A maternal factor affects seed fatty acid biosynthesis and inhibits seed lipid accumulation by targeting seed development and down-regulating a group of genes critical to embryonic development.
Abstract
Fatty acids (FAs) and FA-derived complex lipids play important roles in plant growth and vegetative development and are a class of prominent metabolites stored in mature seeds. The factors and regulatory networks that control FA accumulation in plant seeds remain largely unknown. The role of TRANSPARENT TESTA8 (TT8) in the regulation of flavonoid biosynthesis and the formation of seed coat color is extensively studied; however, its function in affecting seed FA biosynthesis is poorly understood. In this article, we show that Arabidopsis (Arabidopsis thaliana) TT8 acts maternally to affect seed FA biosynthesis and inhibits seed FA accumulation by down-regulating a group of genes either critical to embryonic development or important in the FA biosynthesis pathway. Moreover, the tt8 mutation resulted in reduced deposition of protein in seeds during maturation. Posttranslational activation of a TT8-GLUCOCORTICOID RECEPTOR fusion protein and chromatin immunoprecipitation assays demonstrated that TT8 represses the activities of LEAFY COTYLEDON1, LEAFY COTYLEDON2, and FUSCA3, the critical transcriptional factors important for seed development, as well as CYTIDINEDIPHOSPHATE DIACYLGLYCEROL SYNTHASE2, which mediates glycerolipid biosynthesis. These results help us to understand the entire function of TT8 and increase our knowledge of the complicated networks regulating the formation of FA-derived complex lipids in plant seeds.
Fatty acids (FAs) and FA-derived complex lipids are the major energy reserve substance stored in the seeds of many higher plants. In Arabidopsis (Arabidopsis thaliana), the development of the seed requires the precise regulation of three distinct tissues: the embryo, endosperm, and seed coat. After the occurrence of double fertilization in the embryo sac, the formation of the embryo and endosperm from zygotic tissues is initiated. The accumulation of most seed FAs is physiologically coupled with seed development, which is generally divided into two processes: embryonic morphogenesis (EM) and maturation (Baud and Lepiniec, 2009). EM starts by double fertilization, undergoes a series of programmed cell division and differentiation, and ends at 6 d after pollination (DAP) with the formation of a torpedo-shaped embryo (Goldberg et al., 1994; Mayer and Jürgens, 1998; Jenik et al., 2007). The maturation phase takes place from 7 to 20 DAP and features the accumulation of seed storage reserves, such as seed lipids and proteins, and the acquisition of dormancy and desiccation tolerance (Goldberg et al., 1994; Baud et al., 2002; Fait et al., 2006). During the process of seed development, the endosperm supplies nutrition to the developing embryo. Finally, it is degraded and reduced to merely one cell layer surrounding the embryo, accounting for roughly 10% of the total seed FAs (Penfield et al., 2004; Li et al., 2006; Stone et al., 2008; Baud and Lepiniec, 2009). The seed FA accumulation that mainly occurs in embryonic tissues exhibits a sigmoid pattern, changing slightly at the beginning and end but drastically between 7 and 17 DAP.
The seed coat is derived from the maternal integument and consists of four cell layers, an endothelial layer, a crushed layer forming the inner integument, and an epidermis comprising the outer integument, from inside to outside (Debeaujon et al., 2000). The seed coat not only plays essential roles in nourishing the embryo and protecting it against adverse environmental conditions but also in determining the morphology, dormancy, and longevity of a seed (Mohamedyasseen et al., 1994; Weber et al., 1996; Debeaujon et al., 2000).
Flavonoids are one of the largest groups of secondary metabolites unique to higher plants, biosynthesized from malonyl-CoA and Phe. They can be classified into three major classes, flavonols, anthocyanins, and proanthocyanidins, which are modified from a 15-carbon skeleton. In plants, flavonoid derivatives determine the pigmentation pattern of vegetative organs and seeds (Nesi et al., 2001; Lepiniec et al., 2006) and are involved in multiple functions. For example, they play important roles in protecting plants against a wide range of environmental injuries (Debeaujon et al., 2000; Agati and Tattini, 2010; Agati et al., 2012; Chen et al., 2012b). Seed coat mutants include two major groups. One group, represented by the transparent testa (tt) and transparent testa glabra (ttg) defects in testa flavonoid accumulation, has changes in seed coat color ranging from yellow to pale brown or grayish brown (Shirley et al., 1995; Debeaujon et al., 2000). The second group, represented by glabra2 (gl2), features abnormal testa structures, such as lacking the formation of mucilage (Rerie et al., 1994; Masucci and Schiefelbein, 1996), an important secreta produced by outer seed coat cells and composed primarily of a heterogenous group of acidic polysaccharides that form a gel-like matrix with two key pectins, homogalacturonan and rhamnogalacturonan I (Western et al., 2000; Macquet et al., 2007a, 2007b). The seed FA and flavonoid biosynthesis depends on the precise coordination of the embryo, endosperm, and seed coat. Therefore, the mutants defective in the development of the seed coat could also affect embryonic FA biosynthesis. Recent studies demonstrate that the tt2 mutation resulted in a significant increase of seed FA content (Chen et al., 2012b), and the gl2 mutation produced more seed oil due to increased carbon allocation to the embryo in the absence of seed coat mucilage biosynthesis (Shi et al., 2012).
Previous studies revealed that TT8, which encodes a basic helix-loop-helix domain regulatory protein in the nucleus, is involved in anthocyanin and proanthocyanidin biosynthesis and regulates the expression DIHYDROFLAVONOL 4-REDUCTASE and BANYULS, two genes in the flavonoid biosynthesis pathway (Nesi et al., 2000; Baudry et al., 2006). Despite our knowledge regarding the function of TT8 in flavonoid metabolism, the role of TT8 in seed FA biosynthesis has not been reported. Thus, we designed and performed a series of genetic and molecular experiments to investigate whether TT8 is also involved in FA biosynthesis and to understand the mechanism by which TT8 targets the downstream factors to regulate seed development and FA biosynthesis. Our results clearly show that TT8 inhibits seed FA accumulation by targeting several transcription factors (TFs) regulating seed development in Arabidopsis.
RESULTS
TT8 Changes FA Storage and FA Composition in Mature Seeds
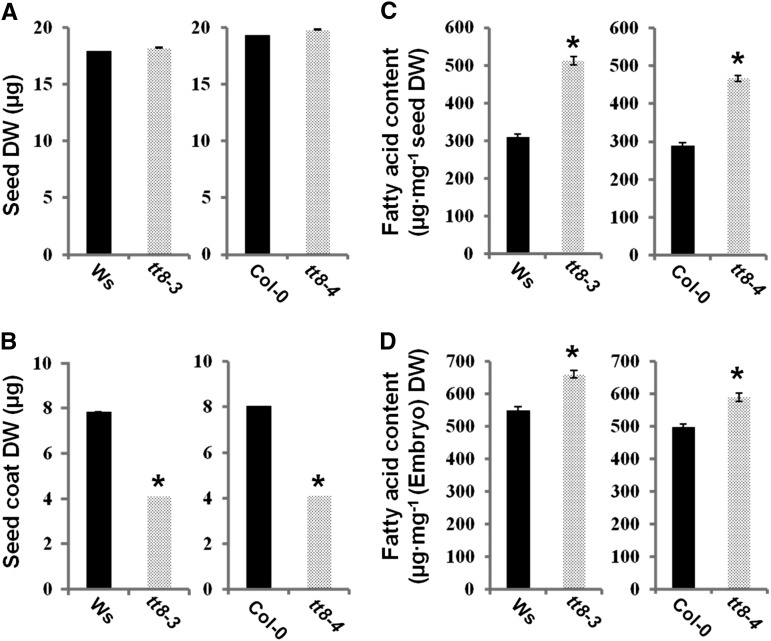
Two transfer DNA insertion mutant lines of the TT8 gene, tt8-3 (Nesi et al., 2000) and tt8-4 (SALK_063334), a newly named allele in this study, were provided by the Arabidopsis Biological Resource Center. The structure of the TT8 gene and the positions of the transfer DNA insertions in the tt8-3 and tt8-4 mutants are shown in Supplemental Figure S1A. Genotyping by PCR (Supplemental Fig. S1B) indicated the presence of the homozygous tt8-4 mutant, which completely lacks the transcript of the TT8 gene, as detected by reverse transcription (RT)-PCR (Supplemental Fig. S1C). The tt8-3 and tt8-4 mutants produce yellow seeds due to the lack of flavonoid deposition in the seed coats. The tt8 mutant seeds are slightly bigger than their respective wild-type controls (Supplemental Fig. S2). However, no obvious differences in embryo size and cell size in the central region of the embryo between tt8-4 and the wild type (Columbia-0 [Col-0]) were observed (Supplemental Fig. S3). The dry weight of tt8 seeds upon maturation was also similar to that of the corresponding wild-type seeds (Fig. 1A). However, the dry weights of tt8-3 and tt8-4 embryos were 40.1% and 39.3% heavier than that of their respective wild-type controls, reflecting the differences in developmental effects. In contrast, the dry weight of seed coats of the tt8 seeds was only 50% of that in the respective wild types (Fig. 1B).
