Abstract
In a widely accepted model, the steroid receptor RNA activator protein (SRA protein; SRAP) modulates the transcriptional regulatory activity of SRA RNA by binding a specific stem-loop of SRA. We first confirmed that SRAP is present in the nucleus as well as the cytoplasm of MCF-7 breast cancer cells, where it is expressed at the level of about 105 molecules/cell. However, our SRAP-RNA binding experiments, both in vitro with recombinant protein and in cultured cells with plasmid-expressed protein and RNA, did not reveal a specific interaction between SRAP and SRA. We determined the crystal structure of the carboxy-terminal domain of human SRAP and found that it does not have the postulated RRM (RNA recognition motif). The structure is a five-helix bundle that is distinct from known RNA-binding motifs and instead is similar to the carboxy-terminal domain of the yeast spliceosome protein PRP18, which stabilizes specific protein-protein interactions within a multisubunit mRNA splicing complex. SRA binding experiments with this domain gave negative results. Transcriptional regulation by SRA/SRAP was examined with siRNA knockdown. Effects on both specific estrogen-responsive genes and genes identified by RNA-seq as candidates for regulation were examined in MCF-7 cells. Only a small effect (~20% change) on one gene resulting from depletion of SRA/SRAP could be confirmed. We conclude that the current model for SRAP function must be re-evaluated; we suggest SRAP may function in a different context to stabilize specific intermolecular interactions in the nucleus.
Keywords: estrogen, nuclear receptor, RNA-binding protein, transcription factor, steroid receptor activator
Introduction
Steroid hormone receptors bind hormones such as estrogen, glucocorticoid or progesterone within the cell and then regulate transcription of steroid-responsive genes. In 1999, an RNA that increased the transcriptional activation of steroid receptors was identified using a yeast two-hybrid screen with the amino-terminal domain of the human progesterone receptor as bait [1]. Hence, the RNA was entitled “steroid receptor RNA activator”, or “SRA”. It was subsequently proposed that SRA asserts similar regulatory effects on transcription in additional biological contexts, such as myogenesis [2] and adipogenesis [3], reviewed recently in [4].
Multiple splice variants of SRA gene transcripts have been identified, in addition to the three that were originally described [5, 6]. Some of them include a candidate open reading frame and two potential start codons for a protein of 224 or 236 amino acids. Translation of this protein from coding variants has been confirmed [7-9], and the protein has been designated “steroid receptor RNA activator protein”, or “SRAP”. The properties of SRAP, including its evolutionary conservation and tissue distribution, as well as its proposed biological activity as a modulator of SRA-dependent transcriptional regulation, have been reviewed [10].
Both SRA RNA and SRAP have been suggested to regulate the transcriptional activity of estrogen receptor (ER) [11-15]. The ratio between the non-coding and coding forms of SRA has been suggested to correlate with the invasion of breast cancer cells [16]. A model for intertwined activities of SRA and SRAP has emerged in which (i) SRA RNA participates as a co-regulator (in most cases reported, a co-activator) of transcription of target genes; (ii) SRAP modulates the activity of SRA by binding a specific stem-loop, designated “stem-loop 7” (STL7), of the RNA; and (iii) the level of SRAP, and by implication the level of its modulation, is controlled by the ratio of coding to noncoding splice variants of the SRA gene transcripts. In essence, this model suggests an intriguing feedback mechanism by which the activity of a “sometimes coding” RNA is self-regulated by the level of modulating protein produced by coding variants of its transcripts. Additionally, other proteins have been suggested to modulate SRA transcriptional regulation, including the SMRT/HDAC1 associated repressor protein (SHARP) [17] and the SRA stem-loop interacting RNA binding protein (SLIRP) [18].
To delineate the molecular basis for this proposed regulatory mechanism, we undertook biochemical, structural, genetic and cell biology studies. To this end, we have expressed and purified recombinant SRAP variants and have carried out protein-RNA binding assays with SRA derivatives. Additionally, we tested in vivo RNA pulldown with SRAP and have searched for evidence of SRA/SRAP-dependent regulation of transcription of specific genes through siRNA knockdown experiments in cells. Our results generally do not corroborate the current model for SRA/SRAP interaction and regulation.
Results
Expression level and cellular localization of SRAP in MCF-7 cells
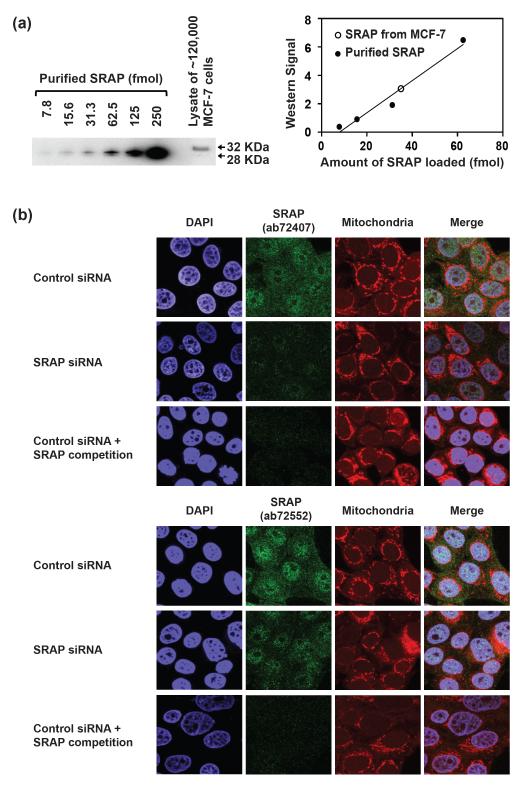
An essential requirement for SRAP to assert an activity as a regulator of transcription is that the protein is expressed and targeted to the cell nucleus. In this context, we measured the expression level and cellular localization of SRAP in the human breast ductal carcinoma cell line, MCF-7. To determine the endogenous expression level of SRAP, we used western blotting, comparing the signal from cell lysates to that of purified recombinant SRAP as a standard (Fig. 1a). (The mobility of endogenous SRAP in MCF-7 on SDS-PAGE differs from that of recombinant SRAP. As discussed in more detail in Materials and Methods and Fig. S1, the MCF-7 SRAP isoform has a V->RL insertion and may in addition be post-translationally modified. The epitope of the antibody used in the western blot lies outside the site of the mutation). Quantification of the western signal demonstrated that there are approximately 1.8 × 105 molecules of SRAP per MCF-7 cell.
Figure 1. SRAP expression level and localization in MCF-7 cells.
(a) SRAP quantitative western blot. Left: Lanes 1-6, purified SRAP (13-236) protein; lane 7, lysate from ~120,000 MCF-7 cells. SRAP was probed with Abcam antibody ab72552. Right: plot of western signal versus amount of SRAP. Open circle, signal from cell lysate. (b) Immunofluorescence analysis of SRAP in MCF-7 cells. SRAP was probed with Abcam antibodies ab72407 (top panels) or ab72552 (bottom panels). The signal specificity was confirmed by knocking down SRAP with pool 2 SRA siRNA (middle rows) as well as by competition with recombinant SRAP (13-236) protein (bottom rows).
Previous studies of SRAP expressed from a plasmid transfected into MCF-7 cells [8] and endogenous SRAP in HeLa cells [19] suggest it localizes to both the nucleus and cytoplasm. To examine the localization of endogenous SRAP in MCF-7 cells, we performed immunofluorescence experiments (Fig. 1b). We used two different rabbit polyclonal antibodies (Abcam ab72407 and ab72552) independently to check for consistency of results (upper and lower panels, Fig. 1b). We observed both nuclear and cytoplasmic localization of SRAP (top rows of figure). The signal was substantially attenuated when SRAP was knocked down with a siRNA pool (pool 2 SRA siRNA, described in more detail below) (middle rows). It was also attenuated when recombinant SRAP protein was added as a competitive inhibitor of antibody binding (bottom rows). While this manuscript was in preparation, another group published results on the localization of endogenous SRAP in MCF-7 cells, using different antibodies [20]; our results are in agreement with theirs.
In summary, SRAP is expressed at substantial levels (of order 105 molecules/cell) in MCF-7 cells. In agreement with the results of others, we find SRAP in both the cytoplasm and the nucleus, supporting the possibility that it asserts a biochemical activity in the nucleus.
Binding of SRA RNA to recombinant SRAP in vitro
It has been proposed that SRAP modulates transcriptional regulation by SRA RNA by binding a specific putative stem-loop of the RNA, designated stem-loop 7 (STL7) in the literature [6]. Therefore, we first attempted to demonstrate a specific SRAP-RNA interaction in vitro with purified components. Recombinant SRAP proteins initiating from both the first start codon (designated SRAP (1-236) for the amino acids it encompasses) and the second start codon (SRAP (13-236)) were expressed in E. coli as carboxy-terminal fusions to maltose binding protein (MBP) and purified after protease cleavage of the MBP tag as described in Materials and Methods. Our data (Fig. S1, and discussed in Materials and Methods) show that the second start codon is used predominantly in vivo; hence, SRAP (13-236) was the primary form used for in vitro experiments, with the primary conclusions being corroborated with experiments using SRAP (1-236).
