Abstract
The lung develops from a very simple outpouching of the foregut into a highly complex, finely structured organ with multiple specialized cell types that are required for its normal physiological function. During both the development of the lung and its remodeling in the context of disease or response to injury, gene expression must be activated and silenced in a coordinated manner to achieve the tremendous phenotypic heterogeneity of cell types required for homeostasis and pathogenesis. Epigenetic mechanisms, consisting of DNA base modifications such as methylation, alteration of histones resulting in chromatin modification, and the action of noncoding RNA, control the regulation of information “beyond the genome” required for both lung modeling and remodeling. Epigenetic regulation is subject to modification by environmental stimuli, such as oxidative stress, infection, and aging, and is thus critically important in chronic remodeling disorders such as idiopathic pulmonary fibrosis (IPF), chronic obstructive pulmonary disease (COPD), bronchopulmonary dysplasia (BPD), and pulmonary hypertension (PH). Technological advances have made it possible to evaluate genome-wide epigenetic changes (epigenomics) in diseases of lung remodeling, clarifying existing pathophysiological paradigms and uncovering novel mechanisms of disease. Many of these represent new therapeutic targets. Advances in epigenomic technology will accelerate our understanding of lung development and remodeling, and lead to novel treatments for chronic lung diseases.
As is true for most tissues, the lung's development, as well as its response to injury, requires the coordinated proliferation, migration, and activation of the different cellular phenotypes already present, as well as phenotypic modulation and reprogramming to generate cells with specialized function. The genome contains the information to direct these events but is insufficient per se. Gene expression must be activated or silenced in a temporally coordinated way in response to developmental or injury/repair signals, and the information passed on to daughter cells as reprogrammed cells divide. This level of control is generally mediated by epigenetic mechanisms. This review will focus on epigenetic mechanisms in lung parenchymal remodeling. Most of the published work in this area is relevant to pulmonary fibrosis, particularly idiopathic pulmonary fibrosis (IPF), and to chronic obstructive pulmonary disease (COPD), although other conditions, such as bronchopulmonary dysplasia (BPD), will be considered as well. Other excellent publications have reviewed epigenetic mechanisms of lung development (48), cancer (14, 32), and airways disease (1, 25, 83).
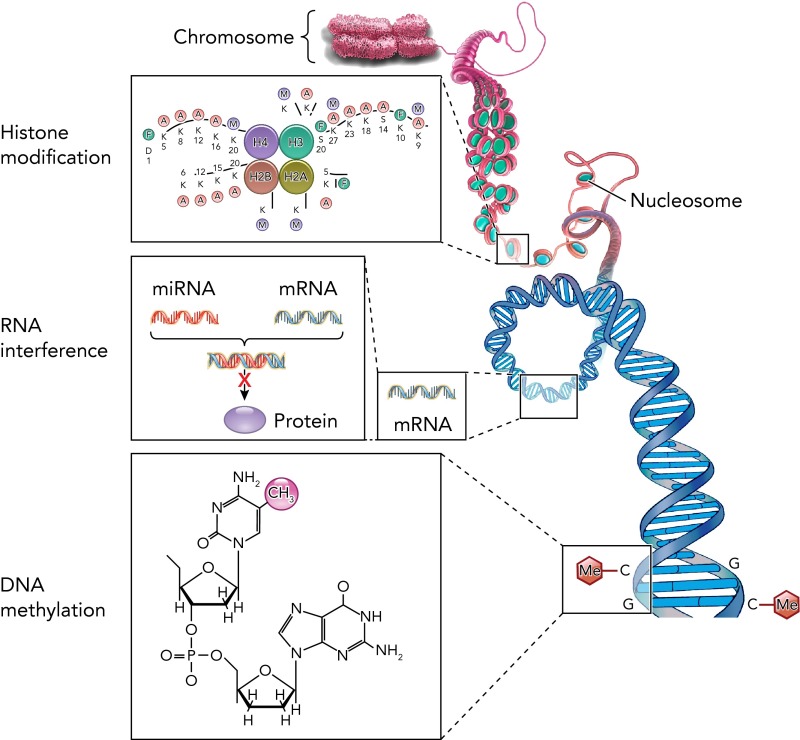
Epigenetic mechanisms are central to reprogramming of cellular phenotypes (38) and are known to be altered in cancer, normal development, and aging, and in responses to the environment. Many of these paradigms are associated with IPF and COPD. Epigenetics is defined as any modification of the genome or of gene expression not resulting from alteration in DNA nucleotide sequence. Many epigenetic alterations are heritable, affecting daughter cells. If an environmental stimulus causes epigenetic changes in the germline, these alterations can be transmitted to subsequent generations. There are three major processes of epigenetic modification: direct DNA methylation, chromatin (histone) modifications, and noncoding RNAs (ncRNAs). A number of epigenetic processes have been identified to play a role in IPF, including the three major epigenetic mechanisms: DNA methylation, histone modifications, and noncoding RNA (see FIGURE 1).
FIGURE 1.
Major epigenetic mechanisms controlling gene expression
Major Mechanisms of Epigenetic Regulation
DNA Methylation
Covalent methylation of the 5′ position of cytosine in the context of cytosine-guanine (CpG) dinucleotides is mediated by DNA methyltransferases (DNMTs) and generally results in tight packing of DNA and histones (heterochromatin), and in the long-term silencing of transcription. DNA methylation may result in gene silencing that can be propagated to daughter cells. Methylation is also responsible for silencing transposons and other parasitic elements; maintaining a normal pattern of genomic methylation is essential for health. The CpG content in the human genome is low (on the order of 1%), but most promoters have areas of high CpG content, often as long stretches of CpG known as “CpG islands.” High CpG-content promoters tend to be unmethylated in “housekeeping” genes. However, in tissue-restricted genes, inactive X chromosomes in females, and in most imprinted genes, CpG islands tend to be methylated. In many types of cancer, there is global hypomethylation resulting in genomic instability, but hypermethylation of promoters in specific genes such as tumor suppressors (15, 77). There are CpGs in areas up to 2 kb away from CpG islands, often referred to as “shores,” which are methylated in a tissue-specific manner (44). Variations on traditionally understood CpG methylation include hydroxymethylcystosine (5hMC) (41, 61, 92) and N6-methyl-adenine (100), methylation within gene bodies, and non-CpG methylation (3, 111), the consequences of which are increasingly being studied.