Figure 1.
Comparison between the tt8 mutant lines and their respective wild types. A, Seed dry weight (DW). B, Seed coat dry weight. C, FA content. D, FA content of the embryo. Data presented are means of three independent biological repeats ± se. Asterisks indicate significant differences (P ≤ 0.05) compared with the wild type.
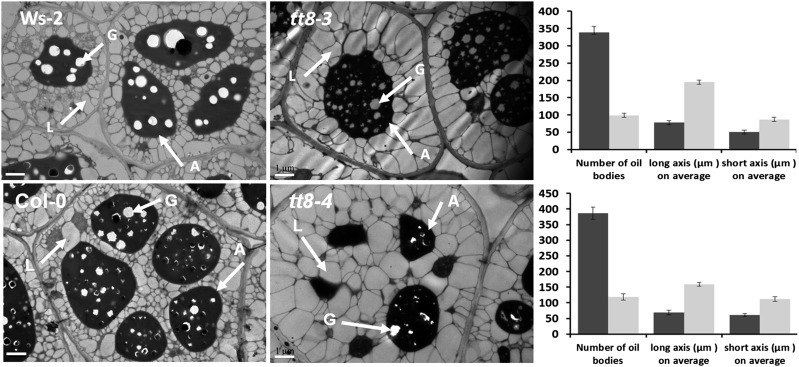
The seed FA content and FA composition in mature tt8 seeds were measured and are shown in Table I and Figure 1C. The FA content of tt8 seeds was 60% higher than that of wild-type seeds. Regardless of which allele mutated, the increase in seed FA content was attributed to the elevated proportion of embryo weight as well as the promoted FA production in the embryos (Fig. 1, B and D). The increase of FA content was accompanied by an alteration in FA composition. In the mature tt8 seeds, the proportions of 16-C and 18-C FAs, with an exception for 18:1, decreased significantly, whereas the proportions of very-long-chain FAs (C ≥ 20), including 20-C, 22-C, and 24-C FAs, increased significantly (Table II). The subcellular structures of the tt8 mutant lines (tt8-3 and tt8-4) were compared with those of Wassilewskija (Ws) and Col-0, respectively (Fig. 2). Figure 2 also indicates the average number of oil bodies that were contained in a cell and the mean lengths of long and short axes of an oil body. It is clear from the figure that there were smaller numbers and larger sizes of oil bodies in the tt8 cells than in the wild-type cells. These results demonstrate that the tt8 mutation results in defective flavonoid accumulation in the seed coat, significant increases in the relative proportions of embryo weight and FA content in seeds, the alteration of the FA composition, and changes in the number and size of oil bodies in seed cells.
Table I. Comparison of FA composition and total FA content (μg mg−1 seed dry weight) in mature seeds between tt8 mutant lines and their respective wild types.
Data reported are means of five independent experiments (five biological repeats) ± se. Asterisks indicate significant differences (P ≤ 0.05).
| Line | 16:0 | 18:0 | 18:1 | 18:2 | 18:3 | 20:0 | 20:1 | 22:0 | 22:1 | 24:0 | Sum |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ws | 29.4 ± 1.1 | 10.2 ± 0.9 | 27.0 ± 1.1 | 115.3 ± 2.3 | 83.1 ± 1.8 | 5.1 ± 0.1 | 39.9 ± 1.1 | 0.8 ± 0.1 | 3.7 ± 0.2 | 0.8 ± 0.1 | 315.1 ± 8.7 |
| tt8-3 | 45.7 ± 1.6* | 15.2 ± 0.6* | 72.8 ± 1.3* | 165.2 ± 2.7* | 126.9 ± 2.3* | 10.1 ± 0.1* | 69.9 ± 1.2* | 1.9 ± 0.8* | 8.2 ± 0.2* | 2.7 ± 0.1* | 518.5 ± 10.9 |
| Col-0 | 28.2 ± 0.9 | 8.5 ± 0.3 | 18.6 ± 0.5 | 109.5 ± 2.5 | 78.2 ± 1.3 | 3.9 ± 0.1 | 36.2 ± 0.6 | 0.8 ± 0.1 | 2.9 ± 0.8 | 0.5 ± 0.1 | 287.3 ± 7.2 |
| tt8-4 | 43.8 ± 1.2* | 12.1 ± 0.4* | 65.9 ± 1.0* | 153.7 ± 3.2* | 117.2 ± 1.3* | 7.4 ± 0.1* | 61.1 ± 1.0* | 1.6 ± 0.1* | 6.6 ± 0.3* | 1.5 ± 0.1* | 471.0 ± 8.6 |
Table II. Comparison of FA composition and total FA content (mol %) in mature seeds between tt8 mutant lines and their respective wild types.
Data reported are means of five independent experiments (five biological repeats) ± se. The up or down arrows indicate an increase or decrease from the respective wild types.
| Line | 16:0 | 18:0 | 18:1 | 18:2 | 18:3 | 20:0 | 20:1 | 22:0 | 22:1 | 24:0 |
|---|---|---|---|---|---|---|---|---|---|---|
| Ws | 9.3 ± 0.8 | 3.3 ± 0.4 | 8.6 ± 0.4 | 36.6 ± 3.8 | 26.36 ± 3.53 | 1.6 ± 0.1 | 12.6 ± 0.6 | 0.3 ± 0.0 | 1.2 ± 0.4 | 0.3 ± 0.0 |
| tt8-3 | 8.8 ± 0.8↓ | 2.9 ± 0.4↓ | 14.0 ± 0.5↑ | 31.9 ± 3.9↓ | 24.5 ± 4.5↓ | 1.9 ± 0.2↑ | 13.5 ± 0.6↑ | 0.4 ± 0.1↑ | 1.6 ± 0.3↑ | 0.5 ± 0.1↑ |
| Col-0 | 9.8 ± 0.8 | 3.0 ± 0.3 | 6.5 ± 0.4 | 38.1 ± 3.6 | 27.2 ± 3.8 | 1.4 ± 0.2 | 12.6 ± 0.5 | 0.3 ± 0.1 | 1.0 ± 0.3 | 0.2 ± 0.0 |
| tt8-4 | 9.3 ± 0.9↓ | 2.6 ± 0.5↓ | 14.0 ± 0.5↑ | 32.6 ± 4.0↓ | 24.9 ± 4.6↓ | 1.6 ± 0.2↑ | 13.0 ± 0.7↑ | 0.3 ± 0.1↑ | 1.4 ± 0.2↑ | 0.3 ± 0.1↑ |
Figure 2.
Subcellular differences in seed embryo cells between the wild types and tt8 mutant lines in pairs. Arrows point to oil bodies (L), aleurone grains (A), and globoids (G). The quantitative data are given in charts beside each pair of mutant and wild-type images. Bars = 1 µm.