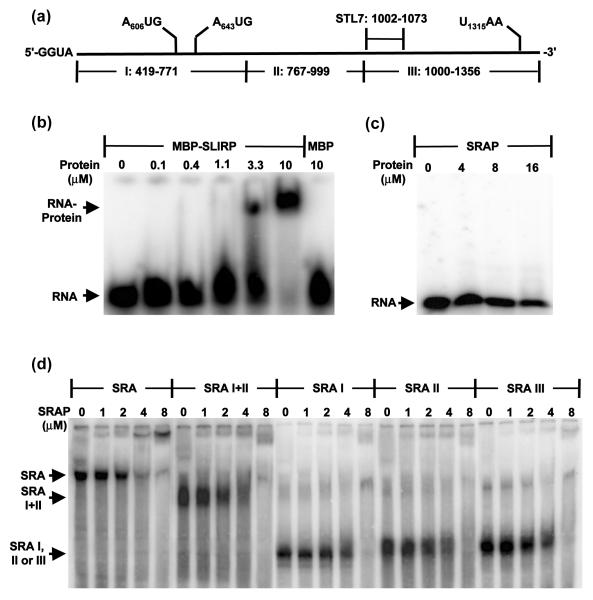
For electrophoretic mobility shift assays (EMSAs), RNAs were synthesized by in vitro transcription, purified and labeled as described in Materials and Methods. Initial efforts to demonstrate binding of SRAP to stem-loop 7 of SRA RNA (Fig. 2a), which has been proposed to be its specific target [6], failed to yield positive results. Hence, a control binding experiment was implemented with the protein SLIRP, which was originally isolated and characterized as a protein that binds STL7 [18]. Using a recombinant MBP-SLIRP fusion that was expressed and purified from E. coli we demonstrated that under the conditions of our experiments, SLIRP binds STL7 with an apparent affinity in the micromolar range (Fig. 2b). Using the same conditions, SRAP showed no apparent binding of STL7 at a concentration of 16 μM (Fig. 2c).
Figure 2. Direct assay for SRA RNA binding by SRAP in vitro.
(a) Schematic diagram of RNA fragments used for EMSA. Numbering corresponds to NCBI reference sequence NM_001035235. (b) Control gel shift of STL-7 RNA by MBP-SLIRP. (c) Gel shift of STL-7 RNA by SRAP. (d) Gel shift of full-length SRA RNA and subfragments thereof by SRAP.
We next attempted binding experiments with a 938-nucleotide construct of SRA RNA, which we refer to as “full-length”, and large fragments thereof (Fig. 2d). Throughout transcription and purification, denaturation of these RNAs was avoided. Loading dyes were excluded from samples that had both protein and RNA to preclude the possibility of dye interference with SRASRAP interaction. Binding to full-length SRA RNA was also checked with SRAP (1-236), the longer putative translation product; no substantial differences from results with SRAP (13-236) were found. These experiments demonstrate a very weak affinity of SRAP for SRA RNA, in the micromolar range. They also show that SRAP binds large, non-overlapping fragments of SRA RNA (fragments I + II, I, II and III) with similar affinities, suggesting a sequence- and site-nonspecific RNA binding activity.
Binding of SRA RNA to SRAP in cultured HEK293T cells
Arguably, a failure to demonstrate a specific interaction between SRAP and SRA RNA in vitro might be due to any of a number of deficiencies in the purified system, such as lack of post-transcriptional modification of the RNA [21], lack of post-translational modification of the protein [22-24], absence of other essential binding partners in a multi-component complex, and lack of assembly chaperones. To address this concern, we co-expressed SRAP and SRA RNA in HEK293T cells in order to include intracellular factors and/or modifications that might be essential for complex formation. In parallel, we carried out experiments with intracellular expression of human telomerase reverse transcriptase (hTERT) and human telomerase RNA (hTR), a system for which successful co-expression and assembly of components into an active protein-RNA complex is well documented ([25] and references therein).
Mimicking the hTERT/hTR precedent, the protein-coding sequence of hTERT was replaced with that of SRAP in a mammalian expression plasmid that utilizes an amino-terminal 3xFLAG tag, and the coding sequence of hTR was replaced with that of SRA RNA, with the modification that the two ATG translational start codons were mutated to TAG in order to preclude expression of SRAP from this plasmid, an approach for which there is a precedent [26].
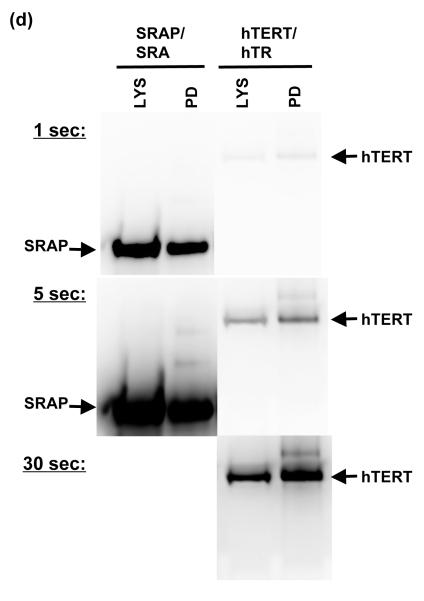
After co-transfection and growth with the alternative protein/RNA plasmid combinations, 3xFLAG-tagged protein (SRAP or hTERT) was adsorbed to anti-FLAG beads. RNA was isolated from both the beads (pull-down) and the residual supernatant from the pull-down. Samples were run on denaturing gels and RNA was detected with northern blots (Fig. 3). In each of three replicate experiments, SRAP failed to pull down any SRA RNA (Fig. 3a). hTERT successfully pulled down the majority of the hTR RNA, while SRAP did not pull down hTR (Fig. 3b). Western blots demonstrated that SRAP was expressed at significant levels in the HEK293T cells under the conditions of our experiments, and that anti-FLAG beads successfully bound it for pull-down (Fig. 3c). Anti-FLAG western blots on samples with equal amounts of SRAP or hTERT material (lysate or pull-down) run on the same gel provided a direct comparison of relative expression levels of the two proteins; SRAP was expressed at substantially higher levels than hTERT in our experiments (Fig. 3d).
Figure 3. Testing for SRAP-SRA RNA binding in human cells by coimmunoprecipitation.
Protein-RNA pairs were expressed in HEK293T cells using a dual plasmid system as described in Materials and Methods. Anti-FLAG peptide beads were used to pull down FLAG-tagged protein (SRAP or hTERT) from cell lysates. RNA (SRA or hTR) was trizol-extracted from lysate (LYS), pull-down beads (PD), and pull-down supernatant (SUPT). For each set of samples, volume of SUPT used (corrected for dilution), and volume of PD (corrected for concentration by centrifugation and resuspension) was approximately equal to volume of LYS used. Samples were run on a denaturing 4% acrylamide gel; ~2 ng each of in vitro transcribed SRA and hTR RNAs were run in separate lanes for reference. (a) Northern blot probed with labeled anti-SRA oligonucleotides. (b) Northern blot from (a) probed additionally with anti-hTR oligonucleotides. (c) Western blot with an anti-FLAG antibody of cell lysate and pull-down samples from SRAP/SRA and SRAP/hTR expression, demonstrating efficient pull-down of SRA protein. Volume of PD (corrected for concentration by centrifugation and resuspension) was approximately equal to volume of LYS. (d) Western blot with an anti-FLAG antibody of cell lysate and pull-down samples from SRAP/SRA and hTERT/hTR expression (equal volumes of each), demonstrating expression level of SRAP relative to that of TERT.
In summary, in these experiments we find no evidence for formation of a stable complex that includes SRAP and SRA RNA in cultured human cells.
Structure and activity of the carboxy-terminal domain of SRAP
It has been noted that the sequence of residues L163LVQEL of SRAP is consistent with an “RNP-2” RNA binding motif found in some RNA recognition motif structural domains [27], leading to the suggestion that the carboxy-terminal domain of SRAP is an RNA binding motif [5]. In support of this suggestion, it was shown that mutating the hexapeptide motif to GGGQEL (mutated residues underlined) impaired SRAP-dependent pull down of SRA RNA in HEK293 cells. To examine this suggestion, we pursued structural and biochemical studies on this domain.
A fragment of human SRAP consisting of residues 105-215, with an amino-terminal V105M mutation, was crystallized. The structure was solved to 2.8 Å resolution by molecular replacement, using an unpublished NMR model of the mouse homolog (PDB ID 2YRU, 85% identical in sequence), as described in Materials and Methods. Correct register of the sequence placement was confirmed by the overlap of sulfur atoms of cysteine and methionine residues of the model with peaks of an anomalous difference Fourier computed using anomalous differences derived from data collected at λ = 1.5 Å (Fig. S2). There are two SRAP fragment protomers per asymmetric unit. They were modeled and refined independently; they superimpose with a root mean square deviation (rmsd) of distances between Cα carbons of 0.4 Å. The protomers of the refined structure superimpose on the NMR model used in molecular replacement with an rmsd difference between Cα carbons of 0.8 Å.
The structure does not comprise an RRM, but rather a five-helix bundle (Fig. 4). The five-helix bundle was noted previously for the NMR structure, although the absence of an RRM was not discussed [10]. For discussion, the helices are designated A-E sequentially. The hexapeptide suggested to be an RNA binding motif lies within helix C (Fig 4a-b). Of the three residues that were mutated to glycine, the side chains of two of them, Leu165 and Val166, are packed in the hydrophobic interior of the protein. Therefore, mutation of these residues would be expected to disrupt the folding of the protein.
Figure 4. Crystal structure of SRAP carboxy terminal domain and its similarity to the structure of PRP18 carboxy-terminal domain.
(a-b) Cartoon drawing of the crystallographic structure of the carboxy-terminal domain of SRAP (residues 105-215). (a) Side view; helices are lettered sequentially. Side chains of the six residues suggested to comprise an RNP-2 motif, L163LVQEL, shown as stick models [6]. The three residues that were mutated to glycines to test RNA binding in the published study are additionally indicated with semitransparent spheres. (b) Top view. (c) Schematic drawing of the domain organization of SRAP and PRP18, indicating the fragments that were crystallized. (d) Cartoon drawing of the SRAP and PRP18 domains illustrating their similarity in structure. Molecular drawings were made with PyMOL (http://www.pymol.org).
