Abstract
Several studies have addressed the importance of various ubiquitin-like (UBL) post-translational modifiers. These UBLs are covalently linked to most, if not all, target protein(s) through an enzymatic cascade analogous to ubiquitylation, consisting of E1 (activating), E2 (conjugating), and E3 (ligating) enzymes. In this report, we describe the identification of a novel ubiquitin-fold modifier 1 (Ufm1) with a molecular mass of 9.1 kDa, displaying apparently similar tertiary structure, although lacking obvious sequence identity, to ubiquitin. Ufm1 is first cleaved at the C-terminus to expose its conserved Gly residue. This Gly residue is essential for its subsequent conjugating reactions. The C-terminally processed Ufm1 is activated by a novel E1-like enzyme, Uba5, by forming a high-energy thioester bond. Activated Ufm1 is then transferred to its cognate E2-like enzyme, Ufc1, in a similar thioester linkage. Ufm1 forms several complexes in HEK293 cells and mouse tissues, revealing that it conjugates to the target proteins. Ufm1, Uba5, and Ufc1 are all conserved in metazoa and plants but not in yeast, suggesting its potential roles in various multicellular organisms.
Keywords: Uba5, ubiquitin, ubiquitin fold, ubiquitin-like protein, Ufm1
Introduction
Protein modification plays a pivotal role in the regulation and expansion of genetic information. In the past two decades, a new type of post-translational protein-modifying system has been identified whose uniqueness is that protein(s) is used as a ligand, that is, modification of protein, by protein, and for protein. A typical system is the ubiquitylation, a modification system in which a single or multiple ubiquitin molecules are attached to a protein, which serves as a signaling player that controls a variety of cellular functions (Hershko and Ciechanover, 1998; Pickart, 2001). Protein ubiquitylation is catalyzed by an elaborate system highly regulated in the cells, which is catalyzed by a sequential reaction of multiple enzymes consisting of activating (E1), conjugating (E2), and ligating (E3) enzymes. E1, which initiates the reaction, forms a high-energy thiolester bond with ubiquitin via adenylation in an ATP-dependent manner. The E1-activated ubiquitin is then transferred to E2 in a thioester linkage. In some cases, E2 can directly transfer the ubiquitin to substrate proteins in an isopeptide linkage; however, E2s mostly requires the participation of E3 to achieve substrate-specific ubiquitylation reaction in the cells. E3s are defined as enzymes required for recognition of specific substrates for ubiquitylation, other than E1 and E2 (Varshavsky, 1997; Bonifacino and Weissman, 1998; Glickman and Ciechanover, 2002).
A set of novel molecules called ubiquitin-like proteins (UBLs) that have structural similarities to ubiquitin has been recently identified (Jentsch and Pyrowolakis, 2000). They are divided into two subclasses: type-1 UBLs, which ligate to target proteins in a manner similar, but not identical, to the ubiquitylation pathway, such as SUMO, NEDD8, and UCRP/ISG15, and type-2 UBLs (also called UDPs, ubiquitin-domain proteins), which contain ubiquitin-like structure embedded in a variety of different classes of large proteins with apparently distinct functions, such as Rad23, Elongin B, Scythe, Parkin, and HOIL-1 (Tanaka et al, 1998; Jentsch and Pyrowolakis, 2000; Yeh et al, 2000; Schwartz and Hochstrasser, 2003).
In this report, we describe a unique human UBL-type modifier named ubiquitin-fold modifier 1 (Ufm1) that is synthesized in a precursor form consisting of 85 amino-acid residues. We also identified the human activating (Uba5) and conjugating (Ufc1) enzymes for Ufm1. Prior to activation by Uba5, the extra two amino acids at the C-terminal region of the human proUfm1 protein are removed to expose Gly whose residue is necessary for conjugation to target molecule(s). Lastly, we show that the mature Ufm1 is conjugated to yet unidentified endogenous proteins, forming ∼28, 38, 47, and 70 kDa complexes in human HEK293 cells and various mouse tissues.
Results
Identification of a novel protein-activating enzyme, Uba5
Our initial plan was to identify the molecule(s) that interacts with human Atg8p homolog GATE16, a type-1 UBL modifier required for autophagy (Klionsky and Emr, 2000; Ohsumi, 2001), using a yeast two-hybrid screening. Please note that the nomenclature of the autophagy-related genes was recently unified as ATG (Klionsky et al, 2003). Among several positive clones, we identified fragments of FLJ23251 (Figure 1A), which encodes a 404-amino-acid protein highly conserved in various multicellular organisms, such as Homo sapiens, Caenorhabditis elegans, Drosophila melanogaster, and Arabidopsis thaliana, but absent in yeasts (Saccharomyces cerevisiae and Schizosaccharomyces pombe) (Figure 1B). The sequence of FLJ23251 in the region containing residues 72–229 is highly homologous to the corresponding regions in Uba1 (i.e., E1 for ubiquitin) and other E1-like proteins for UBLs including the ATP-binding motif (GXGXXG) (Figure 1A and B). We named this protein Uba5, because it is a member of the E1-like enzyme family. Uba5 also has a metal-binding motif conserved in other E1-like enzymes such as Uba2, Uba3, Uba4, and Atg7. Most of E1-like enzymes have an active site Cys residue within the conserved 10–20 amino-acid residues downstream from the metal-binding motif. In the case of Uba5, the Cys250 seems to be the most possible active site Cys residue (Figure 1B). If an active site Cys residue within an E1 and E1-like enzymes is changed to Ser, an O-ester bond instead of a thioester bond is formed with its respective modifier protein and the intermediates become stable even under reducing conditions. Therefore, we mutated Cys250 within Uba5 to Ser and expressed it as a Flag-fused Uba5C250S (Flag-Uba5C250S) or Flag-Uba5 as control in HEK293 cells. As shown in Figure 1C, both Flag-Uba5 and Flag-Uba5C250S were expressed as ∼50 kDa proteins in HEK293 cells. When Flag-Uba5C250S was expressed, an additional band with a higher molecular mass of ∼60 kDa was clearly observed, indicating that Flag-Uba5C250S forms an intermediate complex with an endogenous protein. These results suggest that Uba5 is indeed a novel protein-activating enzyme for a presumptive modifier (see below).
Figure 1.
