Abstract
Extinctions can dramatically reshape biological communities. As a case in point, ancient mass extinction events apparently facilitated dramatic new evolutionary radiations of surviving lineages. However, scientists have yet to fully understand the consequences of more recent biological upheaval, such as the megafaunal extinctions that occurred globally over the past 50 kyr. New Zealand was the world's last large landmass to be colonized by humans, and its exceptional archaeological record documents a vast number of vertebrate extinctions in the immediate aftermath of Polynesian arrival approximately AD 1280. This recently colonized archipelago thus presents an outstanding opportunity to test for rapid biological responses to extinction. Here, we use ancient DNA (aDNA) analysis to show that extinction of an endemic sea lion lineage (Phocarctos spp.) apparently facilitated a subsequent northward range expansion of a previously subantarctic-limited lineage. This finding parallels a similar extinction–replacement event in penguins (Megadyptes spp.). In both cases, an endemic mainland clade was completely eliminated soon after human arrival, and then replaced by a genetically divergent clade from the remote subantarctic region, all within the space of a few centuries. These data suggest that ecological and demographic processes can play a role in constraining lineage distributions, even for highly dispersive species, and highlight the potential for dynamic biological responses to extinction.
Keywords: aDNA, Phocarctos hookeri, New Zealand, priority effects, extinction
1. Introduction
Biologists have long been intrigued by the evolutionary importance of historical contingencies [1,2]. For example, the notion that the extinction of one lineage can directly benefit another remains a central paradigm in evolution and ecology [3,4]. In particular, palaeontologists have inferred that mass extinction events in the geological record can have profound downstream consequences for surviving taxa [5,6]. Despite the extensive attention devoted to the impacts of ancient extinctions [7], our understanding of the biological consequences of more recent upheaval (e.g. worldwide megafaunal extinctions over the past 50 thousand years (kyr) [8]) remains limited.
Human settlement of New Zealand approximately AD 1280 [9] led to the sudden demise of a large component of these islands’ highly endemic and distinctive avifauna, with 40 indigenous bird species going extinct within 200 years of human arrival [10,11]. On the other hand, large dispersive coastal vertebrates such as penguins and pinnipeds have been assumed to have survived New Zealand's human impacts [12], although recent data suggest this may not always have been the case [11]. Specifically, ancient DNA (aDNA) analysis of Megadyptes penguins indicates that an endemic pre-historic mainland species (Megadyptes waitaha) was eliminated soon after human arrival, and subsequently replaced by the subantarctic Megadyptes waitaha antipodes [11]. While such an unexpected finding could readily be dismissed as an intriguing anomaly, we propose that dynamic extinction–recolonization patterns may be a more general phenomenon in impacted ecosystems [13–15]. This finding seems congruent with previously described extinctions that appear to have released dispersive taxa from the biogeographic constraints previously imposed by the presence of sister lineages [8,16]. Here, we test for parallel extinction–recolonization scenarios by conducting ancient and modern DNA analyses of pre-human Holocene fossil, archaeological and extant New Zealand sea lion (Phocarctos) samples, and comparing these temporal genetic data with published penguin datasets [11].
2. Material and methods
(a). Modern DNA extraction and sequencing
Fifty-six modern Phocarctos hookeri skin samples were collected during the 2003–2012 breeding season from pups born at seven breeding colonies throughout the species' modern breeding range (electronic supplementary material, table S1 and figure S1). DNA was extracted from 2 mm2 skin samples following a modified Chelex protocol with 5% and 4 µg proteinase K [17] followed by ethanol precipitation. An approximately 1.2 kbp (excluding primers) fragment of the mitochondrial DNA (mtDNA) control region (CR)/D-loop was amplified using the primers L-NZSL d-loop-tRNA pro (5′ CTCAAGGAAGAGGCAAGAGC 3′) and H-NZSL d-loop tRNA phe (5′ GAAGGGCTAGGACCAAACCT 3′) [18]. PCRs (25 µl) contained 0.4 M of each primer, 1.5 mM MgCl2, 200 µM dNTPs, 0.5 Taq (BIOTAQ, Bioline), 1 × PCR buffer and 2 µl 1 : 20 diluted DNA. PCR thermocyling conditions were 94°C 4 min, 10 cycles of 94°C 20 s, 65°C (decreasing 1°C per cycle), 72° C 90 s, followed by 20 cycles of 94°C 20 s, 55°C 25 s, 72°C 90 s, and a final extension at 72°C 5 min. PCR products were purified using AcroPrep clean up plates (Pall Corporation) following the manufacturer's instructions and sequenced using the primer L-NZSL d-loop tRNA pre using Big Dye terminator technology on an ABI 3730xl (sequencing with H-NZSL d-loop-tRNA phe was not practical owing to repetitive stretches of sequence within CR).
(b). Ancient DNA extraction and sequencing
A total of 97 pre-human Holocene fossil and early archaeological (AD 1280–1450 based on associated archaeological remains) P. hookeri specimens from throughout coastal New Zealand (electronic supplementary material, figure S1) and the Auckland Islands were obtained from museum and university collections, and in situ Holocene fossil deposits [19] (electronic supplementary material, table S2). P. hookeri bones were identified morphologically following Worthy [18,20]. To ensure independence of individual bones, only common elements of the left or right orientation were sampled from an individual deposit, or bones were sampled from different stratigraphic units within the site. All aDNA extractions and PCR set-up were carried out at the University of Otago in a purpose-built aDNA laboratory physically isolated from any other molecular laboratories [21] following strict aDNA procedures to minimize contamination of samples with exogenous DNA [22], including the use of negative controls.