Histone Modifications
DNA in the nucleus is organized into chromatin, together with RNA, histones, and other chromosomal and nuclear proteins. The biochemical composition and physical structure of chromatin have significant effects on transcriptional activity (10). The modification of NH2-terminal tails of histones significantly affects the condensation of chromatin and access to the transcriptional machinery (51). Transcriptionally active, “open” euchromatin generally has hyperacetylated and hypomethylated histones, whereas more inactive heterochromatin tends to be hypoacetylated and hypermethylated. In addition to acetylation and methylation, which have been extensively studied (12), histones can be phosphorylated (5), nitrated (53), ubiqutinylated (101), and SUMOylated (88). Some histone methylation patterns have been extensively characterized; for example, trimethylation of histone 3 at the lysine 4 position (H3K4me3) is often found at transcription start sites and strongly correlates with active transcription, whereas H3K36me3 is often found associated with the gene body of actively transcribed genes. In contrast, H3K9me3 is often associated with the promoter region of repressed genes. However, H3K9me3 enrichment within the gene body is associated with active expression (7, 10). Combinations of histone modifications have been associated with different types of regulatory elements in genome-wide mapping studies; for example, enrichment of H3K4me3, H3K9ac, and H3K27ac is characteristic of active promoters, whereas chromatin regions enriched for H3K4me2/3 and H3K27me3 tend to be associated with silent or “poised” promoters. Active enhancer regions are characterized by H3K4me1/2 and low H3K4me3 (26).
Certain molecular complexes regulate chromatin structure and gene expression by altering the position of the nucleosomes. SWI/SNF, ISWI, NuRD, and INO80 complexes remodel the chromatin architecture of target promoters in an energy-dependent manner, altering the access of the transcriptional machinery to particular loci (30). Many chromatin remodeling complexes contain catalytic subunits such as Brahma (Brm) and Brahma-related gene-1 (Brg1), which have an important role in lung development. Brg1 interacts with Nkx2–1, a critical regulator of lung development, facilitating its binding to the mouse Sftpb promoter, facilitating transcription (13); Brg1 interaction with Smad3 increases the expression of some TGF-β-inducible genes (104). Deletion of Brg1 selectively in lung epithelial cells results in rapid tumor development (13). The three-dimensional structure of chromatin and its remodeling are considered another level of epigenetic regulation, which is beyond the scope of this review (59, 67).
Noncoding RNA
Eukaryotic genomes transcribe large numbers of RNAs that have no coding capacity. In addition to microRNA (miRNA), other forms of ncRNA include Piwi-interacting RNA (piRNA), short interfering RNA (siRNA), and promoter-linked long noncoding RNA (plncRNA), enhancer-linked ncRNA (elncRNA), and long intergenic ncRNA (lincRNA) (17).
Small (∼20–30 nucleotide) ncRNAs, including miRNA, piRNA, and siRNA, are the best characterized and have been found to play major roles in gene regulation at many levels of gene function, including chromatin architecture, RNA processing and stability, and protein translation. In distinction to siRNAs, which target specific mRNAs, miRNAs may target multiple genes, their proteins products, and expression networks. According to miRBase (56), there are currently over 1,800 human miRNAs, which regulate >60% of protein-coding genes (72). In the developing mouse lung, miRNAs are developmentally regulated in a cell type-specific manner (65). Environmental toxins can alter the lung miRNA profile (46, 109). Early life events and exposures can thus modify miRNA profiles during lung development and alter susceptibility to lung diseases throughout the lifespan. Silencing miRNAs in vivo (e.g., using oligonucleotides termed “antagomirs”) is feasible and has been applied in vivo in animal models of lung disease (57, 74), offering the potential of miRNA-based therapeutic strategies for clinical use.
Epigenetic Alterations Associated With Lung Diseases
Idiopathic Pulmonary Fibrosis
Studies have increasingly demonstrated that abnormal DNA methylation patterns alter expression of many genes involved in IPF pathogenesis. DNA methylation leads to silencing of Thy-1 in lung fibroblasts in vitro and in vivo, promoting myofibroblastic differentiation and resistance to apoptosis (85, 86). Thy-1 promoter hypermethylation was confirmed in Thy-1(−) myofibroblasts within the characteristic fibroblastic foci (FF) in IPF, in contrast to the overlying epithelial cells which express Thy-1. Thus DNA methylation can be differentially regulated in distinct cell types within the same pathological lesion. Subsequently, others have found that hypoxia promotes hypermethylation of Thy-1 in lung fibroblasts, indicating that hypoxic modification of DNA methylation can induce myofibroblastic differentiation (82). Other studies have also demonstrated promoter methylation associated with transcriptional silencing of other IPF-associated genes (16, 35, 39). Methyl CpG binding protein 2 (MeCP2) binds to the alpha smooth muscle actin (α-SMA) promoter and alters its expression in fibroblasts (36). In addition, poly(ADP-ribosyl)ation (PARylation), mediated by members of the poly(ADP-ribose) polymerase (PARP) superfamily (principally PARP-1), affects myofibroblast differentiation in IPF by suppressing methylation in the α-SMA gene and modulating the binding of Smad3 to its binding element in the α-SMA promoter (37). It has been hypothesized that epigenetic mechanisms promote fibrosis in multiple tissues by preventing fibroblasts from returning to their resting state once they are activated (103).
Two published studies have measured DNA methylation more globally in the context of IPF. The studies used different controls and different platforms for assessing methylation. One study demonstrated differential methylation of 625 CpG islands between IPF lung tissue and control samples (uninvolved tissue from cancer resections). The majority (91.2%) of the differentially methylated regions (DMRs) were outside promoters (e.g., in intronic, exonic, or intergenic areas) (80). This study reported that methylation patterns in IPF had similarities to both control and lung cancer tissues, and thus might represent an intermediate condition. Another study compared transcriptome (from RNA expression microarray) to DNA methylation (Illumina BeadChip) in lung tissue from IPF subjects and normal controls (84), identifying 835 DMRs. Analysis of genes having twofold difference in expression and statistically significant differences in methylation, in which expression and methylation were inversely correlated, revealed 16 genes, 12 of which had already been described as relevant to fibrotic remodeling in the lung or elsewhere. The study confirmed differences in expression of the remaining genes at the RNA and protein levels in IPF samples. These two genome-wide methylation studies employed different techniques, so there was little overlap between the two studies in the genes found to be differentially methylated and expressed. Both studies carried out pathway analysis of their respective datasets, however, and both found that cellular assembly and organization and cell growth and proliferation were important paradigms regulated by DNA methylation. These studies, although limited by small sample sizes and some technical limitations, nonetheless underscore the importance of alterations of the epigenome in the pathogenesis of IPF and pave the way for more detailed studies using newer sequence-based technologies.