TT8 Controls Seed Starch and Protein Metabolism
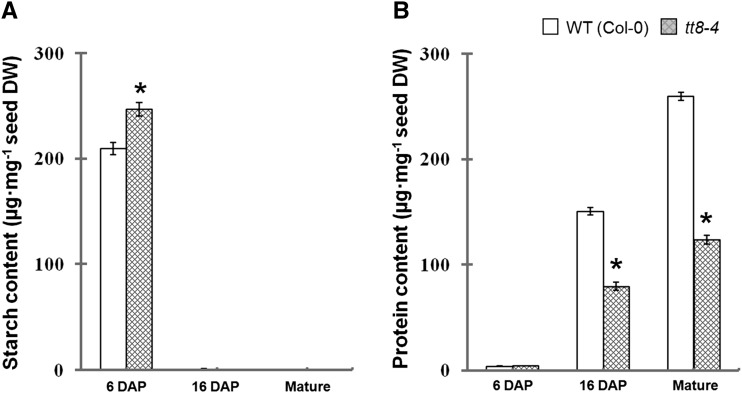
The starch and protein contents of the wild-type (Col-0) and tt8-4 seeds at 6 DAP, 16 DAP, and mature developmental stages were analyzed. As shown in Figure 3A, the starch contents of the wild type (209.38 ± 5.76 µg mg−1 dry weight) and tt8-4 (246.63 ± 6.37 µg mg−1 dry weight) peaked in the developing seeds at 6 DAP. The mutant seeds exhibited significantly more starch than the wild-type seeds. The starch contents of both genotypes decreased considerably during the process of EM. In the seeds at 16 DAP and mature stages, only traces of starch could be detected in both the wild type and the mutant.
Figure 3.
Evaluation of starch (A) and protein (B) contents in developing Arabidopsis seeds between the wild type (WT; Col-0) and the tt8-4 mutant line. The starch and protein contents were analyzed at three different developmental stages, mature seeds and developing seeds at 6 and 16 DAP, based on the dry weight (DW) and seed total protein content. Data presented are means of three independent experiments (three biological repeats) ± se. Asterisks indicate significant differences (P ≤ 0.05) compared with the wild type.
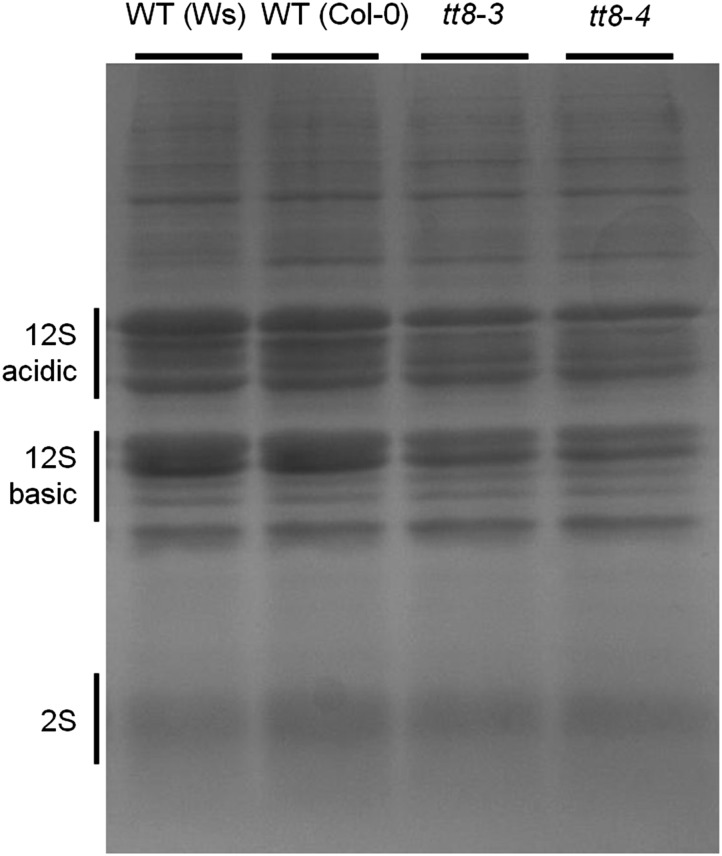
Figure 3B shows the increase of seed total protein contents from 6 DAP to the mature stage in the wild type and the tt8-4 mutant. The seed protein content of the tt8-4 seeds at 6 DAP was slightly higher than that of the wild type at the same stage; however, the tt8-4 seeds at 16 DAP as well as at the mature stage contained lower protein than the wild-type seeds at the same stages. We also analyzed the amounts of proteins such as the 12S and 2S storage proteins in wild-type and tt8 mutant seeds. Fractionation of identical amounts of protein extracts from wild-type and tt8 mutant seeds by SDS-PAGE revealed quantity differences. As shown in Figure 4, the quantities of proteins, including 12S and 2S, in the tt8 mutant seeds were uniformly lower than in the wild-type seeds; however, the overall proportions of individual proteins in the mutant were not significantly changed. Measurement of protein contents in mature seeds confirmed that the tt8 seeds accumulated less protein than the wild-type seeds (Fig. 3B).
Figure 4.
tt8 mutant seeds contain less protein than do wild-type (WT) seeds. Protein extracts from an equal number of wild-type, tt8-3, and tt8-4 mature seeds were fractionated on a 15% SDS polyacrylamide gel and stained.
Effect of the TT8 Mutation on Seed Coat Mucilage
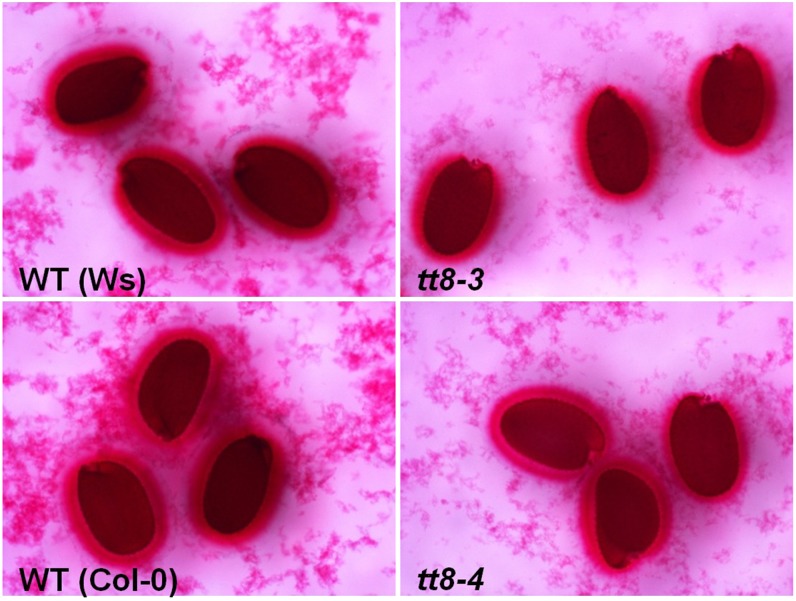
Previous studies showed that TT8 regulates the biosynthesis of seed coat mucilage together with ENHANCER OF GLABRA3 (Zhang et al., 2003). To investigate if the tt8 mutation resulted in less mucilage, we compared the mucilage layers between wild-type and tt8 seeds. The result shows that there was no obvious difference in the thickness of mucilage layers between wild-type and tt8 seeds (Fig. 5).
Figure 5.
Comparison of the mucilage layer attached to the seed coat between wild-type (WT) and tt8 mutant seeds. [See online article for color version of this figure.]