DNA was extracted from 70 to 280 mg of bone (or tooth) powder, following Rohland et al. [23]. Two overlapping fragments (118 and 120 bp, respectively, excluding primers, 189 bp in total) of the mtDNA CR were amplified using primer pairs ‘NZSLCR1f’/‘NZSLCR1r’ and ‘NZSLCR2f’/‘NZSLCR2’ [19]. PCRs (20 µl reaction) contained 0.25 mM of each primer, 1 M of Betaine, 1 × PCR buffer, 4 mM MgCl2, 0.625 mM dNTPs, 2U of Amplitaq Gold (Life Technologies) and 2 µl DNA. PCR thermocycling conditions were 95°C 9 min, 60 cycles of 95°C 20 s, 54°C 30 s, 72°C 30 s, and a final extension at 72°C 4 min. The 189 bp region sequenced contained all 11 variable sites of the approximately 1.2 kbp P. hookeri mtDNA CR. Of these 11 variable sites, nine are parsimony informative. Each PCR was replicated twice. PCR amplification and all downstream procedures were carried out in a modern genetics laboratory. PCR products were purified using ExoSAP (1.5 U ExoI, 1U SAP; GE Healthcare) by incubation at 37°C for 30 min and 80°C for 15 min, then sequenced bidirectionally from independent PCR products using Big Dye terminator technology on an ABI 3730xl.
(c). Ancient DNA authenticity
Each DNA extract was independently amplified at least twice, and the resulting amplicons sequenced bidirectionally from independent PCR products to confirm sequence authenticity [24]. The sea-lion-specific primers did not amplify DNA from any of the extraction and PCR negative controls. If amplification and sequencing could not be replicated, then corresponding sequences were excluded from downstream phylogenetic analysis. In the few instances of inconsistent DNA sequences from an individual, which were consistent with what could be expected for DNA damage (G–A and C–T transitions), the DNA extract was amplified and sequenced bidirectionally at least twice more, and a majority rule consensus was applied to the independent replicates [25]. After sequence authentication, the majority of G–A and C–T transitions were shared by multiple individuals and were geographically concordant within the context of regional phylogeographic structure [26] (i.e. consistent differentiation between subantarctic and mainland specimens) and thus clearly do not represent DNA damage. Additionally, close genetic similarity between archaeological subantarctic and modern subantarctic samples strongly suggests that DNA damage is not a significant factor in our analysis. Only five (M2, M4, M6, M8 and M11; figure 2 and the electronic supplementary material, table S2 and figure S2) sequences had private G–A or C–T changes that were not shared with at least one other DNA sequence, and these are retained in analyses as they show strong phylogenetic similarity with nearby samples. All sequences are deposited in GenBank (accession numbers KJ588766–KJ588778; KJ648152–KJ64854).
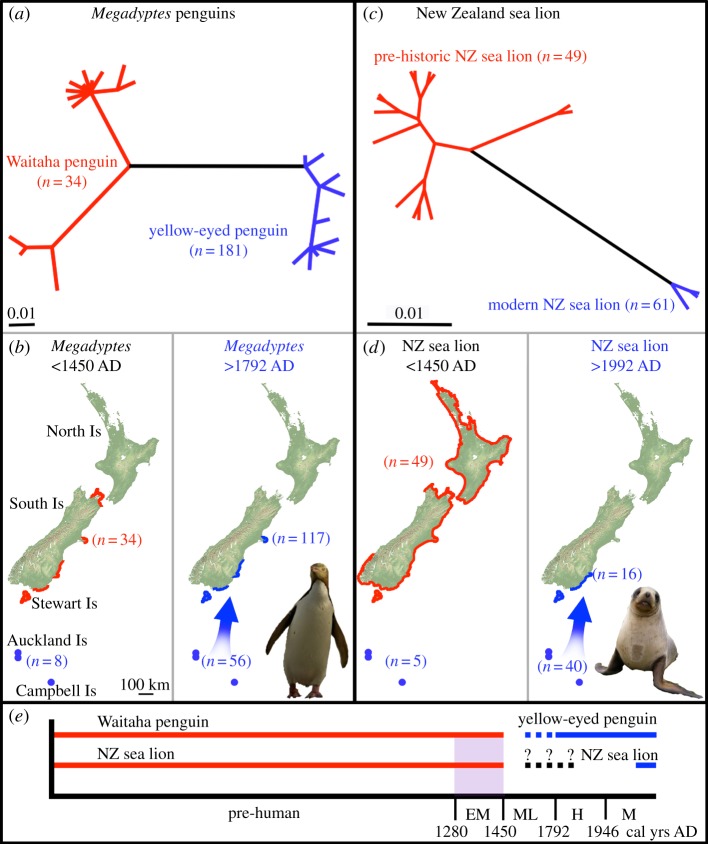
Figure 2.