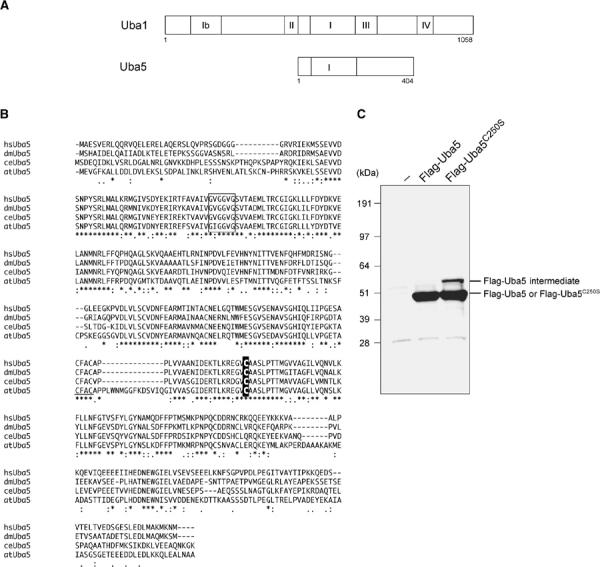
Uba5, a novel E1-like enzyme. (A) Schematic representation of Uba1 and Uba5 in H. sapiens. Uba1 is divided into several domains, including I, Ib, II, III, and IV boxes, which are conserved in other E1-like enzymes, and other regions without obvious similarity, described previously (Komatsu et al, 2001). Note that Uba5 is of a relatively small size and includes the box I and two other parts. The box I region of Uba1 (amino acids 459–611) has 48.4% similarity and 22.3% identity to amino acids 72–229 of Uba5, which includes the conserved ATP-binding motif (GXGXXG). The sequence of Uba5 is available from GenBanK™ under the accession number AK026904. hs, H. sapiens; ce, C. elegans; dm, D. melanogaster; at, A. thaliana. (B) Sequence alignment of hsUba5 and its homologs of other species (dm, NM_132494; ce, NM_058847; at, NM_100414). The amino-acid sequence of hsUba5 is compared by the ClustalW program. Asterisks, identical amino acids; single and double dots, weakly and strongly similar amino acids, respectively, determined by the criteria of ClustalW program. Open box indicates an ATP-binding motif. The putative active site Cys residue is boxed in black. The metal-binding motif is underlined. (C) Identification of the intermediate linked to Uba5 in HEK293 cells. Both Uba5 and Uba5C250S, in which the predicted active site Cys positioned at 250 was changed to Ser by site-directed mutagenesis, were tagged with Flag peptide at N-terminus, resulting in Flag-Uba5 and Flag-Uba5C250S, respectively. Each Flag-Uba5 and Flag-Uba5C250S was expressed in HEK293 cells. The cell lysates were subjected to SDS–PAGE and analyzed by immunoblotting with anti-Flag antibody.
Identification of a novel ubiquitin-fold molecule, Ufm1
Because Uba5 was identified as GATE-16-binding protein, we initially assumed that Uba5 is another GATE-16-activating enzyme, in addition to Atg7. To test this possibility, we examined whether Uba5C250S (the presumptive active site Cys at position 250 was replaced by Ser) forms an intermediate complex with GATE-16 or not. Unexpectedly, we could not identify a stable complex between Uba5C250S and GATE-16 (data not shown). Therefore, we attempted to identify a protein(s) that physically associates with Uba5 in the cells. To do this, Flag-Uba5 was expressed in HEK293 cells, then immunoprecipitated by anti-Flag antibody. The immunoprecipitates were eluted with a Flag peptide, then digested with Lys-C endopeptides (Achromobacter protease I) and the cleaved fragments were directly analyzed using a highly sensitive ‘direct nano-flow LC–MS/MS' system as described in Materials and methods. Following database search, a total of 28 peptides were assigned to MS/MS spectra obtained from four nano-LC–MS/MS analyses for the Flag-Uba5-associated complexes. These peptide data identified three proteins as Uba5-associated components: GATE-16, and hypothetical proteins BM-002 and CGI-126 (excluding the bait protein Uba5 and the background proteins, such as HSP70 and keratins).
One of these identified proteins, BM-002, is an 85-amino-acid protein with a predicted molecular mass of ∼9.1 kDa. This protein is conserved in multicellular organisms, but not in yeasts, like Uba5 (Figure 2A). The human BM-002 has high identity over the species in the central region but has elongated sequences at both N- and C-terminal regions in some species. Although the protein shows no clear overall sequence identity to ubiquitin or other modifiers (Figure 2B), the tertiary structure of BM-002 displays a striking resemblance to human ubiquitin (Figure 2C). The human structure of BM-002 was constructed by a computer-assisted modeling, based on the structure of its C. elegans homolog that has been analyzed previously, as a protein possessing ‘ubiquitin-like fold' with secondary structure elements ordered β–β–α–β–β–α (α-helix and β-sheet) along the sequence (Cort et al, 2002). Thus, we named human BM-002 as Ufm1.
Figure 2.
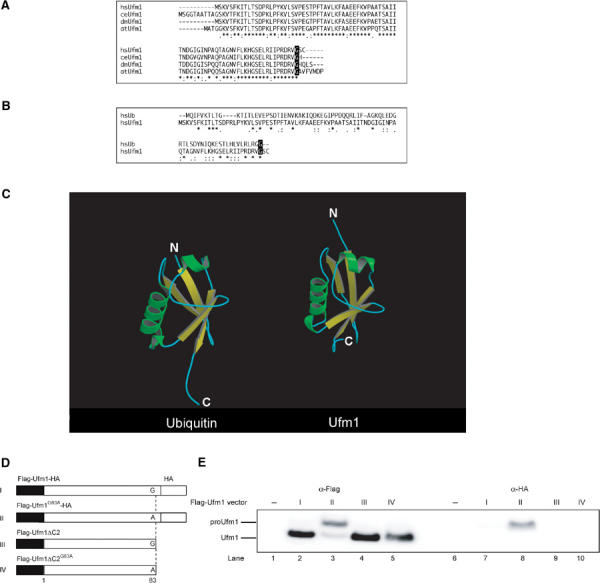
Ufm1, a novel ubiquitin-fold molecule. (A) Sequence alignment of hsUfm1 and its homologs. The sequence of hsUfm1 is available from GenBanK™ under the accession number BC005193 (dm, a coding region of dmUfm1 was found from D. melanogaster genomic sequence; ce, NM_066304; at, NM_106420). The homology analysis was performed as described in Figure 1B. The C-terminal conserved Gly residue is boxed in black. (B) Sequence alignment of hsUbiquitin with hsUfm1. The homology analysis was performed as described in Figure 1B. The C-terminal conserved Gly residue is boxed in black. (C) Structural ribbon of hsUbiquitin and predicted structural ribbon of hsUfm1. α-Helices and β-strands are shown in green and yellow, respectively. The homology model of hsUfm1 was created from the C. elegans Ufm1 structure (Cort et al, 2002) by using MOE program (2003.02; Chemical Computing Group Inc., Montreal, Quebec, Canada). (D) Schematic representation of mammalian expression plasmids for Ufm1 and the derivative mutants. Flag epitope tags at the N-terminus, HA epitope tags at the C-terminus, and putative cleavage site Gly83 residue (vertical dotted lines) are indicated. To construct Ufm1G83A, a single point mutation was introduced into Ufm1, which led to an amino-acid substitution from Gly to Ala at position 83. To construct Ufm1ΔC2, the two C-terminal residues were deleted by PCR. Ufm1ΔC2G83A was also produced by site-directed mutagenesis of Ufm1ΔC2. The ΔC2 mutants were tagged with the Flag epitopes at N-terminus. (E) ProUfm1 processing. HEK293 cells were transfected with Flag-Ufm1-HA, Flag-Ufm1G83A-HA, Flag-Ufm1ΔC2, or Flag-Ufm1ΔC2G83A. The cell lysates were subjected to SDS–PAGE and analyzed by immunoblots with anti-Flag and anti-HA antibodies. ProUfm1 and mature Ufm1 are indicated on the left. The numbers at the top from I to IV are similar to those in (D).
