Abstract
Plant tissue colonization by Trichoderma atroviride plays a critical role in the reduction of diseases caused by phytopathogenic fungi, but this process has not been thoroughly studied in situ. We monitored in situ interactions between gfp-tagged biocontrol strains of T. atroviride and soilborne plant pathogens that were grown in cocultures and on cucumber seeds by confocal scanning laser microscopy and fluorescence stereomicroscopy. Spores of T. atroviride adhered to Pythium ultimum mycelia in coculture experiments. In mycoparasitic interactions of T. atroviride with P. ultimum or Rhizoctonia solani, the mycoparasitic hyphae grew alongside the pathogen mycelia, and this was followed by coiling and formation of specialized structures similar to hooks, appressoria, and papillae. The morphological changes observed depended on the pathogen tested. Branching of T. atroviride mycelium appeared to be an active response to the presence of the pathogenic host. Mycoparasitism of P. ultimum by T. atroviride occurred on cucumber seed surfaces while the seeds were germinating. The interaction of these fungi on the cucumber seeds was similar to the interaction observed in coculture experiments. Green fluorescent protein expression under the control of host-inducible promoters was also studied. The induction of specific Trichoderma genes was monitored visually in cocultures, on plant surfaces, and in soil in the presence of colloidal chitin or Rhizoctonia by confocal microscopy and fluorescence stereomicroscopy. These tools allowed initiation of the mycoparasitic gene expression cascade to be monitored in vivo.
Trichoderma spp. are active ingredients in a variety of commercial biofungicides used to control a range of economically important aerial and soilborne fungal plant pathogens (17, 19). The antagonistic activity of biocontrol Trichoderma strains is attributable to one or more complex mechanisms, including nutrient competition, antibiosis, the activity of cell wall-lytic enzymes, induction of systemic resistance, and increased plant nutrient availability (16, 18, 19, 24, 30, 31, 42). Many studies, primarily in vitro studies, have shed light on the molecular basis of the three-way relationship among the pathogen, the plant, and the biocontrol agent (14, 35). However, the complexities of these interactions have been poorly studied in situ. For example, many of the factors involved in biocontrol are known (32, 34, 51, 55), but the antifungal mechanisms, including mycoparasitism, and the fate of Trichoderma in the soil and on the plant are not well understood. Effective monitoring of biocontrol-related processes in vivo based on the use of vital markers (1, 20, 40) provides a basis for development of new selection methods and improved applications.
The green fluorescent protein (GFP)-encoding gene (gfp) (8) is a powerful tool for monitoring the fate and behavior of bacterial and fungal inoculants in situ (1, 5, 29, 43, 48-50). GFP, unlike other biomarkers (22), does not require any substrate or additional cofactors in order to fluoresce. Even a single cell expressing GFP can be easily seen by epifluorescence microscopy or confocal scanning laser microscopy (CSLM) (23, 47, 48). Zeilinger et al. (55) constructed Trichoderma atroviride biocontrol strains with fusions of gfp to various promoters for constitutive or inducible expression during biocontrol. They found that chitinases were induced by the presence of Rhizoctonia solani and other fungal hosts in coculture experiments and that these strains were particularly useful for in vitro studies of the early phases of the interaction with the fungal host (55). Clearly, these mutants could be very useful in in situ studies of the antagonistic process; e.g., they permitted visualization of the mycoparasitic interactions between Trichoderma and various pathogenic fungi occurring on plant tissues.
Our objective in this study was to use gfp-tagged mutants of T. atroviride to study the in situ interaction of T. atroviride with the plant pathogens Rhizoctonia solani and Pythium ultimum in cocultures and directly on cucumber (Cucumis sativus L.) seeds, on roots, and in sterile soil. In particular, we were interested in the timing of induction of genes encoding chitinases in the presence of the host fungi or chitin. Our working hypothesis was that chitinase gene promoters would be induced in soil and on plant surfaces in the presence of the host fungus, enabling direct visualization of the mycoparasitic gene expression cascade in vivo. The combination of gfp tagging and advanced microscopy for in situ monitoring provides a plethora of new possibilities for studying the complex mechanisms of interactions among fungal antagonists, pathogens, and plants.
MATERIALS AND METHODS
Fungal cultures and growth conditions.
The following T. atroviride mutants (55) of strain P1 were used: the T. atroviride pki1::gfp strain with the gfp gene under control of the constitutive pyruvate kinase promoter; the T. atroviride ech42::gfp strain with gfp under control of the ech42 promoter, which is induced by cell wall oligosaccharides or digested colloidal chitin; and the T. atroviride nagl::gfp strain with gfp under control of the nag1 promoter, which is induced by cell wall oligosaccharides or N-acetylglucosamine. T. atroviride strains were grown on potato dextrose agar (PDA) (Merck, Darmstadt, Germany) supplemented with 100 μg of hygromycin B (Sigma-Aldrich Chemie, Steinheim, Germany) per ml or in SM medium [containing (per liter) 2.8 g of (NH4)2SO4, 0.6 g of urea, 4 g of KH2PO4, 0.6 g of CaCl2 · 2H2O, 0.2 g of MgSO4, 0.01 g of FeSO4 · 7H2O, 0.0028 g of ZnSO4 · H2O, 0.0032 g of CoCl2 · 6H2O, and 1.0 g of sucrose; pH 6.0] supplemented with 100 μg of hygromycin B per ml. P. ultimum Trow var. ultimum HB2 was provided by John Hockenhull (Department of Plant Biology, KVL, Copenhagen, Denmark). R. solani is a pathogenic isolate that was obtained from the Plant Pathology and Biocontrol Unit, The Swedish University of Agricultural Sciences, Uppsala, Sweden. The pathogenic fungal strains were grown on PDA, in potato dextrose broth, on cornmeal agar, or in cornmeal broth (CMB) (Sigma-Aldrich Chemie). All cultures were incubated statically in the dark at 22°C, unless specified otherwise.
Biocontrol assays in vivo.
In vivo tests of the biocontrol ability of T. atroviride were conducted with R. solani. Bean or cucumber seeds were coated with a 10% (wt/vol) aqueous suspension of the adhesive Pelgel (Liphatech, Milwaukee, Wis.) containing 109 spores of T. atroviride wild-type strain P1 per ml, 109 spores of the pki1::gfp strain per ml, 109 spores of the ech42::gfp strain per ml, or 109 spores of the nagl::gfp strain per ml (1 ml of spore suspension/15 g of seeds) and then air dried in an open petri dish overnight in a laminar flow hood.
An R. solani inoculum was prepared in potato dextrose broth liquid cultures that were incubated for 5 days at 25°C with shaking at 150 rpm. The R. solani fungal biomass was collected by using a vacuum in a Miracloth-lined Buchner funnel and rinsed twice with 2 volumes of distilled water. Excess liquid was pressed through the filter, and the biomass was weighed. Three grams of the fungal biomass was homogenized in 100 ml of distilled water and mixed with 1 kg of sterile soil.
Treated bean and cucumber seeds were planted 4 and 3 cm deep in the infested soil, respectively. The pots were kept at 25°C in the light and were watered daily with sterile water. The number of plants that emerged or survived and the plant height were evaluated twice weekly for up to 1 month after planting.