Parallel extinction and re-colonization of Megadyptes penguins and Phocarctos sea lions following human colonization of New Zealand. (a) Bayesian phylogram of Megadyptes [11] penguin mtDNA control region (CR). M. waitaha sequences in red, M. antipodes in blue. (b) Distribution of Megadyptes (M. waitaha: red; M. antipodes: blue) prior to 1450 AD and after 1792 AD [11]. (c) Bayesian phylogram of Phocarctos mtDNA CR. Pre-historic mainland Phocarctos sequences in red, subantarctic and modern Phocarctos in blue. (d) Distribution of Phocarctos (pre-historic New Zealand [19]; red: subantarctic and modern: blue) prior to 1450 AD and after 1992 AD. (e) Extinction and recolonization timeframes for Megadyptes and Phocarctos. Early Maori (EM) represents the period from human colonization approximately 1280 AD to the extinction of avian megafauna approximately 1450 AD (purple shaded bar) [45]; Middle–Late Maori (ML), from 1450 AD until the time of European settlement of New Zealand by sealers in 1792; Historical (H), from 1792 until the end of commercial sealing in 1946; and modern (M) from 1946 onwards.
(d). Phylogenetic analysis
Contiguous sequences were constructed using Geneious v. 6.1.2 [27]. The analysis was restricted to the 189 bp region sequenced for all specimens. Inclusion of partially sequenced specimens, where only one of the two fragments amplified in aDNA extracts, did not change the result (electronic supplementary material, figure S3). The most appropriate model of evolution, as determined by MODELTEST [28] under the Akaike information criterion, was GTR + I + G. Maximum-likelihood (ML) analysis was performed in PAUP* [29] using the full heuristic search option. Parameters for the GTR + I + G model were then re-estimated from the data, and node support was calculated with 10 000 bootstrap replicates. Bayesian trees were estimated with BEAST v. 1.7.4 [30] in two independent runs, using 20 000 000 generations, sampling every 1000th generation and discarding 25% as burn-in. Convergence diagnostics of Bayesian analyses were explored using Tracer [31] and FigTree v. 1.4.0 [32]. The topologies of the ML and Bayesian trees were very similar. A network of ancient and modern haplotypes was constructed using TCS [33] using the 95% parsimony criterion. AMOVAs were performed in Arlequin v. 3.5.1.2 [34] to test for temporal and geographical structure within the mainland lineage.
(e). Radiocarbon dating
Radiocarbon dates on pre-human Holocene fossil P. hookeri remains were sourced from Collins et al. [19] and Worthy [18]. A further two pre-human Holocene fossil P. hookeri bones were radiocarbon-dated (electronic supplementary material, table S3) at Beta Analytic Inc. (USA). Dates are reported as radiocarbon ages, based on Libby T1/2 = 5568 years, uncorrected for calendar variation, in years before present (present is AD 1950). Radiocarbon ages were calibrated using OxCal v. 4.2 [35] and the Marine09 calibration curve [36]. Local ΔR values [37] were applied to the calibrations of radiocarbon dates (electronic supplementary material, table S3). The large geographical ranges of many pinnipeds in their adult stage can make it difficult to select appropriate ΔR values for such taxa [38,39]. To account for this issue, we also applied minimum and maximum New Zealand ΔR values [37] to the adult bones radiocarbon-dated. Calibrated ages are reported as 95.4% confidence calibrated ages in years BC/AD.
(f). Testing demographic scenarios
Approximate Bayesian computation (ABC; [40]) was used to quantitatively test different demographic scenarios against each other. ABC is a model-based analysis method, which calculates the posterior distributions of model parameters using information from prior distributions and extensive simulations rather than calculating the likelihood from the data directly. Bayesian serial Simcoal [41] was used to simulate the temporal DNA data under four different models: (i) a closed single population with constant size, (ii) a closed single population incorporating a bottleneck, (iii) two closed populations followed by re-colonization and (iv) a two closed populations model, in which the subantarctic population remains a constant population size, whereas the mainland NZ population experiences a bottleneck with subsequent recovery (population growth). The BayeSSC input files (showing the prior ranges) can be found in the electronic supplementary material. The number of segregating sites, pairwise differences (between the subantarctic and the mainland NZ populations, and between time layers, pre-human/archaeological and modern), and private haplotypes were selected to summarize the data, which resulted in a total of 30 summary statistics. Nonlinear regression-based machine learning [42] was applied, implemented in the freely available ‘abc’ R package [43], with log transformation in the nonlinear regression step (except for the growth rate in model 4). Tolerance levels of 0.001, 0.002 and 0.004 were used for the parameter estimations. The expected deviance (deviance information criterion, DIC; [44]) was used to infer the best-supported model, implemented in the R package ‘abc’.