Ubiquitin is synthesized in a precursor form that must be processed by de-ubiquitylating enzymes (DUBs) to generate a Gly–Gly sequence at the C-terminus. Similarly, Ufm1 has a single Gly residue conserved across species at the C-terminal region, although the length and sequences of amino acids extending from this Gly residue vary among species. To test whether the C-terminus of Ufm1 is post-translationally cleaved, we constructed an expression vector for Ufm1 tagged at both the N- and C-ends, that is, a Flag epitope at the N-terminus and an HA epitope at the C-terminus (Flag-Ufm1-HA) (Figure 2D). After transfection of Flag-Ufm1-HA into HEK293 cells, the cell lysate was subjected to SDS–PAGE, and Flag-Ufm1-HA was detected by immunoblotting. A 10-kDa protein corresponding to Ufm1 was recognized with anti-Flag antibody, while no appreciable protein was observed with anti-HA antibody (Figure 2E, lanes 2 and 7). The mobility on SDS–PAGE was similar to that of Flag-Ufm1ΔC2 (equivalent to mature Ufm11–83 protein) lacking the C-terminal Ser84 and Cys85 of proUfm1 (Figure 2E, lane 4). These results suggested that the C-terminus of Ufm1 is post-translationally cleaved in the cells, producing mature Ufm1 with the C-terminal Gly83 residue. It is known that the replacement of C-terminal Gly residue of Ub and other UBLs with an Ala residue inhibits the C-terminal processing (Kabeya et al, 2000; Tanida et al, 2003). To examine whether Gly83 of Ufm1 is essential for the cleavage, Gly83 of Flag-Ufm1-HA was mutated to Ala, and expressed in HEK293 cells (Figure 2D, Flag-Ufm1G83A-HA). The mobility of most Flag-Ufm1G83A-HA on SDS–PAGE was apparently slower than that of Flag-Ufm1-HA (Figure 2E, lane 3). This mutant was recognized by immunoblotting with anti-HA antibody as well as anti-Flag antibody, suggesting that mutation Gly83 to Ala confers resistance to its C-terminal cleavage.
Uba5 is an Ufm1-activating enzyme
We next investigated whether Uba5 forms an intermediate complex with Ufm1. We expressed Flag-Uba5 or Flag-Uba5C250S with Myc-tagged Ufm1 (Myc-Ufm1) in HEK293 cells. Myc-tagged Ufm1ΔC3 lacking the C-terminal Gly83 of mature Ufm1 (Myc-Ufm1ΔC3; i.e., deletion form of three residues from precursor Ufm11–85 protein) was used as control. Each cell lysate was prepared and analyzed by immunoblotting with anti-Flag antibody. Flag-Uba5C250S formed an intermediate with an endogenous protein as shown in Figure 1 (Figure 3A, lane 7). When Flag-Uba5C250S was coexpressed with Myc-Ufm1, the intermediate shifted to higher molecular weight (Figure 3A, lane 8). The higher band was not detected when Myc-Ufm1ΔC3 was coexpressed (Figure 3A, lane 9). To verify that the intermediate is indeed the Uba5–Ufm1 complex, Flag-Uba5C250S was immunoprecipitated and blotted with anti-Flag and anti-Myc antibody. Consistent with the above data, a higher sized intermediate was observed when Flag-Uba5C250S was coexpressed with Myc-Ufm1 (Figure 3B, top panel, lane 5), but not alone or with Myc-Ufm1ΔC3 (Figure 3B, top panel, lanes 4 and 6). The intermediate was also recognized by anti-Myc antibody (Figure 3B, lower panel, lane 5), indicating the existence of the Flag-Uba5C250S–Myc-Ufm1 complex. Note that the small-sized intermediate is presumably a complex with an endogenous Ufm1, as mentioned. These results indicate that Uba5 forms an intermediate with Ufm1 and the Gly83 residue of Ufm1 is essential for the formation of the intermediate with Uba5 in vivo.
Figure 3.
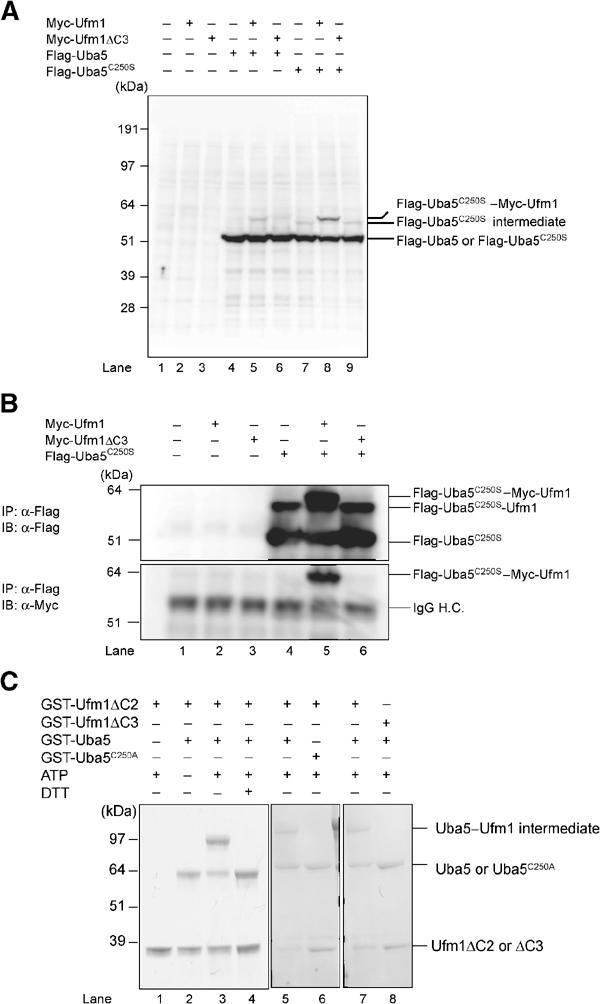
Demonstration that Uba5 is an Ufm1-activating enzyme. (A) Immunoblotting analysis. Each Myc-tagged Ufm1 (Myc-Ufm1) and Myc-Ufm1ΔC3 was expressed alone (lanes 2 and 3, respectively), and coexpressed with Flag-Uba5 (lanes 5 and 6, respectively) or Flag-Uba5C250S (lanes 8 and 9, respectively). Each Flag-Uba5 and Flag-Uba5C250S was also expressed alone (lanes 4 and 7, respectively). The cell lysates were subjected to SDS–PAGE and analyzed by immunoblotting with anti-Flag antibody. The bands corresponding to Flag-Uba5, Flag-Uba5C250S, and Flag-Uba5C250S intermediates are indicated on the right. (B) Immunoblotting analysis after immunoprecipitation. Each Myc-Ufm1 and Myc-Ufm1ΔC3 was expressed alone (lanes 2 and 3, respectively), and coexpressed with Flag-Uba5C250S (lanes 5 and 6, respectively). Flag-Uba5C250S was also expressed alone (lane 4). The cell lysates were immunoprecipitated with anti-Flag antibody. The resulting immunoprecipitates were subjected to SDS–PAGE and analyzed by immunoblotting with anti-Flag and anti-Myc antibodies. The bands corresponding to Flag-Uba5C250S, Flag-Uba5C250S–endogenous Ufm1, and Flag-Uba5C250S–Myc-Ufm1 intermediates are indicated. (C) In vitro activating assay of Ufm1 by Uba5. Purified recombinant GST-Ufm1ΔC2 (2 μg) (lanes 1–7) was incubated for 30 min at 25°C with some of the following: 2 μg of purified recombinant GST-Uba5 (lanes 2–5, 7, and 8), GST-Uba5C250A (lane 6), and 5 mM ATP (lanes 1 and 3–8). Lane 8 was conducted similar to lane 7, except that GST-Ufm1ΔC3 was used instead of GST-Ufm1ΔC2. Reactions were then incubated with SDS loading buffer lacking reducing agent (lanes 1–3 and 5–8) or containing 100 mM DTT (lane 4). The presence or absence of various components is indicated above the lanes. The bands corresponding to free GST-Uba5, GST-Uba5C250A, GST-Ufm1ΔC2 (mature Ufm1), GST-Ufm1ΔC3, and GST-Uba5–GST-Ufm1ΔC2 thioester product are indicated on the right.