For all in vivo biocontrol assays, infected plant material was plated onto acidified PDA to verify the presence of Trichoderma and the pathogenic fungi. For all biocontrol assays we used three or more treatment replicates per experiment, the experiments were repeated on three separate occasions, and the results presented below are the average values obtained from all combined experiments. The percentage of survivors was calculated by dividing the number of plants that emerged by the number of seeds planted. The statistical analyses included a one-way analysis of variance of arcsine-transformed percentage values or raw data obtained from measured growth or plant height, and the significance level was <0.05. Due to poor emergence and deformed growth of control plants (seeds treated with Pelgel or water) in the assays with pathogen-infested soil, it was not possible to include the data from these experiments in the analysis of variance. Therefore, unpaired t tests were conducted with the wild-type and mutant strains to determine differences.
Fungal interactions in cocultures.
Coculture studies of the interaction between the T. atroviride pki1::gfp strain and P. ultimum or R. solani and induction of gfp in the T. atroviride ech42::gfp and nagl::gfp strains by R. solani were performed on glass slides on which 150 μl of 25% PDA was spread flat onto an area that was 25 by 15 by 1.5 mm, as previously described (3, 4, 46). After the medium solidified, a plug (diameter, 2 mm) of P. ultimum or R. solani from the margin of an actively growing culture was inoculated at one end of the slide, and a plug of T. atroviride whose size was similar was inoculated at the opposite end. The slide was incubated with the agar side up at 22°C in the dark in a petri dish sealed with Parafilm that contained a layer of sterilized and premoistened Munktell filter paper (Munktell Filter AB, Grycksbo, Sweden). After 24 to 28 h, the fungal hyphae met. The slides were viewed with a confocal scanning laser microscope (model TCS; Leica, Heidelberg, Germany) with excitation wavelengths of 488 nm (Ar) and 633 nm (HeNe). Emission light was collected in the range from 510 to 560 nm for GFP and in the range from 620 to 660 nm for background fluorescence. A ×20 objective with an instrumental zoom factor of ×1 to ×4 was used. Images were obtained by using Leica confocal software (version 2.477). Three-dimensional rendering of the stack of images was obtained by using the three-dimensional software supplied with the confocal microscope system.
Pretreatment of cucumber seeds for in situ study.
The surfaces of cucumber seeds were lightly brushed with sterilized water by using a small sterile paintbrush. The seeds were then soaked in 70% (vol/vol) ethanol for 2 min and washed three times with sterilized water. The treated seeds were transferred to sterile filter paper to absorb excess water and then disinfected in 2% (wt/vol) sodium hypochlorite (Sigma-Aldrich Chemie) for 2 min (54). The disinfected seeds were washed five times in sterilized water, and excess water was removed by blotting with sterilized filter paper under aseptic conditions. Thirty disinfected seeds and 30 nonsterile seeds (controls) were incubated on six PDA plates (10 seeds/plate) in the light at 28°C for 5 days to determine the efficacy of disinfection and cucumber seed germination. Ten sterilized seeds were ground dry with a mortar and pestle together with 2 g of sterilized silica (diameter, 1 to 1.5 mm; Sigma-Aldrich Chemie). An 80-μl suspension of the macerated seeds was spread onto a PDA plate and incubated at 28°C for 3 days to identify potential endophytes of cucumber seeds.
Preparation of soil for microcosm studies.
The soil utilized was an agricultural soil obtained in Flemingsberg, Sweden (pH 5.5 to 6.5). The soil was autoclaved at 121°C for 1 h on three successive days. A 1-g sample of sterilized soil was added to a sterilized flask containing 20 ml of phosphate-buffered saline (PBS) (8 g of NaCl per liter, 0.2 g of KCl per liter, 1.4 g of Na2HPO4per liter, 0.24 g of KH2PO4 per liter; pH 7.4) with 20 glass beads (diameter, 5 mm; Sigma-Aldrich Chemie). The flask was shaken at 150 rpm for 30 min. Eighty-microliter portions of the soil suspension were spread on PDA plates. The plates were incubated at 28°C for 3 days to determine contamination. Sterile soil was dried in an oven at 65°C for 3 days before it was used.
Preparation of fungal spores or propagules.
A T. atroviride spore suspension was prepared by growing T. atroviride on PDA plates at 22°C for 7 days and suspending spores in 10 ml of PBS by scraping the plate with an inoculating loop. The suspension was filtered through sterilized glass wool (Merck) in a 10-ml syringe. The spores in the spore suspension were counted by using a Bürker-Türk counting chamber (Karl Hecht Assistent KG, Sondheim/Röhm, Germany) and epifluorescence microscopy (Zeiss, Oberkochen, Germany) with a 485-nm excitation filter and emission at 520 nm.
P. ultimum was grown in a flask containing 20 ml of 2% CMB at 28°C with shaking at 150 rpm for 21 days. Propagules (defined as reproductive units capable of producing colonies) were harvested by centrifugation at 1,650 × g for 15 min, washed three times in PBS by centrifugation under the same conditions, and resuspended in PBS. The resuspended propagules were vortexed to detach the sporangia from the majority of the vegetative hyphae, filtered through sterilized glass wool in a 10-ml syringe, and then counted as described above.
Experimental design for confrontation studies on cucumber seeds.
Cucumber seeds were inoculated with a combination of T. atroviride spores and P. ultimum propagules. Three different treatments were used. For treatment A, a P. ultimum propagule suspension was mixed with sterilized soil to obtain 3.1 × 103 propagules/g of soil, and the soil water content was adjusted to 12% (vol/wt) by using sterilized deionized water. Then aliquots (65 g) of the inoculated soil were distributed into petri dishes. Sterilized cucumber seeds were soaked in a Trichoderma spore suspension containing 1.1 × 103 spores/ml for 30 min and then blotted dry with sterilized filter paper under aseptic conditions. Twenty-four inoculated cucumber seeds were planted in two petri dishes (12 seeds/dish) that were sealed with Parafilm, and the seeds were grown at 23°C with a cycle consisting of 10 h of light and 14 h of darkness (45). For treatment B, sterilized cucumber seeds were soaked in a P. ultimum suspension containing 3.1 × 103 propagules/ml, and a T. atroviride spore suspension was mixed with sterilized soil to obtain 103 spores/g of soil. The inoculated seeds were planted in petri dishes filled with inoculated soil as described above. For treatment C, T. atroviride and P. ultimum suspensions were blended together and then mixed with sterilized soil before sterilized cucumber seeds were planted. The concentrations of both spores and propagules and other relative parameters were the same as those described above for the other treatments. For the control, the same volume of PBS that was used for the inoculant suspension was mixed with sterilized soil, and sterilized cucumber seeds were sown in the soil.
Two cucumber seeds were arbitrarily removed from each treatment every 2 days for 20 days in order to observe colonization and coverage by fluorescence stereomicroscopy (model MZ12; Leica AG, Heerbrugg, Switzerland). The colonized cucumber seeds were longitudinally cryosectioned with a cryostat (model CM 3050; Leica, Heidelberg, Germany) to obtain slices that were 20 to 30 μm thick. One drop of Vectashield mounting medium (Vector Laboratories Inc., Burlingame, Calif.) was added to each slice before a coverslip was added and the preparation was examined by CSLM.
Effect of inoculation method on seed surface colonization by T. atroviride.
Front and back images of whole seeds were obtained with a Hamamatsu digital charge-coupled device camera (model C4742-95; Hamamatsu Photonics, Hamamatsu City, Japan) installed on an epifluorescence microscope. A grid consisting of 100 squares was superimposed on four sectors (two upper sectors and two lower sectors) of each seed, and the hyphae crossing the intersection points (except the hyphae on two of the side lines) were counted for five different positions of the grid (for a total of 500 crossing points). The fungal aggregates were recorded by using the same method that was used for the hyphae. The percent coverage of cucumber seeds by T. atroviride was determined as follows: percent coverage = N (1/5), where N is the number of intersection points occupied by fungal hyphae in five views for 100 square grids (39).