EMSA experiments were performed to test whether the carboxy-terminal domain of SRAP binds SRA RNA fragments. Under experimental conditions in which full-length SRAP weakly bound SRA and large subfragments thereof, the fragment SRAP (V105M-215) failed to manifest any binding of SRA RNA fragments I, II, or III at 10 μM protein concentration (Fig. S3).
A search for similar protein structures was undertaken with the DALI server (www.ebi.ac.uk/dali). A single structure, a carboxy-terminal domain of yeast splicing factor PRP18, was found; 99 amino acid residues of the PRP18 domain aligned with the SRAP domain with a rmsd of Cα positions of 2.0 Å and sequence identity of 16% between aligned residues. This similarity has been noted previously [10]. The relationship of the PRP18 domain to that of SRAP, and the relationship of each to its respective parent protein, are summarized in Fig. 4c. The structural similarity is shown in Fig 4d. Both domains are five-helix bundles; the most significant difference between them is an insertion between helices D and E of PRP18, much of which was disordered in that crystal structure.
It is somewhat surprising that only a single structural homolog to the SRAP five-helix bundle was found, in view of the simplicity of the structure. In this context, we can ask what is unique about this particular fold. Arguably the most notable feature, relative to other folds such as four-helix bundles, is the manner in which helix D projects from a “platform” formed by helices C and E. In the discussion section, we consider whether this may have functional significance.
Effects of SRA knockdown on estrogen signaling in MCF-7 cells
Lack of evidence from our experiments in vitro and in cultured cells for a specific, stable complex between SRAP and SRA RNA does not preclude the possibility that SRAP or SRA modulates transcription of specific target genes independent of the other component. Since both SRA and SRAP were suggested to perform regulatory roles in estrogen signaling, we investigated SRA/SRAP modulation of estrogen-dependent transcription in MCF-7 cells (which have only estrogen receptor α (ERα) and are devoid of estrogen receptor β (ERβ) upon depletion of endogenous SRA/SRAP through siRNA knockdown. For these experiments, we utilized knockdowns rather than overexpression out of concern that overexpression may drive interactions that do not occur when the components are present at physiological concentrations.
Our strategy was to use a pool of several siRNAs for knockdown experiments rather than just a single siRNA, since this has been shown to mitigate off-target effects [28]. Initially, we compared the efficacy of two siRNA reagent pools. The first (“pool 1”, Dharmacon Smartpool, D-120329) has been used previously in expression profiling of the SRA gene [29], while the second (“pool 2”, Thermo Scientific ON-TARGETplus), which became available more recently, is designed to further reduce off-target effects by dual-strand modification [30]. A control set of non-target siRNAs was designed for pool 1 by scrambling the sequence of each siRNA; a control set of non-target siRNAs was commercially available for pool 2.
We observed a more complete knockdown of SRA RNA with pool 2, to <10% of control levels, versus pool 1, ~20% of control levels (Fig. 5a). Additionally, we observed that MCF-7 cells transfected with pool 1 SRA siRNA exhibited a significantly slower growth rate (p < 0.05) than cells in the absence of siRNA or transfected with pool 1 control siRNA (Fig. 5b). Following up on this observation, we transfected MCF-7 cells with individual siRNAs of pool 1 SRA siRNA and pool 1 control siRNA. Only one siRNA induced the attenuation of cell growth (Fig. S4), suggesting that this phenotype is an off-target effect. This may correlate with the earlier observation that several cell growth-related genes showed expression level changes when SRA was knocked down with pool 1 SRA siRNA [29]. We did not observe a similar effect on cell growth rate with pool 2 SRA siRNA. Consequently, we used pool 2 SRA siRNA and control siRNA for subsequent experiments.
Figure 5. Knockdown of SRA RNA and SRAP.
In each case, approximately 200,000 MCF-7 cells were transfected with 25 nM of the indicated siRNA pool or no RNA. (a) SRA RNA level, measured by RT-qPCR (primer: SRA-3, with GAPDH mRNA as internal control) 3 days after transfection (n = 1). (b) Cell number, measured 3 days after transfection by flow cytometry (error bars represent standard deviation (SD), n = 2). (c-d) Time dependence of knockdown efficiency of pool 2. MCF-7 cells were transfected with 25 nM corresponding siRNA pool. (c) SRA RNA level, measured by RT-qPCR (error bars represent SD for n = 3; primer: SRA-4, with GAPDH mRNA as internal control). (d) SRAP protein level, measured by western blot (antibody: Abcam ab72552, with GAPDH protein as internal control).
To optimize knockdown conditions, we examined the time dependence of the siRNA treatment (Fig. 5c-d). At all the time points we tested (1-3 days after siRNA transfection), pool 2 SRA siRNA induced efficient knockdown of both SRA RNA (Fig. 5c) and SRAP protein (Fig. 5d), and the knockdown efficiency was stable throughout the three-day time course.
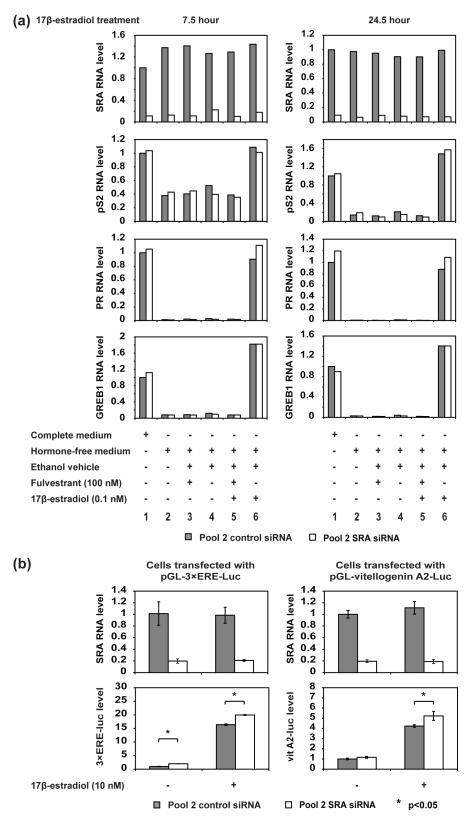
Additionally, to optimize estrogen-dependent gene expression signals within the physiological range of estrogen levels, various 17β-estradiol concentrations were tested. Fulvestrant, an antagonist of estrogen receptors, was used to confirm that the effects observed were caused specifically by 17β-estradiol signaling through estrogen receptors. The results (Fig. S5) show that pS2 (the Trefoil factor 1 gene) and PR (progesterone receptor gene), two endogenous estrogen-responsive genes, were clearly induced by 17β-estradiol treatment when cells were grown in hormone-free media to reduce the background expression level. The level of SRA RNA, however, was not affected by 17β-estradiol. By assessing the gene expression levels of pS2 and PR in a 17β-estradiol dose response experiment (Lane 8-11) compared to expression in unsupplemented medium (Lane 3), we chose a 17β-estradiol concentration of 0.1 nM, the lowest level that induces a substantial increase in mRNA levels, for experiments measuring the expression of endogenous genes.
We then treated cells transfected with pool 2 control siRNA or pool 2 SRA siRNA with 0.1 nM 17β-estradiol and measured the endogenous mRNA level of pS2, PR, and GREB1 (growth regulation by estrogen in breast cancer 1), another estrogen-responsive gene (Fig. 6a). To monitor both early and late responses, we tested different 17β-estradiol treatment times (7.5 hours and 24.5 hours). Single-replicate measurements under each condition revealed a clear and consistent trend. SRA RNA was efficiently knocked down (top panels), and the 3 estrogen-responsive genes exhibited substantially increased mRNA levels in response to 17β-estradiol treatment (compare lane 6 to lanes 2-5). However, depleting SRA had no apparent effect on the responses of the pS2 and GREB1 genes to 17β-estradiol, while the PR RNA levels showed ~20% enhancement. We then monitored the mRNA levels of pS2 and PR over a three day period after siRNA transfection, making measurements in triplicate to verify statistical significance, and found the same trend; PR showed ~20% increase in mRNA level after SRA knockdown, while pS2 showed no significant change (Fig. S6).
Figure 6. Effect of SRA RNA knockdown on estrogen-dependent gene expression.
(a) Knocking down SRA does not significantly affect the transcription of estrogen-dependent genes pS2, PR and GREB1 in MCF-7 cells. Lane 1: MCF-7 cells were grown in complete medium and transfected with 25 nM corresponding siRNA pool. Lanes 2-6: MCF-7 cells were grown in hormone-free medium for 3 days and then transfected with 25 nM corresponding siRNA pool. 24 hours after transfection, the cells were treated with 17β-estradiol with or without fulvestrant for 7.5 hours (left panels) or 24.5 hours (right panels). The RNA level of SRA, pS2, PR and GREB1 was quantified with RT-qPCR with GAPDH mRNA as an internal control (SRA primer: SRA-4) (n = 1).
(b) Knocking down SRA does not significantly affect the transcription of an estrogen-controlled luciferase gene from transfected plasmids. MCF-7 cells were grown in hormone-free medium for 3 days and then transfected with 25 nM corresponding siRNA pool. 23 hours after siRNA transfection, the cells were transfected with luciferase reporter plasmids. 5 hours after the plasmid transfection, cells were treated with 10 nM 17β-estradiol or ethanol vehicle. 18 hours after that, cells were harvested and luciferase activity was measured (error bars represent SD, n = 3).