We subsequently tested whether Uba5 can activate Ufm1 in vitro. The thioester formation assay was performed using recombinant proteins expressed in Escherichia coli. Recombinant GST-tagged Uba5 and mature Ufm1 (Ufm1ΔC2) with exposed C-terminal Gly83 residue were purified, mixed and incubated in the presence of ATP and then subjected to SDS–PAGE at either reducing or nonreducing conditions. GST-Ufm1ΔC3 was used as control. An ∼100 kDa band corresponding to the GST-Ufm1ΔC2–GST-Uba5 intermediate complex was clearly observed when the mixture was applied at nonreducing conditions (Figure 3C, lane 3). This intermediate was not observed when ATP or GST-Uba5 was excluded from the mixture (Figure 3C, lanes 1 and 2), or when the mixture was loaded in the presence of a reducing agent dithiothreitol (DTT) (Figure 3C, lane 4). Furthermore, GST-tagged Uba5C250A mutant, a presumptive active site Cys mutant, could not form the intermediate even at nonreducing conditions (Figure 3C, lane 6). GST-tagged Ufm1ΔC3 was also incapable of forming the intermediate in this reaction (Figure 3C, lane 8). Taken together, we concluded that Uba5 is an Ufm1-activating enzyme and has the active site in Cys250.
Identification of a novel protein-conjugating enzyme, Ufc1
The LC–MS/MS analysis revealed CGI-126 protein as another Uba5 interacting protein. CGI-126 is a protein of 167-amino-acid residues with a predicted molecular mass of 19.4 kDa. This protein is also conserved in multicellular organisms, like Uba5 and Ufm1 (Figure 4A). The C-terminal half of human CGI-126 has a high identity across species as shown in Figure 4A. CGI-126 has a highly conserved region, for example, residues 113–126, with limited similarity to the region of Ubc's that encodes an active site Cys residue capable of forming a thioester bond (Figure 4A). We assumed that this protein may be an E2-like conjugating enzyme for Ufm1 and thus named it Ufm1-conjugating enzyme 1 (Ufc1). If Ufc1 is an authentic E2 enzyme for Ufm1, it is expected to form an intermediate complex with Ufm1 via a thioester linkage. To test this possibility in the same way as Uba5, we mutated the predicted active site Cys residue within Ufc1 (Figure 4A, Cys116) to Ser. We expressed Flag-Ufc1 or Flag-Ufc1C116S (a presumptive active site Cys at position 116 was replaced by Ser) in combination with Myc-Ufm1 or Myc-Ufm1ΔC3 in HEK293 cells. Flag-Ufc1C116S formed a stable intermediate band when coexpressed with Myc-Ufm1 (Figure 4B, lane 8), but not alone or with Myc-Ufm1ΔC3 (Figure 4B, lanes 7 and 9). To ascertain that this is the Flag-Ufc1C116S–Myc-Ufm1 intermediate, Flag-Ufc1C116S was immunoprecipitated and blotted with anti-Myc antibody (Figure 4C). Indeed, Myc-Ufm1, but not Myc-Ufm1ΔC3, formed a complex with Flag-Ufc1C116S (Figure 4C, lanes 5 and 6, top and bottom panels). Note that Flag-Ufc1C116S intermediate with a faster electrophoretic mobility than the Flag-Ufc1C116S–Myc-Ufm1 complex is presumably the intermediate with the endogenous Ufm1 (Figure 4C, lanes 4–6, upper panel). These results indicate that Ufc1 forms an intermediate with Ufm1 in vivo.
Figure 4.
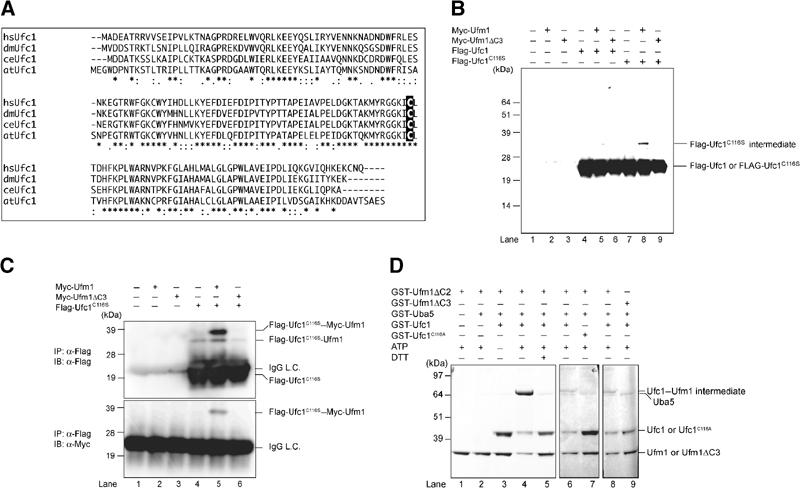
Ufc1, a novel E2-like enzyme. (A) Sequence alignment of hsUfc1 and its homologs. The sequence of Ufc1 is available from GenBanK™ under the accession number BC005187 (dm, NM_137230; ce, NM_066654; at, BT001180). The homology analysis was performed as described in Figure 1B. The putative active site Cys residue is boxed in black. (B) Immunoblotting analysis. Each Myc-tagged Ufm1 (Myc-Ufm1) and Myc-Ufm1ΔC3 was expressed alone (lanes 2 and 3, respectively), and coexpressed with Flag-Ufc1 (lanes 5 and 6, respectively) or Flag-Ufc1C116S (lanes 8 and 9, respectively). Each Flag-Ufc1 and Flag-Ufc1C116S was also expressed alone (lanes 4 and 7, respectively). The cell lysates were subjected to SDS–PAGE and analyzed by immunoblotting with anti-Flag antibody. The bands corresponding to Flag-Ufc1, Flag-Ufc1C116S, and Flag-Ufc1C116S intermediates are indicated on the right. (C) Immunoblotting analysis after immunoprecipitation. Each Myc-Ufm1 and Myc-Ufm1ΔC3 was expressed alone (lanes 2 and 3, respectively), and coexpressed with Flag-Ufc1C116S (lanes 5 and 6, respectively). Flag-Ufc1C116S was also expressed alone (lane 4). The cell lysates were immunoprecipitated with anti-Flag antibody. The resulting immunoprecipitates were subjected to SDS–PAGE and analyzed by immunoblots with anti-Flag and anti-Myc antibodies. The bands corresponding to Flag-Ufc1C116S, Flag-Ufc1C116S–endogenous Ufm1, and Flag-Ufc1C116S–Myc-Ufm1 intermediates are indicated. (D) In vitro thioester bond formation assay of Ufm1 by Ufc1. Purified recombinant GST-Ufm1ΔC2 (2 μg) (lanes 1–8) was incubated for 30 min at 25°C with the following: purified recombinant GST-Uba5 (0.2 μg) (lanes 2–9), GST-Ufc1 (2 μg) (lanes 3–6, 8, and 9), GST-Ufc1C116S (2 μg) (lane 7), and 5 mM ATP (lanes 1, 2, and 4–9). Lane 9 was conducted similar to lane 8, except that GST-Ufm1ΔC3 was used instead of GST-Ufm1ΔC2. Reactions were then incubated with SDS loading buffer lacking reducing agent (lanes 1–4 and 6–9) or containing 100 mM DTT (lane 5). The presence or absence of various components is indicated above the lanes. The bands corresponding to free GST-Ufm1ΔC2 (mature Ufm1), GST-Ufm1ΔC3, GST-Uba5, GST-Ufc1, GST-Ufc1C116S, and GST-Ufc1–GST-Ufm1ΔC2 thioester product are indicated on the right.
