Abstract
Polyclonal antisera to either a synthetic OipA peptide or a recombinant OipA protein detected OipA expression in Helicobacter pylori and correlated with functional oipA status determined by PCR sequence (sensitivity and specificity of >94%). Immunoblotting is a simple and accurate method for detecting expression of the important virulence factor OipA.
The Helicobacter pylori outer inflammatory protein, OipA, is an important virulence factor associated with enhanced interleukin-8 secretion and increased inflammation in vitro as well as the clinically important presentation of peptic ulcer (11, 13, 14). Expression of OipA is regulated by the slipped-strand repair mechanism based on the number of CT dinucleotide repeats in the 5′ region of the oipA gene (switch on = functional and switch off = nonfunctional) (11), such that gene switch status may affect bacterial characteristics such as virulence. Isolates that contain the cag pathogenicity island (CagA is a marker) typically also have oipA with functional status “on” (1, 3, 12, 13). Currently, prediction of the presence of a functional OipA protein requires PCR-based sequencing of the signal region of the gene. However, sequencing of the signal region of the gene does not guarantee that no mutations are present downstream that would prevent production of the protein. The present study was designed to test by immunoblot assay the hypothesis that PCR-based sequencing of the signal-peptide coding region of the oipA gene reliably predicted OipA functional status.
Because full-length clones of the OipA protein proved to be lethal to both bacterial and baculovirus-insect expression systems (Z. Z. Nurgalieva et al., unpublished data), we produced antisera to synthetic peptides and a partial recombinant OipA protein. We used the OipA sequence of strain CA22 isolated from a Korean patient with gastric cancer for construction of synthetic peptides. Synthetic peptides were based on the deduced sequence of the protein and computer algorithms (4-9). Two peptide sequences that were calculated to be amphipathic and hydrophilic with a high surface potential and near a region predicted to function as a T-cell epitope (2) were selected for synthesis. The sequences were KDSTKIANRFAGNGGSG (peptide 56) and DANTLKKVSRHVFRKSSG (peptide 161). Immunogenicity was tested against a recombinant OipA fusion protein using murine serum samples collected after immunization. Only peptide 56 proved to be strongly immunogenic, and it was used to immunize rabbits, resulting in anti-OipA peptide serum (sp-56). Prior to use, nonspecific antibodies were removed from sp-56 antisera by adsorption with an oipA knockout mutant H. pylori strain.
Recombinant OipA antigen was produced as a fusion with an N-terminal MS2-polymerase and a His tag by using the Escherichia coli expression vector pEV40 (10). A selected portion of the oipA gene (45 to 882 bp) was PCR amplified with the primers SO102 (5′-GAGAATTCCACGCTGAAAGGAATGGAT-3′) and SO103 (5′-GATCCTCGAGTCAATAAACGCTCACCACTCTTT-3′) and H. pylori 26695 chromosomal DNA as a template. The PCR fragment (EcoRI/XhoI) was cloned into pEV40a (EcoRI/SalI), resulting in plasmid pSO214a. The fusion protein was expressed by temperature induction of the λPL promoter and purified by Ni2+-nitrilotriacetic acid affinity chromatography according to the manufacturer's protocol (Qiagen, Inc., Valencia, Calif.). The purified recombinant OipA fusion protein was used to immunize a rabbit to obtain the polyclonal antiserum AK282.
Immunoblotting was performed by standard methods. We used a 1:5,000 dilution of anti-recombinant OipA antisera (AK282) and a 1:1,000 dilution of anti-OipA peptide sera (sp-56) (adsorbed) as first antibodies and horseradish peroxidase-conjugated protein A (1:2,000) (Bio-Rad Laboratories, Hercules, Calif.) as the second antibody. Proteins were detected by enhanced chemiluminescence with the ECL system (Amersham Life Sciences, Arlington Heights, Ill.). We also performed an immunoblot assay for CagA with a commercial anti-CagA antibody (1:4,000) (Austral Biologicals, San Ramon, Calif.).