A number of studies have demonstrated alterations in histone modifications that affect the expression of individual genes in the context of IPF, including cyclooxygenase-2 (COX-2), interferon gamma-induced protein 10 (IP-10), monocyte chemoattractant protein-1 (MCP-1), Thy-1, IL-6, PAI-1, αSMA, and Fas (2, 18, 19, 40, 87, 93, 94). Histone deacetylases (HDACs) have been shown to modify lung myofibroblast differentiation (28). Trichostatin A (TSA), an HDAC inhibitor, blocked transforming growth factor β1 (TGF-β1)-mediated α-SMA and α1 type I collagen mRNA induction in normal human lung fibroblasts. In another report, TSA was found to induce Thy-1 expression in Thy-1(−) fibroblasts, as well as induce demethylation of CpG sites in the Thy-1 promoter region (87), demonstrating interaction among the different epigenetic paradigms.
A study of profibrotic fibroblast phenotypes from both a murine model of lung fibrosis and IPF patient-derived fibroblasts demonstrated that altered HDAC2 and HDAC4 expression underlies the differential association of the Fas promoter with acetylated histones, and that treatment of profibrotic fibroblasts with HDAC inhibitors increased Fas expression and restored susceptibility to Fas-mediated apoptosis (40). Another study demonstrated that spiruchostatin A (SpA), a class-I selective HDAC inhibitor, inhibited proliferation and differentiation of IPF fibroblasts (22). The accelerated epithelial senescence observed in IPF can be antagonized by sirtuin (SIRT) 6, a class III HDAC (71). HDAC inhibitors have been proposed as novel therapeutics for fibrotic diseases characterized by fibroblast activation (76). So far, no genome-wide histone modification profiles have been reported in IPF.
A great deal of evidence indicates a number of roles for miRs in the pathogenesis of IPF (20, 21, 62, 63, 70, 73, 74, 78, 106, 107). For example, TGF-β1 upregulates miR-21 in mice following bleomycin-induced lung fibrosis as well as in IPF lung tissue (63). Conversely, TGF-β1 suppresses miR-29 expression, resulting in increased expression of profibrotic target genes and worse fibrosis (20). Interestingly, a majority of miRs increased in IPF are localized to chromosome 14q32, and many are members of the mir-154 family, which modulates the WNT/β-Catenin pathway (70). An interesting recent study of IPF fibroblasts showed that argonaute (AGO)1 and AGO2 [part of the RNA-induced silencing (RISC) miRNA processing complex] were decreased in rapidly progressive IPF compared with normal or slowly progressive IPF biopsies and fibroblasts (73). Another recent study found that expression of the miR-17∼92 cluster is decreased in IPF associated with hypermethylation of the miR-17∼92 promoter (21). This cluster is important in lung development and lung epithelial homeostasis (66, 98). Remarkably, methylation of the miR 17∼92 promoter correlated inversely with lung function, showing a direct correlation of an epigenetic alteration and a physiological measurement in lung fibrosis (21). See Table 1 for additional miRs implicated in IPF pathogenesis. The interaction between miRNAs and DNA methylation machinery appears to be important in regulating fibroblast phenotypes and fibrosis in other tissues as well (90). A re-analysis of published studies of miRs and expression arrays in IPF indicated that the TGF-β1, Wnt, sonic hedgehog, p53, and vascular endothelial growth factor pathways are regulated by miRNA networks in IPF (75). It has been hypothesized that, in IPF, the aberrant expression of miRs that regulate or are regulated by TGF-β1 leads to release of inhibitions on the TGF-β1 pathway and the formation of feed-forward loops (74).
Table 1.
Selected microRNAs implicated in IPF
| MicroRNAs | Observed Change | Physiological Consequence | Reference |
|---|---|---|---|
| miR-199a-5p | Upregulated in the bleomycin model and in samples from IPF patients | Key effector of TGF-β signaling in lung fibroblasts by regulating caveolin 1 | 62 |
| miR-145 | Increased in TGF-β1-treated lung fibroblasts and in the lungs of patients with IPF compared with normal human lungs | In lung fibroblasts, increased SMA-α expression, enhanced contractility, and promoted formation of focal and fibrillar adhesions; activation of latent TGF-β1 | 107 |
| miR-155 | Mouse model of lung fibrosis showed that miR-155 expression level was correlated with the degree of lung fibrosis | Participate in lung epithelial-mesenchymal interactions by binding to and decreasing the release of keratinocyte growth factor induced by IL-1β or TNF-α in human normal pulmonary fibroblasts | 78 |
| miR-200 family members | Reversing the fibrogenic activity of pulmonary fibroblasts from mice with experimental pulmonary fibrosis and from patients with IPF | Inhibited the TGF-β1-induced epithelial-mesenchymal transition of alveolar epithelial cells | 106 |
Shown are selected microRNAs implicated in IPF; see text for others.
Chronic Obstructive Pulmonary Disease
Epigenetic alterations in asthma and chronic obstructive pulmonary disease (COPD) have been expertly reviewed recently (50, 108). In both of these very highly prevalent disorders, epigenetic regulation of inflammatory responses, resulting primarily from exposure to environmental stimuli, plays a prominent role in alteration of the cellular phenotypes that drives pathogenesis. In common diseases such as these, where genetic variation has been found to account for only a small portion of the risk, epigenetic alteration is likely a key link between genetic susceptibility and disease expression (9).
As is the case for IPF, a number of recent studies have begun to address epigenetic alterations more globally in COPD (epigenomics). The first of these measured genome-wide DNA methylation in two large family cohorts, one of which included nonsmokers with COPD, using DNA from white blood cells (79). A total of 3,565 CpG sites were statistically significantly differentially methylated (P value of <0.05) genome-wide in the initial cohort. Interestingly, the top-ranked differentially methylated gene based on both diagnosis and severity (FEV1) was SERPINA1, the gene for alpha-1 antitrypsin, which is arguably the best characterized monogenic cause of COPD. Using more stringent statistical criteria in both cohorts and association with severity, 349 DMRs were identified, 95% of which were hypomethylated and 70% of which were outside of CpG islands. Gene ontology (GO) analysis indicated overrepresentation of immune and inflammatory system pathways, responses to stress and external stimuli, as well as wound-healing and coagulation cascades among these 349 DMRs (79). Similar to the epigenomic studies in IPF, this study identified differential methylation of a large number of genes with either known association with COPD or significant biological plausibility, underscoring the robustness of this type of analysis. A limitation is that the changes were not measured in the lung, the target tissue in COPD. A subsequent study by the same group used a more extensive methylation platform in the same patient cohorts to analyze the effects of corticosteroid use in COPD (99). Using a combination of statistical models, a total of 511 sites were significantly associated with current steroid use. Pathway analysis indicated significant enrichment for intrinsic membrane components, hemostasis and coagulation, cellular ion homeostasis, leukocyte and lymphocyte activation and chemotaxis, protein transport, and responses to nutrients.