It has previously been reported that overexpression of SRA RNA in HeLa cells from a human cytomegalovirus-driven expression vector enhances the luciferase signal from a hormone-responsive luciferase reporter plasmid approximately tenfold over its level in the absence of SRA [1, 31]. To look for similar behavior in MCF-7 cells, we used two different plasmids with the firefly luciferase gene under the control of estrogen-responsive promoters: pGL3-3×ERE-Luc, which has 3 copies of the classic vitellogenin ERE (Estrogen Response Element), and pGL3-vitellogenin A2-Luc, which has the endogenous vitellogenin A2 promoter sequence −331 to −87. We asked whether depletion of SRA RNA from endogenous levels would result in a reduction of estrogen-dependent luciferase signal in MCF-7 cells. In experiments with these plasmids, we treated MCF-7 cells with a higher level of 17β-estradiol (10 nM) than in the previous experiment in order to increase the estrogen-induced signal, as well as the potential change in signal resulting from SRA knockdown. Both plasmids showed a substantially elevated luciferase signal in response to 17β-estradiol treatment (Fig. 6b); however, knocking down SRA RNA did not attenuate the estrogen-dependent luciferase expression. On the contrary, SRA RNA knockdown resulted in a small (~10-15%) but statistically significant increase in luciferase expression. This increase is similar to the percentage increase we described above for PR mRNA in response to SRA knockdown (Fig. 6a).
In these experiments, we have searched for evidence for SRA/SRAP modulation of estrogen-dependent transcription of three specific genes and from specific hormone-dependent promoters when SRA is depleted from endogenous levels through siRNA knockdown. The effects we observe range from negligible to relatively small enhancements of the level of transcription; they are substantially smaller and opposite in sign from what would be expected from previous reports [1, 26, 31].
Attempts to identify SRA-regulated genes with RNA-seq
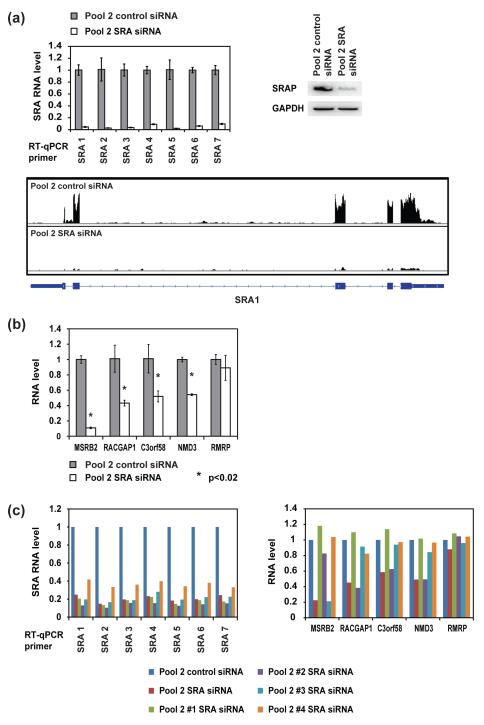
Since we were unable to detect accentuated regulation of estrogen-responsive gene expression by SRA/SRAP in the specific cases we examined, we undertook a genome-wide screen for SRA-regulated genes by deep sequencing. Although a conceptually similar experiment using microarrays was described previously [29], our identification of a siRNA pool that gave improved knockdown of SRA without affecting cell growth provided the incentive for new experiments. Cells were grown in normal medium, rather than hormone-free medium supplemented with 17β-estradiol, in order to more closely approximate physiological conditions, and were harvested 32 hours after transfection. RT-qPCR and western blotting confirmed that both SRA and SRAP were successfully depleted (Fig. 7a). Because SRA RNA has alternative splicing isoforms, we tested seven different RT-qPCR primers spanning the entire SRA core region [1]; all the primers revealed efficient knockdown.
Figure 7. RT-qPCR examination of the top candidates for SRA/SRAP-regulated genes identified with RNA-seq.
(a) Verification of SRA knockdown in the RNA-seq samples. MCF-7 cells were transfected with 25 nM corresponding siRNA pool. Cells were harvested 32 hours after transfection. Top left: SRA RNA level, measured by RT-qPCR (error bars represent SD, n = 3, with GAPDH mRNA as internal control). Top right: SRAP protein level, measured by western blot (antibody: Abcam ab72407, with GAPDH protein as internal control). Bottom: RNA-seq data at the locus of SRA also confirm the knockdown. (b) RT-qPCR verification of the expression level differences of the candidate genes at 32 hours after transfection with 25 nM corresponding siRNA pool, with GAPDH mRNA as internal control (error bars represent SD, n = 3). (c) The expression level changes of the candidate genes are not consistent when SRA is knocked down with individual siRNAs in pool 2. MCF-7 cells were transfected with 25 nM of the indicated siRNA. 32 hours after transfection, RNA levels of SRA and the candidate genes were examined with RT-qPCR, with GAPDH mRNA as internal control (n = 1).
157 million reads were generated from triplicate samples of the pool 2 control siRNA group (46 million from replicate 1, 62 million from replicate 2, 49 million from replicate 3); 151 million reads were generated from triplicate samples of the pool 2 SRA siRNA group (57 million from replicate 1, 29 million from replicate 2, 65 million from replicate 3). The reads were aligned to the human genome with the program tophat2.0.4 in conjunction with bowtie2.0.0. Expression level differences of each gene were analyzed with cuffdiff. Criteria used to select top candidates for SRA-regulated genes were: cutoff of q value ≤ 0.05, fold change ≤ 0.5 or ≥ 2, and reads per kilobase per million mapped reads (RPKM) ≥ 1. We only identified 6 genes that met all the conditions listed above (Table 2 and supplementary file); as expected, one of these was SRA. Examination of the distribution of total reads at the SRA locus further confirmed the nearly complete knockdown of this gene product (Fig. 7a, bottom panel). We then used RT-qPCR to examine the RNA level of the other 5 genes (Fig. 7b). Except for RMRP, the expression level changes of the other 4 genes were confirmed and the fold change corresponded well with that obtained from the RNA-seq results. We then tested whether these genes were true SRA targets by knocking down SRA with the individual siRNAs in pool 2 SRA siRNA (Fig. 7c). RT-qPCR analysis of SRA RNA level showed that each of the 4 siRNAs knocked down SRA efficiently. However, when we examined the RNA level of the other 5 candidate genes, in each case only one of the four individual siRNAs induced gene expression changes similar to that induced by the whole pool. Therefore, the observed gene expression changes do not correlate rigorously with knockdown of SRA; they are likely to be off-target effects of individual siRNAs [32]. We do not observe any changes in gene expression that can be unequivocally attributed to depletion of SRA/SRAP.
Table 2.
Top candidate SRA-regulated genes in MCF-7 cells identified with RNA-seq.
| Gene Name | RPKM* of Control |
RPKM of SRA Knockdown |
Fold Change (SRA Knockdown/ Control) |
p Value | q Value |
|---|---|---|---|---|---|
| SRA1 | 18.8303 | 1.60853 | 0.085 | 0 | 0 |
| MSRB2 | 17.6106 | 2.69954 | 0.153 | 0 | 0 |
| RACGAP1 | 55.6141 | 21.8603 | 0.393 | 0 | 0 |
| C3orf58 | 3.1492 | 1.53036 | 0.486 | 2.75E-07 | 0.000126 |
| NMD3 | 137.413 | 67.1504 | 0.489 | 0 | 0 |
| RMRP | 10.5995 | 26.0199 | 2.455 | 3.8E-06 | 0.00124 |
RPKM: Reads per kilo base pair per million reads.
Discussion
Re-evaluation of SRAP as an SRA-binding protein
The contemporary model for intertwined activities of SRAP and SRA states that splice variants of SRA act as mRNAs to produce SRAP, which in turn modulates SRA-dependent transcriptional regulation when its carboxy-terminal domain binds a specific stem-loop, STL7, of SRA RNA ([4, 10] and references therein). We utilized several approaches to evaluate this model. Working with purified reagents in vitro, we find binding of SRA RNA and fragments thereof by recombinant SRAP has the characteristics of sequence-nonspecific RNA binding [33]. We found no binding specificity for STL7 of SRA. To address the possibility that cellular factors may be required for assembly of a SRAP-SRA complex, we co-expressed SRAP and SRA in HEK293T cells. However, pull-down of SRAP from cell lysates failed to carry any significant amount of SRA RNA. The carboxy-terminal domain of SRAP is a five-helix bundle (PDB ID 2YRU and this work) that has no structural similarity to known RNA-binding structural domains and in our hands does not bind RNA. Of the three amino acid residues whose simultaneous mutation to glycine were reported to abolish SRA binding [6], the side chains of two are buried and inaccessible for interactions. Our results lead us to conclude that the current model for the function of SRAP needs to be re-evaluated.
We have examined the published accounts of a specific SRAP-SRA interaction to see how the discrepancy between previous results and ours might be resolved. Evidence to support the model that SRAP specifically binds STL7 of SRA comes from experiments in which (i) full-length or deletion mutants of in vitro transcribed RNA were mixed with RNase-treated, immunoprecipitated endogenous SRAP from cell lysates, and (ii) presence of SRA RNA was monitored by RT-PCR [6]. In these experiments, of the four SRA deletions tested, only the deletion of STL7 resulted in a loss of signal attributed to bound RNA. Our experiments that argue against SRAP binding specifically to STL7 differ from these in that we measured protein-RNA binding directly using purified components.
Experiments have been reported in which SRAP (endogenous or plasmid-expressed) was immunoprecipitated from cell lysates, and bound SRA was monitored by RT-PCR amplification [6]. In view of the concern that RT-PCR may amplify a minor component, we utilized northern blots to test for stoichiometric levels of binding of SRA to SRAP in cells, with negative results. Our results still allow the possibility that, in absence of a direct interaction, SRA and SRAP may co-localize in larger molecular complexes in some cases.
