Abstract
Differentiating radiation necrosis (a radiation induced treatment effect) from recurrent brain tumors (rBT) is currently one of the most clinically challenging problems in care and management of brain tumor (BT) patients. Both radiation necrosis (RN), and rBT exhibit similar morphological appearance on standard MRI making non-invasive diagnosis extremely challenging for clinicians, with surgical intervention being the only course for obtaining definitive “ground truth”. Recent studies have reported that the underlying biological pathways defining RN and rBT are fundamentally different. This strongly suggests that there might be phenotypic differences and hence cues on multi-parametric MRI, that can distinguish between the two pathologies. One challenge is that these differences, if they exist, might be too subtle to distinguish by the human observer. In this work, we explore the utility of computer extracted texture descriptors on multi-parametric MRI (MP-MRI) to provide alternate representations of MRI that may be capable of accentuating subtle micro-architectural differences between RN and rBT for primary and metastatic (MET) BT patients. We further explore the utility of texture descriptors in identifying the MRI protocol (from amongst T1-w, T2-w and FLAIR) that best distinguishes RN and rBT across two independent cohorts of primary and MET patients. A set of 119 texture descriptors (co-occurrence matrix homogeneity, neighboring gray-level dependence matrix, multi-scale Gaussian derivatives, Law features, and histogram of gradient orientations (HoG)) for modeling different macro and micro-scale morphologic changes within the treated lesion area for each MRI protocol were extracted. Principal component analysis based variable importance projection (PCA-VIP), a feature selection method previously developed in our group, was employed to identify the importance of every texture descriptor in distinguishing RN and rBT on MP-MRI. PCA-VIP employs regression analysis to provide an importance score to each feature based on their ability to distinguish the two classes (RN/rBT). The top performing features identified via PCA-VIP were employed within a random-forest classifier to differentiate RN from rBT across two cohorts of 20 primary and 22 MET patients. Our results revealed that, (a) HoG features at different orientations were the most important image features for both cohorts, suggesting inherent orientation differences between RN, and rBT, (b) inverse difference moment (capturing local intensity homogeneity), and Laws features (capturing local edges and gradients) were identified as important for both cohorts, and (c) Gd-C T1-w MRI was identified, across the two cohorts, as the best MRI protocol in distinguishing RN/rBT.
Keywords: Radiation necrosis, recurrent disease, primary brain tumors, metastatic brain tumors, texture analysis, gradient orientations, treatment evaluation, MRI
1. INTRODUCTION
One of the most significant unsolved problems in treatment and management of brain tumors is differentiating radiation necrosis (RN),1 a radiation induced effect, from brain tumor recurrence (rBT)2 following radiation therapy. The current standard treatment regimen for patients with brain tumors (BT) comprises of surgical resection followed by adjunctive radiation and chemotherapy. Such aggressive treatment, although has shown to significantly improve median survival in BT patients, has also resulted in up to a threefold increase in radiation induced effects such as RN1 for both primary as well as metastatic (MET) brain tumor patients. RN, an irreversible radiation effect, that manifests after 6-9 months post-chemo-radiation treatment is characterized by life-threatening complications such as edema, severe neuropsychological symptoms, and usually mimics signs of rBT on MRI. Consequently, as many as 20 to 40% of all patients are subjected to multiple radiological studies, brain lesion biopsies, and even resections for what is ultimately deemed RN. Unfortunately, both these manifestations (RN and rBT) have significantly different treatment regimens and could be potentially fatal if not identified in time. The ability to reliably distinguish RN from rBT early could have immediate clinical implications in determining prognosis, guiding subsequent therapy, and improving patient outcome.1 There is hence a significant need to identify non-invasive imaging based markers that can reliably distinguish patients with rBT from RN to identify appropriate treatment regimens and thus avoid unnecessary and potentially harmful surgical interventions.
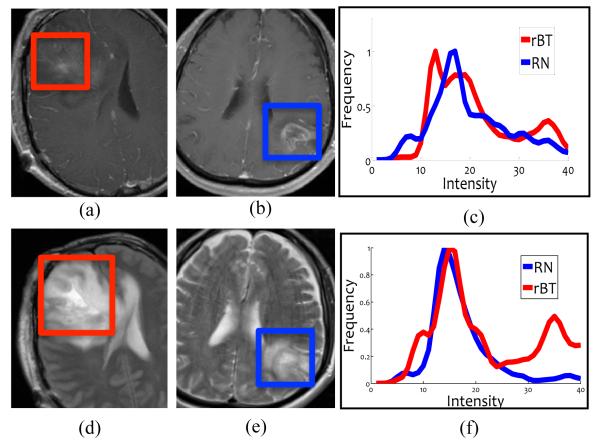
Due to its high resolution and clear definition of tumor margins, structural MRI is routinely used in the clinical setting to follow and monitor patients with brain tumors; however, it suffers from limitations in differentiating RN from rBT.3 Figures 1 (a) and (b) show a T1-w MRI image for rBT and RN respectively and the corresponding intensity histogram plots for RN (blue) and rBT (red) in 1 (c). Figures 1 (d) and (e) similarly show the two pathologies for T2-w and the corresponding intensity histogram plots in 1 (f). The similarity and overlap of intensities across RN and rBT for both T1-w as well as T2-w MRI is apparent, demonstrating the poor separability of RN and rBT using original MR intensities. Recently, a few studies have identified visual (qualitative) descriptors as “swiss cheese”, “soap bubble enhancement”, and “moving wave-front effect” based on their appearance on MRI as corresponding to RN.4 However these investigations have used image characteristics that are subjectively assessed and qualitatively defined and, therefore, potentially have inter-observer variability. Moreover, qualitatively defined features may not be able to capture the subtle localized differences across different pathologies with similar overall appearance.5
Figure 1.