To confirm that Ufc1 is indeed an E2-like enzyme that conjugates with Ufm1 via a thioester linkage, we conducted an in vitro Ufm1 conjugation assay. Recombinant GST-Uba5, GST-Ufc1, and GST-Ufm1ΔC2 were mixed and incubated in the presence of ATP. GST-Ufc1C116A mutant and GST-Ufm1ΔC3 were used as negative controls. Under nonreducing conditions, an ∼70 kDa band corresponding to GST-Ufm1ΔC2–GST-Ufc1 intermediate was observed (Figure 4D, lane 4). This product was not formed at reducing conditions, or when any of the components was omitted from the reaction (Figure 4D, lanes 1–3 and 5). GST-tagged Ufc1C116A mutant could not form the intermediate, suggesting that Cys116 is indeed the active site (Figure 4D, lane 7). GST-Ufm1ΔC3 was again unable to form the intermediate complex in this reaction (Figure 4D, lane 9). Taken together, we concluded that Ufc1 functions as an Ufm1-conjugating enzyme and has the active site in Cys116.
Conjugation of Ufm1 to cellular protein(s)
We next examined whether Ufm1 conjugates to the target protein(s) in cells. To this end, we expressed Flag- and 6xHis-tagged Ufm1 constructs in HEK293 cells, and purified them under denaturing conditions by Ni2+ beads. The resulting precipitates were then analyzed by immunoblotting with anti-Flag antibody. When FlagHis-Ufm1-HA (proUfm1) or FlagHis-Ufm1ΔC2 (mature form) was expressed, several proteins with sizes of about 28, 38, and 47 kDa were detected, in addition to the 10 kDa corresponding to free FlagHis-Ufm1ΔC2 (Figure 5A, lanes 2 and 4). These bands were not detected by FlagHis-Ufm1G83A-HA and FlagHis-Ufm1ΔC3, suggesting that both C-terminal cleavage and C-terminal Gly residue are required for the conjugation reaction (Figure 5A, lanes 3 and 6). Moreover, these protein bands were resistant to reducing agents, such as DTT and β-mercaptoethanol. These results indicate that Ufm1 is covalently attached to some target proteins, probably through an isopeptide bond between the C-terminal Gly83 of Ufm1 and a Lys residue in the cellular proteins. It is of note that FlagHis-Ufm1G83A mutant with exposed C-terminal Ala instead of Gly can conjugate to target proteins (Figure 5A, lane 5), consistent with the previous report on ubiquitin and SUMO (Hodgins et al, 1992; Kamitani et al, 1997). Since C-terminal Gly to Ala mutation confers resistance to the Ufm1 processing, the conjugates with FlagHis-Ufm1G83A mutant may be more stable than those with FlagHis-Ufm1ΔC2 (Figure 5A, compare lanes 4 and 5). These results suggest that the Ufm1 conjugation is also a reversible reaction.
Figure 5.
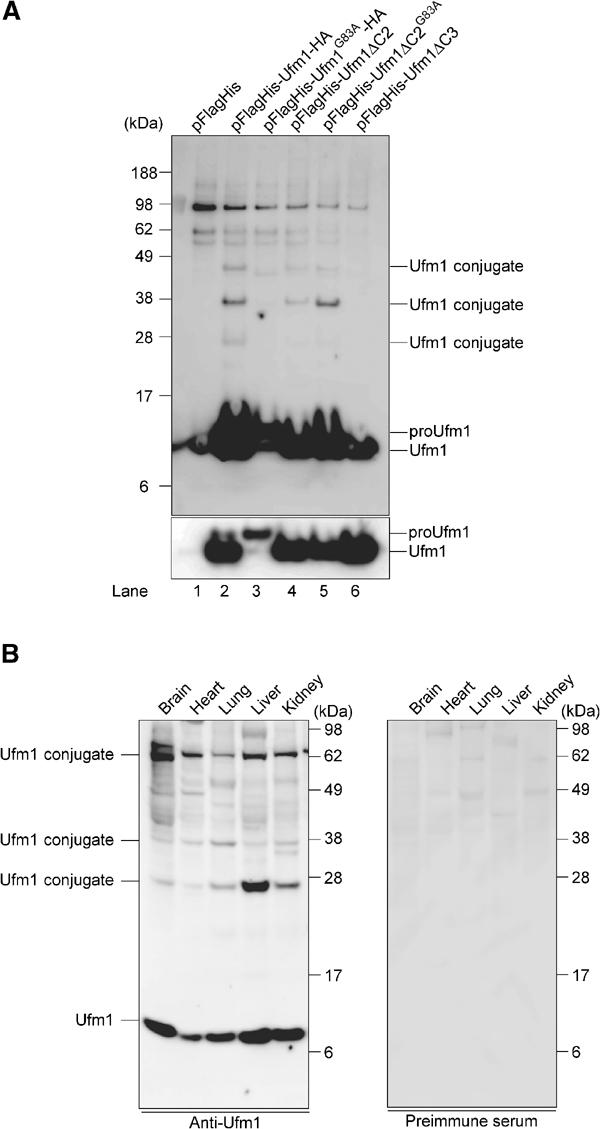
Formation of a covalent protein conjugate(s) with Ufm1 in HEK293 cells and mouse tissues. (A) Ufm1 conjugates in human HEK293 cells. HEK293 cells were transfected with FlagHis-Ufm1-HA, FlagHis-Ufm1G83A-HA, FlagHis-Ufm1ΔC2, FlagHis-Ufm1ΔC2G83A, or FlagHis-Ufm1ΔC3 expression plasmids. These cells were lysed under denaturing conditions, and the lysates were precipitated with Ni2+ beads. The precipitates were subjected to SDS–PAGE and analyzed by immunoblotting with anti-Flag antibody. The bottom panel shows the short exposure of the upper panel. The bands corresponding to mature Ufm1, proUfm, and Ufm1 conjugates are indicated on the right. (B) Ufm1 conjugates in various mouse tissues. Homogenates from mouse tissues as indicated were prepared and subjected to SDS–PAGE and analyzed by immunoblotting with anti-Ufm1 serum (left panel) or preimmune serum (right panel). The bands corresponding to Ufm1 and conjugates between Ufm1 and target proteins are indicated on the left.
We further investigated the expression of Ufm1 and its conjugated proteins in mouse tissues using anti-Ufm1 serum. Ufm1 was widely expressed in all tissues examined, such as brain, heart, lung, liver, and kidney (Figure 5B, left panel). In addition, several bands with striking similarity to proteins detected in HEK293 cells were observed. These bands were not detected by preimmune or preabsorbed antisera (Figure 5B, right panel), suggesting that they are likely the Ufm1 conjugates. Although the intensity of each band varied among tissues and HEK293 cells, 28 and 38 kDa proteins were commonly detected. The 70-kDa band observed in all tissues was also detected faintly in HEK293 cells (Figure 5A, lane 5). The 47-kDa band observed in HEK293 cells was not clear. These protein bands were resistant to reducing agents, such as DTT and β-mercaptoethanol, indicating that Ufm1 covalently attaches to cellular proteins like other Ubl proteins. The targets of Ufm1 appeared to be common in a variety of tissues. These results suggest the universal roles of Ufm1 in the regulation of cellular function in multicellular organisms.