Landmark studies demonstrated an imbalance of HDAC and HAT activity in COPD and asthma, with reduction in HDAC2 activity and increased HAT activity (45). Subsequent studies have shown a critical role for HDAC/HAT imbalance in mediating steroid resistance and established histone-modifying enzymes as important therapeutic targets in inflammatory airway disease (6, 24). Other HDACs, such as HDAC7, may play a role in the abnormal adaptation to hypoxia seen in COPD (95). Oxidant stress, such as that associated with cigarette smoke, environmental toxicants, and aging, modifies chromatin remodeling, which can significantly alter gene expression. The interaction of oxidant stress with HDACs and histone acetyltransferases (HATs), with subsequent effects on inflammation, autophagy, and senescence in the context of COPD, has been very well reviewed recently (91).
Muscle dysfunction is a common complication of COPD, which affects exercise tolerance, quality of life, and survival. Recently, several interesting studies have shown interaction among epigenetic mechanisms, including miRNAs and chromatin modification, which lead to muscle dysfunction in COPD and may offer novel therapeutic targets (8).
Other Parenchymal Lung Diseases
The etiology of chronic lung disease of prematurity, also known as bronchopulmonary dysplasia (BPD), is complex, multifactorial, and incompletely understood, but is thought to result from a very immature, developing lung subjected to inflammation, hypoxic, or hyperoxic stress and ventilator-associated trauma. The lung in BPD is characterized by inhibited alveolar development (IAD), with “simplified” alveoli and abnormal vascular remodeling, resulting from disordered reprogramming of lung cell phenotypes and abnormal cellular communication. Because epigenetic mechanisms are critical to normal lung development and many of the etiological mechanisms of BPD have epigenetic effects, it follows that epigenetic studies may clarify much of the pathophysiology of this disorder and offer new therapeutic targets.
Gender affects susceptibility to BPD. One study has demonstrated gender-specific interaction of methyl CpG binding protein 2 (MeCP2) with the PPARγ promoter in a rat model of intrauterine growth retardation (IUGR), resulting in differential effects of PPARγ on impaired alveolarization of the lung (49). In a preterm lamb model of BPD using mechanical ventilation, inhibition of histone deacetylation significantly improved alveolar formation in the lung associated with changes in specific histone modifications, suggesting that mechanical ventilation can cause epigenetic alterations that can be reversed pharmacologically with therapeutic benefit (29). Newborn mice exposed to hyperoxia show inhibited alveolarization associated with decreased HDAC1 and HDAC2 expression and increased p53 and p21; drugs that affect the activity of histone deacetylases may thus confer protection against hyperoxia-induced alveolar hypoplasia (64).
The expression of numerous genes involved in the pathogenesis of pulmonary hypertension (PH) is epigenetically regulated (4, 102); a number of studies in experimental models of PH have described epigenetic alterations that could lead to novel therapeutic targets (54). Histone demethylases alter the contractile and proliferative phenotype of fetal sheep pulmonary artery smooth muscle cells (PASMC) in vitro (105). HDAC inhibitors reverse PH in mouse pups resulting from maternal undernutrition (81), as well as improving hypoxia-induced PH in a rat model and altering epigenetically mediated changes in the expression profiles of adventitial fibroblasts and inflammatory cells in pulmonary arteries of calves with PH (60, 110). The role of miRNA as pathophysiological mediators and therapeutic targets in PH has been recently reviewed elsewhere (27, 68).
Future Perspectives
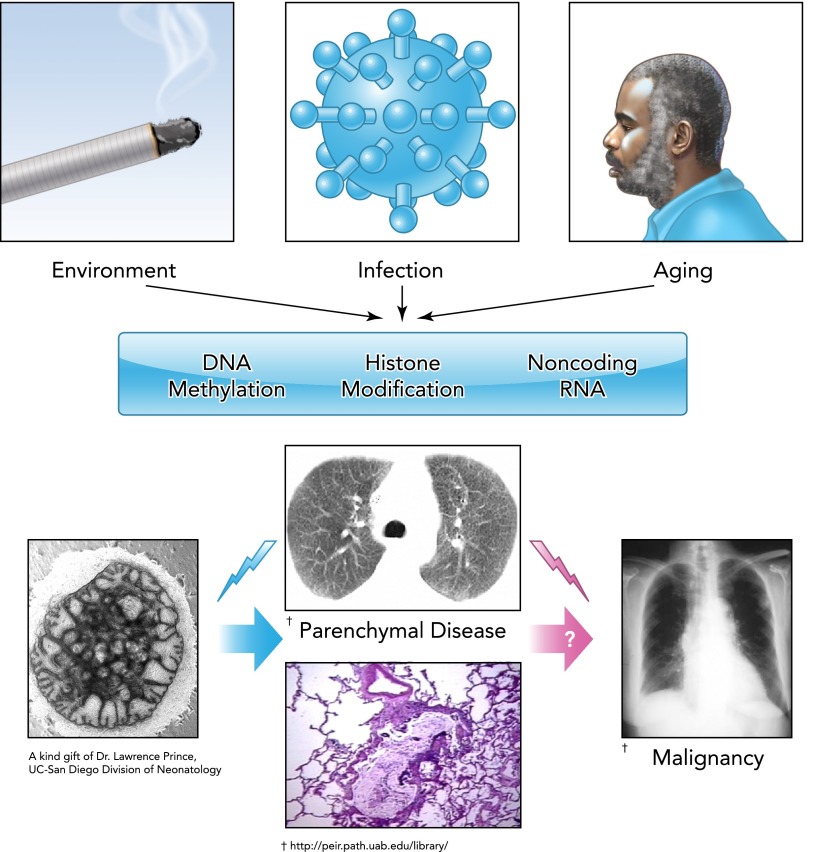
There is an emerging view based on published studies to date of cooperative interaction among the genome, the environment, and the epigenetic machinery in conditions resulting in lung remodeling, such as IPF, COPD, BPD, and PH (see FIGURE 2). Increasingly, interactions among the different epigenetic mechanisms have been shown to accelerate remodeling. Hypomethylated CpG in bacterial and viral DNA may promote rapid progression of pulmonary fibrosis via TLR9-dependent myofibroblast differentiation (69). Because clinical progression in IPF could result from aberrant processing of miRNAs (73), it has been hypothesized that activation of pulmonary fibroblasts by hypomethylated DNA leads to aberrant miR processing, promoting rapid progression of fibrosis in the lung (34).