Induction of T. atroviride mutants.
Ground R. solani hyphae were prepared by growing R. solani in a 250-ml flask containing 50 ml of 0.2% (wt/vol) CMB at 28°C with shaking at 150 rpm for 14 days. The hyphae were washed five times with sterile double-distilled water in a sterile beaker with stirring and were harvested by centrifugation at 6,000 × g for 15 min. Pelleted hyphae were ground in a mortar and pestle, dried overnight at 80°C, and then sieved with a 60-mesh sieve. The sieved hyphal powder was sterilized by autoclaving and stored at 4°C until it was used.
Colloidal chitin was prepared by dissolving 50 g of crab shell chitin (Sigma) in 150 ml of 12 N HCl. Ice-cold sterile distilled water (200 ml) was added, and the pH was adjusted to 7.0 with 5 N NaOH. The suspension was centrifuged at 9,000 × g for 10 min, the supernatant was discarded, and 0.2 M phosphate buffer (pH 7.0) was added. The suspension was mixed well and centrifuged again at 9,000 × g for 10 min. After the supernatant was discarded, sterile distilled water was added to obtain a chitin concentration of 4% (wt/vol). Finally, the suspension was sterilized by autoclaving and stored at 4°C until it was used.
Spore suspensions of the T. atroviride ech42::gfp and nagl::gfp mutants were inoculated to obtain a concentration of 103 spores ml−1 into 125-ml flasks containing 30 ml of SM medium with 100 μg of hygromycin B ml−1, 0.1% sucrose, and various concentrations of sterile colloidal chitin or the R. solani ground hyphae stock (0, 0.125, 0.25, 0.5, 1.5, and 2.5 mg ml−1). The cultures were grown at 28°C with shaking at 150 rpm. Samples (1 ml) of the cultures were taken at zero time and after 24, 48, 72, 96, and 120 h of incubation and were centrifuged at 9,000 × g for 10 min. The hyphal pellets were resuspended in 400 μl of sterile double-distilled water and then transferred to separate wells of a 96-well microtiter plate and incubated at 4°C for 30 min. Fluorescence was measured with a microtiter plate spectrofluorometer (BMG LabTechnologies, Offenburg, Germany) by using an excitation wavelength of 485 nm and an emission wavelength of 520 nm. After this, each sample suspension was transferred to a tared plastic weighing dish and dried at 65°C to a constant weight, and the weight of the hyphae was determined with an accuracy of 10−4 g.
Induction of T. atroviride mutants in soil microcosms.
Microcosms in petri dishes contained 65 g of sterile soil (relative moisture content, 65%) blended with 6.5 × 104 spores of the T. atroviride ech42::gfp or nagl::gfp strain. R. solani was grown on PDA at 22°C until sclerotia that were 2 mm in diameter were formed. R. solani sclerotia were inoculated into the soil in the petri dishes with 8 mm between the sclerotia and were covered with 2 mm of soil. The petri dishes were sealed with Parafilm, covered with black cloth, and incubated at 22°C. Soil samples were taken daily for 1 week after inoculation and examined by confocal microscopy.
RESULTS
In vivo biocontrol activity of gfp-tagged mutants.
Transformation with the gfp gene did not affect the biocontrol ability of T. atroviride strain P1 (Table 1). Both the emergence (percentage of survival) and the growth of bean plants from antagonist-treated seeds were significantly greater than the emergence and the growth observed for the control treatments (Pelgel or water alone) in soil containing a pathogen (P < 0.001). There were no differences among any of the seed treatments, T. atroviride strains, or controls if the soil did not contain a pathogen (Table 1).
TABLE 1.
Effects of T. atroviride wild-type strain P1 and gfp-tagged strains on plant survival and plant height of emerged bean seedlings 14 days after planting
| Seed treatment |
R. solani-infested soil
|
Soil without pathogen
|
||
|---|---|---|---|---|
| % Survivala | Height (cm)b | % Survival | Height (cm) | |
| Wild type P1 + Pelgel | 71 | 6.6 ± 3.6 | 100 | 19 ± 2.8 |
| pki1::gfp + Pelgel | 76 | 5.6 ± 5.0 | 100 | 17 ± 2.9 |
| ech42::gfp + Pelgel | 81 | 4.3 ± 2.8 | 100 | 18 ± 2.8 |
| nag1::gfp + Pelgel | 86 | 12.0 ± 6.0 | 100 | 18 ± 1.4 |
| Pelgel alone | 37 | NA | 100 | 18 ± 1.1 |
| Water | 30 | NA | 100 | 18 ± 1.9 |
The percentage of survival was determined by determining the number of emergent plants during the evaluation period. Analysis of variance for arcsine-transformed values: P1 versus pki1::gfp, ech42::gfp, or nag1::gfp mutant, not significantly different; P1, pki1::gfp mutant, ech42::gfp mutant, or nag1::gfp mutant versus Pelgel or water alone, significantly different (P < 0.001). Seed treatments were replicated three times per experiment, and the experiments were repeated on three separate occasions. Data from all experiments were combined for the analysis.
Unpaired t test for plant height: for soil treated with pathogen, P1 versus pki1::gfp, ech42::gfp or nag1::gfp mutant, not significantly different: for soil with no pathogen, P1, pki1::gfp mutant, ech42::gfp mutant, or nag1::gfp mutant versus Pelgel or water alone, not significantly different. NA, not applicable (plants in poor condition).
Mycoparasitism by T. atroviride in cocultures.
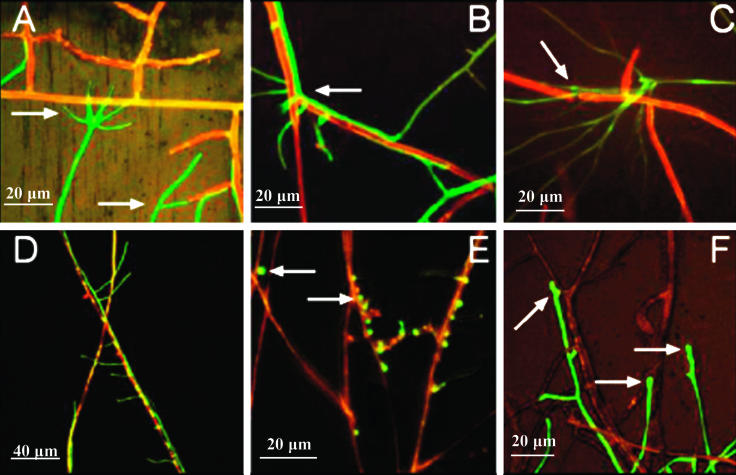
gfp-tagged hyphae were easily seen due to their green fluorescence, whereas the pathogenic fungal hyphae were either weakly autofluorescent (red or yellow) or nonfluorescent (brown). With R. solani, T. atroviride formed a cluster of branches immediately before contact that grew towards the host hyphae (Fig. 1A). Subsequently, the mycoparasite aligned with R. solani hyphae (Fig. 1B), which often broke during an attack in a manner not observed with the other fungal hosts tested (Fig. 1C). With P. ultimum, T. atroviride grew alongside the P. ultimum hyphae and branched toward adjacent hyphae (Fig. 1D). T. atroviride spores adhered to the host hyphae (Fig. 1E) and germinated after attachment. Then the young T. atroviride hyphae extended toward and parasitized the host mycelium (Fig. 1F). The confrontation with P. ultimum also was characterized by growth of T. atroviride alongside the host hyphae and subsequent coiling around them. The specialized, appressorium-like structures that formed in the presence of the host mycelium and were previously described by Chet et al. (11) with R. solani were visualized during parasitism of P. ultimum (data not shown). In addition, the hyphal tips of T. atroviride swelled and formed papilla-like structures not only during contact with the host hyphae but also before they touched the host mycelium (Fig. 1F).