TT8 Acts Maternally to Influence Seed FA
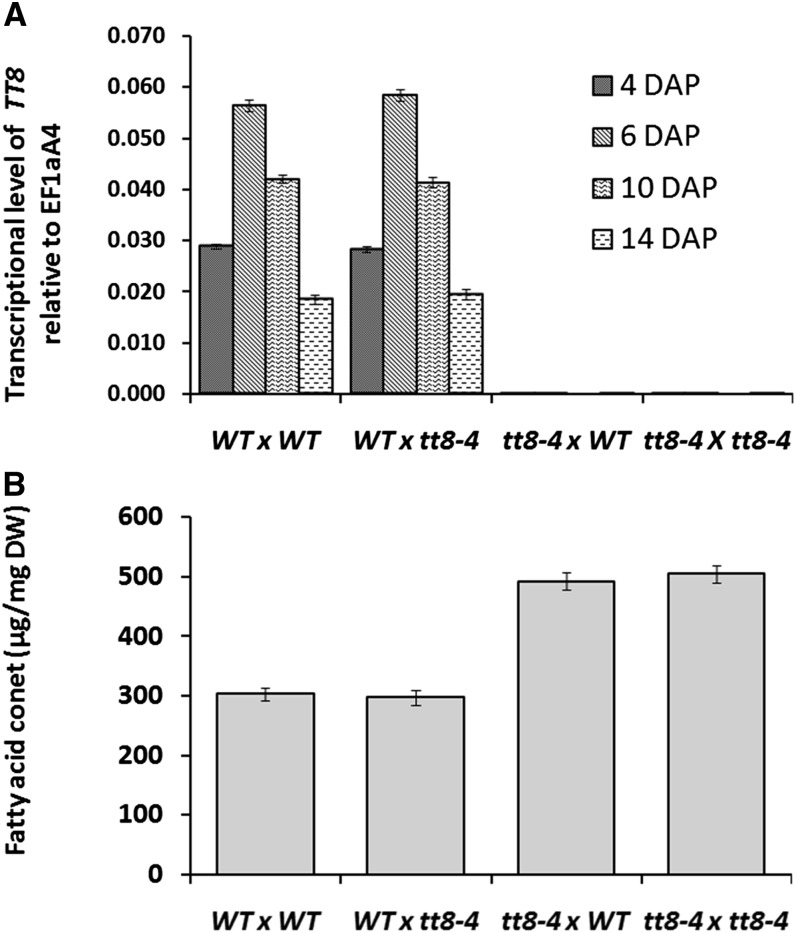
Previous studies indicate that TT8 regulates the biosynthesis of flavonoids in the seed coat, which is a maternal tissue. To understand whether TT8 acts maternally or zygotically in affecting FA biosynthesis, we made reciprocal crosses between the wild type and the tt8-4 mutant and measured the expression of TT8 as well as FAs in the respective F1 seeds. Table III shows the homozygous or heterozygous status of the TT8 gene in the embryo, endosperm, and seed coat tissues of various F1 hybrids derived from reciprocal crossings. In the F1 seeds derived from the crossings wild type × wild type and wild type × tt8-4, the levels of TT8 expression were quite similar, peaking at 6 DAP and then decreasing at 10 and 14 DAP. In contrast, in the F1 seeds originating from the crossings tt8-4 × wild type and tt8-4 × tt8-4, the expression of TT8 could hardly be detected (Fig. 6A). Moreover, high seed oil accumulations were measurable only in the F1 seeds from the crossings where tt8-4 was used as a maternal parent, indicating that the tt8 mutation had a maternal effect on seed FA accumulation (Fig. 6B).
Table III. Status of the TT8 allele in the embryo, endosperm, and seed coat tissues of F1 seeds derived from Col-0, tt8-4, and the reciprocal crossings between Col-0 and tt8-4.
Plus and minus signs indicate dominant TT8 and recessive tt8 status, respectively.
Figure 6.
Maternal effects of the tt8-4 mutation on TT8 expression in embryonic tissues (A) and seed FA accumulation (B). DW, Dry weight; WT, wild type.
TT8 Down-Regulates a Number of Genes Involved in Embryonic Development and FA Synthesis during Seed Development
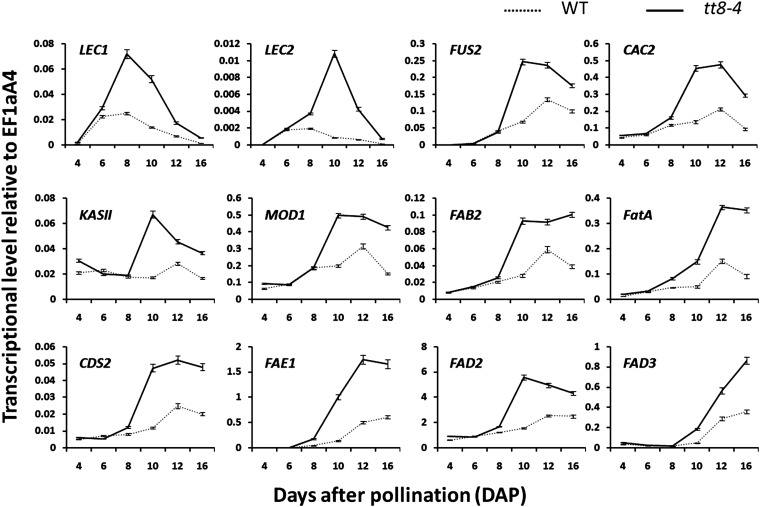
The expression of the TFs regulating seed development, such as LEAFY COTYLEDON1 (LEC1), LEC2, and FUSCA3 (FUS3), and the enzymes that are important in the FA biosynthesis pathway, including ACETYL CO-ENZYME A CARBOXYLASE BIOTIN CARBOXYLASE SUBUNIT (CAC2; At5g35360), BETA-KETOACYL-ACP SYNTHETASE2 (KASII; At1g74960), MOSAIC DEATH1 (MOD1; At2g05990), FATTY ACID BIOSYNTHESIS2 (FAB2; At2g43710), FATA ACYL-ACP THIOESTERASE (FatA; At3g25110), CYTIDINEDIPHOSPHATE DIACYLGLYCEROL SYNTHASE2 (CDS2; At4g22340), FATTY ACID ELONGATION1 (FAE1; At4g34520), FATTY ACID DESATURASE2 (FAD2; At3g12120), and FAD3 (At2g29980), was investigated by performing RT-quantitative PCR (qPCR) using the RNAs extracted from seeds at 4, 6, 8, 10, 12, and 16 DAP (Fig. 7). Relative to the wild type, the tt8 mutation resulted in about 2-fold higher LEC1 and LEC2 transcripts at early stages of seed development. The mutation did not significantly promote the relative expression of FUS3 from 4 to 8 DAP but resulted in 3.5-fold higher transcripts at 10 DAP. As detailed in Figure 7, the tt8-4 mutation did not lead to significant changes in the expression of genes encoding enzymes in the FA biosynthesis pathway at the developmental stages before 10 DAP but led to significant increases at 10 DAP and afterward. We also investigated the effect of tt8-4 on the expression of WRINKLED1 (WRI1) and ABSCISIC ACID3 (ABI3), two other TFs regulating seed development, and of CAC3 (At2g38040), BCCP2 (At5g15530), and PIS2 (At4g38570), other enzymes in the FA biosynthesis pathway. However, there were no significant expression differences in these genes between the wild type and tt8-4 (Supplemental Fig. S4). Taken together, the tt8-4 mutation promoted a class of genes regulating either embryonic development or FA synthesis during seed development.
Figure 7.
Comparison of the transcription levels of genes either regulating seed development or important in the FA synthesis pathway at different developmental stages between wild-type (WT) and tt8-4 seeds. RNA samples were extracted from the developing seeds at 4, 6, 8, 10, 12, and 16 DAP. The gene transcripts were measured by RT-qPCR using EF1aA4 (At5g60390) as the internal standard. Values are means of two replicates carried out on cDNA dilutions obtained from two independent RNA extractions.
TT8 Directly Targets LEC1, LEC2, FUS3, and CDS2 in Arabidopsis Seedlings
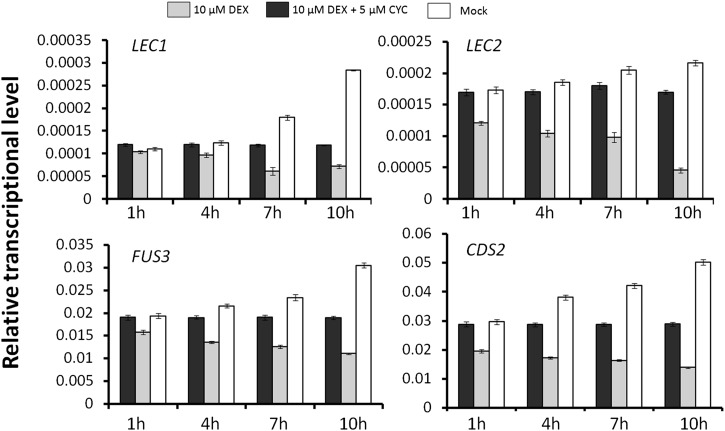
To further elucidate whether the TFs and enzymes mentioned above are the direct downstream targets of TT8, an inducible TT8 gene fused with the steroid-binding domain of glucocorticoid receptor (35S:TT8-GR) was constructed and transferred into the wild type and then crossed with tt8-4 mutant plants. Treatment of tt8-4 plants harboring 35S:TT8-GR 20 d after sowing with the mammalian steroid hormone analog dexamethasone (DEX) rescued the yellow seed coat phenotype of the tt8-4 mutant (Supplemental Fig. S5). Thus, DEX treatment induced high levels of TT8 activity. RT-qPCR was performed using the RNA samples from the transgenic seedlings (the tt8-4 plants harboring the 35S:TT8-GR construct) 15 d after sowing mock treated or treated with 10 µm DEX or 10 µm DEX plus 5 µm cycloheximide (CYC) for 1, 4, 7, and 10 h. Figure 8 shows that DEX treatment for 1 h or longer resulted in continuous reduction of LEC1, LEC2, FUS3, and CDS2 RNA levels relative to the mock-treated controls. To determine whether 35S:TT8-GR directly or indirectly represses these four genes, we repeated DEX applications to the 35S:TT8-GR seedlings of the tt8-4 background in the presence of the protein synthesis inhibitor CYC, because the induction of TT8-GR activity by DEX does not require protein synthesis. The results showed that CYC strongly removed the repression of these genes exerted by DEX treatment, especially at 4 and 7 h after combined treatment with DEX and CYC (Fig. 8), suggesting that LEC1, LEC2, FUS3, and CDS2 could be the direct targets of TT8.