Function of the SRA system (SRA/SRAP) in estrogen-dependent transcription in MCF-7 cells
Lack of a specific, stable association of SRAP with SRA does not rule out the possibility that SRA/SRAP functions as a transcriptional regulator. To test this, we have looked for changes in gene expression in MCF-7 cells consequent to siRNA knockdown of SRA, which depletes both non-coding RNA functions of SRA and the production of SRAP. First, focusing on specific estrogen-responsive genes, we found that mRNA levels increased in response to 17β-estradiol treatment as expected, but the mRNA levels were mostly insensitive to SRA/SRAP depletion, with the largest changes being ~15-20%. Then, in a genome-wide search for SRA/SRAP-dependent genes, we identified several genes whose expression levels changed in response to the siRNA knockdown. The genes identified do not overlap with the set of genes previously identified as sensitive to SRA siRNA knockdown by microarray analysis [29]. Further, we found that in each case we examined, the expression response was due to a single siRNA of the pool 2 siRNAs, indicating a probable off-target effect. The largest effects we see in response to SRA/SRAP depletion are relatively modest, ~15-20%. This contrasts with the reports of several investigators [1, 26, 31] in which overexpression of SRA from transfected plasmids resulted in as much as a tenfold enhancement of transcriptional activity. Our approach differs from theirs in that we looked for changes of expression in response to SRA/SRAP depletion from endogenous levels, rather than in response to overexpression above endogenous levels. Our concern about overexpression experiments is their potential to drive weak interactions that would not occur to any substantial extent at wild-type levels of macromolecules; thus, transcriptional changes caused by overexpression of SRA could be real but limited to the overexpression situation.
We acknowledge that our siRNA knockdown method does not allow separation-of-function observations on SRAP protein and SRA RNA independently. Previous separation-of-function studies monitoring the signal from hormone-responsive luciferase reporter plasmids have given divergent results. In one study [31], mutagenesis of the SRA sequence in a manner that would disrupt predicted RNA secondary structures while leaving the translated protein sequence unchanged identified several RNA stem-loops whose putative disruption resulted in attenuation of hormone-dependent coactivation. A later study using a similar approach [26] reported enhanced activation at EREs when RNA secondary structures were disrupted but translated protein sequence was unchanged. In a reciprocal experiment in which RNA structure was not disrupted but protein expression was abolished by mutagenizing start codons, signals from human progesterone receptor and pS2 EREs were unchanged, while the signal from a Xenopus vitellogenin ERE was enhanced. Hence, a consistent pattern for separable effects of SRA RNA versus SRAP protein has not emerged from previous studies. We have not attempted to separate the effect of SRA depletion from the effect of SRAP depletion with our knockdown methodology.
We recognize a number of caveats with our conclusions, which inevitably are drawn from data from the selected set of experimental conditions we have explored. In our RNA-seq experiments, we looked for changes occurring at 32 hours post-transfection; there is a possibility of acute responses that are ameliorated by 32 hours or effects that occurred on a longer time scale. In this regard, we have observed time-dependent effects; for example, we examined mRNA levels of pS2 and PR at different times after siRNA transfection and found PR up-regulated a few percent on days 2 and 3 (Fig. S6). However, this effect is relatively subtle. For RNA-seq we chose a cutoff for substantial effects on gene expression of < 0.5-fold and > 2-fold. We recognize that genes with more modest responses to SRA knockdown could be biologically meaningful.
Additionally, previous studies by Klinge et al. have shown that regulation of estrogen receptor activity depends on both the specific sequence of the ERE and the flanking DNA sequences [14]; we have explored a limited set of ERE sequences. Notably, in the studies by Klinge et al. it was found that overexpression of SRA had no significant effect on the estrogen-stimulated transcriptional activity of ERα on pS2 ERE, but did inhibit the activity on PR ERE. This is in agreement with our results from SRA knockdown, which showed no effect on pS2 but a slight enhancement of PR transcriptional activity when SRA was depleted.
Does PRP18 similarity suggest alternative functions for SRAP?
If SRAP is not a modulator of SRA-dependent transcriptional regulation, what is its function? SRAP is expressed at relatively high levels in cells, and several independent studies show it is partitioned in part to the nucleus ([8, 19, 20] and this work). Arguably, an intriguing hint may come from the structural similarity of the SRAP carboxy-terminal domain to that of the yeast PRP18 protein—intriguing because, although the 5-helix bundle is a very simple fold, these are the only two proteins in the protein structure database that share it. Unfortunately, the sequence similarity between these two domains is too low to allow BLAST searches to “link” them through homologous sequences, so that tracing a biological function through a path of sequence relatedness is not an option.
PRP18 is involved in the second step of yeast mRNA splicing. Specifically, genetic and mutagenesis studies have demonstrated that the carboxy-terminal domain interacts with the protein Slu7 at a weakly conserved face of the domain, and with the protein component of U5 snRNP through the highly conserved loop region linking helices D and E [34, 35]. PRP18 is thought to orient U5 snRNP in a manner that aligns an RNA loop of U5 snRNP with the exon junction; PRP18 thereby guides a specific RNA-RNA interaction through direct protein-protein interactions. Reasoning by analogy, it is interesting to consider whether the five-helix bundle found in both SRAP and PRP18 is a protein domain utilized for specific protein-protein interactions.
In summary, we do not find convincing evidence that SRAP interacts with SRA RNA to regulate transcription. SRAP resides in part in the nucleus ([8, 19, 20] and this work), suggesting it is likely to have a nuclear function. The structural similarity of its carboxy-terminal domain to that of PRP18 suggests it may be involved in the assembly of multisubunit complexes through specific protein-protein interactions. The (S/T)P-rich central domain suggests that its activity may be regulated by phosphorylation. Indeed, proteomic mass spectrometry experiments provide evidence for phosphorylation by proline-directed serine, threonine kinases (consensus target sequence (S/T)P) in this domain [22-24]. The specific targets of SRAP, and by implication, its biological function, remain to be found.
Materials and Methods
Reagents and plasmids
LB-based autoinduction medium with trace metal ions for E. coli growth was obtained from Formedium, United Kingdom (AIMLB0210). 17β-estradiol and fulvestrant (Sigma) were dissolved in ethanol and DMSO, respectively.
SRAP and SLIRP protein coding sequences that were codon-optimized for expression in E. coli were synthesized by GenScript and provided in pUC57 plasmids. Standard cloning methods were used to construct plasmids for expression of recombinant SRAP and SLIRP or subfragments thereof in E. coli as either (a) carboxy terminal fusions to maltose binding protein (MBP) in tandem with a tobacco etch virus (TEV) protease cleavage site, or (b) amino terminal fusions to the intein-chitin binding domain of the pTWIN1 vector (New England Biolabs). Plasmid pRK1043 for expressing TEV protease as a MBP-TEV[S219V]-Arg5 protease fusion in E. coli [36], referred to as MBP-TEV protease in this article, was obtained from Addgene.
A coding sequence for hSRA (nucleotides 419-1356 of GenBank sequence NM_001035235) preceded by the T7 promoter and KpnI recognition sequence (used in making 5′ deletions), and with a SapI site at the 3′ end, was synthesized by GenScript and provided in plasmid pUC57. The endogenous SapI site of pUC57 was subsequently removed from the vector.
To construct plasmids for transient transfections in human cells, the native coding sequence for human SRAP (amino acid residues 1-236) with an amino-terminal 3xFLAG tag, and the coding sequence for SRA as described above, with the two ATG ‘start’ codons for SRAP protein mutated to TAG ‘stop’ codons, were cloned into vectors provided by J. Lingner [37] and used for similar coexpression with human telomerase RNA and reverse transcriptase protein [25].
pGL3-vitellogenin A2-Luc (which has the vitellogenin A2 promoter region, −331 to −87 [38], inserted upstream of a minimal TATA box-containing promoter driving firefly luciferase expression) was constructed as follows. Xenopus laevis genomic DNA was amplified with the sense primer: 5’GGGGTACCGGTCACAGTGACCTG and the antisense primer: 5’AACTGCAGACTGTTATCAGGTTGTGTAAAC. The amplicon was inserted between the KpnI and PstI sites of p3APP-Luc, a derivative of pGL3-basic containing the TATA box sequence: 5’AGGGTATATAAT [39]. pGL3-3×ERE-Luc (which has 3 copies of the classic vitellogenin ERE consensus sequence, TTATAAAGTCCACGCGCAACCCGCGGACTCACCAT, followed by a TATA box and the E1a minimal promoter driving firefly luciferase expression) and pRL-TK (a control plasmid for luciferase-based assays which has the thymidine kinase promoter driving renilla luciferase expression) were contributed by Dr. Leslie Leinwand, University of Colorado.
Protein expression and purification
Expression plasmids for MBP or intein fusions of SRAP or SLIRP were transformed into E. coli BL21(DE3) cells, grown on autoinduction media at 37 °C, and harvested by centrifugation. Chromatography was done at 4 °C. For MBP fusions, cells were resuspended in “buffer A” (50 mM Tris-Cl, 0.1 M NaCl, 0.4 mM EDTA, 1 mM DTT, pH 7.9) and lysed by sonication. Centrifugation, followed by precipitation of nucleic acids by addition of polyethyleneimine (PEI) to a concentration of 0.1% (w/v) and a second centrifugation, yielded a lysate that was applied directly to an amylose affinity column. Protein was eluted with buffer A + 1 M α-methyl-glucose, concentrated, and purified by FPLC gel filtration on a Superdex-75 column (GE Healthcare) equilibrated with buffer A. We found that expression of full-length SRAP in E. coli consistently yielded, in addition to full-length product, shorter variants as a substantial fraction of the product. Purification and analysis by mass spectrometry of these variants revealed them to be truncations that terminated within the central, proline-rich region of SRAP, presumably as a result of premature translation termination.