3. Results
(a). Phylogenetic analysis
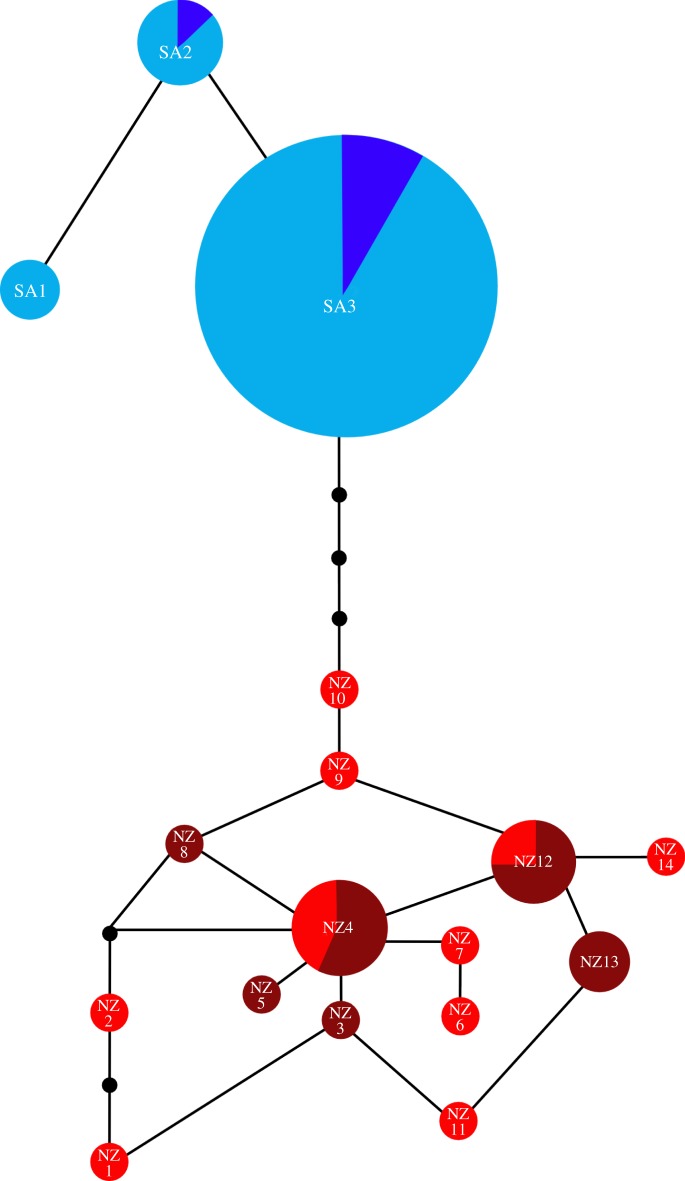
DNA was successfully amplified and sequenced from 56 modern and 54 aDNA P. hookeri specimens (figure 1 and the electronic supplementary material, figure S2). Fourteen of the 54 aDNA extracts amplified and sequenced for only one primer pair, resulting in 40 full aDNA sequences included in genetic analyses (electronic supplementary material, table S2). The inclusion of partial sequences did not significantly alter the phylogenetic results (electronic supplementary material, figure S3); therefore, only full sequences were used in downstream analyses. Phylogenetic analyses of modern and aDNA sequence data reveal a clear genetic distinction between modern versus ancient mainland New Zealand Phocarctos (figure 2 and the electronic supplementary material, figures S2 and S3). Fourteen haplotypes were identified from aDNA mainland New Zealand samples, and two from pre-historic Auckland Islands samples (figure 2 and the electronic supplementary material, table S2). There is compelling evidence for genetic turnover in mainland New Zealand: all pre-historic mainland samples cluster together in a distinct clade not detected in modern mainland and subantarctic samples. Specifically, all pre-human and archaeological samples from mainland New Zealand belong to a now-extinct evolutionary lineage that is highly distinct (mtDNA CR divergence 2.1–4.6%; table 1) from the newly colonized ‘subantarctic’ lineage that replaced it (figure 1). This distinction is comparable to the genetic distance between the now extinct M. waitaha and M. antipodes following a similar extinction and replacement scenario (mtDNA CR divergence 2.2–4.2%; [11]). By contrast, there is no evidence of genetic turnover in the subantarctic, with two of the three modern Auckland Islands haplotypes also detected in pre-historic subantarctic (Enderby Island) samples (figure 2 and the electronic supplementary material, figure S2). These data confirm that the subantarctic lineage was present on Enderby Island at the time of Polynesian occupancy approximately 650 yr BP [45]. Results from AMOVA indicate a lack of significant temporal and spatial genetic differentiation among pre-historic mainland samples, but show major mtDNA differentiation between pre-historic mainland and subantarctic specimens (table 2). Additionally, genetic diversity observed within extant P. hookeri and Megadyptes is considerably lower than that detected within the now extinct ‘mainland’ lineages of these taxa (table 1 and figure 2). Moreover, the P. hookeri haplotype network (figure 2) does not show the star-like pattern expected to characterize a recent population expansion, nor does it show extensive 'gaps’ that might be expected following a population decline.
Figure 1.
Haplotype network of mainland (red) and subantarctic (blue) Phocarctos hookeri. Haplotype frequency is indicated by circle area, black dots represent hypothetical intermediate haplotypes. Pie charts within each haplotype represent the proportion of samples of Holocene, archaeological or modern age represented by that haplotype. Naturally deposited Holocene fossils from mainland New Zealand are indicated by dark red, archaeological mainland NZ specimens are light red, archaeological subantarctic specimens are dark blue and modern subantarctic specimens are light blue.
Table 1.
Within-lineage genetic variation in Megadyptes spp. and P. hookeri. Genetic data for Megadyptes spp. from [11], GenBank accession numbers FJ391944–FJ391968. n, sample size, h, haploype diversity, π, nucleotide diversity.
| taxa | lineage | na | h | π |
|---|---|---|---|---|
| P. hookeri | ‘mainland’ | 49 | 0.872 | 0.007 |
| ‘subantarctic’ | 61 | 0.339 | 0.0001 | |
| Megadyptes | M. waitaha | 74 | 0.834 | 0.009 |
| M. antipodes | 111 | 0.547 | 0.004 |
aSample size (n) based on aDNA only for ‘mainland’ P. hookeri and M. waitaha and modern and aDNA for ‘subantarctic’ P. hookeri and M. antipodes.
Table 2.