FIG. 1.
CSLM images of confrontations between cocultures of the T. atroviride pkil::gfp mutant and R. solani (A, B, and C) and between cocultures of the T. atroviride pkil::gfp mutant and P. ultimum (D, E, and F). The host hyphae are reddish (R. solani) or reddish brown (P. ultimum), whereas the hyphae of the T. atroviride pkil::gfp mutant are green. The left arrow in panel A indicates a conidium, and the right arrow in panel A and the arrow in panel B indicate branching in the T. atroviride hyphae. In panel C the arrow indicates a breakage point in the R. solani hyphae. In panel E the arrows indicate green fluorescent T. atroviride spores deposited on P. ultimum hyphae. In panel F the arrows indicate papilla-like structures in the T. atroviride hyphae. The images were obtained with a ×20 objective.
Effect of inoculation method on colonization of the cucumber seed surface by T. atroviride.
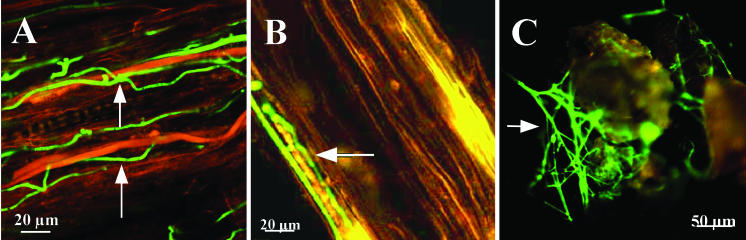
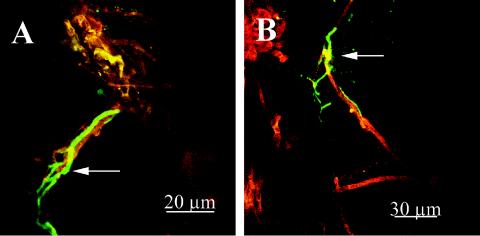
The method of inoculation of T. atroviride and P. ultimum onto cucumber seeds affected the level of coverage of the seed surface by T. atroviride. When P. ultimum was mixed with sterilized soil into which seeds that had been soaked in a T. atroviride spore suspension were planted (treatment A), T. atroviride hyphae appeared 2 days after planting (Table 2). If T. atroviride spores were mixed with sterilized soil before seeds that had been soaked in a P. ultimum suspension were planted (treatment B) or if sterilized cucumber seeds were sown in soil that was inoculated with the same concentrations of T. atroviride spores and P. ultimum propagules (treatment C), colonization by the biocontrol agent was not observed until 4 days after planting. Colonization by T. atroviride increased for up to 14 days after planting for treatment A and for up to 16 days for treatments B and C. After this, the percent coverage stabilized or slightly decreased for all treatments (Table 2). The gfp-tagged Trichoderma colonized the outer layer of the roots of the cucumber seedlings (Fig. 2A and B), as well as the soil inoculated with the biocontrol agent and the pathogen together (treatment C) (Fig. 2C).
TABLE 2.
Colonization of the cucumber seed surface by T. atroviride, expressed as seed coveragea
| Time (days) | Seed coverage (%)
|
||
|---|---|---|---|
| Treatment A | Treatment B | Treatment C | |
| 0 | 0 | 0 | 0 |
| 2 | 0.20 ± 0.12 | 0 | 0 |
| 4 | 1.7 ± 0.48 | 0.53 ± 0.67 | 0.13 ± 0.67 |
| 6 | 5.3 ± 0.73 | 2.4 ± 0.64 | 2.6 ± 0.70 |
| 8 | 7.7 ± 3.0 | 4.1 ± 0.98 | 3.3 ± 0.75 |
| 10 | 15 ± 1.3 | 9.8 ± 0.53 | 9.8 ± 1.1 |
| 12 | 15 ± 1.2 | 13 ± 0.87 | 13 ± 0.64 |
| 14 | 17 ± 0.90 | 13 ± 0.61 | 13 ± 0.33 |
| 16 | 16 ± 0.71 | 15 ± 0.98 | 14 ± 0.46 |
| 18 | 15 ± 1.0 | 14 ± 1.4 | 14 ± 1.5 |
| 20 | 16 ± 1.8 | 14 ± 1.3 | 13 ± 1.00 |
The data are means ± standard errors for three seeds. The treatment A cucumber seeds inoculated with T. atroviride were planted in P. ultimum-infested soil; in treatment B cucumber seeds infested with P. ultimum were planted in T. atroviride-inoculated soil; and in treatment C both P. ultimum and T. atroviride were inoculated together into sterile soil before cucumber seeds were planted.
FIG. 2.
Images of the T. atroviride pki1::gfp mutant colonizing a cucumber root (A and B) and soil particles (C) in sterilized soil inoculated with P. ultimum after 3 days of incubation. The initial concentrations of P. ultimum and T. atroviride added to the soil were 3.1 × 103 propagules g−1 and 3 × 103 spores g−1, respectively. Cucumber seeds were also planted at the time of inoculation. The images in panels A and B were taken with a CSLM with a ×20 objective. The arrows indicate green fluorescent T. atroviride hyphae alongside P. ultimum hyphae (yellow or reddish brown) on the outer surface of a cucumber root (reddish brown). The image in panel C was taken with a fluorescence steromicroscope by using a ×6.9 objective and shows green fluorescent T. atroviride hyphae (arrow) adhering to soil particles (brownish or yellow).
Interaction between T. atroviride and P. ultimum on cucumber seeds.
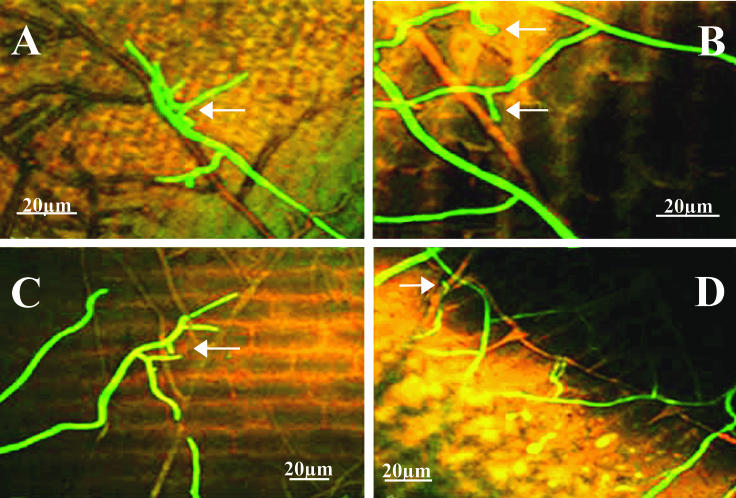
P. ultimum was identifiable as reddish brown hyphae by CSLM. Direct mycoparasitism of P. ultimum by T. atroviride was observed on the cucumber seed surfaces during seed germination (i.e., 8 days after planting) and for all three inoculation treatments. The morphological changes of the mycoparasite and the relative phases of attack by T. atroviride were similar to those observed in cocultures. On the cucumber seed surface, T. atroviride hyphae grew alongside the P. ultimum mycelium (Fig. 3A). The papilla-like swelling of T. atroviride hyphal tips was observed when the hyphae touched the host and also before contact (Fig. 3B). After the first contact with the host was established, T. atroviride formed new hyphal branches that further extended toward the P. ultimum mycelium (Fig. 3C), and eventually the branches coiled around the host mycelium on the cucumber seed surface (Fig. 3D). Growth of T. atroviride hyphae on the cucumber seeds apparently was stimulated by the presence of the host. The hyphae were more concentrated in regions of the seed surface colonized by P. ultimum mycelium.