Subcellular localization of Ufm1 in HeLa cells
We finally examined the subcellular distribution of Ufm1 in HeLa cells. Immunocytochemical analysis using anti-Ufm1 serum revealed that Ufm1 was predominantly localized in the nucleus and diffusely in the cytoplasm (Figure 6A). These staining patterns were not observed when anti-Ufm1 serum had been preadsorbed with excess amounts of recombinant Ufm1 protein or preimmune serum was used instead of anti-Ufm1 serum (Figure 6B). Moreover, Ufm1 localization in the cytoplasm and nucleus was similar to the localization of exogenously expressed GFP-tagged Ufm1 in HeLa cells (data not shown). In the nucleus, strong immunoreactivity to anti-Ufm1 serum was observed as a dot-like structure. Although such dots-like structures were detected by preimmune serum, those intensities were weak. Thus, some of these dot-like structures may represent conjugates of Ufm1.
Figure 6.
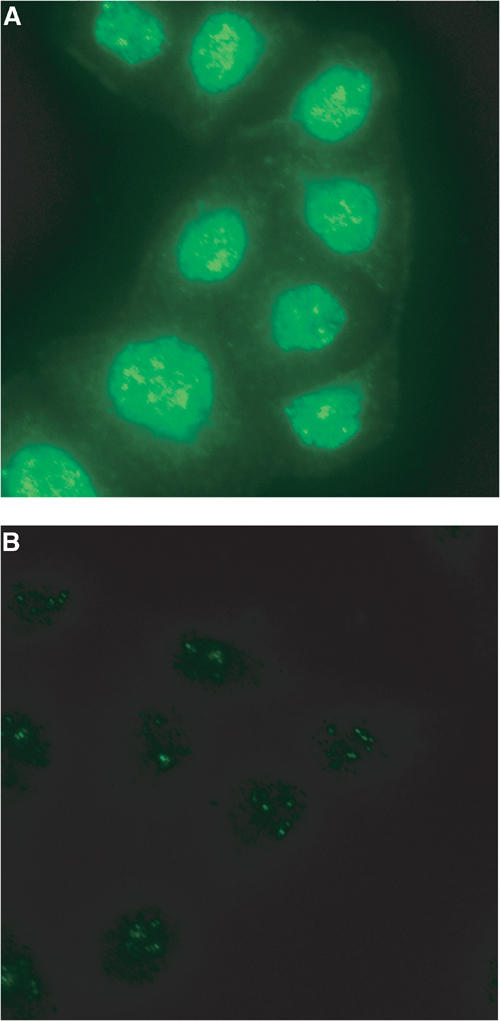
Intracellular distribution of Ufm1 in HeLa cells. (A) HeLa cells were seeded on coverslips 24 h before fixation for immunostaining. Ufm1 was detected with anti-Ufm1 serum and visualized with Alexa 488 nm anti-rabbit antibody. (B) Immunocytochemical analysis was conducted as for (A), except that preimmune serum was used. Cells were observed using a fluorescence microscope. Magnification, × 400.
Discussion
In the present study, we reported that Ufm1 acts as a new post-translational UBL modifier, based on the following criteria: (1) It is a small protein of 9.1 kDa with a ubiquitin-fold structure. (2) Ufm1 is synthesized in a precursor form, and the extra amino-acid residues at the C-terminal side need to be processed to expose the Gly residue. (3) The C-terminal processing and exposure of glycine residue are essential to the formation of Ufm conjugates in the cells. (4) Ufm1 has specific E1-like (Uba5) and E2-like (Ufc1) enzymes for activation and conjugation, respectively. Intriguingly, many UBL modifiers are evolutionarily conserved from yeast to human, except interferon-inducible UBL modifiers, such as UCRP/ISG15, Fat10, and Fau1/MNSFβ (Nakamura et al, 1995; D'Cunha et al, 1996; Liu et al, 1999). Ufm1, Uba5, and Ufc1 found in the present study are conserved in various multicellular organisms (Figures 1B, 2A and 4A), but not in both budding and fission yeasts, suggesting that they all have been generated by coevolution.
We identified Uba5 as an E1 enzyme for Ufm1. This enzyme is relatively small compared to Uba1, that is, an E1 for ubiquitin (Figure 1A). In the in vitro assay, the recombinant Uba5 protein formed a thioester linkage with recombinant Ufm1 (Figure 3C) and transferred the activated Ufm1 to recombinant Ufc1 (an E2 enzyme) (Figure 4D), indicating that Uba5 can activate Ufm1 as a single molecule. This is in marked contrast to other E1s such as Uba2 and Uba3, which retain obvious similarities to the C-terminal half of Uba1 but require the formation of heterodimer complexes with respective partner molecules, AOS1 and APP-BP1, respectively, with similarities to the N-terminal half of Uba1 (Johnson et al, 1997; Liakopoulos et al, 1998; Osaka et al, 1998). Another E1-like enzyme, Uba4 that activates Urm1, is of similar size to Uba5 (Furukawa et al, 2000), but it remains unknown whether Uba4 acts as a single molecule or needs a partner subunit. The homology of Uba5 to Uba1 is less than those of Uba2 and Uba3, except their ThiF domain conserved in E1s, and thus it is likely that Uba5 may uniquely activate Ufm1, differing from other E1s such as Uba1, Uba2/AOS1, and Uba3/APP-BP1. Thus, although the structure of APP-BP1/Uba3 heterodimer is determined and the mechanism by which E1s activate their cognate UBLs was proposed (Walden et al, 2003a, 2003b), the weak homology of Uba5 with other E1s hampered the computer-assisted structural analysis. To clarify this issue, structural analysis of Uba5 is required. This issue is currently under investigation in our laboratories. So far, most E1-like enzymes activate single species of UBL protein, although Atg7 is exception, which can activate both Atg8 and Atg12 (Mizushima et al, 1998; Tanida et al, 1999; Ichimura et al, 2000). A total of 10 E1-like enzymes can be identified in the human genome by computer analysis. Considering the limited number of E1-like proteins, it is possible that some E1-like proteins can activate a distinct set of UBL proteins. Whether or not Uba5 is capable of activating proteins other than Ufm1 remains to be clarified.
There are more than a dozen of E2 family genes in human genomes. In the budding yeast, 13 different E2s, namely Ubc1–Ubc13, have been documented and functionally characterized. Functionally, most of them catalyze the conjugation of ubiquitin, except that Ubc9 and Ubc12 are for SUMO and NEDD8/Rub1, respectively (Johnson and Blobel, 1997; Lammer et al, 1998; Osaka et al, 1998). In addition, in the autophagic pathway, Atg3 and Atg10 are both E2 enzymes for Atg8 and Atg12, respectively, but they do not have obvious sequence similarities to known Ubc's, except for a short region encompassing an active Cys residue (Shintani et al, 1999; Ichimura et al, 2000). Similarly, Ufc1 is a unique E2-like enzyme with no obvious sequence homology with other E2s, except approximately 10 amino-acid residues encompassing the active site Cys residue.