AMOVA results assessing hierarchical genetic variation between regional and temporal sample groupings.
| groups | variance | % variation | FST | P(FST) |
|---|---|---|---|---|
| north island versus south island | −0.01455 | −2.97 | −0.02970 | 0.54839 ± −0.01336 |
| north NZ versus south NZa | −0.00729 | −1.47 | −0.01471 | 0.43988 ± −0.01620 |
| archaeological versus naturalb | −0.02431 | −4.97 | −0.04975 | 0.99218 ± −0.00280 |
| mainland NZ versus subantarcticc | 0.11435 | 25.61 | 0.25605 | 0.00000 ± 0.00000 |
aNorth NZ versus South NZ sample groupings corresponding to northern and southern clade distributions within Megadyptes waitaha [11].
bBones sampled from archaeological (post-1280 AD) or natural (pre-1280 AD) deposits.
cSubantarctic samples include modern sequences.
The recent recolonization of P. hookeri ended a 150-year period during which no sea lions bred on mainland New Zealand [46,47]. Temporal turnover in mainland New Zealand is highlighted by the finding that the recently colonized ‘subantarctic’ lineage now occupies terrain that was inhabited by the extinct ‘mainland’ lineage until just a few centuries ago. Despite extensive sampling of pre-historic samples (49 specimens successfully sequenced from 18 mainland localities), there is no indication that the distinctive subantarctic lineage was present on New Zealand's mainland until after the extinction of the endemic mainland clade.
(b). Radiocarbon dating
In addition to eight previously published radiocarbon dates from mainland New Zealand P. hookeri bones [19], a further two P. hookeri bones from natural deposits in Northland (North Island) and Enderby Island (Auckland Islands) were radiocarbon dated in order to confirm the suspected pre-human age of these deposits, and the long-term presence of P. hookeri on mainland New Zealand prior to human arrival [19]. Along with the previously published radiocarbon dates [19], these new dates provide further evidence for the presence of P. hookeri on mainland New Zealand during the Holocene, prior to human arrival (electronic supplementary material, figure S4 and table S3).
(c). Testing demographic scenarios
Four different demographic scenarios were compared using an ABC framework. Model 3 (recolonization model) had the highest support values, using both rejection and nonlinear regression model comparisons (electronic supplementary material, table S4). Interestingly, support values were highest for model 4 using the nonlinear regression method and accepting 4000 estimates (electronic supplementary material, table S4). However, the rejection step accepting 4000 estimates still favours model 3 (electronic supplementary material, table S4), providing support for model 3 in five out of six model comparisons. Parameter estimations for model 3 are provided in the electronic supplementary material, table S5.
4. Discussion
Spatio-temporal genetic analyses of Megadyptes and Phocarctos reveal strikingly similar patterns of turnover. These extinctions of genetically distinctive, endemic mainland lineages coincide with the arrival of humans approximately 1280 AD. [9]. Additionally, the fact that the remains of both of these endemic coastal taxa are abundant in early Maori middens (ca 1280–1450 AD), but are absent from later archaeological sites (ca 1450–1792 AD), is consistent with rapid extinction owing to over-hunting (electronic supplementary material, figure S4), as also occurred for iconic mainland terrestrial megafauna such as moa [10,48]. While temporal shifts in haplotype/haplogroup frequencies could potentially be caused by genetic drift alone, the loss of New Zealand's distinctive mainland sea lion mtDNA lineage is clearly related to the complete elimination of this pre-historic endemic population (electronic supplementary material, table S4).
Numerous drivers have been proposed to explain megafaunal extinctions in different parts of the world [8]. For instance, Holocene shifts inferred for Antarctic vertebrate populations have been linked to climatic fluctuations [39]. In mainland New Zealand, however, the distribution of Phocarctos at the time of human arrival spanned a latitudinal range of some 13° (1500 km; electronic supplementary material, figure S1) [19], encompassing a broad thermal envelope, such that minor climatic fluctuations [49] occurring around the time of human arrival are unlikely to explain the near-simultaneous extinctions of these lineages over such a wide spatial and thermal range. Indeed, genetic divergence levels and fossil data [19] (electronic supplementary material, figure S4) imply that Phocarctos persisted in mainland New Zealand throughout the Late Glacial (ca 14–11.6 kya) and Holocene (11.6 kya–present). By contrast, the archaeological and genetic evidence for over-hunting and extinction of New Zealand's megafauna during the early period of Polynesian settlement (ca 1280–1450 AD) is compelling [48,50].
Regardless of underlying causes of extinction, the dramatic temporal turnover detected for Phocarctos and Megadyptes populations raises the intriguing possibility that subantarctic lineages of both taxa may have been formerly excluded from establishing in mainland New Zealand by the presence of their endemic mainland counterparts. While, in the case of Megadyptes, three pre-historic M. antipodes were detected in New Zealand's mainland archaeological record, and interpreted as non-breeding vagrants from the subantarctic [11], as yet no ‘subantarctic’ lineage Phocarctos have been detected in pre-historic mainland samples. Based on our present sampling of 49 pre-historic mainland individuals, however, we cannot categorically reject the possibility that the mainland had occasional, low frequency (e.g. less than 0.02) ‘subantarctic’ P. hookeri vagrants or breeders during pre-historic times. Further sampling of pre-historic material is required to test for such individuals. More broadly, these findings suggest that lineage extinction–replacement scenarios might be more common than is currently recognized [1–7]. For instance, dynamic biological responses to extinction have also been suggested for continental megafauna of the Northern Hemisphere [13]. In such cases, extinction–replacement scenarios are thought to reflect the retraction and/or expansion of lineages as a result of a changing climate and habitat preferences, over timeframes far exceeding those of the present study [51–53]. Additional examples of apparent lineage replacement come from New Zealand's terrestrial avifauna, which is now dominated by self-introduced, post-human arrivals following the elimination of their original endemic relatives (e.g. Porphyrio; Circus) [54]. Over ancient timeframes, New Zealand's palynological record provides strong evidence for biotic turnover (extinction–replacement) associated with the Oligocene drowning [55]. We predict that future studies will reveal additional cases of lineage extinction and replacement in response to human impacts, especially across recently colonized ecosystems (e.g. islands of the Pacific Ocean) [56,57]. Furthermore, additional cases of extinction and replacement are likely to be evident within New Zealand's coastal fauna. We hypothesize that the apparently recent establishment of an Australian blue penguin (Eudyptula minor) lineage, for example, may reflect similar extinction–replacement dynamics [58,59].