(a) and (b) show representative T1-w MRI images while (d) and (e) show representative T2 images for RN and rBT respectively. Histograms of T1-w (c) and T2-w MRI (f) intensities within the lesion area for RN (blue) and rBT (red). Note the similarity of RN and rBT on (a) and (b), (d) and (e) and the overlap in histograms for T1-w and T2-w MRI.
Interestingly, recent studies have reported that the physiological pathways leading to the development of RT and rBT are fundamentally different.1,6 This strongly suggests that there might be phenotypic differences and hence cues on multi-parametric MRI, that can distinguish between the two pathologies.7 Over the last two decades, texture descriptors have shown substantial utility in quantifying morphological information for computer aided analysis for a myriad of diseases and tumor types.8–10 However, to our knowledge, the utility of texture descriptors, in the context of differentiating RN and rBT, has not yet been explored in detail. In this work, we explore the applicability of texture descriptors for evaluating subtle morphological differences across RN, rBT towards addressing the following questions:
Are there specific texture descriptors that can distinguish RN from rBT for primary brain tumor patients?
Are there specific texture descriptors that could differentiate RN and rBT for MET brain tumor patients? Is there a commonality between the texture features identified as important for primary and MET patients?
Could we quantify relative importance of standard-of-care MRI protocols (Gd-C T1-w, T2, FLAIR) in distinguishing RN and rBT for primary and MET patients?
The remainder of the paper is organized as follows. Section 2 discusses the previous work and novel contributions. In Section 3, we provide methodological details of this work. Experimental results are presented in Section 4. We provide concluding remarks in Section 5.
2. PREVIOUS WORK AND NOVEL CONTRIBUTIONS
The existing research on identifying differences between RN and rBT on MRI has primarily focused on reporting qualitative clues regarding the location, shape, and surrounding pathologies of the two types. The qualitative features that have been reported in the literature as distinctive of RN and rBT include, (1) origin near the primary tumor site, (2) contrast-agent enhancement, (3) vasogenic edema, (4) growth over time, and (5) mass effect.1,11 Features such as conversion from a non-enhancing to an enhancing lesion after radiation therapy, lesions appearing distant from the primary resection site, corpuscallosum or peri-ventricular white matter involvement, “Swiss cheese” and “soap bubble” shape patterns have been suggested to characterize RN.1 However, some studies have reported contradictory results regarding the validity of these qualitative findings in distinguishing RN and rBT on MRI.5
Apart from the visual descriptors, a few small cohort studies have displayed promise in utilizing semi-quantitative measures obtained via magnetic resonance spectroscopy (MRS), perfusion-and diffusion-weighted MR imaging, and Positron emission tomography (PET) to successfully distinguish the two morphologies. For e.g., Taylor et al.12 found that MRS reliably identified 5 of 7 patients with rBT and 4 of 5 patients with RN. Others however have found that MRS reliably distinguished pure rBT from pure RN but not where mixed specimens were involved.13 Tsuyuguchi et al.14 found that methionine PET had a sensitivity of 78% and a specificity of 100% for detecting rBT. However, Belohlvek et al.15 with a different agent, fluorodeoxyglucose, found PET to be insensitive but specific in distinguishing RN and rBT. Unfortuantely, none of these investigations have yet been identified to be clearly superior to the other modalities in terms of diagnostic sensitivity or specificity.16 Additionally, to our knowledge, none of these existing methods have explored the utility of computerized quantitative descriptors to obtain alternate representations of the lesions for differentiating RN and rBT on MRI.