Figure 8.
Induced TT8 activity can transcriptionally repress the expression of LEC1, LEC2, FUS3, and CDS2. RT-qPCR results were determined from RNA samples of tt8-4 35S:TT8-GR seedlings 15 d after sowing mock treated or treated with 10 µm DEX or 10 µm DEX plus 5 µm CYC for 1, 4, 7, and 10 h. The transcriptional level of the mock-treated samples at each time point was set to 1, and EF1aA4 was used as the internal control. Values are means of three biological repeats, and error bars indicate se.
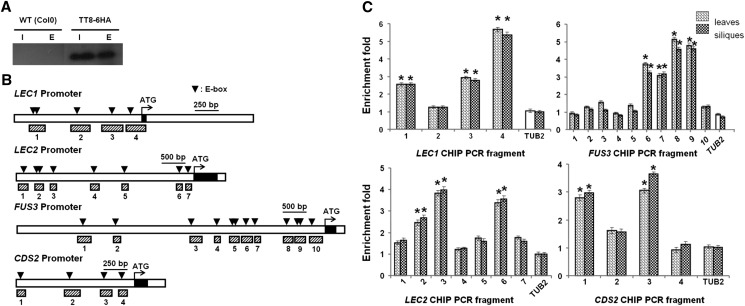
To further elucidate the molecular mechanism by which TT8 affects the expression of LEC1, LEC2, FUS3, and CDS2, chromatin immunoprecipitation (ChIP) assays were performed to test whether TT8 could bind to the promoter regions of these four genes using 35S:TT8-6HA transgenic plants (HA for human influenza hemagglutinin epitope) in which yellow seed coat was restored (Supplemental Fig. S5). The TT8 protein displays the typical features of a TF, with a basic helix-loop-helix signature at its C terminus, which recognizes the binding consensus site CANNTG (Abe et al., 1997; Massari and Murre, 2000). The abundance of the TT8-6HA protein in nuclear extracts (input) of 35S:TT8-6HA rosette leaves and siliques and the corresponding immunoprecipitated fractions (eluate) used for ChIP assays was comparable to that in the wild type after fixation (Fig. 9A). ChIP assays showed that TT8-6HA was associated with the promoter regions near fragments 1, 3, and 4 of LEC1, fragments 2, 3, and 6 of LEC2, fragments 6, 7, 8, and 9 of FUS3, and fragments 1 and 3 of CDS2. Additionally, fragments 4, 3, 8, and 3 of LEC1, LEC2, FUS3, and CDS2, respectively, showed the highest enrichment folds (Fig. 9, B and C). These data strongly support the hypothesis that these four genes are directly targeted by TT8.
Figure 9.
ChIP enrichment test showing the binding of TT8-6HA to the LEC1, LEC2, FUS3, and CDS2 promoters. A, Western-blotting results showing the TT8-6HA band. I, Input, E, eluate. B, The promoter structures of LEC1, LEC2, FUS3, and CDS2. The upstream region and the first intron of the genes are represented by white boxes, while the first exon is represented by the black box. The arrowheads at the top indicate the sites containing putative E-boxes on the promoters of these four genes above. Hatched boxes represent the DNA fragments amplified in ChIP assays. C, ChIP assay results in rosette leaves and siliques. EF1aA4 was used as an internal control. Significant differences in comparison with the enrichment of a TUBULIN BETA CHAIN2 (TUB2) fragment are indicated with asterisks. Error bars denote se.
DISCUSSION
The factors and the regulatory network that control FA biosynthesis in plant seeds are still largely unknown and require further investigation. In this study, we discover the novel function of TT8 in regulating seed FA accumulation in Arabidopsis. We demonstrate that the tt8 mutation drastically increases seed FA content (about 64%; Tables I and II; Fig. 1C), in particular the seed embryo FA content (Fig. 1D), and also changes the proportion of various FA species (Tables I and II). The significant increase of FA content in the tt8 seeds was accompanied by reductions of seed coat proportion (Fig. 1B) and protein deposition in embryos (Figs. 3B and 4) and by an elevation of starch synthesis at early seed developmental stages (Fig. 3A). The tt8 embryos are heavier than wild-type embryos, reflecting the differences in developmental effects from metabolic outcomes. TT8 acts maternally to affect seed FA accumulation (Fig. 6). Furthermore, it directly binds to the regulatory regions of LEC1, LEC2, FUS3, and CDS2 and down-regulates the expression of these genes (Figs. 7–9). On the other hand, it indirectly inhibits the expression of a number of genes, including CAC2, KASII, MOD1, FAB2, FatA, FAE1, FAD2, and FAD3, that catalyze various steps in the FA biosynthesis pathway (Fig. 6).
In Arabidopsis, TT8, together with TT2 and TTG1, synergistically regulates flavonoid biosynthesis (Nesi et al., 2000, 2001; Debeaujon et al., 2003; Lepiniec et al., 2006). The reduction of flavonoids in the seed coat caused yellowish tt8 seeds (Nesi et al., 2000; Supplemental Fig. S2), a reduction of seed coat weights, and heavier embryos based on similar seed weight (Fig. 1, A and B). Consequently, mature tt8-4 seeds yield higher FAs in seeds and embryos (Fig. 1, C and D). Nevertheless, there were no obvious differences in the sizes of embryo cells, embryos (Supplemental Fig. S3), and seeds (Supplemental Fig. S1) between the wild type and tt8 mutants.
In this study, we observed that tt8 mutants have larger oil bodies (Fig. 2). However, it was difficult to determine a causal relationship between oil body size/number and the oil content. In one of our previous publications, we analyzed the possible functions of a group of lipases, named seed FA reducers, to diminish the size of oil bodies by hydrolyzing triacylglycerol (Chen et al., 2012a). We also noticed that large oil bodies sometimes are correlated with lower seed oil content. For example, the disruption of OLEOSIN by RNA interference results in unusually larger oil bodies that correlate with lower seed lipid content (Siloto et al., 2006). Moreover, some rapeseed (Brassica napus) cultivars with low oil content have oil bodies similar in size to those in cultivars with high oil content (Hu et al., 2009).
Seed starch accumulates transiently during EM in Arabidopsis (Baud et al., 2002). Some studies proposed that starch accumulation strengthens a seed as a principal sink before EM starts, and starch degradation serves the synthesis of storage compounds after EM ends (Norton and Harris, 1975; daSilva et al., 1997; Periappuram et al., 2000). By contrast, seed protein is deposited mainly during the seed maturation stage, reflected by the high expression of a class of genes encoding the seed storage proteins in embryos (Pang et al., 1988; Baud et al., 2002). The storage proteins of Arabidopsis seeds consist mainly of a 12S globulin (cruciferin) and a 2S albumin (arabin; Heath et al., 1986). It is believed that the patterns of starch and/or protein accumulation in developing seeds affect seed FA biosynthesis remarkably (Mu et al., 2008; Chen et al., 2012b). Our study demonstrates that the amount of seed starch accumulation at 6 DAP is negatively correlated with the amount of protein deposition at late stages (16 DAP or the mature phase; Figs. 3 and 4), suggesting that the tt8-4 mutation supplies more carbohydrates for the biosynthesis of FAs, which otherwise may be used for the production of proteins.