Fractions with the desired protein fusion were pooled, and either used as MBP fusions in subsequent experiments (for SLIRP), or cleaved with MBP-TEV protease overnight at 4 °C. Protease-cleaved fusions were subsequently applied to an amylose column (which bound both the MBP-TEV protease and uncleaved MBP fusion protein), and the flow-through, which contained protein without MBP tag, was collected and chromatographed on Superdex-75 equilibrated with “storage buffer” (20 mM MOPS, 80 mM NaCl, pH 7.0) as a final purification step, and stored frozen.
Cells with intein fusions were resuspended in “buffer B” (20 mM Tris-Cl, 0.5 M NaCl, 0.4 mM EDTA, pH 8.0), lysed by sonication, centrifuged and treated with PEI as described above for MBP fusions. The lysate was applied to a chitin column, washed extensively with buffer B, then washed with “buffer C” (20 mM Tris-Cl, 0.1 M NaCl, 0.4 mM EDTA, pH 8.0); intein cleavage was effected by equilibrating the column with buffer C plus 50 mM DTT overnight. Protein was eluted from the chitin column and chromatographed on Superdex-75 equilibrated with storage buffer, and stored frozen.
There are two candidate start codons present in protein-coding splice variants of SRA RNA [8]. Hence, both candidate forms of SRAP, starting at Met1 and Met13, were expressed and purified. With the MBP-SRAP fusions used, TEV protease cleavage leaves a glycine at the amino-terminus of the product. The two products are designated SRAP (1-236) and SRAP (13-236), with the amino-terminal glycine implied. Additionally, candidate mRNAs for SRAP identify three isoforms, differing in translation product by a T50I mutation (SRA2) and a V110RL insertion/mutation (SRA3) relative to wild type (SRA1) [8]. However, we have expressed only the wild-type versions of SRAP. To compare the apparent size of endogenous SRAP directly with purified protein, we performed western blots on endogenous SRAP in HeLa and HEK293T cell lysates, using recombinant SRAP proteins as molecular size standards (Fig. S1a). The endogenous SRAP in HeLa and HEK293T cells aligns with the shorter form of the protein, SRAP (13-236). MCF-7 cell lysates revealed an endogenous form of SRAP with an apparent mobility closer to that of SRAP (1-236) (Fig. S1b). We cloned and sequenced the SRA cDNA from MCF-7 cells, and found it to be the SRA3 form, with an insertion and mutation relative to wild-type SRAP. Additionally, the results of RNA-seq experiments (Fig. S1c) demonstrate predominant use of the second start codon, M13, in this cell line. This is consonant with results of sequencing of 40 clones of SRA isoforms from myogenic cells, which showed a distribution of transcription start sites in which coding transcripts favored the second start codon [6]. We conclude that the second start codon is used predominantly in vivo. Discrepancies in mobility from recombinant standards or molecular weight standards on gels, observed by others in breast cancer cells [8] and HeLa cells[7], as well as by us in this work, may be due to both the differences in size of the different isoforms and to post-translational modification of SRAP, which has been suggested by several studies [22-24]. We have not directly addressed the presence or influence of post-translational modifications of SRAP in this work.
MBP-TEV[S219V]-Arg5 protease was expressed and purified as recommended (Dr. David Waugh, http://irp.nih.gov/pi/david-waugh).
RNA transcription and purification
RNA was transcribed by T7 RNA polymerase from DNA templates using standard conditions [40]. For STL7 RNA, a 72-mer consisting of nucleotides 1002-1073 of GenBank sequence NM_001035235 was transcribed from a PCR-generated template; 2′-O-methyl deoxyoligonucleotides were incorporated in the two 5′- sites of the 3′ PCR primer in order to suppress random nucleotide addition beyond the end of the template by the polymerase [41]. Full-length SRA RNA and large (>200 nucleotide) fragments thereof were transcribed from plasmids that had been digested with SapI to define the 3′ terminus. Since the plasmid used for transcription incorporated a KpnI site following the T7 promoter, the transcribed RNAs included a 5′ GGUA(CC) in addition to the native SRA RNA sequences, where in some cases one or both of the CC nucleotides in parenthesis were endogenous to SRA.
STL7 RNA was purified by gel filtration on Superdex-75. Larger fragments of SRA RNA were purified by gel filtration on either Superdex-200 or Sephacryl-300.
Electrophoretic Mobility Shift Assays (EMSA)
For use as EMSA substrates, purified RNAs were dephosphorylated using calf intestinal phosphatase, purified by phenol extraction and ethanol precipitation, 5′- 32P-labeled using T4 polynucleotide kinase, and then purified on G-25 spin columns. STL7 RNA was heated to 90 °C for three minutes and then snap-cooled on ice; heating and snap-cooling was avoided with larger RNAs, due to the potential for misfolding. Trace amounts of RNA (< 1 nM) were equilibrated with protein in a binding buffer consisting of 20 mM MOPS, 80 mM KCl, 0.1 mg/ml BSA, 10 μg/ml polyA, 5% ficoll, pH 7.0. Samples were equilibrated at room temperature for >30 min, then loaded onto native acrylamide gels and electrophoresed in TBE at 4 °C. RNA-only samples included xylene cyanol for tracking; tracking dye was excluded from samples with protein to avoid the possibility of interference with RNA binding. After electrophoresis, gels were dried and exposed on phosphorimager plates by standard methods.
Cell Culture
MCF-7 cells were a gift from Dr. Leslie Leinwand. The cells were routinely grown in complete medium (high glucose DMEM with 10% fetal bovine serum, 2 mM Glutamax-1, 100 units/ml penicillin and 100 μg/ml streptomycin) at 37 °C, 5% CO2. If cells were treated with 17β-estradiol, they were grown in hormone-free medium (high glucose, phenol-red free DMEM with 10% charcoal-stripped fetal bovine serum, 2 mM Glutamax-1, 100 units/ml penicillin and 100 μg/ml streptomycin) for at least 3 days before the experiment. Plasmids were transfected into cells with Lipofectamine® 2000 Reagent (Invitrogen) according to the manufacturer’s instructions.
HEK293T cells were grown and transfected with plasmids as described previously [25].
RNA pull-downs and northern blots
HEK293T cells were transiently transfected with pairs of protein and RNA expression plasmids; cells were grown and lysates extracted as described previously [25]. Pull-down of 3x-FLAG-tagged SRAP or hTERT was effected by agitating lysates with anti-FLAG agarose beads overnight at 4 °C. RNA was extracted from lysates, beads, or supernatants from bead pull-downs with TRIzol (Ambion) following the manufacturer’s instructions. Redissolved RNA samples were run on denaturing 4% acrylamide gels (7 M urea, TBE), RNA was transferred to Hybond-N+ membranes (GE Healthcare RPN303B) in 1X TBE at 1.0 amp, 1.5 h, and then crosslinked to the membrane using UV irradiation. A combination of three different deoxyoligonucleotides hybridizing to different sites of SRA RNA, and two different oligonucleotides hybridizing to different sites of hTR RNA, were 5′- 32P-labeled and purified as described above. Hybridization was effected in ‘RapidHyb’ buffer (GE Healthcare UK NIF939) at 55 °C for 2 h. Blots were then washed twice with 2× SSC + 0.1% SDS, twice with 0.2× SSC + 0.1% SDS, and exposed to phosphorimager plates.
Western blots
Proteins were heat-denatured at 95 °C for 5 min in sample buffer, separated on polyacrylamide gels (NuPAGE Bis-Tris, Invitrogen) by SDS–PAGE, and transferred to HyBond-ECL membranes (GE Healthcare RPN78D) using standard protocols. The following antibodies were used at specified dilutions: rabbit polyclonal anti-SRA1 antibody (Abcam, ab72407, 1:5000), rabbit polyclonal anti-SRA1 antibody (Abcam, ab72552, 1:5000), mouse monoclonal GAPDH (A-3) antibody (Santa Cruz, sc-137179, 1:1000). Secondary antibodies, peroxidase-AffiniPure donkey anti-rabbit IgG (H+L) (Jackson, 711-035-152, 1:5000) and peroxidase-AffiniPure donkey anti-mouse IgG (H+L) (Jackson, 715-035-150, 1:5000), were used to reveal primary antibodies with SuperSignal® West Pico Chemiluminescent Substrate (Thermo Scientific). Anti-FLAG antibody (Sigma A8592), used at 1:5000 dilution, is horseradish peroxidase (HRP) conjugated and requires no secondary antibody. The signals were quantified with a FluorChem HD2 imaging system (Alpha Innotech).
siRNA transfection
Pool 1 SRA siRNA (SRA Smartpool, Dharmacon, D-120329) has been described [29]. Pool 1 nontarget siRNA was designed by scrambling the siRNA sequences in pool 1 SRA siRNA. Sequences of these siRNAs are listed in Table S1. Pool 2 SRA siRNA is ON-TARGETplus siRNA pool for human SRA1 (Thermo Scientific, L-027192-00-0005 and LU-027192-00-0002). Pool 2 Control siRNA is ON-TARGETplus Non-targeting control pool (Thermo Scientific, D-001810-10-05).
For siRNA transfection, corresponding siRNAs were reverse-transfected into 200,000-300,000 MCF-7 cells with Lipofectamine® RNAiMAX Reagent (Invitrogen) according to the manufacturer’s instructions.