FIGURE 2.
Epigenetic alterations related to environmental exposures and aging affect the trajectory from early lung development to advanced age
Similar mechanisms may be involved in the development of pulmonary malignancies.
Epigenomic technology is advancing rapidly as cost is decreasing. Next-generation sequencing (NGS)-based mRNA analysis is making array-based transcriptomics a thing of the past. Advances such as ion semiconductor sequencing are putting NGS within reach of many laboratories. NGS-based analysis is very well suited to ncRNA, with the capacity to identify previously undescribed miRNAs, as well as other ncRNAs, which are biologically relevant and not incorporated into array-based techniques. NGS-based analysis of DNA methylation and histone modifications on an unbiased, whole-genome level have been published but are still beyond the scope of most laboratories. These genome-wide, single-base level approaches generate massive datasets with the burden of storage and analysis. In silico analysis of existing data offers a wealth of information without additional experimentation (42, 47, 58). However, as NGS costs continue to decrease, it may become more cost-effective to re-sequence samples than to archive massive datasets. The developing field of systems biology offers coordinated analysis of multiple “-omic” datasets to identify emergent systems (43).
Epigenetic therapies are beyond the scope of this review but are in preclinical and clinical trials for many diseases (11, 31, 33, 52, 55, 97). Most of the current epigenetic modifiers are DNMT inhibitors and HDAC inhibitors, but additional chromatin modifications can be targeted by a number of small molecule inhibitors. The future challenges in epigenetic therapeutics are increasing specificity and minimizing off-target effects. Antisense oligonucleotides can be used to specifically target miRNAs (23, 89, 96). Targeting multiple miRNAs simultaneously may be necessary in complex pathologies, such as pulmonary fibrosis and COPD. It is possible that a personalized miRNA targeting approach may need to be developed for individual patients.
Notwithstanding the challenges ahead, epigenomics has become an integral part of our understanding of the complex process of tissue remodeling in multiple lung diseases and is likely to provide many breakthroughs that should improve quality and quantity of life for millions.
Footnotes
J. S. Hagood receives funding from the National Heart, Lung, and Blood Institute (grants HL-082818 and HL-111169) and from the Pulmonary Fibrosis Foundation, and is the sole author of this review.
No conflicts of interest, financial or otherwise, are declared by the author(s).
Author contributions: J.S.H. prepared figures; J.S.H. drafted manuscript; J.S.H. edited and revised manuscript; J.S.H. approved final version of manuscript.
References
- 1.Adcock IM, Ford P, Ito K, Barnes PJ. Epigenetics and airways disease. Respir Res 7: 21, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akhtar S, Coward WR, Feghali-Bostwick CA, Jenkins G, Knox A, Pang L. Molecular mechanisms of increased expression of the chemokine monocyte chemoattractant. Protein-1 (mcp-1) in idiopathic pulmonary fibrosis (ipf). Am J Respir Crit Care Med 183: A3466, 2011 [Google Scholar]
- 3.Arand J, Spieler D, Karius T, Branco MR, Meilinger D, Meissner A, Jenuwein T, Xu G, Leonhardt H, Wolf V, Walter J. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLos Genet 8: e1002750, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archer SL, Marsboom G, Kim GH, Zhang HJ, Toth PT, Svensson EC, Dyck JR, Gomberg-Maitland M, Thebaud B, Husain AN, Cipriani N, Rehman J. Epigenetic attenuation of mitochondrial superoxide dismutase 2 in pulmonary arterial hypertension: a basis for excessive cell proliferation and a new therapeutic target. Circulation 121: 2661–2671, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee T, Chakravarti D. A peek into the complex realm of histone phosphorylation. Mol Cell Biol 31: 4858–4873, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnes PJ. Corticosteroid resistance in patients with asthma and chronic obstructive pulmonary disease. J Allergy Clin Immunol 131: 636–645, 2013 [DOI] [PubMed] [Google Scholar]
- 7.Barnes PJ. Targeting the epigenome in the treatment of asthma and chronic obstructive pulmonary disease. Proc Am Thor Soc 6: 693–696, 2009 [DOI] [PubMed] [Google Scholar]
- 8.Barreiro E, Sznajder JI. Epigenetic regulation of muscle phenotype and adaptation: a potential role in COPD muscle dysfunction. J Appl Physiol 114: 1263–1272, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet 20: 350–358, 2004 [DOI] [PubMed] [Google Scholar]
- 10.Black JC, Whetstine JR. Chromatin landscape: methylation beyond transcription. Epigenetics 6: 9–15, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolden JE, Shi W, Jankowski K, Kan CY, Cluse L, Martin BP, MacKenzie KL, Smyth GK, Johnstone RW. HDAC inhibitors induce tumor-cell-selective pro-apoptotic transcriptional responses. Cell Death Dis 4: e519, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet 43: 559–599, 2009 [DOI] [PubMed] [Google Scholar]
- 13.Cao Y, Vo T, Millien G, Tagne JB, Kotton D, Mason RJ, Williams MC, Ramirez MI. Epigenetic mechanisms modulate thyroid transcription factor 1-mediated transcription of the surfactant protein B gene. J Biol Chem 285: 2152–2164, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carvalho RH, Haberle V, Hou J, van Gent T, Thongjuea S, van Ijcken W, Kockx C, Brouwer R, Rijkers E, Sieuwerts A, Foekens J, van Vroonhoven M, Aerts J, Grosveld F, Lenhard B, Philipsen S. Genome-wide DNA methylation profiling of non-small cell lung carcinomas. Epigenetics Chromatin 5: 9, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C, Yin N, Yin B, Lu Q. DNA methylation in thoracic neoplasms. Cancer Lett 301: 7–16, 2011 [DOI] [PubMed] [Google Scholar]
- 16.Cisneros J, Hagood J, Checa M, Ortiz-Quintero B, Negreros M, Herrera I, Ramos C, Pardo A, Selman M. Hypermethylation-mediated silencing of p14(ARF) in fibroblasts from idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol 303: L295–L303, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costa FF. Non-coding RNAs: meet thy masters. Bioessays 32: 599–608, 2010 [DOI] [PubMed] [Google Scholar]