Seed mucilage production in Arabidopsis is part of a remarkable differentiation process during which the epidermal cells of the mature ovule grow, rearrange their cytoplasm, synthesize and secrete mucilage, and form a secondary cell wall (Western et al., 2000). Several mutants associating with abnormal seed coat development, including tt2 and gl2, demonstrated an elevated seed FA content but a reduced mucilage layer in comparison with wild-type plants (Shi et al., 2012). It is supposed that seed coat mucilage and seed FAs are the competing sinks for the limiting photosynthates imported into developing seeds (Shi et al., 2012). In our study, we did not observe a significant reduction in mucilage layer in tt8-4 seeds (Fig. 5), suggesting that the tt8-4 seeds yield higher FAs without any cost in mucilage biosynthesis.
The comparison of FAs in the F1 seeds generated from reciprocal crossings between wild-type and tt8-4 plants indicates a clear maternal effect of the TT8 function. Only the F1 embryos derived from a crossing in which tt8-4 was used as the maternal parent exhibited a higher FA yield (Fig. 6). There could be a close association between the yield of FA in embryonic tissues and the amount of flavonoids in the surrounding seed coat, because flavonoids are considered to be inhibitors of FA biosynthesis, repressing the expression of FabG and FabI, which encode important reductases involved in chain elongation during FA biosynthesis (Zhang and Rock, 2004). They could possibly permeate from a seed coat to an adhering embryo, inhibiting the accumulation of FAs in the embryonic tissues. Another interpretation could be an epigenetic modification such as gene imprinting, since it seems that the expression of the TT8 allele demonstrated a clear parent-of-origin effect (i.e. that the TT8 allele derived from the paternal parent was not expressed; Fig. 6A; Grossniklaus et al., 1998; Adams et al., 2000; Gehring et al., 2004; Jiang and Köhler, 2012).
Seed FA biosynthesis starts by the formation of malonyl-CoA from the precursor acetyl-CoA catalyzed by acetyl-CoA carboxylase (ACCase), which is the key switch that controls FA flux, acting as a sensor or a gating system to monitor the overall flux of FA biosynthesis (Mu et al., 2008). ACCase is composed of three subunits, CAC2, CAC3, and BCCP2. CAC2 was up-regulated in developing tt8-4 seeds at 10 and 12 DAP (Fig. 7). More active ACCase should lead to an elevation of FAs in the tt8 seeds. On the other hand, the genes catalyzing the rest of the critical steps in the FA synthetic pathway, such as KAS (which transfers acetyl-CoA and malonyl-ACP to 3-ketoacyl-ACP), MOD1 (which encodes an enoyl-ACP reductase), FAB2 (which is a stearoyl-ACP desaturase), FatA (which encodes acyl-ACP thioesterase; Moreno-Pérez et al., 2012), CDS2 (which catalyzes 1,2-diacylglycerol-3-phosphate into CDP-diacylglycerol), FAD2 (which serves for the synthesis of polyunsaturated lipids; Okuley et al., 1994), and FAD3 (which is essential for the formation of the FA 18:3; Shah et al., 1997), were also moderately up-regulated at various stages and drastically peaked at 10 DAP in the tt8 developing seeds (Fig. 7). These up-regulated genes would be responsible for the elevated seed FA accumulation and the changed proportions of various FA species in the tt8 seeds.
FA accumulation is determined at multiple levels, of which transcriptional regulation is essential for controlling the FA biosynthesis pathway (Ohlrogge and Jaworski, 1997; Millar et al., 2000). The key TFs controlling FA accumulation mainly include WRI1, LEC1, LEC2, FUS3, and ABI3, which have a large number of downstream target genes. In Arabidopsis, LEC1 shares extensive sequence similarity with the HAPLESS3 subunit of a CCAAT-binding TF, and the increased expression of the gene elevates many FA biosynthetic genes involved in condensation, chain elongation, and desaturation (Lee et al., 2003; Mu et al., 2008). LEC2 and FUS3 are B3 domain TFs that play essential roles in the regulation of seed maturation (Luerssen et al., 1998; Stone et al., 2001; To et al., 2006). The LEC2 protein induces many maturation traits, including auxin activity, and the expression of other seed development regulators such as FUS3 and ABI3, which function redundantly in many aspects, including seed storage protein accumulation, chlorophyll degradation, and abscisic acid responses (Luerssen et al., 1998; To et al., 2006; Baud et al., 2007). These TFs function in unique and overlapping ways in regulating embryonic development in Arabidopsis (Wang et al., 2007; Santos-Mendoza et al., 2008; Baud and Lepiniec, 2009). In this study, the tt8 mutation did not cause significant changes in the expression levels of WRI1 and ABI3 (Supplemental Fig. S4) but did cause significant changes in the expression levels of LEC1, LEC2, and FUS3, in particular, a remarkable increase of FUS3 expression at the late stage of seed development (Fig. 7), which was consistent with the TT8 expression pattern (Nesi et al., 2000; Xu et al., 2013).
By posttranslational activation of TT8-GR, we further demonstrated the repression of LEC1, LEC2, FUS3, and CDS2 by induced TT8 activity (Fig. 8). Moreover, down-regulation of these four genes by DEX treatment of TT8-GR seedlings was not affected by CYC, indicating that the repression of these four genes by TT8 is independent of protein synthesis (Fig. 8). ChIP assays using specific HA antibodies further revealed in vivo TT8 binding to the cis-regulatory regions of these genes (Fig. 9), thus suggesting that TT8 acts as a direct regulator repressing LEC1, LEC2, and FUS3, which have a large number of downstream targets during seed development, and CDS2, which mediates glycerolipid biosynthesis.
Taken together, we show that the tt8 mutation remarkably increased the FA contents of seeds, altered the FA composition, and lowered seed protein accumulation during seed maturation. TT8 acted maternally to affect seed FA accumulation, and it repressed the expression of LEC1, LEC2, FUS3, and CDS2 by binding to the cis-regulatory regions of these genes and indirectly down-regulated a number of other genes, such as CAC2 (At5g35360), KASII (At1g74960), MOD1 (At2g05990), FAB2 (At2g43710), FatA (At3g25110), CDS2 (At4g22340), FAE1 (At4g34520), FAD2 (At3g12120), and FAD3 (At2g29980), which encode enzymes catalyzing various steps of FA biosynthesis. For decades, yellow-seeded Brassica species oilseeds have been interesting to breeders. This work enriches our knowledge of why yellow seeds accumulate more FAs in Arabidopsis, a model plant belonging to the family Cruciferae that includes major Brassica species oilseeds.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All the transgenic Arabidopsis (Arabidopsis thaliana) plants are of the Col-0 background. tt8-3 and tt8-4 are of the Ws and Col-0 backgrounds, respectively. The primers used for the tt8-4 genotyping are listed in Supplemental Table S1. The Arabidopsis plants were grown with a light cycle of 16 h of light and 8 h of dark. The light tubes were turned off at 11 pm and turned on at 7 am, and the tissue samples were collected around 1 pm. Other growth conditions are given in our previous publications (Chen et al., 2012a, 2012b).
Dissection of Seeds
For the separation of seed coat from embryo, we soaked the seeds in tap water at 4°C for at least 1 h and then carefully detached the embryos from seed coats using a pair of forceps. The seed coats actually came off with endosperm layers. The detached embryos were further observed using a microscope to determine their sizes and the cell shapes/cell numbers of the epidermal layer. The seed coats were dried at room temperature and then weighed.
Determination of FAs, Seed Starch, and Protein Content
The extraction and analysis of FAs in the seed and seed embryo were carried out as described before (Poirier et al., 1999; Chen et al., 2012b). The quantification of the seed starch and protein was determined as described previously by Chen et al. (2012b).