RT-qPCR
RNA was extracted from cells with TRIzol (Ambion). cDNA were prepared using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). RT-qPCR was performed on the LightCycler® 480 Real-Time PCR System (Roche), with iQ™ SYBR® Green Supermix (Bio-Rad) or Lightcycler® 480 Probes Master (Roche), according to specific primers. Sequences of the primers used are listed in Table S2. The signal of the examined gene products was normalized to the signal of GAPDH mRNA for each reaction.
Flow cytometry
MCF-7 cells were trypsinized and separated to single cells with 70 μm cell strainers. The cell numbers were measured with the MoFlow flow cytometer (DakoCytomation; Fort Collins, CO) with a single 488 nm laser. Data were analyzed using Summit Software (DakoCytomation).
Luciferase assay
Cells were harvested and tested for firefly and renilla luciferase signals using the Dual-Luciferase® Reporter Assay System (Promega) according to the manufacturer’s instructions. The signal was examined with the Synergy2 microplate reader (BioTek).
Immunofluorescence
MCF-7 cells were reverse-transfected with corresponding siRNAs and seeded on coverslips in 12-well dishes. Right before harvest, the cells were incubated in DMEM containing 200 nM MitoTracker® Red CMXRos (Invitrogen) for 30 min. Then medium was removed and all the subsequent steps were performed at room temperature. The cells were fixed with 3.7% formaldehyde in PBS for 10 min and permeablized with 0.5% Triton-X 100 for 5 min. Then the cells were incubated with either Abcam antibody ab72407 (1:200) or ab72552 (1:100), described above, for 1 h. Cells were then incubated with Alexa Fluor® 488 goat anti-rabbit (H+L), highly cross-adsorbed (Invitrogen A11034, 1:500) for 30 min in the dark. Coverslips were mounted on microscope slides using Vectashield mounting medium with DAPI (Vector Laboratories), sealed with transparent nail polish and stored in the dark at 4 °C until imaging. Images were obtained using a Nikon A1R confocal microscope and analyzed with NIS-Elements AR and ImageJ software.
RNA-seq
Total RNA was purified from MCF-7 cells treated with pool 2 Control siRNA or pool 2 SRA siRNA (3 replicates for each group). cDNA libraries were prepared and double-stranded cDNA was fragmented using DNase I according to Illumina specifications, prior to adaptor ligation. Sequencing libraries were amplified and sequenced using an Illumina HiSeq 2000 sequencer. RNA-seq reads were aligned to the human genome (hg19 build) using tophat version 2.0.4 (http://tophat.cbcb.umd.edu), in conjunction with bowtie version 2.0.0 (http://bowtiebio.sourceforge.net). Expression level differences for specific genes were analyzed by cuffdiff. RNA-seq data have been deposited in the NCBI Gene Expression Omnibus data repository.
Protein crystallization and structure determination
A fragment consisting of residues 105-215 of SRAP was expressed as an intein fusion and purified as described above. Protein was concentrated to 20 mg/ml, dialyzed into 1.0 M malonate(Na+), 0.1 M MOPS, pH 6.8, and crystallized in hanging drops by gradually increasing the malonate concentration in the precipitant wells from 1.4 M to 2.0 M. Crystals were stabilized in 2.4M malonate(Na+), 0.1 M MOPS, pH 6.5, and flash-frozen in the same solution, which served as a cryoprotectant.
Crystallographic data were collected on beamline 8.2.2 of the Advanced Light Source (ALS). Native data were collected to 2.8 Å resolution at a wavelength λ = 1.000 Å; a separate dataset was also collected on a second crystal to 4.0 Å resolution at a wavelength λ = 1.500 Å, using inverse beam strategy, to measure significant anomalous scattering of sulfur atoms from cysteine and methionine. Data were processed with mosflm [42] and HKL2000 [43]. Standard crystallographic computations were implemented with CCP4 [44, 45] and PHENIX [46]; model building was done with COOT [47].
The crystals are tetragonal, space group P43212, with two molecules per asymmetric unit (Table 1). An initial molecular replacement solution was found using an unpublished NMR structure of a fragment of mouse SRAP (PDB ID 2YRU), which has 85% sequence identity to the human protein fragment used here, as a search model. Correct placement of the sequence in the model was confirmed with an anomalous difference Fourier using the data collected at λ = 1.500 Å and model phases; peaks in the anomalous Fourier align with sulfurs of methionines and cysteines (Fig. S2). Successive cycles of model building and refinement yielded a model with Rcryst = 0.224, Rfree = 0.267; the model includes residues 109-210 of each protomer; 9 residues of each protomer are disordered in the final model.
Table 1.
Crystallographic data collection and refinement statistics for native data.
| Data Collection | |
| Wavelength (Å) | 1.000 |
| Resolution range (last shell) (Å) | 50.0-2.8 (2.85-2.80) |
| Observations (total/unique) | 71,967/9695 |
| Redundancy | 7.4 (8.0) |
| Completeness (%) | 99.9 (100.0) |
| <I/σ(I)>* | 17.1 (6.3) |
| Rsym ± | 0.059 (0.295) |
| Refinement | |
| Resolution range (last shell) (Å) | 39.5-2.80 (3.21-2.80) |
| Rcryst § | 0.224 (0.265) |
| Rfree | 0.267 (0.388) |
| Number of reflections (working set) | 9150 |
| Number of reflections (test set) | 479 |
| Number of protein atoms | 3295 |
| Average B value, all atoms (Å2) | 57.4 |
| Rmsd# bond length (Å) | 0.005 |
| Rmsd angles (°) | 0.089 |
Spacegroup P432121, a = 85.90 Å, c = 100.41 Å.
<I/σ(I)> = intensity divided by standard deviation of intensity, averaged over all measurements.
Rsym = Σ∣Ihkl - <Ihkl>∣/Σ<Ihkl> where Ihkl = single value of measured intensity of hkl reflection, and <Ihkl> = mean of all measured values intensity of hkl reflection.
Rcryst = Σ∣Fobs-Fcalc∣/ΣFobs where Fobs = observed structure factor amplitude and Fcalc = structure factor calculated from model. Rfree is computed in the same manner as Rcryst, using the test set of reflections.
Rmsd is root mean square deviation from ideal value.
PDB and GEO accession codes
Coordinates and structure factors have been deposited with the Research Collaboratory for Structural Biology with the identifier 4NBO. RNA-seq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus data repository (http://www.ncbi.nlm.nih.gov/geo/), accession identifier GSE52385.
Supplementary Material
Research Highlights.
[3 to 5, <86 characters, including spaces, each; first should state “background or context of the question”; one to three to describe principal results; last should “conclude on a clear description of the conceptual advance and significance of the work”]
Current model: SRAP modulates SRA RNA, a transcriptional regulator, by binding SRA.
SRAP-SRA binding experiments in vitro and in cells do not show specific binding.
Crystal structure of C-terminal domain of SRAP shows it is similar to PRP18.
siRNA knockdowns of SRA/SRAP show only minor effect on transcription.
Data do not support current model for SRAP activity; alternative are discussed.