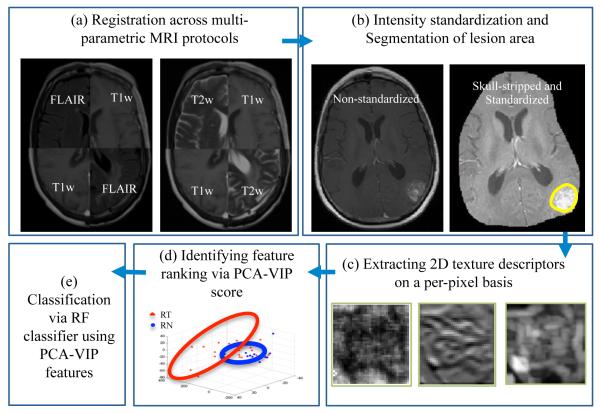
Figure 2 illustrates an overview of our framework. In Step 1, different MRI protocols are aligned in the same frame of reference, Gd-T1-w MRI in our case. In Step 2, skull stripping is first performed to remove background intensities that might affect feature extraction and classification across MRI protocols. Intensities across different MRI protocols are then aligned via intensity standardization to enable quantitative evaluation of MR parameters across patient studies, while ensuring tissue specific meaning to the parameters being compared. After intensity standardization, manual segmentation by a hand held annotation tool is performed by an expert radiologist to localize the lesion area. In Step 3, texture descriptors capturing information regarding orientations, heterogeneity, edge, spots, ripples, wave effects for RN and rBT (Table 1) on a per-pixel are extracted for every MRI protocol. Principal component analysis based variable importance projection (PCA-VIP),17 a feature ranking method previously developed in our group, is then employed to rank different texture descriptors in the order of their performance in distinguishing RN and rBT. A random forest (RF) classifier is employed as a final step to train a classifier using the best performing PCA-VIP texture descriptors to evaluate their performance in distinguishing RN and rBT.
Figure 2.
Overview of the methodology and overall workflow. In Step 1, registration of different MRI protocols (T2-w, FLAIR) is performed to bring them the same frame of reference (T1-w MRI). In Step 2, pre-processing via intensity standardization and segmentation of lesion area is performed, while in Step 3, different 2D texture features are extracted on a per-pixel basis. PCA-VIP is performed in Step 4 on the texture descriptors to rank the features based on their ability to distinguish RN and rBT, and finally in Step 5, a random forest classifier is trained on the best performing features (identified via PCA-VIP) to distinguish RN and rBT for two independent cohorts of primary and MET patients.
Table 1.
Biological relevance and significance of the texture features explored in this work in distinguishing RN/rBT on multi-parametric MRI.
| Feature set | Significance | Biological relevance in distinguishing RN/rBT |
|---|---|---|
| Laws Energy (25) | Filter masks extract level, edges, waves, ripples, spot patterns |
Appearance of ROI (wavefront, soap bubble) |
| Haralick Texture (13) | Statistics of gray-level co- occurrence matrices such as angular second moment, con trast and difference entropy |
Structural Heterogeneity |
| Laplacian pyramids (24) | Multi-resolution filters capture edges at different levels |
Prominent edges between RN, rBT appearance |
| Gradient orientations (57) | Intensity orientation captures prominent direction of intensity change |
Cellular activity, local entropy |
3. METHODOLOGY
3.1 Notation
We denote as a 3D grid for Gd-contrast (Gd-C) T1-w MRI protocol. The remaining MRI protocols are registered to to obtain, , where fβ(c) is the associated intensity at every voxel location c on a 3D grid Cβ, β ∈ {T2, FLAIR}. Texture feature descriptors are denoted as , where ϕ denotes the feature operator, and β denotes the MRI protocol, β ∈ {T1, T2, FLAIR}. The PCA-VIP score corresponding to every feature is denotes as πϕ,β, while the combined PCA-VIP score for every MRI protocol is denoted as πβ.
3.2 Co-registration of different MP-MRI protocols
A 3D affine transformation with 12 degrees of freedom, encoding rotation, translation, shear, and scale, was employed via the 3D Slicer software 4.1. (http://www.slicer.org/) to accurately align every MRI protocol with reference to Gd-C T1-w MRI, which yielded a registered 3D volume, , for every β, β ∈ {T2, FLAIR}. During registration, the 3D volume is appropriately resampled and interpolated, in order to account for varying voxel sizes and resolutions between different MRI protocols. Note that all the different MP-MRI acquisitions are aligned to frame of reference to enable per-voxel quantitative comparisons across different protocols (Figure 2(a)).
3.3 Pre-processing of MRI protocols
Pre-processing involves skull stripping, bias field correction, and intensity standardization of MRI images across different studies. Skull stripping is performed via an open-source automated BrainSuite tool (http://brainsuite.org/). We then correct the MRI protocols for known acquisition based intensity artifacts; bias field inhomogeneity and intensity nonstandardness.
3.3.1 Bias field inhomogeneity correction
The bias-field artifact manifests as a smooth variation of signal intensity across the structural MRI, and has been shown to significantly affect computerized image analysis algorithms such as the automated classification of tissue regions.18 Bias field artifacts were corrected for by means of the popular N3 algorithm,18 which incrementally de-convolves smooth bias field estimates from acquired image data, resulting in a bias-field corrected image.