FIG. 3.
CSLM images of cucumber seed cryosections prepared 10 days (A), 12 days (B), 14 days (C), and 18 days (D) after planting in sterile soil inoculated with P. ultimum at a concentration of 3.1 × 103 propagules g−1. The arrow in panel A indicates an aggregation of T. atroviride hyphae on P. ultimum hyphae. The arrows in panel B indicate papilla-like swelling of T. atroviride hyphal tips. The arrow in panel C indicates branching T. atroviride hyphae extending towards P. ultimum hyphae. The arrow in panel D indicates coiling of T. atroviride hyphae around P. ultimum hyphae. All images were taken with a ×20 objective.
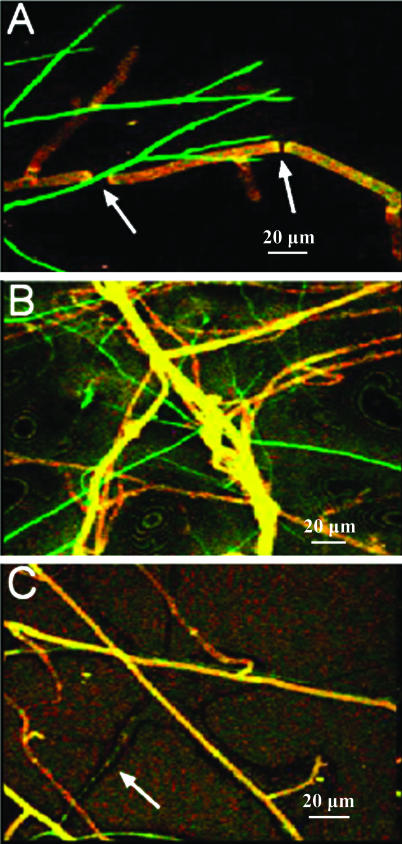
Induction of expression of T. atroviride mutants by R. solani in cocultures.
Induction of expression of chitinase genes resulted in a GFP fluorescent phenotype in both Trichoderma mutants (the ech::gfp and nag1::gfp mutants) 2 days after inoculation when the Trichoderma hyphae contacted R. solani hyphae (Fig. 4). The nagl::gfp mutant (Fig. 4A) was more intensely green fluorescent than the ech42::gfp mutant (Fig. 4B) upon contact with R. solani hyphae in PDA. The fluorescence intensity of both mutants became fainter as hyphal growth and sporulation continued, and 2 to 3 days after contact, the older hyphae lost green fluorescence. Prior to contact of the hyphae only weak autofluorescence was observed, which was comparable to the autofluorescence of the wild-type T. atroviride strain (Fig. 4C).
FIG. 4.
CSLM images of induction of chitinase genes in the T. atroviride nag1::gfp mutant (A) and the T. atroviride ech42::gfp mutant (B) by R. solani hyphae. In panels A and B the T. atroviride hyphae are green fluorescent due to contact with the R. solani hyphae, and the R. solani hyphae are reddish yellow. The arrows in panel A indicate breakage points in the R. solani hyphae. (C) Coculture of R. solani and wild-type T. atroviride (no gfp) (arrow) for comparison. All images were taken with a ×20 objective.
Induction of expression of T. atroviride mutants by colloidal chitin or ground R. solani hyphae.
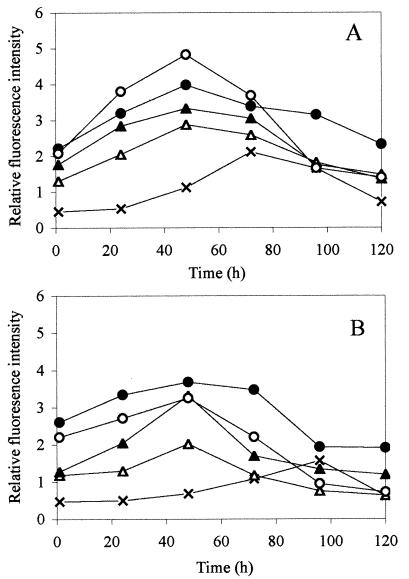
The T. atroviride mutants were incubated in the presence of colloidal chitin or ground R. solani hyphae, and fluorescence was monitored over time. The greatest fluorescence intensity appeared 48 h after inoculation of either the nagl::gfp or ech42::gfp mutant into SM medium supplemented with colloidal chitin or ground R. solani (Fig. 5). Up to the time of maximum intensity (48 h), the increase in fluorescence intensity was proportional to the concentration of colloidal chitin or ground R. solani for both mutants (the results for the highest concentration and an intermediate concentration of each are shown in Fig. 5). The fluorescence intensity of the nag1::gfp mutant in SM medium was greater than that of the ech42::gfp mutant after induction with colloidal chitin (Fig. 5), which is consistent with the confrontation studies performed with the T. atroviride mutants and R. solani (Fig. 4). However, the difference between the mutants was less obvious when they were induced by R. solani hyphae (Fig. 5). The relative fluorescence intensity increased over time for the controls without added hyphae or chitin as well, but to a much lesser extent, and the greatest fluorescence intensity was observed after 72 or 96 h of incubation (Fig. 5). The fluorescence observed in the controls was autofluorescence, not induction of gfp gene expression, and it was similar to that seen in wild-type T. atroviride strains that lacked the gfp gene (data not shown).
FIG. 5.
Relative fluorescence intensity, expressed per 0.1 mg (dry weight) of Trichoderma hyphae, upon chitinase gene induction of the T. atroviride nag1::gfp mutant (A) or the T. atroviride ech42::gfp mutant (B) in the presence of ground chitin at concentrations of 0.5 mg ml−1 (▵) and 2.5 mg ml−1 (○) or in the presence of R. solani hyphae at concentrations of 0.5 mg ml−1 (▴) and 2.5 mg ml−1 (•). Controls (×) consisted of T. atroviride mutant strains with no chitin or R. solani hyphae added. Each data point represents the mean of triplicate samples, which did not vary more than 10% from the mean.
Microcosm studies of inducible expression of T. atroviride mutants.
Hyphae of both T. atroviride mutants were observed growing in soil 2 days after inoculation. Three days after inoculation, GFP-fluorescing hyphae of the nagl::gfp mutant were observed growing alongside R. solani hyphae (Fig. 6A). After 4 days, the gfp gene of the ech::gfp mutant was expressed when the mutant was in contact with R. solani hyphae (Fig. 6B). More T. atroviride hyphae interacted with R. solani hyphae during the rest of the 7-day sampling period. On the seventh day the Trichoderma strain began to sporulate, and the spores were green fluorescent (data not shown). After sporulation the green fluorescence of the Trichoderma hyphae began to fade.