Acknowledgements
We thank Etienne Leygue (University of Manitoba, Canada) for SRA expression plasmids; Arthur Zaug (University of Colorado) for performing transfections and making cell extracts; James Huntley and Steven Gao (University of Colorado) for performing the next-generation sequencing; Theresa Nahreini for assistance with cell culture and sorting; Robin Dowell, Phillip Richmond and Mary Allen for help with RNA-seq data analysis; Rebecca Hoffman for producing the western blots shown in Fig. S1a; Leslie Leinwand, Emily Pugach and members of the Cech lab for assistance and discussion. T.R.C. is an investigator of the Howard Hughes Medical Institute. L.X. is supported by NIH T32 GM08759. K.B.B. is supported by AHA 13POST14410014. Crystallographic data were collected at the Berkeley Center for Structural Biology, which is supported in part by the National Institutes of Health NIGMS, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Abbreviations
- BSA
bovine serum albumin
- EMSA
electrophoretic mobility shift assay
- ER
estrogen receptor
- ERE
estrogen-responsive element
- hTERT
human telomerase reverse transcriptase
- hTR
human telomerase RNA
- Luc
luciferase
- MBP
maltose binding protein
- MOPS
3-(N-morpholino) propanesulfonic acid
- PNK
T4 polynucleotide kinase
- pS2
Trefoil factor 1
- PR
progesterone receptor
- GREB1
growth regulation by estrogen in breast cancer 1
- RRM
RNA recognition motif
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- SD
standard deviation
- SHARP
SMRT/HDAC1 associated repressor protein
- SLIRP
SRA stem-loop interacting RNA binding protein
- SRA
steroid receptor RNA activator
- SRAP
steroid receptor RNA activator protein
- STL7
stem-loop 7
- TEV
tobacco etch virus
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O’Malley BW. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 2.Caretti G, Lei EP, Sartorelli V. The DEAD-box p68/p72 proteins and the noncoding RNA steroid receptor activator SRA: eclectic regulators of disparate biological functions. Cell Cycle. 2007;6:1172–1176. doi: 10.4161/cc.6.10.4228. [DOI] [PubMed] [Google Scholar]
- 3.Xu B, Gerin I, Miao H, Vu-Phan D, Johnson CN, Xu R, Chen XW, Cawthorn WP, MacDougald OA, Koenig RJ. Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity. PLoS One. 2010;5:e14199. doi: 10.1371/journal.pone.0014199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colley SM, Leedman PJ. Steroid Receptor RNA Activator - A nuclear receptor coregulator with multiple partners: Insights and challenges. Biochimie. 2011;93:1966–1972. doi: 10.1016/j.biochi.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Hube F, Guo J, Chooniedass-Kothari S, Cooper C, Hamedani MK, Dibrov AA, Blanchard AA, Wang X, Deng G, Myal Y, Leygue E. Alternative splicing of the first intron of the steroid receptor RNA activator (SRA) participates in the generation of coding and noncoding RNA isoforms in breast cancer cell lines. DNA Cell Biol. 2006;25:418–428. doi: 10.1089/dna.2006.25.418. [DOI] [PubMed] [Google Scholar]
- 6.Hube F, Velasco G, Rollin J, Furling D, Francastel C. Steroid receptor RNA activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. Nucleic Acids Res. 2011;39:513–525. doi: 10.1093/nar/gkq833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chooniedass-Kothari S, Emberley E, Hamedani MK, Troup S, Wang X, Czosnek A, Hube F, Mutawe M, Watson PH, Leygue E. The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett. 2004;566:43–47. doi: 10.1016/j.febslet.2004.03.104. [DOI] [PubMed] [Google Scholar]
- 8.Emberley E, Huang GJ, Hamedani MK, Czosnek A, Ali D, Grolla A, Lu B, Watson PH, Murphy LC, Leygue E. Identification of new human coding steroid receptor RNA activator isoforms. Biochem Biophys Res Commun. 2003;301:509–515. doi: 10.1016/s0006-291x(02)03070-x. [DOI] [PubMed] [Google Scholar]
- 9.Kawashima H, Takano H, Sugita S, Takahara Y, Sugimura K, Nakatani T. A novel steroid receptor co-activator protein (SRAP) as an alternative form of steroid receptor RNA-activator gene: expression in prostate cancer cells and enhancement of androgen receptor activity. Biochem J. 2003;369:163–171. doi: 10.1042/BJ20020743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper C, Vincett D, Yan Y, Hamedani MK, Myal Y, Leygue E. Steroid Receptor RNA Activator bi-faceted genetic system: Heads or Tails? Biochimie. 2011;93:1973–1980. doi: 10.1016/j.biochi.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Coleman KM, Lam V, Jaber BM, Lanz RB, Smith CL. SRA coactivation of estrogen receptor-alpha is phosphorylation-independent, and enhances 4-hydroxytamoxifen agonist activity. Biochem Biophys Res Commun. 2004;323:332–338. doi: 10.1016/j.bbrc.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 12.Cooper C, Guo J, Yan Y, Chooniedass-Kothari S, Hube F, Hamedani MK, Murphy LC, Myal Y, Leygue E. Increasing the relative expression of endogenous non-coding Steroid Receptor RNA Activator (SRA) in human breast cancer cells using modified oligonucleotides. Nucleic Acids Res. 2009;37:4518–4531. doi: 10.1093/nar/gkp441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deblois G, Giguere V. Ligand-independent coactivation of ERalpha AF-1 by steroid receptor RNA activator (SRA) via MAPK activation. J Steroid Biochem Mol Biol. 2003;85:123–131. doi: 10.1016/s0960-0760(03)00225-5. [DOI] [PubMed] [Google Scholar]
- 14.Klinge CM, Jernigan SC, Mattingly KA, Risinger KE, Zhang J. Estrogen response element-dependent regulation of transcriptional activation of estrogen receptors alpha and beta by coactivators and corepressors. J Mol Endocrinol. 2004;33:387–410. doi: 10.1677/jme.1.01541. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe M, Yanagisawa J, Kitagawa H, Takeyama K, Ogawa S, Arao Y, Suzawa M, Kobayashi Y, Yano T, Yoshikawa H, Masuhiro Y, Kato S. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor alpha coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. Embo J. 2001;20:1341–1352. doi: 10.1093/emboj/20.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Chooniedass-Kothari S, Hamedani MK, Troup S, Hube F, Leygue E. The steroid receptor RNA activator protein is expressed in breast tumor tissues. Int J Cancer. 2006;118:1054–1059. doi: 10.1002/ijc.21425. [DOI] [PubMed] [Google Scholar]
- 17.Shi Y, Downes M, Xie W, Kao HY, Ordentlich P, Tsai CC, Hon M, Evans RM. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 2001;15:1140–1151. doi: 10.1101/gad.871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatchell EC, Colley SM, Beveridge DJ, Epis MR, Stuart LM, Giles KM, Redfern AD, Miles LE, Barker A, MacDonald LM, Arthur PG, Lui JC, Golding JL, McCulloch RK, Metcalf CB, Wilce JA, Wilce MC, Lanz RB, O’Malley BW, Leedman PJ. SLIRP, a small SRA binding protein, is a nuclear receptor corepressor. Mol Cell. 2006;22:657–668. doi: 10.1016/j.molcel.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 19.Yan Y, Skliris GP, Penner C, Chooniedass-Kothari S, Cooper C, Nugent Z, Blanchard A, Watson PH, Myal Y, Murphy LC, Leygue E. Steroid Receptor RNA Activator Protein (SRAP): a potential new prognostic marker for estrogen receptor-positive/node-negative/younger breast cancer patients. Breast Cancer Res. 2009;11:R67. doi: 10.1186/bcr2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan Y, Penner CC, Skliris GP, Cooper C, Nugent Z, Blanchard A, Hamedani MK, Wang X, Myal Y, Murphy LC, Leygue E. Steroid receptor RNA activator protein (SRAP) expression as a prognostic factor in ER+ human breast tumors. J Cancer Res Clin Oncol. 2013;139:1637–1647. doi: 10.1007/s00432-013-1485-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao X, Patton JR, Ghosh SK, Fischel-Ghodsian N, Shen L, Spanjaard RA. Pus3p- and Pus1p-dependent pseudouridylation of steroid receptor RNA activator controls a functional switch that regulates nuclear receptor signaling. Mol Endocrinol. 2007;21:686–699. doi: 10.1210/me.2006-0414. [DOI] [PubMed] [Google Scholar]
- 22.Brill LM, Xiong W, Lee KB, Ficarro SB, Crain A, Xu Y, Terskikh A, Snyder EY, Ding S. Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell. 2009;5:204–213. doi: 10.1016/j.stem.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Hoof D, Munoz J, Braam SR, Pinkse MW, Linding R, Heck AJ, Mummery CL, Krijgsveld J. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell. 2009;5:214–226. doi: 10.1016/j.stem.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 25.Zaug AJ, Crary SM, Jesse Fioravanti M, Campbell K, Cech TR. Many disease-associated variants of hTERT retain high telomerase enzymatic activity. Nucleic Acids Res. 2013;41:8969–8978. doi: 10.1093/nar/gkt653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chooniedass-Kothari S, Vincett D, Yan Y, Cooper C, Hamedani MK, Myal Y, Leygue E. The protein encoded by the functional steroid receptor RNA activator is a new modulator of ER alpha transcriptional activity. FEBS Lett. 2010;584:1174–1180. doi: 10.1016/j.febslet.2010.02.024. [DOI] [PubMed] [Google Scholar]
- 27.Kielkopf CL, Lucke S, Green MR. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 2004;18:1513–1526. doi: 10.1101/gad.1206204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myers JW, Chi JT, Gong D, Schaner ME, Brown PO, Ferrell JE. Minimizing off-target effects by using diced siRNAs for RNA interference. J RNAi Gene Silencing. 2006;2:181–194. [PMC free article] [PubMed] [Google Scholar]
- 29.Foulds CE, Tsimelzon A, Long W, Le A, Tsai SY, Tsai MJ, O’Malley BW. Research resource: expression profiling reveals unexpected targets and functions of the human steroid receptor RNA activator (SRA) gene. Mol Endocrinol. 2010;24:1090–1105. doi: 10.1210/me.2009-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, Marshall W, Khvorova A, Linsley PS. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanz RB, Razani B, Goldberg AD, O’Malley BW. Distinct RNA motifs are important for coactivation of steroid hormone receptors by steroid receptor RNA activator (SRA) Proc Natl Acad Sci U S A. 2002;99:16081–16086. doi: 10.1073/pnas.192571399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 33.Kowalczykowski SC, Paul LS, Lonberg N, Newport JW, McSwiggen JA, von Hippel PH. Cooperative and noncooperative binding of protein ligands to nucleic acid lattices: experimental approaches to the determination of thermodynamic parameters. Biochemistry. 1986;25:1226–1240. doi: 10.1021/bi00354a006. [DOI] [PubMed] [Google Scholar]
- 34.Bacikova D, Horowitz DS. Mutational analysis identifies two separable roles of the Saccharomyces cerevisiae splicing factor Prp18. RNA. 2002;8:1280–1293. doi: 10.1017/s1355838202023099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bacikova D, Horowitz DS. Genetic and functional interaction of evolutionarily conserved regions of the Prp18 protein and the U5 snRNA. Mol Cell Biol. 2005;25:2107–2116. doi: 10.1128/MCB.25.6.2107-2116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 37.Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. Embo J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker P, Germond JE, Brown-Luedi M, Givel F, Wahli W. Sequence homologies in the region preceding the transcription initiation site of the liver estrogen-responsive vitellogenin and apo-VLDLII genes. Nucleic Acids Res. 1984;12:8611–8626. doi: 10.1093/nar/12.22.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hua X, Liu X, Ansari DO, Lodish HF. Synergistic cooperation of TFE3 and smad proteins in TGF-beta-induced transcription of the plasminogen activator inhibitor-1 gene. Genes Dev. 1998;12:3084–3095. doi: 10.1101/gad.12.19.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milligan JF, Uhlenbeck OC. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 41.Kao C, Zheng M, Rudisser S. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA. 1999;5:1268–1272. doi: 10.1017/s1355838299991033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leslie AGW, Powell HR. In: Processing Diffraction Data with Mosflm. Read RJ, Sussman JL, editors. Vol. 245. 2007. pp. 41–51. (NATO Science Series). [Google Scholar]
- 43.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 44.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr. 2011;67:D235–D242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.