3.3.2 Intensity standardization
A second artifact termed intensity nonstandardness refers to the issue of MR image “intensity drift” across different imaging acquisitions; both between different patients as well as for the same patient at different imaging instances. Intensity nonstandardness results in MR image intensities lacking tissue-specific numeric meaning within the same MRI protocol, for the same body region, or for images of the same patient obtained on the same scanner.19 Correcting for this artifact hence enables quantitative evaluation of MR parameters across patient studies, while ensuring tissue specific meaning to the parameters being compared. Every MRI protocol, is quantitated by correcting for intensity drift between different patient studies.19 The ROI was then manually segmented on by an expert radiologist via a hand-annotation tool in 3D Slicer.
3.4 Texture feature extraction of MP-MRI
A total of 119 texture features were extracted from each of , β ∈ {T1, T2, FLAIR} on a per-voxel basis. These features are obtained by (1) calculating responses to various filter operators, and (2) computing gray level intensity co-occurrence statistics, as follows,
Haralick texture features: Haralick texture features10 are based on quantifying the spatial gray-level co-occurrence within local neighborhoods around each pixel in an image, stored in the form of matrices. A total of 13 Haralick texture descriptors were calculated from each of , β ∈ {T1, T2, FLAIR} based on statistics derived from the corresponding co-occurrence matrices.
Laws texture features: Laws features use 5 × 5 separable masks20 that are symmetric or anti-symmetric to extract level (L), edge (E), spot (S), wave (W), and ripple (R) patterns to detect various types of textures on an image. The convolution of these masks with every , β ∈ {T1, T2, FLAIR} resulted in a total of 25 distinct laws features for every MRI protocol.
Laplacian pyramids: Laplacian pyramids allows to capture multi-scale representations via a set of band pass filters.21,22 First, the original image is convolved with a Gaussian kernel. The Laplacian is then computed as the difference between the original image and the low pass filtered image. The resulting image is then sub-sampled by a factor of two, and the filter sub-sample operation is repeated recursively. This process is continued to obtain a set of band-pass filtered images (since each is the difference between two levels of the Gaussian pyramid). A total of 24 filtered image representations were obtained from each of , β ∈ {T1, T2, FLAIR}.
- Histogram of gradient (HoG) orientations: For every c ∈ C, gradients along the X and Y directions are computed as,23
Here, and are the gradient magnitudes along the X and the Y axes respectively denoted by f(c)X and f(c)Y . Once the gradient magnitudes along the two coordinate axes are calculated, the gradient orientation θ of every c ∈ C is calculated as(1)
After obtaining the gradient orientations of all the points of interest, they are binned into histograms that span 0 to 360°. The entire histogram is divided into twenty bins, each encompassing 18°. The feature vector consists of the binned histogram values in the form of 20×1 vectors.(2)
Feature extraction results in feature scenes , where fϕ,β (c) is the feature value at location c ∈ C when feature operator ϕ is applied to scene , β ∈ {T1, T2, FLAIR}, resulting in a total of 120 texture feature scenes (including original intensity) corresponding to each of , β ∈ {T1, T2, FLAIR}.
3.5 Feature ranking via PCA-VIP
Once feature scenes are identified for every β, PCA-VIP scheme is used to rank each of the feature sets based on their ability to distinguish RN and rBT. PCA-VIP quantifies the contributions of individual features to regression or classification on an embedding obtained via principal components analysis. PCA-VIP score for every feature scene, ϕϕ,β, is computed as follows:
| (3) |
where m is the number of features in the original, high-dimensional feature space; h is the number of retained features in the low-dimensional embedding space; the ti are the principal components; the pi are the loadings, estimated by (denotes pseudo-inverse); and the bi are the coefficients that solve the regression equation y = Tb˕, where y is a vector of class labels. The degree to which a feature contributes to classificationin the PCA transformed space is directly proportional to the square of its PCA-VIP score. Thus, features with PCA-VIP scores near 0 have little predictive power, and the features with the highest PCA-VIP scores contribute the most to class discrimination in the embedding space.
The importance of a feature subset is quantified by summing the squared PCA-VIP scores associated with each of the features in , as follows:
| (4) |
We obtain a combined πβ for every β by summing PCA-VIP values over ϕ for every , β ∈ {T1, T2, FLAIR}, to obtain the relative importance of every MRI protocol in distinguishing RN and rBT.