Microscopy Analysis
To observe the seed coat color, mature seeds of the wild type and tt8 mutants were randomly selected and photographed using an Olympus SZ 61 stereomicroscope. The seeds stained with Ruthenium Red were inspected with a compound light microscope (Nikon SMZ1500) and photographed. The embryo size and embryo cell size were measured as follows (Ohto et al., 2005). Twenty mature dried seeds of each line were imbibed for 1 h and dissected using the microscope to isolate mature embryos. Embryos were incubated in a buffer (50 mm sodium phosphate, pH 7, 10 mm EDTA, 1% [v/v] Triton X-100, and 1% [v/v] dimethyl sulfoxide) at 37°C overnight, fixed with 10% (v/v) formalin, 5% (v/v) acetic acid, 45% (v/v) ethanol, and 0.01% (v/v) Triton X-100 for 45 min, and then rehydrated using a graded series of ethanol (50%, 70%, 80%, 90%, 95%, and 100% [v/v]) for about 5 min each. Embryos were then treated for 1 h in Hoyer’s solution (chloral hydrate:water:glycerol, 3:0.8:0.4). The cleared embryo and embryo cell sizes were observed and measured using an Olympus compound microscope (Nikon SMZ1500) with differential interference contrast optics. Transmission electron microscopy observation was performed as described previously (Chen et al., 2012a, 2012b).
Protein Analysis by SDS-PAGE
We used the method to analyze different kinds of seed total protein described by Ohto et al. (2005). In brief, 15 mature dried seeds from wild-type plants or tt8 mutants were homogenized with 30 µL of extraction buffer (100 mm Tris-HC1, pH 8, 0.5% [v/v] SDS, 10% [v/v] glycerol, and 20% [v/v] 2-mercaptoethanol) using a microglass pestle and mortar, followed by boiling for 5 min and centrifugation for 10 min at highest speed at 4°C (Naito et al., 1988). Fifteen microliters of each extract was used for SDS-PAGE.
Ruthenium Red Staining for Seed Coat Mucilage
Dry seeds from each line were randomly selected and shaken in water for 1 h and then stained in a 0.01% (w/v) Ruthenium Red solution.
RT-PCR and RT-qPCR
Flowers on primary shoots were tagged with different colored threads to indicate the days after pollination, and the seeds from the tagged siliques were harvested for RNA extraction. Young leaves at the rosette stage were utilized for RNA extraction to identify the transgenic plants. The total RNA of different tissues was isolated using the RNeasy Plant Mini Kit (Qiagen) following the manufacturer’s instructions. First-strand complementary DNA (cDNA) was synthesized in a 10-µL reaction solution containing about 1.5 µg of total RNA reverse transcribed by the ThermoScript RT-PCR system (Invitrogen). Diluted aliquots of the reverse-transcribed cDNAs were used as templates in RT-qPCR containing the SYBR Green PCR Master Mix (Applied Biosystems). RT-qPCR assays were performed in triplicate on the 7900HT Fast Real-Time PCR system (Applied Biosystems). All the primers for the semiquantitative RT-PCR and RT-qPCR analysis are listed in Supplemental Table S2.
Generation of Transgenic Plants
To construct 35S:TT8-6HA, the TT8 cDNA was amplified with the primers TT8_P1_XhoI (5′-ATTCTCGAGATGGATGAATCAAGTATTATTCC-3′) and TT8-6HA_P2_SpeI (5′-GGACTAGTTAGATTAGTATCATGTATTAT-3′). The PCR products were digested by XhoI and SpeI and then cloned into pGreen-35S-6HA to obtain an in-frame fusion of TT8-6HA under the control of the 35S promoter (Liu et al., 2008). Similarly, the TT8 cDNA was also amplified with the primers TT8_P1_XhoI and TT8-GR_P2_ClaI (5′-CCATCGATTAGATTAGTATCATGTATTAT-3′) and then cloned into pGreen-35S-GR to obtain an in-frame fusion of TT8-GR under the control of the 35S promoter (Yu et al., 2004). These constructs were transformed into Agrobacterium tumefaciens strain GV3101, which was then used for the transformation of Arabidopsis wild-type (Col-0) plants via floral dip (Clough and Bent, 1998). The transgenic plants were selected by basta until the T3 homozygous transgenic plants were generated, and then RT-qPCR and western blotting were applied to further verify the transgenic plants. The confirmed 35S:TT8-6HA and 35S:TT8-GR transgenic plants were independently crossed with tt8-4 to check whether the yellow seed coat of tt8-4 can be rescued in the two transgenic plants.
GR Induction
For the induction of TT8:GR, seedlings at 15 d after sowing were collected 1, 4, 7, and 10 h after a single mock, DEX, or DEX plus CYC treatment. The concentrations of DEX and CYC were 10 and 5 μm, respectively.
ChIP Assay
The ChIP assays were performed according to Xi et al. (2010). For each ChIP assay, three independent experiments were performed using rosette leaves or siliques collected separately. DNA enrichment was examined by real-time qPCR in triplicate as described previously (Liu et al., 2008). The enrichment of a TUB2 genomic fragment was used as a negative control. Primer pairs used for the ChIP enrichment test are listed in Supplemental Table S3.
Statistical Analysis
A completely randomized block design with at least three biological replicates for each experiment was applied. The relative amounts of RNAs were calculated using the method of Livak and Schmittgen (2001). Data were classified with Win-Excel and analyzed via ANOVA using the SPSS (version 8.0) statistical package. Comparisons between the treatment means were made using Tukey’s test at P ≤ 0.05.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Identification of tt8 mutant lines.
Supplemental Figure S2. Microscopic observation of tt8 mature seeds.
Supplemental Figure S3. The embryo size and cell size are similar to those of the wild type.
Supplemental Figure S4. Comparison of the transcription levels of WRI1, ABI3, CAC3, and BCCP2 genes at different developmental stages between wild-type and tt8-4 seeds.
Supplemental Figure S5. Confirmation of the functional transgenic plants of 35S:TT8-GR and 35S:TT8-6HA.
Supplemental Table S1. Primers used for genotyping of the tt8-4 mutants.
Supplemental Table S2. Primers used for RT-PCR and RT-qPCR analyses.
Supplemental Table S3. Primers used for the ChIP assay.
Supplementary Material
Acknowledgments
We thank Mei Li for technical assistance.
