google Scholar]
- 116.Hjelmeland AB, et al. Loss of phosphatase and tensin homologue increases transforming growth factor beta-mediated invasion with enhanced SMAD3 transcriptional activity. Cancer research. 2005;65:11276–11281. doi: 10.1158/0008-5472.CAN-05-3016. [DOI] [PubMed] [Google Scholar]
- 117.Mehenni H, et al. LKB1 interacts with and phosphorylates PTEN: a functional link between two proteins involved in cancer predisposing syndromes. Human molecular genetics. 2005;14:2209–2219. doi: 10.1093/hmg/ddi225. [DOI] [PubMed] [Google Scholar]
- 118.Ahmed SF, et al. The chaperone-assisted E3 ligase C terminus of Hsc70-interacting protein (CHIP) targets PTEN for proteasomal degradation. The Journal of biological chemistry. 2012;287:15996–16006. doi: 10.1074/jbc.M111.321083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gonzalez-Santamaria J, et al. Regulation of the tumor suppressor PTEN by SUMO. Cell death & disease. 2012;3:e393. doi: 10.1038/cddis.2012.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Herlevsen M, et al. A novel model to identify interaction partners of the PTEN tumor suppressor gene in human bladder cancer. Biochemical and biophysical research communications. 2007;352:549–555. doi: 10.1016/j.bbrc.2006.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Freeman DJ, et al. PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase-dependent and -independent mechanisms. Cancer cell. 2003;3:117–130. doi: 10.1016/s1535-6108(03)00021-7. [DOI] [PubMed] [Google Scholar]
- 122.Zhou M, et al. PTEN reverses MDM2-mediated chemotherapy resistance by interacting with p53 in acute lymphoblastic leukemia cells. Cancer research. 2003;63:6357–6362. [PubMed] [Google Scholar]
- 123.Meuillet EJ, et al. Thioredoxin-1 binds to the C2 domain of PTEN inhibiting PTEN's lipid phosphatase activity and membrane binding: a mechanism for the functional loss of PTEN's tumor suppressor activity. Archives of biochemistry and biophysics. 2004;429:123–133. doi: 10.1016/j.abb.2004.04.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PTEN interacting proteins and the PTEN domain to which they bind. References to the papers that describe these interactions are listed.