4. EXPERIMENTAL RESULTS
4.1 Dataset Description
A total of 20 primary (10 RN, and 10 rBT) and 22 MET (10 RN and 12 rBT) patient studies were retrospectively acquired between 9 months to 2 years post-chemo-radiation with 3 Tesla MP-MRI. Gd-C T1-w, T2-w, and FLAIR protocols were acquired as a part of the routine standard of care imaging at different time-points. Both cohorts of patients (primary and MET) were histologically confirmed on biopsy samples either with RN (>80% RN) and rBT (>80% tumor) by an expert pathologist. The MP-MRI acquisitions for the time-point immediately before the biopsy was identified and used in this work for identification of RN and rBT for the two cohorts.
4.2 Experiment 1: Ranking performance of texture descriptors in distinguishing RN, rBT for primary BT patients
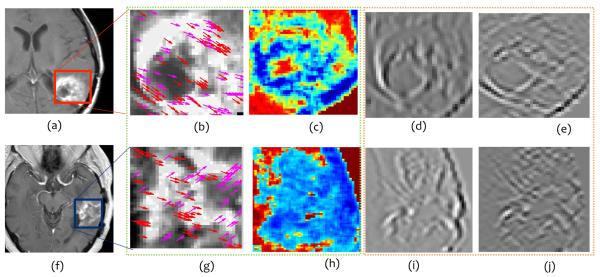
Figure 3 shows the top 2 best (Figures 3 (b), (c) for rBT and (g),(h) for RN) and worst (Figures 3 (d), (e) for rBT, and (i), (j) for RN) performing texture descriptors, outlined in green and orange respectively, on Gd-C T1-w MRI for primary BT patients. The top three texture descriptors for each of the MRI protocols (Gd-C T1-w, T2-w, and FLAIR) with corresponding PCA-VIP scores are listed in Table 2. It is interesting to note that HoG features capturing the dominant orientations in X and Y directions (at orientation range 72° – 107° for Gd-C T1-w, and 180° – 215° for T2-w MRI) were identified as the best performing texture descriptors for primary BT patients. This suggests that there are some fundamental orientation differences between RN and rBT that may not be appreciable by visual inspection of original MR intensities. Inverse difference moment which captures heterogeneity in the lesion, in laplacian space, was also consistently picked up as an important feature for all three MRI protocols. This feature in laplacian space is potentially emphasizing the edges thereby making heterogeneity between RN and rBT more prominent in the alternate representation space (Figures 3(c), and (h) respectively). The worst performing features were identified as Laws features (S5L5 (spots and level filter), and L5E5 (level and edge filter)) for Gd-C T1-w MRI suggesting the poor discriminability based on spot, waves, and ripple characteristics, that have previously been identified as qualitative descriptors (“soap bubble”, “swiss cheese” effect) to distinguish the two pathologies.
Figure 3.
Representative T1-w MR images for rBT (a) and RN (f). Figures 3 (b), (c) and (g), (h) outlined in green, represent top performing features (HoG (red, magenta arrows show top 2 prominent directions), and Laplacian inverse moment (red shows more heterogeneity)) for rBT, RN respectively. Figures 3 (d), (e) and (i), (j), outlined in orange, represent the worst performing features (S5L5 and L5E5 (laplacian) Laws features) for rBT and RN respectively for primary BT patients.
Table 2.
Top 3 texture descriptors and their PCA-VIP scores listed for T2-FLAIR, T2-w, and T1-w MRI protocols for primary BT patients. HoG and inverse difference moment were consistently identified as the best performing features in distinguishing RN and rBT for primary BT patients for the three protocols.
| MRI protocol | Texture descriptor | π ϕ,β |
|---|---|---|
| Gd-C T1-w MRI | HoG (72° – 107°) | 2.15 |
| Laplacian (Inverse difference moment) | 2.026 | |
| Laws (E5L5) | 1.621 | |
|
| ||
| T2-w MRI | Inverse difference moment | 2.21 |
| HoG (180° – 215°) | 1.58 | |
| Laplacian (Inverse difference moment) | 1.42 | |
|
| ||
| FLAIR MRI | Laplacian (Inverse difference moment) | 2.47 |
| Correlation | 1.84 | |
| Inverse difference moment | 1.83 | |
4.3 Experiment 2: Ranking performance of texture descriptors in distinguishing RN and rBT patients for metastasis BT patients
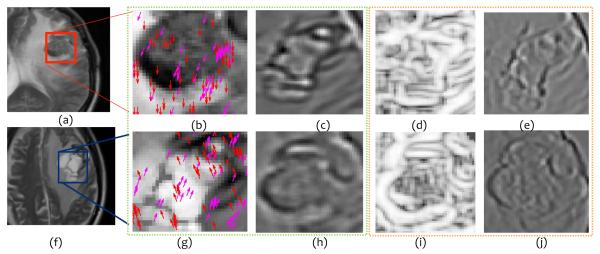
Figure 4 shows the top 2 best (Figures 4 (b), (c) for rBT and (g),(h) for RN) and worst (Figures 4 (d), (e) for rBT, and (i), (j) for RN) performing texture descriptors, outlined in green and orange respectively, on Gd-C T1-w MRI for MET BT patients. The top three texture descriptors for each of the MRI protocols (Gd-C T1-w, T2-w, and FLAIR) for MET patients with corresponding PCA-VIP are listed in Table 3. Similar to primary BT cases, HoG features were identified as the most discriminatory feature for each of the three protocols in distinguishing RN and rBT for MET patients. This reaffirms our hypothesis that there exists different orientation directions between RN and rBT for both primary as well as MET patients. Similarly, Laws features quantifying wave and level filter, and wave and ripple filter, were identified as the worst performing features for MET patients. Interestingly, however, Laws features quantifying edge and level filter, were identified as important in distinguishing RN and rBT for Gd-C T1-w as well as FLAIR MRI for MET patients, suggesting that edge and gradient characteristics are important in distinguishing the two pathologies.