OipA status was determined in 150 clinical isolates (60 from Japan and 90 from the United States). H. pylori isolates from Japan and the United States were used to ensure that slight sequence differences in oipA sequences between strains from Asia and Western countries did not influence the results (1, 11). We studied 105 isolates with oipA gene status “on” and 45 with status “off” as determined by PCR-based sequencing of the signal region of the oipA as previously described (11). The strains were selected to consist of equal numbers from patients with gastritis only, duodenal ulcer, and gastric cancer. The protocols under which the strains were obtained were approved by local ethics committees, and written informed consent was obtained. Controls consisted of isogenic oipA and cagE mutant strains (11).
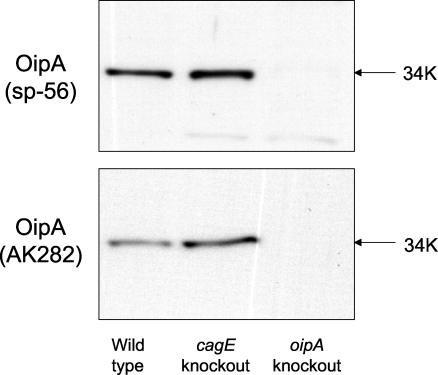
The antisera to both the OipA peptide (sp-56) and the recombinant OipA protein recognized a protein with a molecular weight of approximately 34,000 in both the wild-type strain (Fig. 1, lane 1) and in cagE knockout mutants (lane 2), which is consistent with the predicted size of OipA. The protein band was absent in the corresponding oipA knockout mutant (lane 3).
FIG. 1.
Western blotting analysis of OipA in H. pylori wild-type and mutant strains. Total cell lysates of H. pylori strains were probed with anti-OipA peptide sera (sp-56) raised against the synthetic OipA peptide (A) or antiserum (AK282) raised against a partial recombinant OipA protein (B). OipA protein was expressed in the wild-type strain (lane 1) and cagE knockout strain (lane 2) but not in the oipA knockout strain (lane 3). Based on the size, the lower band seen in Fig. 1A (sp-56) is thought to be nonspecific.
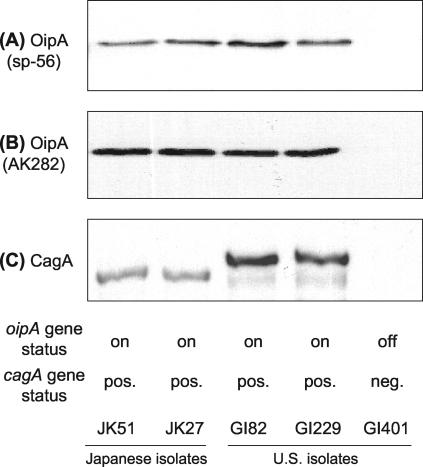
Immunoblot analysis of H. pylori isolates with oipA “on” status detected the OipA protein in 94% of isolates with sp-56 antiserum and 99% of isolates with AK282 antiserum. Neither antiserum detected OipA in H. pylori isolates with “off” status (Fig. 2A and B). In six (4%) instances sp-56 results and in one (0.6%) instance AK282 results were inconsistent with the results of PCR-based sequencing. These results were confirmed three times. We sequenced the entire oipA gene in all six isolates, and sequence analyses revealed no insertions or deletions that would produce frameshifts in the oipA gene. The isolates were all from the United States. Comparison of the sequences of the sp-56 region between isolates that were and were not detected by immunoblot did not show amino acid substitutions in the gene. Mutations in the promoter region preventing expression of a functional open reading frame were not excluded.
FIG. 2.
Western blotting analysis of OipA and CagA in clinical isolates. Total cell lysates of five H. pylori strains were probed with anti-OipA peptide sera (sp-56) raised against the synthetic OipA peptide (A), antiserum (AK282) raised against a recombinant OipA protein (B), or antibody raised against a recombinant CagA protein (Austral Biologicals) (C). OipA and CagA proteins were expressed in Japanese strains (lanes 1 and 2) and U.S. strains (lane 3 and 4). No band was detected from the lysates of the U.S. strain with oipA status “off” and without the cagA gene (lane 5).
In agreement with previous studies (1, 13), CagA protein was detected in 103 of 105 oipA “on” strains and in 3 of 45 oipA “off” strains (Fig. 2C). The results for oipA gene status were identical to those for OipA protein status in all five cases in which there were inconsistent results between CagA and the oipA gene status.