Glossary
- FA
fatty acid
- EM
embryonic morphogenesis
- DAP
days after pollination
- RT
reverse transcription
- Col-0
Columbia-0
- Ws
Wassilewskija
- TF
transcription factor
- qPCR
quantitative PCR
- DEX
dexamethasone
- CYC
cycloheximide
- ChIP
chromatin immunoprecipitation
- cDNA
complementary DNA
Footnotes
This work was supported by the Natural Science Foundation of China (grant nos. 31171463 and 31371542), the Chinese Ministry of Education (grant no. 20130101110077), and the Zhejiang Provincial Bureau for Science and Technology (grant no. 2013C32004).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. (1997) Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams S, Vinkenoog R, Spielman M, Dickinson HG, Scott RJ. (2000) Parent-of-origin effects on seed development in Arabidopsis thaliana require DNA methylation. Development 127: 2493–2502 [DOI] [PubMed] [Google Scholar]
- Agati G, Azzarello E, Pollastri S, Tattini M. (2012) Flavonoids as antioxidants in plants: location and functional significance. Plant Sci 196: 67–76 [DOI] [PubMed] [Google Scholar]
- Agati G, Tattini M. (2010) Multiple functional roles of flavonoids in photoprotection. New Phytol 186: 786–793 [DOI] [PubMed] [Google Scholar]
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C. (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40: 151–160 [Google Scholar]
- Baud S, Lepiniec L. (2009) Regulation of de novo fatty acid synthesis in maturing oilseeds of Arabidopsis. Plant Physiol Biochem 47: 448–455 [DOI] [PubMed] [Google Scholar]
- Baud S, Mendoza MS, To A, Harscoët E, Lepiniec L, Dubreucq B. (2007) WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J 50: 825–838 [DOI] [PubMed] [Google Scholar]
- Baudry A, Caboche M, Lepiniec L. (2006) TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J 46: 768–779 [DOI] [PubMed] [Google Scholar]
- Chen MX, Du X, Zhu Y, Wang Z, Hua SJ, Li ZL, Guo WL, Zhang GP, Peng JR, Jiang LX. (2012a) Seed Fatty Acid Reducer acts downstream of gibberellin signalling pathway to lower seed fatty acid storage in Arabidopsis. Plant Cell Environ 35: 2155–2169 [DOI] [PubMed] [Google Scholar]
- Chen MX, Wang Z, Zhu YN, Li ZL, Hussain N, Xuan LJ, Guo WL, Zhang GP, Jiang LX. (2012b) The effect of TRANSPARENT TESTA2 on seed fatty acid biosynthesis and tolerance to environmental stresses during young seedling establishment in Arabidopsis. Plant Physiol 160: 1023–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- daSilva PMFR, Eastmond PJ, Hill LM, Smith AM, Rawsthorne S. (1997) Starch metabolism in developing embryos of oilseed rape. Planta 203: 480–487 [Google Scholar]
- Debeaujon I, Léon-Kloosterziel KM, Koornneef M. (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122: 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, Caboche M, Lepiniec L. (2003) Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell 15: 2514–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fait A, Angelovici R, Less H, Ohad I, Urbanczyk-Wochniak E, Fernie AR, Galili G. (2006) Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol 142: 839–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Choi Y, Fischer RL. (2004) Imprinting and seed development. Plant Cell (Suppl) 16: S203–S213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB, de Paiva G, Yadegari R. (1994) Plant embryogenesis: zygote to seed. Science 266: 605–614 [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB. (1998) Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280: 446–450 [DOI] [PubMed] [Google Scholar]
- Heath JD, Weldon R, Monnot C, Meinke DW. (1986) Analysis of storage proteins in normal and aborted seeds from embryo-lethal mutants of Arabidopsis thaliana. Planta 169: 304–312 [DOI] [PubMed] [Google Scholar]
- Hu Z, Wang X, Zhan G, Liu G, Hua W, Wang H. (2009) Unusually large oilbodies are highly correlated with lower oil content in Brassica napus. Plant Cell Rep 28: 541–549 [DOI] [PubMed] [Google Scholar]
- Jenik PD, Gillmor CS, Lukowitz W. (2007) Embryonic patterning in Arabidopsis thaliana. Annu Rev Cell Dev Biol 23: 207–236 [DOI] [PubMed] [Google Scholar]
- Jiang H, Köhler C. (2012) Evolution, function, and regulation of genomic imprinting in plant seed development. J Exp Bot 63: 4713–4722 [DOI] [PubMed] [Google Scholar]
- Lee HS, Fischer RL, Goldberg RB, Harada JJ. (2003) Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc Natl Acad Sci USA 100: 2152–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M. (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57: 405–430 [DOI] [PubMed] [Google Scholar]
- Li YH, Beisson F, Pollard M, Ohlrogge J. (2006) Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Liu C, Chen H, Er HL, Soo HM, Kumar PP, Han JH, Liou YC, Yu H. (2008) Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135: 1481–1491 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta CT) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Luerssen H, Kirik V, Herrmann P, Miséra S. (1998) FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J 15: 755–764 [DOI] [PubMed] [Google Scholar]
- Macquet A, Ralet MC, Kronenberger J, Marion-Poll A, North HM. (2007a) In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiol 48: 984–999 [DOI] [PubMed] [Google Scholar]
- Macquet A, Ralet MC, Loudet O, Kronenberger J, Mouille G, Marion-Poll A, North HM. (2007b) A naturally occurring mutation in an Arabidopsis accession affects a β-d-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. Plant Cell 19: 3990–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari ME, Murre C. (2000) Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 20: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. (1996) Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell 8: 1505–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U, Jürgens G. (1998) Pattern formation in plant embryogenesis: a reassessment. Semin Cell Dev Biol 9: 187–193 [DOI] [PubMed] [Google Scholar]
- Millar AA, Smith MA, Kunst L. (2000) All fatty acids are not equal: discrimination in plant membrane lipids. Trends Plant Sci 5: 95–101 [DOI] [PubMed] [Google Scholar]
- Mohamedyasseen Y, Barringer SA, Splittstoesser WE, Costanza S. (1994) The role of seed coats in seed viability. Bot Rev 60: 426–439 [Google Scholar]
- Moreno-Pérez AJ, Venegas-Calerón M, Vaistij FE, Salas JJ, Larson TR, Garcés R, Graham IA, Martínez-Force E. (2012) Reduced expression of FatA thioesterases in Arabidopsis affects the oil content and fatty acid composition of the seeds. Planta 235: 629–639 [DOI] [PubMed] [Google Scholar]
- Mu J, Tan H, Zheng Q, Fu F, Liang Y, Zhang J, Yang X, Wang T, Chong K, Wang XJ, et al. (2008) LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol 148: 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito S, Dubé PH, Beachy RN. (1988) Differential expression of conglycinin α′ and β subunit genes in transgenic plants. Plant Mol Biol 11: 109–123 [DOI] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L. (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12: 1863–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13: 2099–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton G, Harris JF. (1975) Compositional changes in developing rape seed (Brassica napus L.). Planta 123: 163–174 [DOI] [PubMed] [Google Scholar]
- Ohlrogge JB, Jaworski JG. (1997) Regulation of fatty acid synthesis. Annu Rev Plant Physiol Plant Mol Biol 48: 109–136 [DOI] [PubMed] [Google Scholar]
- Ohto MA, Fischer RL, Goldberg RB, Nakamura K, Harada JJ. (2005) Control of seed mass by APETALA2. Proc Natl Acad Sci USA 102: 3123–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J. (1994) Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6: 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang PP, Pruitt RE, Meyerowitz EM. (1988) Molecular cloning, genomic organization, expression and evolution of 12S seed storage protein genes of Arabidopsis thaliana. Plant Mol Biol 11: 805–820 [DOI] [PubMed] [Google Scholar]
- Penfield S, Rylott EL, Gilday AD, Graham S, Larson TR, Graham IA. (2004) Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1. Plant Cell 16: 2705–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periappuram C, Steinhauer L, Barton DL, Taylor DC, Chatson B, Zou JT. (2000) The plastidic phosphoglucomutase from Arabidopsis: a reversible enzyme reaction with an important role in metabolic control. Plant Physiol 122: 1193–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Ventre G, Caldelari D. (1999) Increased flow of fatty acids toward β-oxidation in developing seeds of Arabidopsis deficient in diacylglycerol acyltransferase activity or synthesizing medium-chain-length fatty acids. Plant Physiol 121: 1359–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerie WG, Feldmann KA, Marks MD. (1994) The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev 8: 1388–1399 [DOI] [PubMed] [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L. (2008) Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J 54: 608–620 [DOI] [PubMed] [Google Scholar]
- Shah S, Xin ZG, Browse J. (1997) Overexpression of the FAD3 desaturase gene in a mutant of Arabidopsis. Plant Physiol 114: 1533–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Katavic V, Yu YY, Kunst L, Haughn G. (2012) Arabidopsis glabra2 mutant seeds deficient in mucilage biosynthesis produce more oil. Plant J 69: 37–46 [DOI] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Siloto RMP, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM. (2006) The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Braybrook SA, Paula SL, Kwong LW, Meuser J, Pelletier J, Hsieh TF, Fischer RL, Goldberg RB, Harada JJ. (2008) Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. Proc Natl Acad Sci USA 105: 3151–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ. (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F. (2006) A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18: 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Guo JH, Lambert KN, Lin Y. (2007) Developmental control of Arabidopsis seed oil biosynthesis. Planta 226: 773–783 [DOI] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Wobus U. (1996) Controlling seed development and seed size in Vicia faba: a role for seed coat-associated invertases and carbohydrate state. Plant J 10: 823–834 [Google Scholar]
- Western TL, Skinner DJ, Haughn GW. (2000) Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol 122: 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi WY, Liu C, Hou XL, Yu H. (2010) MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 22: 1733–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu WJ, Grain D, Le Gourrierec J, Harscoët E, Berger A, Jauvion V, Scagnelli A, Berger N, Bidzinski P, Kelemen Z, et al. (2013) Regulation of flavonoid biosynthesis involves an unexpected complex transcriptional regulation of TT8 expression, in Arabidopsis. New Phytol 198: 59–70 [DOI] [PubMed] [Google Scholar]
- Yu H, Ito T, Wellmer F, Meyerowitz EM. (2004) Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat Genet 36: 157–161 [DOI] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao MZ, Payne CT, Lloyd A. (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zhang YM, Rock CO. (2004) Evaluation of epigallocatechin gallate and related plant polyphenols as inhibitors of the FabG and FabI reductases of bacterial type II fatty-acid synthase. J Biol Chem 279: 30994–31001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.