Figure 4.
Representative T1-w MR images for rBT (a) and RN (f). Figures 4 (b) , (c) and (g), (h), outlined in green, represent the top performing features (HoG (red, magenta arrows show top 2 prominent directions), and Laws E5L5 (edge and level filter) ) for rBT and RN respectively. Figures 4 (d), (e) and (i), (j), outlined in orange, represent the worst performing (W5L5 (wave and level filter in laplacian pyramid space) and W5R5 (wave and ripple filter)) features for rBT and RN respectively for MET patients.
Table 3.
Top 3 texture descriptors and their PCA-scores listed for T1-w, T2-w, and FLAIR MRI protocols for MET patients. HoG features were consistently identified as best performing features across all 3 MRI protocols for distinguishing RN and rBT for MET patients.
| MRI protocol | Texture descriptor | π ϕ,β |
|---|---|---|
| Gd-C T1-w MRI | HoG (0-35) | 1.98 |
| Laws (E5L5) | 1.78 | |
| Gabor (Θ = 135, ʌ = 32) | 1.51 | |
|
| ||
| T2-w MRI | HoG (108-143) | 1.71 |
| Sum Entropy | 1.49 | |
| Sum Variance | 1.39 | |
|
| ||
| FLAIR MRI | HoG (72-107) | 1.91 |
| Laws (E5L5) | 1.88 | |
| Laws (L5E5) | 1.63 | |
4.4 Experiment 3: Identifying the MRI protocol that best separates RN from rBT
Table 4 demonstrates the combined PCA-VIP scores, πβ (obtained using equation 4), along with the mean area under curve (AUC) values, ββ, obtained for every β, β ∈ {T1, T2, FLAIR} for the two cohorts, primary and MET. The top 10 texture descriptors identified based on their PCA-VIP scores for the two cohorts were identified. A random forest classifier was then trained using these top features via a three-fold cross validation strategy, for each of the 3 protocols for each of the two cohorts independently. The ββ values were reported as the mean AUC over 25 iterations of 3-fold cross validation. The quantitative results based on πβ and ββ identified Gd-C T1-w MRI as the best performing feature set in distinguishing RN and rBT for both primary and MET cohorts. These findings resonate with the clinical findings as Gd-C T1-w MRI is the current standard-of-care MRI protocol that is routinely employed by clinicians to make a distinction for RN and rBT. T2-w MRI for primary and FLAIR for MET patients was identified as the second best protocol after Gd-C T1-w MRI.
Table 4.
PCA-VIP scores (πβ) and mean AUC values (μβ) for each of the three MRI protocols for both primary and MET patients. Gd-C T1-w MRI was identified as the best performing MRI protocol both in terms of πβ and μβ, from amongst T2-w MRI and FLAIR protocols.
| Cohort | Protocol | π β | μ β |
|---|---|---|---|
| Primary | Gd-C T1-w MRI | 2.89 | 0:74 ± 0:06 |
| T2-w MRI | 2.55 | 0:62 ± 0:2 | |
| FLAIR | 2.46 | 0:50 ± 0:2 | |
|
| |||
| MET | Gd-C T1-w MRI | 2.73 | 0:71 ± 0:2 |
| T2-w | MRI 2.70 | 0:48 ± 0:20 | |
| FLAIR | 2.69 | 0:55 ± 0:24 | |
5. CONCLUDING REMARKS
Accurately distinguishing radiation necrosis, a radiation induced effect, from recurrent brain tumor is a challenging clinical problem due to the apparent similarities in symptoms and appearance of the two pathologies on traditional MRI. In this work, we investigated the utility of computer extracted texture descriptors on multi-parametric MRI to reliably distinguishing RN and rBT for two independent cohorts of primary and metastatic brain tumor patients. The first objective was to employ texture descriptors for distinguishing RN from rBT for primary brain tumor patients. The texture descriptors were utilized, in the second objective, to distinguish RN and rBT for metastatic patients. The third objective was to quantify the relative importance of standard-of-care MRI protocols (Gd-C T1-w, T2-w, FLAIR) in distinguishing RN and rBT for both primary and metastatic patients. Computerized texture descriptors provided quantitative assessment to distinguish RN from rBT that appeared accurate in capturing subtle architectural details that may not be appreciable on original MR intensities. Our preliminary results based on the two cohorts of primary and metastatic brain tumor patients indicate that,
HoG features at different orientations were identified as the most important features for both primary and metastatic cohorts. This suggests some inherent direction orientation differences between RN, rBT.