This study reinforces the hypothesis that processes such as competitive exclusion and high-density blocking [59] can potentially play important roles in mediating biological distributions and biodiversity patterns over a wide range of temporal and spatial scales. The apparent disconnect between dispersal ability per se and realized distributions may help to explain the paradox of spatial structuring of intrinsically dispersive biotas [60].
Acknowledgements
We thank Auckland Museum (Brian Gill), University of Auckland Department of Anthropology (Rod Wallace and Melinda Allen), Museum of New Zealand Te Papa Tongarewa (Alan Tennyson), Canterbury Museum, and Otago University Department of Anthropology and Archaeology (Phil Latham) for bone samples, and the Department of Conservation (Jim Fyfe) for modern P. hookeri samples. Thank you to Ken Miller for assistance in figure preparation. Funding was provided by a Royal Society of New Zealand Marsden Grant (UOO1112) and the Allan Wilson Centre for Molecular Ecology and Evolution. Modern P. hookeri samples were collected with funding from a Department of Conservation (DOC) Grant (DOC Inv 4219) with approval for sample collection from the DOC Animal Ethics Committee (Approvals AEC 200 2009, AEC 159, AEC 174, AEC 232).
References
- 1.Gould SJ. 1989. Wonderful Life. New York, NY: Norton. [Google Scholar]
- 2.Erwin DH. 2001. Lessons from the past: biotic recoveries from mass extinctions. Proc. Natl Acad. Sci. USA 98, 5399–5403. ( 10.1073/pnas.091092698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wyles JS, Kunkel JG, Wilson AC. 1983. Birds, behavior, and anatomical evolution. Proc. Natl Acad. Sci. USA 80, 4394–4397. ( 10.1073/pnas.80.14.4394) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sloan RE, Rigby JK, Van Valen LM, Gabriel D. 1986. Gradual dinosaur extinction and simultaneous ungulate radiation in the Hell Creek formation. Science 232, 629–633. ( 10.1126/science.232.4750.629) [DOI] [PubMed] [Google Scholar]
- 5.Hallam A, Wignall PB. 1997. Mass extinctions and their aftermath. Oxford, UK: Oxford University Press. [Google Scholar]
- 6.Meredith RW, et al. 2011. Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334, 521–524. ( 10.1126/science.1211028) [DOI] [PubMed] [Google Scholar]
- 7.Bininda-Emonds ORP, et al. 2007. The delayed rise of present-day mammals. Nature 446, 507–512. ( 10.1038/nature/05634) [DOI] [PubMed] [Google Scholar]
- 8.Lorenzen ED, et al. 2011. Species-specific responses of Late Quaternary megafauna to climate and humans. Nature 479, 359–364. ( 10.1038/nature10574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilmshurst JM, Anderson AJ, Higham TFG, Worthy TH. 2008. Dating the late prehistoric dispersal of polynesians to New Zealand using the commensal Pacific rat. Proc. Natl Acad. Sci. USA 105, 7676–7680. ( 10.1073/pnas.0801507105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tennyson AJD, Martinson P. 2007. Extinct birds of New Zealand, p. 180 Wellington, New Zealand: Te Papa Press. [Google Scholar]
- 11.Boessenkool S, Austin JJ, Worthy TH, Scofield P, Cooper A, Seddon PJ, Waters JM. 2009. Relict or colonizer? Extinction and range expansion of penguins in southern New Zealand. Proc. R. Soc. B 276, 815–821. ( 10.1098/rspb.2008.1246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith IWG. 2005. Retreat and resilience: fur seals and human settlement in New Zealand. In The exploitation and cultural importance of marine mammals (ed. Monks GG.). Oxford, UK: Oxbrow Books. [Google Scholar]
- 13.Barnes I, Matheus P, Shapiro B, Jensen D, Cooper A. 2002. Dynamics of Pleistocene population extinctions in Beringian brown bears. Science 295, 2267–2270. ( 10.1126/Science.1067814) [DOI] [PubMed] [Google Scholar]