In assessing the biological roles of the Ufm1-modifying system, characterization of the target molecule(s) is of particular importance. Regarding this issue, we identified several putative proteins that are conjugated with Ufm1 in human HEK293 cells and various mouse tissues. It is noteworthy that the sizes of these bands (28, 38, 47 kDa) increase by 10 kDa, which is consistent with the size of Ufm1. Considering that several Ubl modifiers can attach to target proteins as a polymer, it is possible that these bands correspond to multi- or poly-Ufm1 conjugates. In fact, Ufm1 has six Lys residues. Whether Ufm1 is conjugated to several distinct proteins or multiple Lys residues in a single target or polymerized in a single Lys residue awaits future study. Unfortunately, we could not identify the protein, and detailed analysis of the cellular function of Ufm1 conjugation awaits future study. It was recently reported that Uba5 is induced by certain reagents that induce stress in the endoplasmic reticulum (ER), a so-called ‘unfolded protein response' (Harding et al, 2003). However, we could not observe the induction of Uba5, Ufc1, and Ufm1 by treatment with various compounds known to induce ER stress in mammalian cells (data not shown). In addition, exposure to other stresses including high temperature or heavy metals also did not induce the appearance of obvious new conjugation band(s) of Ufm1, by immunoblot analysis. Further studies on the biological roles of the Ufm1 conjugation pathway are under investigation in our laboratories.
Materials and methods
DNA construction
The cDNA encoding human Uba5 was obtained by PCR from human liver cDNA with the Uba5-s5′ primer (5′-CGGAGGGATCCCCATGGCGGAGTCTGTGGAG-3′) and the Uba5-r3′ primer (5′-CAGTCCTCGAGCTACATATTCTTCATTTT-3′). It was then subcloned into pcDNA3 vector (Invitrogen, San Diego, CA). A point mutation for Cys at position 250 to Ser or Ala was generated by PCR-based site-directed mutagenesis. The Flag tag was introduced at the N-terminus of Uba5 or Uba5C250S. Similarly, cDNA encoding human Ufm1 was amplified by PCR from human liver cDNA with the Ufm1-s5′ primer (5′-TTCCGGGATCCCCATGTCGAAGGTTTCCTTT-3′) and the Ufm1-r3′ primer (5′-AGTAGCTCGAGTTAACAACTTCCAACACGAT-3′), and subcloned into pcDNA3 vector. The Flag, FlagHis, or Myc tags were introduced at the N-terminus of Ufm1. The HA tag was introduced at the C-terminus of Ufm1. The C-terminal deletion mutants of Ufm1 named Ufm1ΔC2 and Ufm1ΔC3, encoding amino acids 1–83 and 1–82, respectively, were generated by PCR. A point mutation for Gly at position 83 to Ala of Ufm1 and Ufm1ΔC2 (Ufm1G83A and Ufm1ΔC2G83A, respectively) was generated by PCR-based site-directed mutagenesis. The cDNA encoding human Ufc1 was obtained by PCR from human liver cDNA with the Ufc1-s5′ primer (5′-GCCCTGGATCCAGATGGCGGATGAAGCCACG-3′) and the Ufc1-r3′ primer (5′-TTCTCGAGTCATTGGTTGCATTTCTCTT-3′). It was then subcloned into pcDNA3 vector. A point mutation for Cys at position 116 to Ser or Ala was generated by PCR-based site-directed mutagenesis. The Flag tag was introduced at the N-terminus of Ufc1 and Ufc1C116S. To express GST-fused Ufm1ΔC2, Ufm1ΔC3, Uba5, Uba5C250A, Ufc1, and Ufc1C116A in E. coli, these cDNAs were subcloned into pGEX-6p vector (Amersham Biosciences). All mutations mentioned above were confirmed by DNA sequencing.
Cell culture and transfection
Media and reagents for cell culture were purchased from Life Technologies (Grand Island, NY). HEK293 cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS), 5 U/ml penicillin, and 50 μg/ml streptomycin. HEK293 cells at subconfluence were transfected with the indicated plasmids using Fugene 6 reagent (Roche Molecular Biochemicals, Mannheim, Germany). Cells were analyzed at 20–24 h after transfection.
Immunological analysis
For immunoblot analysis, cells were lysed with ice-cold TNE buffer (10 mM Tris–HCl, pH 7.5, 1% Nonidet P-40, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), and protease inhibitors) and the lysates were separated by SDS–PAGE (12% gel or 4–12% gradient gel) and transferred to a polyvinylidene difluoride (PVDF) membrane. Mouse monoclonal anti-Flag antibody (M2; Sigma Chemical Co., St Louis, MO), anti-HA antibody (F7; Santa Cruz Biotechnology, Santa Cruz, CA), and rabbit polyclonal anti-Myc antibody (N14; Santa Cruz) were used for immunodetection. Development was performed by the Western lighting detection methods.
For immunoprecipitation analysis, cells were lysed by 200 μl of TNE, and the lysate was then centrifuged at 10 000 g for 10 min at 4°C to remove debris. In the next step, 800 μl of TNE and 30 μl of M2-agarose (Sigma) were added to the lysate, and the mixture was mixed under constant rotation for 12 h at 4°C. The immunoprecipitates were washed five times with ice-cold TNE. The complex was boiled for 10 min in SDS sample buffer in the presence of β-mercaptoethanol to elute proteins and centrifuged at 10 000 g for 10 min at 4°C. The supernatant was subjected to SDS–PAGE, transferred to PVDF membrane, and analyzed by immunoblots with anti-Flag (M2) or anti-Myc (N14) antibody.
For purification of 6xHis-tagged proteins under denaturing conditions, cells were lysed by 1 ml of denaturing lysis buffer (8 M urea, 0.1 M NaH2PO4, and 0.01 M Tris–HCl, pH 8.0) in the presence of 20 mM N-ethylmaleimide as an inhibitor of isopeptidases, and the lysate was sonicated briefly and then centrifuged at 10 000 g for 10 min at room temperature to remove debris. Then, 30 μl of Ni-NTA Superflow (QIAGEN) was added to the lysate, and the mixture was shaken under constant rotation for 30 min at room temperature. The precipitates were washed five times with denaturing wash buffer (8 M urea, 0.1 M NaH2PO4, and 0.01 M Tris–HCl, pH 5.9). To elute proteins, elution buffer (8 M urea, 0.1 M NaH2PO4, and 0.01 M Tris–HCl, pH 4.5) was added to the complex, and the mixture was centrifuged at 10 000 g for 10 min at room temperature. The resulting supernatant was subjected to SDS–PAGE, transferred to PVDF membrane, and analyzed by immunoblots with anti-Flag (M2).
Freshly isolated tissues from mice were homogenized in lysis buffer (50 mM Tris–HCl, pH 7.5, 1% SDS, 5 mM EDTA, and 10 mM β-mercaptoethanol) using potter-Elvehjem homogenizer. The homogenate was centrifuged at 10 000 g for 10 min to remove debris. The resulting supernatant was subjected to SDS–PAGE, transferred to PVDF membrane, and analyzed by immunoblotting with anti-Ufm1 or preimmune serum. The anti-Ufm1 polyclonal antibody was raised in rabbits using the recombinant protein produced in E. coli as an antigen.
In vitro thioester formation assay
Recombinant GST-Ufm1ΔC2, GST-Ufm1ΔC3, GST-Uba5, GST-Uba5C250A, GST-Ufc1, and GST-Ufc1C116A (tagged N-terminally with GST) were produced in E. coli and recombinant proteins were purified by chromatography on glutathione sepharose 4B (Amersham Biosciences). After elution of proteins from the beads, the preparations were dialyzed against 50 mM BisTris (pH 6.5), 100 mM NaCl, 10 mM MgCl2, and 0.1 mM DTT (reaction buffer). Most thioester formation reactions contained reaction buffer with 4 μg GST-Ufm1ΔC2 or GST-Ufm1ΔC3 and some of the following: 5 mM ATP, 2 or 0.2 μg GST-Uba5 or GST-Uba5C250A, and 4 μg GST-Ufc1 or GST-Ufc1C116A. Reactions were incubated for 30 min at 25°C and stopped by the addition of SDS-containing loading buffer either lacking reducing agent or containing 100 mM DTT, followed by a 10 min incubation at 37°C, SDS–PAGE (4–12% acrylamide gradient) and Coomassie brilliant blue staining.