Inverse difference moment (capturing local intensity homogeneity), and Laws features (capturing local edges and gradients) were identified as important in distinguishing RN and rBT for primary brain tumor patients. Laws features (local edges and gradients) was identified as important features in distinguishing RN and rBT for MET patients.
Gd-C T1-w MRI was identified as the best feature based on PCA-VIP scores and AUC values, followed by T2-w and FLAIR for primary BT cases. Gd-C T1-w was similarly identified as important in distinguishing RN and rBT for metastatic cases, followed by FLAIR and T2-w MRI. These findings are consistent with clinical findings as Gd-C T1-w MRI is the modality of choice for visually distinguishing RN and rBT.
Previous work has reported sensitivity and specificity values in the range of 70-95% using advanced imaging modalities such as MRS, PET, and perfusion-MRI.12,14,15 However, these advanced modalities may not be reproducible24 or widely available across diagnostic centers in the US. Additionally, the reported findings have largely been qualitatively investigated over a small cohort of studies.
The presented work is the first approach at employing computerized texture descriptors to investigate histologically-proven RN and rBT over two independent cohorts of 20 primary and 22 MET patients. Although the results are promising, a limitation of our study was the ground truth. In the absence of pure RN and rBT cases, the studies with > 80% of RN or rBT presence identified by a neuropathologist were considered as ground truth. Future work will include obtaining pure RN and rBT studies to understand the fundamental biological significance of the texture descriptors (specially orientation differences) that have shown promise in distinguishing the two pathologies, and their role in predicting patient outcome. We will also investigate the differences in texture descriptors across primary and metastatic brain tumors to understand the manifestation of RN over the two cohorts.
ACKNOWLEDGMENTS
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award numbers R01CA136535-01, R01CA140772-01, and R21CA167811-01; the National Institute of Diabetes and Digestive and Kidney Diseases under award number R01DK098503-02, the DOD Prostate Cancer Synergistic Idea Development Award (PC120857); the QED award from the University City Science Center and Rutgers University, the Ohio Third Frontier Technology development Grant, and the Coulter foundation award (RES508406). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- [1].Siu A, Wind J, Iorgulescu J, Chan T, Yamada Y, Sherman J. Radiation necrosis following treatment of high grade gliomaa review of the literature and current understanding. Acta Neurochir. 2012;154:191–201. doi: 10.1007/s00701-011-1228-6. [DOI] [PubMed] [Google Scholar]
- [2].Nordon A, Drappatz J, Wen P. Novel anti-angiogenic therapies for malignant gliomas. Lancet Neurol. 2008;7(12):1152–1160. doi: 10.1016/S1474-4422(08)70260-6. [DOI] [PubMed] [Google Scholar]
- [3].Macdonald D, et al. Response criteria for phase ii studies of malignant glioma. J Clin Oncol. 1990;8:1277–80. doi: 10.1200/JCO.1990.8.7.1277. [DOI] [PubMed] [Google Scholar]
- [4].Kumarand A, Leeds N, GN GF. Malignant gliomas: Mr imaging spectrum of radiation therapy- and chemotherapy-induced necrosis of the brain after treatment. Radiology. 2000;217(2):377384. doi: 10.1148/radiology.217.2.r00nv36377. [DOI] [PubMed] [Google Scholar]
- [5].Mullins M, Barest G, et al. P. S. Radiation necrosis versusglioma recurrence: conventional mr imaging clues to diagnosis. AJNR Am J Neuroradiol. 2005;26(8):19671972. [PMC free article] [PubMed] [Google Scholar]
- [6].Tofilon P, Fike J. The radioresponse of the central nervous system: A dynamic process. Radiation research. 2000;153:357–370. doi: 10.1667/0033-7587(2000)153[0357:trotcn]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- [7].Verma N, Cowperthwaite M, Burnett M, Markey M. Differentiating tumor recurrence from treatment necrosis: a review of neuro-oncologic imaging strategies. Neuro Oncology. 2013;5(5):515–534. doi: 10.1093/neuonc/nos307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Moura D, Lpez MG. An evaluation of image descriptors combined with clinical data for breast cancer diagnosis. IJCARS. 2013;8(4):561–74. doi: 10.1007/s11548-013-0838-2. [DOI] [PubMed] [Google Scholar]