- 14.Dalen L, Nystrom V, Valdiosera C, Germonpre M, Sablin M, Turner E, Angerbjorn A, Arsuaga JL, Gotherstrom A. 2007. Ancient DNA reveals lack of postglacial habitat tracking in the arctic fox. Proc. Natl Acad. Sci. USA 104, 6726–6729. ( 10.1073/pnas.0701341104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leonard JA, Vilà C, Fox-Dobbs K, Koch PL, Wayne RK, Van Valkenburgh B. 2007. Megafaunal extinctions and the disappearance of a specialized wolf ecomorph. Curr. Biol. 17, 1146–1150. ( 10.1016/j.cub.2007.05.072) [DOI] [PubMed] [Google Scholar]
- 16.Roca AL, Georgiadis N, Pecon-Slattery J, O'Brien SJ. 2001. Genetic evidence for two species of elephant in Africa. Science 293, 1473–1477. ( 10.1126/Science.1059936) [DOI] [PubMed] [Google Scholar]
- 17.Walsh SP, Metzger DA, Higuchi R. 1991. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10, 506–513. [PubMed] [Google Scholar]
- 18.Worthy TH. 1992. Fossil bones of Hooker's sea lions in New Zealand caves. N.Z. J. Nat. Sci. 19, 32–39. [Google Scholar]
- 19.Collins CJ, Rawlence NJ, Worthy TH, Scofield RP, Tennyson AJD, Smith IWG, Knapp M, Waters JM. 2013 Pre-human New Zealand sea lion (Phocarctos hookeri) rookeries on mainland New Zealand. J. Royal Soc. N.Z. 44, 1–16. ( 10.1080/03036758.2013.828761) [DOI] [Google Scholar]
- 20.Worthy TH. 1994. Holocene breeding of sea lions (Phocarctos hookeri) at Delaware Bay, Nelson, South Island, New Zealand. N.Z. J. Nat. Sci. 21, 61–70. [Google Scholar]
- 21.Knapp M, Clarke AC, Horsburgh KA, Matisoo-Smith EA. 2012. Setting the stage: building and working in an ancient DNA laboratory. Ann. Anat. 194, 3–6. ( 10.1016/j.aanat.2011.03.008) [DOI] [PubMed] [Google Scholar]
- 22.Cooper A, Poinar HN. 2000. Ancient DNA: Do it right or not at ALL. Science 289, 1139 ( 10.1126/science.289.5482.1139b) [DOI] [PubMed] [Google Scholar]
- 23.Rohland N, Siedel H, Hofreiter M. 2010. A rapid column-based ancient DNA extraction method for increased sample throughput. Mol. Ecol. Resour. 10, 677–683. ( 10.1111/J.1755-0998.2009.02824.X) [DOI] [PubMed] [Google Scholar]
- 24.Hofreiter M, Serre D, Poinar H, Kuch M, Paabo S. 2001. Ancient DNA. Nat. Rev. Genet. 2, 353–359. ( 10.1038/35072071) [DOI] [PubMed] [Google Scholar]
- 25.Brotherton P, Endicott P, Sanchez JJ, Beaumont M, Barnett R, Austin J, Cooper A. 2007. Novel high-resolution characterization of ancient DNA reveals C > U-type base modification events as the sole cause of post mortem miscoding lesions. Nucleic Acids Res. 35, 5717–5728. ( 10.1093/nar/gkm588) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood JR, Wilmshurst JM, Wagstaff SJ, Worthy TH, Rawlence NJ, Cooper A. 2012. High-resolution coproecology: using coprolites to reconstruct the habits and habitats of New Zealand's extinct upland moa (Megalapteryx didinus). PLoS ONE 7, e40025 ( 10.1371/journal.pone.0040025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drummond A, et al. 2011. Geneious version 6.1.2 created by Biomatters See http://www.geneious.com.
- 28.Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14, 817–818. ( 10.1093/Bioinformatics/14.9.817) [DOI] [PubMed] [Google Scholar]
- 29.Swofford DL. 2003. PAUP* phylogenetic analysis using parsimony (* and other methods) See http://www.paup.csit.fst.edu/.
- 30.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. ( 10.1093/Molbev/Mss075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rambaut A, Drummond A. 2007. Tracer v. 1.4 See http://www.beast.bio.ed.ac.uk/Tracer. [DOI] [PMC free article] [PubMed]
- 32.Rambaut A. 2009. FigTree. v. 1.4.0 ed See http://www.tree.bio.ed.ac.uk/software/figtree/.