- 18.Coward WR, Watts K, Feghali-Bostwick CA, Jenkins G, Pang L. Repression of IP-10 by interactions between histone deacetylation and hypermethylation in idiopathic pulmonary fibrosis. Mol Cell Biol 30: 2874–2886, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coward WR, Watts K, Feghali-Bostwick CA, Knox A, Pang L. Defective histone acetylation is responsible for the diminished expression of cyclooxygenase 2 in idiopathic pulmonary fibrosis. Mol Cell Biol 29: 4325–4339, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, Thannickal VJ, Cardoso WV, Lu J. miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol 45: 287–294, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dakhlallah D, Batte K, Wang Y, Cantemir-Stone CZ, Yan P, Nuovo G, Mikhail A, Hitchcock CL, Wright VP, Nana-Sinkam SP, Piper MG, Marsh CB. Epigenetic regulation of miR-17∼92 contributes to the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med 187: 397–405, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies ER, Haitchi HM, Thatcher TH, Sime PJ, Kottmann RM, Ganesan A, Packham G, O'Reilly KM, Davies DE. Spiruchostatin A inhibits proliferation and differentiation of fibroblasts from patients with pulmonary fibrosis. Am J Respir Cell Mol Biol 46: 687–694, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res 34: 2294–2304, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dekker FJ, Haisma HJ. Histone acetyl transferases as emerging drug targets. Drug Discov Today 14: 942–948, 2009 [DOI] [PubMed] [Google Scholar]
- 25.Durham A, Chou PC, Kirkham P, Adcock IM. Epigenetics in asthma and other inflammatory lung diseases. Epigenomics 2: 523–537, 2010 [DOI] [PubMed] [Google Scholar]
- 26.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grant JS, White K, Maclean MR, Baker AH. MicroRNAs in pulmonary arterial remodeling. Cell Mol Life Sci 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo W, Shan B, Klingsberg RC, Qin X, Lasky JA. Abrogation of TGF-beta1-induced fibroblast-myofibroblast differentiation by histone deacetylase inhibition. Am J Physiol Lung Cell Mol Physiol 297: L864–L870, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamvas A, Deterding R, Balch WE, Schwartz DA, Albertine KH, Whitsett JA, Cardoso WV, Kotton DN, Kourembanas S, Hagood JS. Diffuse lung disease in children: summary of a scientific conference. Pediatr Pulmonol. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res 21: 396–420, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatzimichael E, Crook T. Cancer epigenetics: new therapies and new challenges. J Drug Delivery 2013: 529312, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herceg Z, Vaissiere T. Epigenetic mechanisms and cancer: an interface between the environment and the genome. Epigenetics 6: 804–819, 2011 [DOI] [PubMed] [Google Scholar]
- 33.Ho AS, Turcan S, Chan TA. Epigenetic therapy: use of agents targeting deacetylation and methylation in cancer management. Onco Targets Ther 6: 223–232, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hogaboam CM, Murray L, Martinez FJ. Epigenetic mechanisms through which Toll-like receptor-9 drives idiopathic pulmonary fibrosis progression. Proc Am Thorac Soc 9: 172–176, 2012 [DOI] [PubMed] [Google Scholar]
- 35.Hu B, Gharaee-Kermani M, Wu Z, Phan SH. Epigenetic regulation of myofibroblast differentiation by DNA methylation. Am J Pathol 177: 21–28, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu B, Gharaee-Kermani M, Wu Z, Phan SH. Essential role of MeCP2 in the regulation of myofibroblast differentiation during pulmonary fibrosis. Am J Pathol 178: 1500–1508, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu B, Wu Z, Hergert P, Henke CA, Bitterman PB, Phan SH. Regulation of myofibroblast differentiation by poly(ADP-ribose) polymerase 1. Am J Pathol 182: 71–83, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang K, Fan G. DNA methylation in cell differentiation and reprogramming: an emerging systematic view. Regen Med 5: 531–544, 2010 [DOI] [PubMed] [Google Scholar]
- 39.Huang SK, Fisher AS, Scruggs AM, White ES, Hogaboam CM, Richardson BC, Peters-Golden M. Hypermethylation of PTGER2 confers prostaglandin E2 resistance in fibrotic fibroblasts from humans and mice. Am J Pathol 177: 2245–2255, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang SK, Scruggs AM, Donaghy J, Horowitz JC, Zaslona Z, Przybranowski S, White ES, Peters-Golden M. Histone modifications are responsible for decreased Fas expression and apoptosis resistance in fibrotic lung fibroblasts. Cell Death Dis 4: e621, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLos One 5: e8888, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huopaniemi I, Suvitaival T, Nikkila J, Oresic M, Kaski S. Multivariate multi-way analysis of multi-source data. Bioinformatics 26: i391–i398, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hyduke DR, Lewis NE, Palsson BO. Analysis of omics data with genome-scale models of metabolism. Mol Bio Sys 9: 167–174, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash JB, Sabunciyan S, Feinberg AP. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nature Gen 41: 178–186, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ito K, Ito M, Elliott WM, Cosio B, Caramori G, Kon OM, Barczyk A, Hayashi S, Adcock IM, Hogg JC, Barnes PJ. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. N Engl J Med 352: 1967–1976, 2005 [DOI] [PubMed] [Google Scholar]
- 46.Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J 23: 806–812, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jordan R, Patel S, Hu H, Lyons-Weiler J. Efficiency analysis of competing tests for finding differentially expressed genes in lung adenocarcinoma. Cancer Inform 6: 389–421, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joss-Moore LA, Albertine KH, Lane RH. Epigenetics and the developmental origins of lung disease. Mol Genet Metab 104: 61–66, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joss-Moore LA, Wang Y, Ogata EM, Sainz AJ, Yu X, Callaway CW, McKnight RA, Albertine KH, Lane RH. IUGR differentially alters MeCP2 expression and H3K9Me3 of the PPARgamma gene in male and female rat lungs during alveolarization. Birth Defects Res A Clin Mol Teratol 91: 672–681, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kabesch M, Adcock IM. Epigenetics in asthma and COPD. Biochimie 94: 2231–2241, 2012 [DOI] [PubMed] [Google Scholar]
- 51.Karlic R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. Histone modification levels are predictive for gene expression. Proc Natl Acad Sci USA 107: 2926–2931, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karras JG, Sun G, Tay J, Jackson AL. Reflections on microRNAs in chronic pulmonary disease: looking into the miR-ror and crystal ball. Inflammation Allergy Drug Targets 12: 88–98, 2013 [DOI] [PubMed] [Google Scholar]