- [9].Nanni L, Brahnam S, Ghidoni S, Menegatti E, Barrier T. Different approaches for extracting information from the co-occurrence matrix. PLOS ONE. 2013;8(12):e83554. doi: 10.1371/journal.pone.0083554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Viswanath S, Bloch N, Chappelow J, Toth R, Rofsky N, Genega E, Lenkinski R, Madabhushi A. Central gland and peripheral zone prostate tumors have significantly different quantitative imaging signatures on 3 tesla endorectal, t2-w mr imagery. JMRI. 2012;36(1):213–24. doi: 10.1002/jmri.23618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chan Y, Leung S, et al. A. K. Late radiation injury to the temporal lobes: morphologic evaluation at mr imaging. Radiology. 1999;213(3):800807. doi: 10.1148/radiology.213.3.r99dc07800. [DOI] [PubMed] [Google Scholar]
- [12].Taylor J, Langston J, Reddick W, Kingsley P, Ogg R, Pui M, Kun L, 3rd, J. J, Chen G, Ochs J, Sanford R, Heideman R. Clinical value of proton magnetic resonance spectroscopy for differentiating recurrent or residual brain tumor from delayed cerebral necrosis. Int J Radiat Oncol Biol Phys. 1996;36:1251–1261. doi: 10.1016/s0360-3016(96)00376-8. [DOI] [PubMed] [Google Scholar]
- [13].Dowling C, Bollen A, Noworolski S, McDermott M, Barbaro N, Day M, Henry R, Chang S, Dillon W, Nelson S, Vigneron D. Preoperative proton mr spectroscopic imaging of brain tumors: Correlation with histopathologic analysis of resection specimens. AJNR Am J Neuroradiol. 2001;22:604612. [PMC free article] [PubMed] [Google Scholar]
- [14].Tsuyuguchi N, Sunada I, Iwai Y, Yamanaka K, Tanaka K, Takami T, Otsuka Y, Sakamoto S, Ohata K, Goto T, M MH. Methionine positron emission tomography of recurrent metastatic brain tumor and radiation necrosis after stereotactic radiosurgery: Is a differential diagnosis possible? J Neurosurgery. 2003;98:10561064. doi: 10.3171/jns.2003.98.5.1056. [DOI] [PubMed] [Google Scholar]
- [15].Belohlvek O, Simonov G, Kantorov I, Jr, J. N, Lisck R. Brain metastases after stereotactic radiosurgery using the leksell gamma knife: Can fdg pet help to differentiate radionecrosis from tumour progression? Eur J Nucl Med Mol Imaging. 2003;30:96100. doi: 10.1007/s00259-002-1011-2. [DOI] [PubMed] [Google Scholar]
- [16].Gilbert M. Cerebral radiation necrosis. Neurologist. 2003;9:180188. doi: 10.1097/01.nrl.0000080951.78533.c4. [DOI] [PubMed] [Google Scholar]
- [17].Ginsburg S, Tiwari P, Kurhanewicz J, Madabhushi A. Variable ranking with pca: Finding multiparametric mr imaging markers for prostate cancer diagnosis and grading. Prostate Cancer Imaging. Image Analysis and Image-Guided Interventions Lecture Notes in Computer Science. 2011;6963:146–157. [Google Scholar]
- [18].Sled J, Zijdenbos A, Evans A. A nonparametric method for automatic correction of intensity nonuniformity in mri data. IEEE Transactions on Medical Imaging. 1998;17(1):8797. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- [19].Madabhushi A, Udupa J. New methods of mri intensity standardization via generalized scale. Med. Phy. 2006;33(9):3426–34. doi: 10.1118/1.2335487. [DOI] [PubMed] [Google Scholar]
- [20].Laws K. Ph.D. Dissertation. University of Southern California; 1980. Textured image segmentation. [Google Scholar]
- [21].Burt P, Adelson E. The laplacian pyramid as a compact image code,” Communications. IEEE Transactions on 31. 1983 Apr;:532–540. [Google Scholar]
- [22].Prasanna P, Jain S, Bhagat N, Madabhushi A. Decision support system for detection of diabetic retinopathy using smartphones. [Pervasive Computing Technologies for Healthcare (PervasiveHealth), 2013 7th International Conference on].May, 2013. pp. 176–179. [Google Scholar]
- [23].Dalal N, Triggs B. Histograms of oriented gradients for human detection. Computer Vision and Pattern Recognition, 2005. CVPR 2005; IEEE Computer Society Conference on], 1.Jun, 2005. pp. 886–893. [Google Scholar]
- [24].Marshall I, Wardlaw J, Cannon J, Slattery J, Sellar R. Reproducibility of metabolite peak areas in 1h mrs of brain. Magn Reson Imaging. 1996;14(3):281–92. doi: 10.1016/0730-725x(95)02084-7. [DOI] [PubMed] [Google Scholar]