FIG. 6.
CSLM images of induction of chitinase genes after 3 days of incubation of the T. atroviride nag1::gfp mutant (A) and after 4 days of incubation of the T. atroviride ech42::gfp mutant (B) upon contact with R. solani hyphae (yellow or reddish brown) in sterile soil. The arrows indicate T. atroviride hyphae. Both images were taken with a ×20 objective.
DISCUSSION
The mycoparasitic mode of action of Trichoderma spp. against fungal plant pathogens has been studied extensively in two-culture assays and under other in vitro conditions (9, 10, 12, 21, 31). This study, performed in situ, allowed direct observation of the development of a mycoparasitic attack that occurred in sterile soil and on plant roots and seeds, conditions in which the physical effects of the complex soil matrix and plant tissues were taken into account. The wild-type strain and the gfp-tagged mutants of T. atroviride strain P1 had similarly high levels of biocontrol activity, indicating that transformation and expression of the gfp gene did not reduce the biocontrol capability of the fungus.
Our study revealed both the in vivo occurrence of and the sequence order for the various phases and gene expression of a necrotrophic parasitic interaction between T. atroviride and P. ultimum. The chemotactic growth of T. atroviride toward the host and the coiling around the host hyphae were the most common observations. We found, for instance, that development of the helix-shaped hyphae by the mycoparasite (44) occurred not only in the presence but also in the absence of direct contact with the host (data not shown).
Previous in vitro studies have shown that Trichoderma harzianum hyphae grow and branch directly towards their host (9). Here we found that in situ the branching of T. atroviride hyphae is an active, probably chemotactic, response to the presence of the host. We also observed papilla-like structures at the T. atroviride hyphal tips, which occurred both in the presence and in the absence of direct contact with P. ultimum. Bartnicki-Garcia et al. (2) speculated that papilla formation may be caused by exudates released from the host mycelium capable of displacing the Spitzenkörper (a phase-dark body found at the tip of elongating hyphae of higher fungi), which results in the apex becoming rounded and increasing in diameter. Since the fungal tip is an active growing area and very sensitive to many types of disturbances and stimuli, an alternative explanation may be that these morphological alterations are due to the effect of osmotic pressure changes (13). Our results support the hypothesis that papilla formation can occur due to environmental factors other than contact with host fungi. Alternatively, exudates released from the host mycelium could diffuse and induce distant papilla formation in Trichoderma.
In coculture experiments, T. atroviride spores adhered to the hyphae of P. ultimum, where they germinated and parasitized the host. Adhesion of fungal spores to the host surface is generally thought to be a necessary step for germination of the spores of a fungal mycoparasite and establishment of a successful parasitic interaction (25, 27, 28). There may be specific compounds released from the host hyphae that induce germination of T. atroviride spores and induce the later steps of mycoparasitism (12, 52, 53). This process may be quite complex. For instance, studies of Cochliobolus heterostrophus (6) showed that adhesion of fungal spores to leaves and artificial surfaces is accomplished through a variety of passive and active mechanisms.
The plant seed surface usually is a microbe-rich habitat in which multiple interactions among the germinating seeds, soil pathogens, and antagonists occur. Our in situ study showed that mycoparasitism of P. ultimum by T. atroviride takes place on the seed surface. We used sterile conditions so that the nontagged fungal hosts could be specifically identified in soil and on plant surfaces. On the seed surface, the mycoparasite usually formed hyphal branches that grew towards the host and resulted in intense mycelial growth around the host mycelium. This active growth may have been supported by the production of extracellular enzymes capable of releasing cell wall components that provided nutrients and/or further stimulated host colonization (4, 12, 33, 35, 52, 53).
The inoculation method affected colonization of the cucumber seed surface by T. atroviride. Rapid and extensive coverage of the surface was observed with seeds pretreated with T. atroviride spores (treatment A). Early colonization by a biocontrol agent often is required to fill the critical niches and to effectively compete against pathogenic fungi (38). Thus, seed coating with bacterial and fungal biocontrol agents often is utilized or required to control aggressive, rapidly growing soilborne pathogens, such as P. ultimum and R. solani (15, 38, 41).
Colonization by T. atroviride was observed during all growth stages of the young cucumber plant. Before cucumber seeds germinated, the hyphae colonized the seed surfaces. Subsequently, T. atroviride colonization extended to the cucumber radicle (Fig. 2), and this organism colonized the rhizosphere of the young cucumber root. This active colonization process may be related to the ability of T. atroviride to suppress P. ultimum- or R. solani-caused diseases (Table 1). Although we found no evidence of the presence of the mycoparasite inside the seeds, we cannot exclude the possibility that there is a direct relationship between the biocontrol agent and the plant.
We observed induction of the biocontrol-related ech42 and nag-1 genes during mycoparasitism by fusing the promoters to gfp (7, 31, 55). The transformants were activated by the presence of the host, chitin, and chitoligomers (37, 55) and fluoresced during the early phase of the interaction. This interaction occurred during coculture in vitro with R. solani in medium containing colloidal chitin or R. solani hyphal fragments (55) and in situ in the soil and in the presence of the host (Fig. 6). This is the first observation of in vivo expression of a fungal biocontrol-related gene, a phenomenon that has been predicted by various molecular studies (51; see reference 26 for a review) but has never been observed microscopically previously. GFP fluorescence was detectable within 24 h after T. atroviride started to colonize the soil, indicating that induction of both nag1 and ech42 is a rather early event during the interaction with R. solani and that both endo- and exochitinases may be used by T. atroviride to mycoparasitize the living host rather than to simply degrade dead hyphae.
The data presented here and in other studies clearly indicate that biocontrol- or mycoparasitism-related promoters associated with vital markers, such as GFP or DsRed (40), can be effectively used to study microbial interactions. For instance, it is possible to discern patterns of gene induction and to observe fungal interactions in vivo that occur in the soil and around the plant (36). This methodology may provide a way to monitor biocontrol activity (20) and the plant-Trichoderma interaction, thereby improving the selection of useful strains and the effectiveness of biopesticide and biofertilizer treatments.
In this work, gfp tagging was effective for monitoring in situ interactions between T. atroviride and other microbes grown in cocultures or on a plant (1, 23). In our experiments with cucumber, we observed a direct, mycoparasitic interaction between T. atroviride P1 and P. ultimum on the seed surface, which of course does not rule out the possible involvement of other antagonistic mechanisms (e.g., antibiosis, competition for nutrients or space, induction of resistance in the plant, etc.). On the basis of our CSLM-based study, we concluded that direct mycoparasitism and colonization of plant roots have roles in the biocontrol of P. ultimum by T. atroviride. In addition, we observed the presence and colonization of the tagged Trichoderma in the soil without killing the microorganisms. Such in situ monitoring studies of fungal antagonists should improve both our understanding of the ecology and the agricultural applications of these useful microbes.
Acknowledgments
This work was supported by a grant from the Swedish Council for Engineering Science to J.K.J. and by grants from the following sources to M.L.: FIRB CNR, PON, EU “TRICHOEST,” EU FAIR 98PL-4140, and MIPAF.