- 53.Khan MA, Dixit K, Jabeen S, Moinuddin, Alam K. Impact of peroxynitrite modification on structure and immunogenicity of H2A histone. Scand J Immunol 69: 99–109, 2009 [DOI] [PubMed] [Google Scholar]
- 54.Kim GH, Ryan JJ, Marsboom G, Archer SL. Epigenetic mechanisms of pulmonary hypertension. Pulm Circ 1: 347–356, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koch MW, Metz LM, Kovalchuk O. Epigenetics and miRNAs in the diagnosis and treatment of multiple sclerosis. Trends Mol Med 19: 23–30, 2013 [DOI] [PubMed] [Google Scholar]
- 56.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39: D152–D157, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438: 685–689, 2005 [DOI] [PubMed] [Google Scholar]
- 58.Kumar M, Ahmad T, Sharma A, Mabalirajan U, Kulshreshtha A, Agrawal A, Ghosh B. Let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. J Allergy Clin Immunol 128: 1077–1085, 2011 [DOI] [PubMed] [Google Scholar]
- 59.Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev 21: 175–186, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li M, Riddle SR, Frid MG, El Kasmi KC, McKinsey TA, Sokol RJ, Strassheim D, Meyrick B, Yeager ME, Flockton AR, McKeon BA, Lemon DD, Horn TR, Anwar A, Barajas C, Stenmark KR. Emergence of fibroblasts with a proinflammatory epigenetically altered phenotype in severe hypoxic pulmonary hypertension. J Immunol 187: 2711–2722, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q, Lee CW, Hu D, Lian BQ, Kleffel S, Yang Y, Neiswender J, Khorasani AJ, Fang R, Lezcano C, Duncan LM, Scolyer RA, Thompson JF, Kakavand H, Houvras Y, Zon LI, Mihm MC, Jr, Kaiser UB, Schatton T, Woda BA, Murphy GF, Shi YG. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 150: 1135–1146, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lino Cardenas CL, Henaoui IS, Courcot E, Roderburg C, Cauffiez C, Aubert S, Copin MC, Wallaert B, Glowacki F, Dewaeles E, Milosevic J, Maurizio J, Tedrow J, Marcet B, Lo-Guidice JM, Kaminski N, Barbry P, Luedde T, Perrais M, Mari B, Pottier N. miR-199a-5p Is upregulated during fibrogenic response to tissue injury and mediates TGFbeta-induced lung fibroblast activation by targeting caveolin-1. PLos Genet 9: e1003291, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med 207: 1589–1597, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Londhe VA, Sundar IK, Lopez B, Maisonet TM, Yu Y, Aghai ZH, Rahman I. Hyperoxia impairs alveolar formation and induces senescence through decreased histone deacetylase activity and up-regulation of p21 in neonatal mouse lung. Pediatr Res 69: 371–377, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu J, Qian J, Chen F, Tang X, Li C, Cardoso WV. Differential expression of components of the microRNA machinery during mouse organogenesis. Biochem Biophys Res Commun 334: 319–323, 2005 [DOI] [PubMed] [Google Scholar]
- 66.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17–92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol 310: 442–453, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol 13: 436–447, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meloche J, Paulin R, Provencher S, Bonnet S. Therapeutic Potential of microRNA modulation in pulmonary arterial hypertension. Curr Vasc Pharmacol. In press [DOI] [PubMed] [Google Scholar]
- 69.Meneghin A, Choi ES, Evanoff HL, Kunkel SL, Martinez FJ, Flaherty KR, Toews GB, Hogaboam CM. TLR9 is expressed in idiopathic interstitial pneumonia and its activation promotes in vitro myofibroblast differentiation. Histochem Cell Biol 130: 979–992, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Milosevic J, Pandit K, Magister M, Rabinovich E, Ellwanger DC, Yu G, Vuga LJ, Weksler B, Benos PV, Gibson KF, McMillan M, Kahn M, Kaminski N. Profibrotic role of miR-154 in pulmonary fibrosis. Am J Respir Cell Mol Biol 47: 879–887, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Minagawa S, Araya J, Numata T, Nojiri S, Hara H, Yumino Y, Kawaishi M, Odaka M, Morikawa T, Nishimura SL, Nakayama K, Kuwano K. Accelerated epithelial cell senescence in IPF and the inhibitory role of SIRT6 in TGF-beta-induced senescence of human bronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol 300: L391–L401, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nana-Sinkam SP, Hunter MG, Nuovo GJ, Schmittgen TD, Gelinas R, Galas D, Marsh CB. Integrating the MicroRNome into the study of lung disease. Am J Respir Crit Care Med 179: 4–10, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oak SR, Murray L, Herath A, Sleeman M, Anderson I, Joshi AD, Coelho AL, Flaherty KR, Toews GB, Knight D, Martinez FJ, Hogaboam CM. A micro RNA processing defect in rapidly progressing idiopathic pulmonary fibrosis. PLos One 6: e21253, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, Gibson KF, Konishi K, Yousem SA, Singh M, Handley D, Richards T, Selman M, Watkins SC, Pardo A, Ben-Yehudah A, Bouros D, Eickelberg O, Ray P, Benos PV, Kaminski N. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 182: 220–229, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pandit KV, Milosevic J, Kaminski N. MicroRNAs in idiopathic pulmonary fibrosis. Transl Res 157: 191–199, 2011 [DOI] [PubMed] [Google Scholar]
- 76.Pang M, Zhuang S. Histone deacetylase: a potential therapeutic target for fibrotic disorders. J Pharmacol Exp Ther 335: 266–272, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pfeifer GP, Rauch TA. DNA methylation patterns in lung carcinomas. Semin Cancer Biol 19: 181–187, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pottier N, Maurin T, Chevalier B, Puissegur MP, Lebrigand K, Robbe-Sermesant K, Bertero T, Lino Cardenas CL, Courcot E, Rios G, Fourre S, Lo-Guidice JM, Marcet B, Cardinaud B, Barbry P, Mari B. Identification of keratinocyte growth factor as a target of microRNA-155 in lung fibroblasts: implication in epithelial-mesenchymal interactions. PLos One 4: e6718, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qiu W, Baccarelli A, Carey VJ, Boutaoui N, Bacherman H, Klanderman B, Rennard S, Agusti A, Anderson W, Lomas DA, DeMeo DL. Variable DNA methylation is associated with chronic obstructive pulmonary disease and lung function. Am J Respir Crit Care Med 185: 373–381, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rabinovich EI, Kapetanaki MG, Steinfeld I, Gibson KF, Pandit KV, Yu G, Yakhini Z, Kaminski N. Global methylation patterns in idiopathic pulmonary fibrosis. PLos One 7: e33770, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rexhaj E, Bloch J, Jayet PY, Rimoldi SF, Dessen P, Mathieu C, Tolsa JF, Nicod P, Scherrer U, Sartori C. Fetal programming of pulmonary vascular dysfunction in mice: role of epigenetic mechanisms. Am J Physiol Heart Circ Physiol 301: H247–H252, 2011 [DOI] [PubMed] [Google Scholar]