Protein identification by LC–MS/MS analysis
The Uba5-associated complexes were digested with Achromobacter protease I and the resulting peptides were analyzed using a nanoscale LC–MS/MS system as described previously (Natsume et al, 2002). The peptide mixture was applied to a Mightysil-PR-18 (1 μm particle, Kanto Chemical) frit-less column (45 mm × 0.150 mm ID) and separated using a 0–40% gradient of acetonitrile containing 0.1% formic acid over 30 min at a flow rate of 50 nl/min. Eluted peptides were sprayed directly into a quadropole time-of-flight hybrid mass spectrometer (Q-Tof Ultima, Micromass, Manchester, UK). MS and MS/MS spectra were obtained in a data-dependent mode. Up to four precursor ions above an intensity threshold of 10 counts/s were selected for MS/MS analyses from each survey scan. All MS/MS spectra were searched against protein sequences of Swiss Prot and RefSeq (NCBI) using batch processes of Mascot software package (Matrix Science, London, UK). The criteria for match acceptance were the following: (1) When the match score was 10 over each threshold, identification was accepted without further consideration. (2) When the difference of score and threshold was lower than 10, or when proteins were identified based on a single matched MS/MS spectrum, we manually confirmed the raw data prior to acceptance. (3) Peptides assigned by less than three y series ions and peptides with +4 charge state were all eliminated regardless of their scores.
Immunofluorescence
HeLa cells grown on glass coverslips were fixed in 4% paraformaldehyde (PFA) in PBS for 15 min, and permeabilized with 0.2% (vol/vol) Triton X-100 in PBS for 30 min. After permeabilization, the cells were blocked for 30 min with 5% (vol/vol) normal goat serum in PBS, incubated for 1 h at 37°C with anti-Ufm1 serum or preimmune serum, washed with PBS, and incubated for 30 min with Alexa 488 nm anti-rabbit antibodies (Molecular Probes). The coverslips were washed and mounted on slides. Fluorescence images were obtained using a fluorescence microscope (DMIRE2; Leica) equipped with a cooled charge-coupled device camera (CTR MIC; Leica). Pictures were taken using Leica Qfluoro software (Leica).
Acknowledgments
We thank T Mizushima (Nagoya University) for the computer-assisted structural modeling of human Ufm1. This work was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Bonifacino JS, Weissman AM (1998) Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu Rev Cell Dev Biol 14: 19–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cort JR, Chiang Y, Zheng D, Montelione GT, Kennedy MA (2002) NMR structure of conserved eukaryotic protein ZK652.3 from C. elegans: a ubiquitin-like fold. Proteins 48: 733–736 [DOI] [PubMed] [Google Scholar]
- D'Cunha J, Knight E Jr, Haas AL, Truitt RL, Borden EC (1996) Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc Natl Acad Sci USA 93: 211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K, Mizushima N, Noda T, Ohsumi Y (2000) A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J Biol Chem 275: 7462–7465 [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A (2002) The ubiquitin–proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82: 373–428 [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11: 619–633 [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hodgins RR, Ellison KS, Ellison MJ (1992) Expression of a ubiquitin derivative that conjugates to protein irreversibly produces phenotypes consistent with a ubiquitin deficiency. J Biol Chem 267: 8807–8812 [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y (2000) A ubiquitin-like system mediates protein lipidation. Nature 408: 488–492 [DOI] [PubMed] [Google Scholar]
- Jentsch S, Pyrowolakis G (2000) Ubiquitin and its kin: how close are the family ties? Trends Cell Biol 10: 335–342 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G (1997) Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem 272: 26799–26802 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G (1997) The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J 16: 5509–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19: 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani T, Nguyen HP, Yeh ET (1997) Preferential modification of nuclear proteins by a novel ubiquitin-like molecule. J Biol Chem 272: 14001–14004 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Cregg JM, Dunn WA Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y (2003) A unified nomenclature for yeast autophagy-related genes. Dev Cell 5: 539–545 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD (2000) Autophagy as a regulated pathway of cellular degradation. Science 290: 1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Tanida I, Ueno T, Ohsumi M, Ohsumi Y, Kominami E (2001) The C-terminal region of an Apg7p/Cvt2p is required for homodimerization and is essential for its E1 activity and E1–E2 complex formation. J Biol Chem 276: 9846–9854 [DOI] [PubMed] [Google Scholar]
- Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, Goebl M, Estelle M (1998) Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev 12: 914–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos D, Doenges G, Matuschewski K, Jentsch S (1998) A novel protein modification pathway related to the ubiquitin system. EMBO J 17: 2208–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC, Pan J, Zhang C, Fan W, Collinge M, Bender JR, Weissman SM (1999) A MHC-encoded ubiquitin-like protein (FAT10) binds noncovalently to the spindle assembly checkpoint protein MAD2. Proc Natl Acad Sci USA 96: 4313–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y (1998) A protein conjugation system essential for autophagy. Nature 395: 395–398 [DOI] [PubMed] [Google Scholar]
- Nakamura M, Xavier RM, Tsunematsu T, Tanigawa Y (1995) Molecular cloning and characterization of a cDNA encoding monoclonal nonspecific suppressor factor. Proc Natl Acad Sci USA 92: 3463–3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume T, Yamauchi Y, Nakayama H, Shinkawa T, Yanagida M, Takahashi N, Isobe T (2002) A direct nanoflow liquid chromatography–tandem mass spectrometry system for interaction proteomics. Anal Chem 74: 4725–4733 [DOI] [PubMed] [Google Scholar]
- Ohsumi Y (2001) Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2: 211–216 [DOI] [PubMed] [Google Scholar]
- Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, Kawashima S, Tanaka K, Kato S (1998) A new NEDD8-ligating system for cullin-4A. Genes Dev 12: 2263–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Schwartz DC, Hochstrasser M (2003) A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem Sci 28: 321–328 [DOI] [PubMed] [Google Scholar]
- Shintani T, Mizushima N, Ogawa Y, Matsuura A, Noda T, Ohsumi Y (1999) Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J 18: 5234–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Suzuki T, Chiba T (1998) The ligation systems for ubiquitin and ubiquitin-like proteins. Mol Cells 8: 503–512 [PubMed] [Google Scholar]
- Tanida I, Komatsu M, Ueno T, Kominami E (2003) GATE-16 and GABARAP are authentic modifiers mediated by Apg7 and Apg3. Biochem Biophys Res Commun 300: 637–644 [DOI] [PubMed] [Google Scholar]
- Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E (1999) Apg7p/Cvt2p: a novel protein-activating enzyme essential for autophagy. Mol Biol Cell 10: 1367–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A (1997) The ubiquitin system. Trends Biochem Sci 22: 383–387 [DOI] [PubMed] [Google Scholar]
- Walden H, Podgorski MS, Huang DT, Miller DW, Howard RJ, Minor DL Jr, Holton JM, Schulman BA (2003a) The structure of the APPBP1–UBA3–NEDD8–ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol Cell 12: 1427–1437 [DOI] [PubMed] [Google Scholar]
- Walden H, Podgorski MS, Schulman BA (2003b) Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature 422: 330–334 [DOI] [PubMed] [Google Scholar]
- Yeh ET, Gong L, Kamitani T (2000) Ubiquitin-like proteins: new wines in new bottles. Gene 248: 1–14 [DOI] [PubMed] [Google Scholar]