REFERENCES
- 1.Bae, Y. S., and G. R. Knudsen. 2000. Cotransformation of Trichoderma harzianum with beta-glucuronidase and green fluorescent protein genes provides a useful tool for monitoring fungal growth and activity in natural soils. Appl. Environ. Microbiol. 66:810-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartnicki-Garcia, S., D. D. Bartnicki, G. Gierz, R. Lopez-Franco, and C. E. Bracker. 1995. Evidence that Spitzenkorper behavior determines the shape of a fungal hypha: a test of the hyphoid model. Exp. Mycol. 19:153-159. [DOI] [PubMed] [Google Scholar]
- 3.Benhamou, N., and I. Chet. 1993. Hyphal interactions between Trichoderma harzianum and Rhizoctonia solani: ultrastructure and gold cytochemistry of the mycoparasitic process. Phytopathology 83:1062-1071. [Google Scholar]
- 4.Benhamou, N., and I. Chet. 1997. Cellular and molecular mechanisms involved in the interaction between Trichoderma harzianum and Pythium ultimum. Appl. Environ. Microbiol. 63:2095-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloemberg, G. V., G. A. O'Toole, B.-J. J. Lugtenberg, and R. Kolter. 1997. Green fluorescent protein as a marker for Pseudomonas spp. Appl. Environ. Microbiol. 63:4543-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, E. J., and R. J. Howard. 1994. Adhesion of fungal spores and germlings to host plant surfaces. Protoplasma 181:202-212. [Google Scholar]
- 7.Brunner, K., C. K. Peterbauer, R. L. Mach, M. Lorito, S. Zeilinger, and C. P. Kubicek. 2003. The Nag1 N-acetylglucosaminidase of Trichoderma atroviride is essential for chitinase induction by chitin and of major relevance to biocontrol. Curr. Genet. 43:289-295. [DOI] [PubMed] [Google Scholar]
- 8.Chalfie, M., Y. Tu, G. Euskirchen, W. W. Ward, and D. C. Prasher. 1994. Green fluorescent protein as a marker for gene expression. Science 263:802-805. [DOI] [PubMed] [Google Scholar]
- 9.Chet, I. 1987. Trichoderma applications, mode of action and potential as a biocontrol agent of soilborne plant pathogenic fungi, p. 137-160. In I. Chet (ed.), Innovative approaches to plant disease control. John Wiley & Sons, New York, N.Y.
- 10.Chet, I., N. Benamou, and S. Haran. 1998. Mycoparasitism and lytic enzymes, p. 153-171. In G. E. Harman and C. P. Kubicek (ed.), Trichoderma and Gliocladium, vol. 2. Taylor and Francis Ltd., London, United Kingdom.
- 11.Chet, I., G. E. Harman, and R. Baker. 1981. Trichoderma hamatum: its hyphal interactions with Rhizoctonia solani and Phythium spp. fungal parasites. Microb. Ecol. 7:29-38. [DOI] [PubMed] [Google Scholar]
- 12.Cortes, C., A. Gutierrez, V. Olmedo, J. Inbar, I. Chet, and A. Herrera-Estrella. 1998. The expression of genes involved in parasitism by Trichoderma harzianum is triggered by a diffusible factor. Mol. Gen. Genet. 260:218-225. [DOI] [PubMed] [Google Scholar]
- 13.Deacon, J. W. 1997. Fungal interaction: Mechanism, relevance and practical exploitation, p. 205-223. In J. W. Deacon (ed.), Modern mycology. Blackwell Science, London, United Kingdom.
- 14.Elad, Y., I. Chet, P. Boyle, and Y. Henis. 1983. Parasitism of Trichoderma spp. on Rhizoctonia solani and Sclerotium rolfsii—scanning electron microscopy and fluorescence microscopy. Fungi, structure, soilborne plant pathogens, biological control. Phytopathology 73:85-88. [Google Scholar]
- 15.Filonow, A. B., and J. M. Dole. 1999. Biological control of Pythium damping-off and root rot of greenhouse-grown geraniums and poinsettias. Proc. Okla. Acad. Sci. 79:29-32. [Google Scholar]
- 16.Haran, S., H. Schickler, A. Oppenheim, and I. Chet. 1996. Differential expression of Trichoderma harzianum chitinases during mycoparasitism. Phytopathology 86:980-985. [Google Scholar]
- 17.Harman, G. E. 2000. Myths and dogmas of biocontrol: changes in perceptions derived from research on Trichoderma harzianum T-22. Plant Dis. 84:377-393. [DOI] [PubMed] [Google Scholar]
- 18.Harman, G. E., I. Chet, and R. Baker. 1981. Factors affecting Trichoderma hamatum applied to seeds peas, radishes as a biocontrol agent parasite of fungi. Phytopathology 71:569-572.
- 19.Harman, G. E., and C. P. Kubicek. 1998. Trichoderma and Gliocladium, vol. 1 and 2. Taylor and Francis Ltd., London, United Kingdom.
- 20.Hassan, M., G. Corkidi, E. Galindo, C. Flores, and L. Serrano-Carreon. 2002. Accurate and rapid viability assessment of Trichoderma harzianum using fluorescence-based digital image analysis. Biotechnol. Bioeng. 80:677-684. [DOI] [PubMed] [Google Scholar]
- 21.Inbar, J., and I. Chet. 1995. The role of recognition in the induction of specific chitinases during mycoparasitism by Trichoderma harzianum. Microbiology 141:2823-2829. [DOI] [PubMed] [Google Scholar]
- 22.Jansson, J. K. 2002. Antibiotic, chromogenic, and luminescent markers for bacteria, p. 604-614. In C. J. Hurst, R. L. Crawford, G. R. Knudsen, M. J. McInerney, and L. D. Stetzenback (ed.), Manual of environmental microbiology, 2nd ed. ASM Press, Washington, D.C.
- 23.Jansson, J. K. 2003. Marker and reporter genes: illuminating tools for environmental microbiologists. Curr. Opin. Microbiol. 6:310-316. [DOI] [PubMed] [Google Scholar]
- 24.Jeffries, P. 1995. Biology and ecology of mycoparasitism. Can. J. Bot. 73:1284-1290. [Google Scholar]
- 25.Kubicek, C. P. 1987. Involvement of a conidial endoglucanase and a plasma-membrane-bound beta-glucosidase in the induction of endoglucanase synthesis by cellulose in Trichoderma reesei. J. Gen. Microbiol. 133:1481-1487. [DOI] [PubMed] [Google Scholar]
- 26.Kubicek, C. P., R. L. Mach, C. K. Peterbauer, and M. Lorito. 2001. Trichoderma chitinases: from genes to biocontrol. J. Plant Pathol. 83:11-23. [Google Scholar]
- 27.Kubicek, C. P., G. Muhlbauer, M. Klotz, E. John, and E. M. Kubicek-Pranz. 1988. Properties of a conidial-bound cellulase enzyme system from Trichoderma reesei. J. Gen. Microbiol. 134:1215-1222. [Google Scholar]
- 28.Kuo, K., and H. C. Hoch. 1996. Germination of Phyllosticta ampelicida pycnidiospores: prerequisite of adhesion to the substratum and the relationship of substratum wettability. Fungal Genet. Biol. 20:18-29. [DOI] [PubMed] [Google Scholar]
- 29.Lorang, J. M., R. P. Tuori, J. P. Martinez, T. L. Sawyer, R. S. Redman, J. A. Rollins, T. J. Wolpert, K. B. Johnson, R. J. Rodriguez, M. B. Dickman, and L. M. Ciuffetti. 2001. Green fluorescent protein is lighting up fungal biology. Appl. Environ. Microbiol. 67:1987-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lorito, M. 1998. Chitinolytic enzymes and their genes, p. 73-99 In G. E. Harman and C. P. Kubicek (ed.), Trichoderma and Gliocladium, vol. 2. Taylor and Francis, London, United Kingdom.