- 82.Robinson CM, Neary R, Levendale A, Watson CJ, Baugh JA. Hypoxia-induced DNA hypermethylation in human pulmonary fibroblasts is associated with Thy-1 promoter methylation and the development of a pro-fibrotic phenotype. Respir Res 13: 74, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Salam MT, Zhang Y, Begum K. Epigenetics and childhood asthma: current evidence and future research directions. Epigenomics 4: 415–429, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sanders YY, Ambalavanan N, Halloran B, Zhang X, Liu H, Crossman DK, Bray M, Zhang K, Thannickal VJ, Hagood JS. Altered DNA Methylation Profile in Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med 186: 525–535, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sanders YY, Cisneros J, Nuovo G, Selman M, Hagood JS. Epigenetic regulation mediates thy-1 gene silencing and induction of the myofibroblast phenotype. Proc Am Thorac Soc 5: 363–364, 2008 [Google Scholar]
- 86.Sanders YY, Pardo A, Selman M, Nuovo GJ, Tollefsbol TO, Siegal GP, Hagood JS. Thy-1 promoter hypermethylation: a novel epigenetic pathogenic mechanism in pulmonary fibrosis. Am J Respir Cell Mol Biol 39: 610–618, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sanders YY, Tollefsbol TO, Varisco BM, Hagood JS. Epigenetic regulation of thy-1 by histone deacetylase inhibitor in rat lung fibroblasts. Am J Respir Cell Mol Biol 45: 16–23, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci USA 100: 13225–13230, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stenvang J, Petri A, Lindow M, Obad S, Kauppinen S. Inhibition of microRNA function by antimiR oligonucleotides. Silence 3: 1, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun X, He Y, Huang C, Ma TT, Li J. The epigenetic feedback loop between DNA methylation and microRNAs in fibrotic disease with an emphasis on DNA methyltransferases. Cell Signal 25: 1870–1876, 2013 [DOI] [PubMed] [Google Scholar]
- 91.Sundar IK, Yao H, Rahman I. Oxidative stress and chromatin remodeling in chronic obstructive pulmonary disease and smoking-related diseases. Antioxid Redox Signal 18: 1956–1971, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tang X, Peng R, Phillips JE, Deguzman J, Ren Y, Apparsundaram S, Luo R, Bauer CM, Fuentes ME, Demartino JA, Tyagi G, Garrido R, Hogaboam CM, Denton CP, Holmes AM, Kitson C, Stevenson CS, Budd DC. Assessment of Brd4 inhibition in idiopathic pulmonary fibrosis lung fibroblasts and in vivo models of lung fibrosis. Am J Pathol 183: 470–479, 2013 [DOI] [PubMed] [Google Scholar]
- 94.Tang X, Peng R, Ren Y, Apparsundaram S, Deguzman J, Bauer CM, Hoffman AF, Hamilton S, Liang Z, Zeng H, Fuentes ME, Demartino JA, Kitson C, Stevenson CS, Budd DC. BET bromodomain proteins mediate downstream signaling events following growth factor stimulation in human lung fibroblasts and are involved in bleomycin-induced pulmonary fibrosis. Mol Pharmacol 83: 283–293, 2013 [DOI] [PubMed] [Google Scholar]
- 95.To M, Yamamura S, Akashi K, Charron CE, Barnes PJ, Ito K. Defect of adaptation to hypoxia in patients with COPD due to reduction of histone deacetylase 7. Chest 141: 1233–1242, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.van Rooij E. The art of microRNA research. Circ Res 108: 219–234, 2011 [DOI] [PubMed] [Google Scholar]
- 97.van Rooij E, Purcell AL, Levin AA. Developing microRNA therapeutics. Circ Res 110: 496–507, 2012 [DOI] [PubMed] [Google Scholar]
- 98.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132: 875–886, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wan ES, Qiu W, Baccarelli A, Carey VJ, Bacherman H, Rennard SI, Agusti A, Anderson WH, Lomas DA, DeMeo DL. Systemic steroid exposure is associated with differential methylation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 186: 1248–1255, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wion D, Casadesus J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nature Rev Microbiol 4: 183–192, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wright DE, Wang CY, Kao CF. Histone ubiquitylation and chromatin dynamics. Front Biosci 17: 1051–1078, 2012 [DOI] [PubMed] [Google Scholar]
- 102.Wu X, Chang MS, Mitsialis SA, Kourembanas S. Hypoxia regulates bone morphogenetic protein signaling through C-terminal-binding protein 1. Circ Res 99: 240–247, 2006 [DOI] [PubMed] [Google Scholar]
- 103.Wynn TA. Integrating mechanisms of pulmonary fibrosis. J Exp Med 208: 1339–1350, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xi Q, He W, Zhang XH, Le HV, Massague J. Genome-wide impact of the BRG1 SWI/SNF chromatin remodeler on the transforming growth factor beta transcriptional program. J Biol Chem 283: 1146–1155, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang Q, Lu Z, Singh D, Raj JU. BIX-01294 treatment blocks cell proliferation, migration and contractility in ovine foetal pulmonary arterial smooth muscle cells. Cell Prolif 45: 335–344, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang S, Banerjee S, de Freitas A, Sanders YY, Ding Q, Matalon S, Thannickal VJ, Abraham E, Liu G. Participation of miR-200 in pulmonary fibrosis. Am J Pathol 180: 484–493, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yang S, Cui H, Xie N, Icyuz M, Banerjee S, Antony VB, Abraham E, Thannickal VJ, Liu G. miR-145 regulates myofibroblast differentiation and lung fibrosis. FASEB J 27: 2382–2391, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yao H, Rahman I. Role of histone deacetylase 2 in epigenetics and cellular senescence: implications in lung inflammaging and COPD. Am J Physiol Lung Cell Mol Physiol 303: L557–L566, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhai Y, Zhong Z, Chen CY, Xia Z, Song L, Blackburn MR, Shyu AB. Coordinated changes in mRNA turnover, translation, and RNA processing bodies in bronchial epithelial cells following inflammatory stimulation. Mol Cell Biol 28: 7414–7426, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhao L, Chen CN, Hajji N, Oliver E, Cotroneo E, Wharton J, Wang D, Li M, McKinsey TA, Stenmark KR, Wilkins MR. Histone deacetylation inhibition in pulmonary hypertension: therapeutic potential of valproic acid and suberoylanilide hydroxamic acid. Circulation 126: 455–467, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ziller MJ, Muller F, Liao J, Zhang Y, Gu H, Bock C, Boyle P, Epstein CB, Bernstein BE, Lengauer T, Gnirke A, Meissner A. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLos Genet 7: e1002389, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]