Although there are reports that the genomic structures of many H. pylori genes, including oipA, differ between Asian and Western H. pylori strains (1, 11), there are few data to suggest phenotypic differences. We found that anti-OipA immunoblots detected OipA equally well in both Asian and Western strains, with sensitivities of >94% and specificities of 100%. The synthetic peptides and partial recombinant OipA proteins are currently being tested as antigens to examine the relationship between anti-OipA antibody in patients' sera and oipA functional status and disease presentation.
The high sensitivity and specificity of detection of OipA by immunoblotting provide a convenient and rapid method to detect the OipA status of clinical isolates without the time and expense of DNA sequencing.
Acknowledgments
This work was supported in part by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs (D. Y. Graham and M. E. Conner), and by Public Health Service grants DK56338 (which funds the Texas Gulf Coast Digestive Diseases Center) and AI24998 (M. E. Conner). This study was also supported by DFG (OD 21/1-1) (S. Odenbreit).
We thank Judith M. Ball, Department of Pathobiology, Texas A & M University, for invaluable assistance in constructing the synthetic peptides.
REFERENCES
- 1.Ando, T., R. M. Peek, D. Pride, S. M. Levine, T. Takata, Y.-C. Lee, K. Kusugami, A. van der Ende, E. J. Kuipers, J. G. Kusters, and M. J. Blaser. 2002. Polymorphisms of Helicobacter pylori HP0638 reflect geographic origin and correlate with cagA status. J. Clin. Microbiol. 40:239-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berzofsky, J. A., K. B. Cease, J. L. Cornette, J. L. Spouge, H. Margalit, I. J. Berkower, M. F. Good, L. H. Miller, and C. DeLisi. 1987. Protein antigenic structures recognized by T cells: potential applications to vaccine design. Immunol. Rev. 98:9-52. [DOI] [PubMed] [Google Scholar]
- 3.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou, P. Y., and G. D. Fasman. 1974. Prediction of protein conformation. Biochemistry 13:222-245. [DOI] [PubMed] [Google Scholar]
- 5.Chou, P. Y., and G. D. Fasman. 1978. Prediction of the secondary structure of proteins from their amino acid sequence. Adv. Enzymol. Relat. Areas Mol. Biol. 47:45-148. [DOI] [PubMed] [Google Scholar]
- 6.Hopp, T. P., and K. R. Woods. 1981. Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl. Acad. Sci. USA 78:3824-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 8.Margalit, H., J. L. Spouge, J. L. Cornette, K. B. Cease, C. Delisi, and J. A. Berzofsky. 1987. Prediction of immunodominant helper T cell antigenic sites from the primary sequence. J. Immunol. 138:2213-2229. [PubMed] [Google Scholar]
- 9.Parker, J. M., D. Guo, and R. S. Hodges. 1986. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry 25:5425-5432. [DOI] [PubMed] [Google Scholar]
- 10.Pohlner, J., J. Kramer, and T. F. Meyer. 1993. A plasmid system for high-level expression and in vitro processing of recombinant proteins. Gene 130:121-126. [DOI] [PubMed] [Google Scholar]
- 11.Yamaoka, Y., D. H. Kwon, and D. Y. Graham. 2000. A M(r) 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 97:7533-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamaoka, Y., M. Kita, T. Kodama, S. Imamura, T. Ohno, N. Sawai, A. Ishimaru, J. Imanishi, and D. Y. Graham. 2002. Helicobacter pylori infection in mice: role of outer membrane proteins in colonization and inflammation. Gastroenterology 123:1992-2004. [DOI] [PubMed] [Google Scholar]
- 13.Yamaoka, Y., S. Kikuchi, H. M. El-Zimaity, O. Gutierrez, M. S. Osato, and D. Y. Graham. 2002. Importance of Helicobacter pylori OipA in clinical presentation, gastric inflammation, and mucosal interleukin-8 production. Gastroenterology 123:414-424. [DOI] [PubMed] [Google Scholar]
- 14.Zambon, C. F., D. Basso, F. Navaglia, G. Germano, N. Gallo, M. Milazzo, E. Greco, P. Fogar, S. Mazza, F. Di Mario, G. Basso, M. Rugge, and M. Plebani. 2002. Helicobacter pylori virulence genes and host IL-1RN and IL-1β genes interplay in favouring the development of peptic ulcer and intestinal metaplasia. Cytokine 18:242-251. [DOI] [PubMed] [Google Scholar]