- 33.Clement M, Posada D, Crandall KA. 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659. ( 10.1046/j.1365-294x.2000.01020.x) [DOI] [PubMed] [Google Scholar]
- 34.Excoffier L, Laval G, Schneider S. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. 1, 47–50. [PMC free article] [PubMed] [Google Scholar]
- 35.Bronk Ramsey C. 2009. Bayesian analysis of radiocarbon dates. Radiocarbon 51, 337–360. [Google Scholar]
- 36.Reimer PJ, et al. 2009. Intcal09 and marine09 radiocarbon age calibration curves, 0–50,000 years cal BP. Radiocarbon 51, 1111–1150. [Google Scholar]
- 37.Petchey F, Anderson A, Hogg A, Zondervan A. 2008. The marine reservoir effect in the Southern Ocean: an evaluation of extant and new ΔR values and their application to archaeological chronologies. J. Royal Soc. N.Z. 38, 104–114. ( 10.1080/03014220809510559) [DOI] [Google Scholar]
- 38.Yoneda M, Hirota M, Uchida M, Uzawa K, Tanaka A, Shibata Y, Morita M. 2001. Marine radiocarbon reservoir effect in the western North Pacific observed in archaeological fauna. Radiocarbon 43, 465–471. [Google Scholar]
- 39.de Bruyn M, Hall BL, Chauke LF, Baroni C, Koch PL, Hoelzel AR. 2009. Rapid response of a marine mammal species to holocene climate and habitat change. PLoS Genet. 5, e1000554 ( 10.1371/Journal.Pgen.1000554) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beaumont MA, Zhang W, Baldin DJ. 2002. Approximate Bayesian computation in population genetics. Genetics 162, 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderson CNK, Ramakrishnan U, Chan YL, Hadly EA. 2005. Serial SimCoal: a population genetics method for data from multiple populations and points in time. Bioinformatics 21, 1733–1734. ( 10.1093/bioinformatics/bti154) [DOI] [PubMed] [Google Scholar]
- 42.Blum MGB, François O. 2010. Non-linear regression models for approximate Bayesian computation. Stat. Comput. 20, 63–73. ( 10.1007/s11222-009-9116-0) [DOI] [Google Scholar]
- 43.Csilléry K, François O, Blum MGB. 2011. abc: an R package for approximate Bayesian computation (ABC). Methods Ecol. Evol. 3, 475–479. ( 10.1111/j.2041-210X.2011.00179.X) [DOI] [Google Scholar]
- 44.François O, Lava G. 2011. Deviance information criteria for model selection in approximate Bayesian computation. (http://www.arxiv.org/abs/1105.0269)
- 45.Anderson A. 2005. Subpolar settlement in South Polynesia. Antiquity 79, 791–800. [Google Scholar]
- 46.Childerhouse S, Gales N. 1998. Historical and modern distribution and abundance of the New Zealand sea lion Phocarctos hookeri. N.Z. J. Zool. 25, 1–16. ( 10.1080/03014223.1998.9518131) [DOI] [Google Scholar]
- 47.McConkey S, Heinrich S, Lalas C, McConnell H, McNally N. 2002. Pattern of immigration of New Zealand sea lions Phocarctos hookeri to Otago, New Zealand: Implications for management. Aust. Mammol. 24, 107–116. ( 10.1071/AM02107) [DOI] [Google Scholar]
- 48.Rawlence NJ, Metcalf J, Wood JR, Worthy TH, Austin J, Cooper A. 2012. The effect of climate and environmental change on the megafaunal moa of New Zealand in the absence of humans. Quat. Sci. Rev. 50, 141–153. ( 10.1016/j.quascirev.2012.07.004) [DOI] [Google Scholar]
- 49.Wanner H, et al. 2008. Mid- to Late Holocene climate change: an overview. Quat. Sci. Rev. 27, 1791–1828. ( 10.1016/j.quascirev.2008.06.013) [DOI] [Google Scholar]
- 50.Holdaway RN, Jacomb C. 2000. Rapid extinction of the moas (Aves : Dinorinthiformes): model, test, and implications. Science 287, 2250–2254. ( 10.1126/science.287.5461.2250) [DOI] [PubMed] [Google Scholar]
- 51.Leonard JA, Wayne R, Cooper A. 2000. Population genetics of Ice Age brown bears. Proc. Natl Acad. Sci. USA 97, 1651–1654. ( 10.1073/pnas.040453097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brace S, et al. 2012. Serial population extinctions in a small mammal indicate Late Pleistocene ecosystem instability. Proc. Natl Acad. Sci. USA 109, 20 532–20 536. ( 10.1073/pnas.1213322109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hofreiter M, Muenzel S, Conard NJ, Pollack J, Slatkin M, Weiss G, Paabo S. 2007. Sudden replacement of cave bear mitochondrial DNA in the late Pleistocene. Curr. Biol. 17, R122–R123. ( 10.1016/J.Cub.2007.01.026) [DOI] [PubMed] [Google Scholar]
- 54.Holdaway R, Worthy TH, Tennyson AJD. 2001. A working list of breeding bird species of the New Zealand region at first human contact. N.Z. J. Zool. 28, 119–187. ( 10.1080/03014223.2001.9518262) [DOI] [Google Scholar]
- 55.Landis CA, Campbell HJ, Begg JG, Mildernhall DC, Paterson AM, Trewick SA. 2008. The Waipounamu erosion surface: questioning the antiquity of the New Zealand land surface and terrestrial fauna and flora. Geol. Mag. 145, 173–197. ( 10.1017/S0016756807004268) [DOI] [Google Scholar]
- 56.Duncan RP, Boyer AG, Blackburn TM. 2013. Magnitude and variation of prehistoric bird extinctions in the Pacific. Proc. Natl Acad. Sci. USA 110, 6436–6441. ( 10.1073/pnas.1216511110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Russell JC, Miller SD, Harper GA, MacInnes HE, Wylie MJ, Fewster RM. 2010. Survivors or reinvaders? Using genetic assignment to identify invasive pests following eradication. Biol. Invas. 12, 1747–1757. ( 10.1007/s10530-009-9586-1) [DOI] [Google Scholar]
- 58.Banks JC, Mitchell AD, Waas JR, Paterson AM. 2002. An unexpected pattern of molecular divergence within the blue penguin (Eudyptula minor) complex. Notornis 49, 29–38. [Google Scholar]
- 59.Waters JM, Fraser CI, Hewitt GM. 2013. Founder takes all: density-dependent processes structure biodiversity. Trends Ecol. Evol. 28, 78–85. ( 10.1016/j.tree.2012.08.024) [DOI] [PubMed] [Google Scholar]
- 60.Hallatschek O, Hersen P, Ramanathan S, Nelson DR. 2007. Genetic drift at expanding frontiers promotes gene segregation. Proc. Natl Acad. Sci. USA 104, 19 926–19 930. ( 10.1073/pnas.0710150104) [DOI] [PMC free article] [PubMed] [Google Scholar]