- 31.Lorito, M., V. Farkas, S. Rebuffat, B. Bodo, and C. P. Kubicek. 1996. Cell wall synthesis is a major target of mycoparasitic antagonism by Trichoderma harzianum. J. Bacteriol. 178:6382-6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorito, M., C. K. Hayes, A. Di Pietro, S. L. Woo, and G. E. Harman. 1994. Purification, characterization, and synergistic activity of a glucan 1,3-beta-glucosidase and an N-acetyl-beta-glucosaminidase from Trichoderma harzianum. Phytopathology 84:398-405. [Google Scholar]
- 33.Lorito, M., R. L. Mach, P. Sposato, J. Strauss, C. K. Peterbauer, and C. P. Kubicek. 1996. Mycoparasitic interaction relieves binding of the Cre1 carbon catabolite repressor protein to promoter sequences of the ech42 (endochitinase-encoding) gene in Trichoderma harzianum. Proc. Natl. Acad. Sci. USA 93:14868-14872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorito, M., F. Scala, A. Zoina, and S. L. Woo. 2001. Enhancing biocontrol of fungal pests by exploiting the Trichoderma genome, p. 248-259. In J. Gressel and M. Vurro (ed.), Enhancing biocontrol agents and handling risks. IOS Press, Amsterdam, The Netherlands.
- 35.Lorito, M., and S. L. Woo. 1998. Advances in understanding the antifungal mechanism(s) of Trichoderma and new applications for biological control, p. 73-80. In B. Duffy, U. Rosenberger, and G. Défago (ed.), Molecular approaches in biological control, vol. 21. IOBC WPRS Bulletin/Bulletin OILB SROP, Dijon, France.
- 36.Lutz, M. P., G. Feichtinger, G. Defago, and B. Duffy. 2003. Mycotoxigenic Fusarium and deoxynivalenol production repress chitinase gene expression in the biocontrol agent Trichoderma atroviride P1. Appl. Environ. Microbiol. 69:3077-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mach, R. L., C. K. Peterbauer, K. Payer, S. Jaksits, S. L. Woo, S. Zeilinger, C. M. Kullnig, M. Lorito, and C. P. Kubicek. 1999. Expression of two major chitinase genes of Trichoderma atroviride (T. harzianum P1) is triggered by different regulatory signals. Appl. Environ. Microbiol. 65:1858-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin, F. N., and J. E. Loper. 1999. Soilborne plant diseases caused by Pythium spp.: ecology, epidemiology, and prospects for biological control. Crit. Rev. Plant Sci. 18:111-181. [Google Scholar]
- 39.Meese, R. J., and P. A. Tomich. 1992. Dots on the rocks, a comparison of percent cover estimation methods. J. Exp. Mar. Biol. Ecol. 165:59-73. [Google Scholar]
- 40.Mikkelsen, L., S. Sarrocco, M. Lubeck, and D. F. Jensen. 2003. Expression of the red fluorescent protein DsRed-Express in filamentous ascomycete fungi. FEMS Microbiol. Lett. 223:135-139. [DOI] [PubMed] [Google Scholar]
- 41.Moulin, F., P. Lemanceau, and C. Alabouvette. 1994. Pathogenicity of Pythium species on cucumber in peat-sand rockwool and hydroponics. Eur. J. Plant Pathol. 100:3-17. [Google Scholar]
- 42.Naseby, D. C., J. A. Pascual, and J. M. Lynch. 2000. Effect of biocontrol strains of Trichoderma on plant growth, Pythium ultimum populations, soil microbial communities and soil enzyme activities. J. Appl. Microbiol. 88:161-169. [DOI] [PubMed] [Google Scholar]
- 43.Ramos, C., L. Molbak, and S. Molin. 2000. Bacterial activity in the rhizosphere analyzed at the single-cell level by monitoring ribosome contents and synthesis rates. Appl. Environ. Microbiol. 66:801-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sherwood-Higham, J., W. Y. Zhu, C. A. Devine, G. W. Gooday, N. A. Gow, and D. W. Gregory. 1994. Helical growth of hyphae of Candida albicans. J. Med. Vet. Mycol. 32:437-445. [DOI] [PubMed] [Google Scholar]
- 45.Sivan, A., and I. Chet. 1989. The possible role of competition between Trichoderma harzianum and Fusarium oxysporum on rhizosphere colonization. Phytopathology 79:198-203. [Google Scholar]
- 46.Siwek, K., A. R. Harris, and E. S. Scott. 1997. Mycoparasitism of Pythium ultimum by antagonistic binucleate Rhizoctonia isolates in agar media on capsicum seeds. J. Phytopathol. 145:417-423. [Google Scholar]
- 47.Tombolini, R., and J. K. Jansson. 1998. Monitoring of GFP-tagged bacterial cells. Methods Mol. Biol. 102:285-298. [DOI] [PubMed] [Google Scholar]
- 48.Tombolini, R., D. J. Van der Gaag, B. Gerhardson, and J. K. Jansson. 1999. Colonization pattern of the biocontrol strain Pseudomonas chlororaphis MA 342 on barley seeds visualized by using green fluorescent protein. Appl. Environ. Microbiol. 65:3674-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Unge, A., and J. Jansson. 2001. Monitoring population size, activity, and distribution of gfp-luxAB-tagged Pseudomonas fluorescence SBW25 during colonization of wheat. Microb. Ecol. 41:290-300. [DOI] [PubMed] [Google Scholar]
- 50.Unge, A., R. Tombolini, L. Molbak, and J. K. Jansson. 1999. Simultaneous monitoring of cell number and metabolic activity of specific bacterial populations with a dual gfp-luxAB marker system. Appl. Environ. Microbiol. 65:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woo, S. L., B. Donzelli, F. Scala, R. Mach, G. E. Harman, C. P. Kubicek, G. Del Sorbo, and M. Lorito. 1999. Disruption of the ech42 (endochitinase-encoding) gene affects biocontrol activity in Trichoderma harzianum P1. Mol. Plant-Microbe Interact. 12:419-429. [Google Scholar]
- 52.Woo, S. L., V. Fogliano, L. Mach, R. Ciliento, C. P. Kubicek, and M. Lorito. 2000. Host-derived molecules that activate the mechanism of biocontrol in Trichoderma spp., p. 89. In Proceedings of the 5th Congress of the European Foundation for Plant Pathology (EFPP 2000), Taormina-Giardini Naxos, Italy, 18 to 22 September 2000.
- 53.Woo, S. L., F. Scala, and M. Lorito. 2000. Host-derived molecules that activate the mechanism of biocontrol in Trichoderma spp., p. 25. In Proceedings of the Meeting on Methods To Monitor Microbial Inoculants To Improve Their Success, Wageningen, The Netherlands.
- 54.Yedidia, I., N. Benhamou, and I. Chet. 1999. Induction of defense responses in cucumber plants (Cucumis sativus L.) by the biocontrol agent Trichoderma harzianum. Appl. Environ. Microbiol. 65:1061-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeilinger, S., C. Galhaup, K. Payer, S. L. Woo, R. L. Mach, C. Fekete, M. Lorito, and C. P. Kubicek. 1999. Chitinase gene expression during mycoparasitic interaction of Trichoderma harzianum with its host. Fungal Genet. Biol. 26:131-140. [DOI] [PubMed] [Google Scholar]