Abstract
Membrane biofouling is widely acknowledged as the most frequent adverse event in wastewater treatment systems resulting in significant loss of treatment efficiency and economy. Different strategies including physical cleaning and use of antimicrobial chemicals or antibiotics have been tried for reducing membrane biofouling. Such traditional practices are aimed to eradicate biofilms or kill the bacteria involved, but the greater efficacy in membrane performance would be achieved by inhibiting biofouling without interfering with bacterial growth. As a result, the search for environmental friendly non-antibiotic antifouling strategies has received much greater attention among scientific community. The use of quorum quenching natural compounds and enzymes will be a potential approach for control of membrane biofouling. This approach has previously proven useful in diseases and membrane biofouling control by triggering the expression of desired phenotypes. In view of this, the present review is provided to give the updated information on quorum quenching compounds and elucidate the significance of quorum sensing inhibition in control of membrane biofouling.
Keywords: quorum quenching, quorum sensing inhibition, natural compounds, wastewater treatment, biofouling, membrane bioreactor.
Introduction
Wastewater treatment has become an issue of global concern as scientific community strives for ways to keep environment clean. Different conventional as well as advanced treatment processes such as activated sludge, rotating biological contactor or sequencing batch reactor and membrane bioreactor (MBR) have been in use to treat industrial and municipal wastewaters. Among them, MBR is now accepted as a technology of choice for various wastewater treatments. MBR is state-of-the-art high quality wastewater treatment technology consisted of common bioreactors with membrane filtration units for biomass retention 1. Recent technological innovations and significant membrane cost reduction have allowed the MBRs to become an established treatment system for industrial and municipal wastewaters 2. MBRs have emerged as an effective solution to transform wastewaters into high quality effluent suitable for discharge into surface waterways or to be reclaimed for irrigation purpose. The other advantages of MBR include small footprint, modular design, easy retrofit and ease of operation with high automation potential 2, 3. As a result the use of MBR for treating domestic sewage, landfill leachate, hospital wastewater, restaurant wastewater, petrochemical wastewater and high-concentration industrial wastewater has significantly increased and the widening of application areas will occur in future 4. To date, the MBR wastewater treatment technology is being successfully applied at number of locations around the world. It is expected that MBR use for wastewater treatment will be sustained in future and further acceleration will depend on its better performance. The major players involved in the commercial production of MBR unit include CNC-Siemens, Zenon-GE, Mitsubishi-Rayon, Tory, Kubota, Motimo etc 4.
Despite numerous advantages of MBR over conventional wastewater treatment processes, there are some challenges faced by technology that may restrict its wider applications. These are; pretreatment of wastewater to remove hairs, lint, fibrous materials and other debris, high operational costs due to the use of anti-fouling strategies applied to the system, lack of long-term performance, clogging of aerators and so on 5, 1. One of the major drawback and process limitation of MBR is clogging of membrane surface during the filtration. This phenomenon termed as 'membrane fouling' is the coverage of membrane surfaces by deposition of soluble and particulate materials onto and into the membrane which lead to a loss in permeability. The fouling of membrane initiated with the attachment of soluble microbial products (SMP), bacteria and other colloidal particles onto the membrane surface. The early bacterial attachment, subsequent growth and colonization across the overall membrane surface result into biofilm formation. The developed biofilms are highly heterogeneous and consist of both water-permeable and non-permeable substances. Biofilms lead to several adverse effects on MBR performance such as significant reduction in hydraulic performance, transmembrane pressure (TMP) increase, loss of system productivity, shorten membrane lifespan and increased operation cost; which makes biofouling becomes one of the most challenging issues facing further MBR developments.
To make the MBR as an option of choice for wastewater treatment, it is necessary to overcome the problem of membrane biofouling. Various perspectives including causes, characteristics, mechanism of fouling and methods of control have been investigated previously 6. More recent, different physicochemical and biological strategies have been attempted to control membrane biofouling. Most strategies for reducing MBR biofouling are focused on the physical cleaning of membrane surfaces, modification of existing membranes and incorporation of antibiotics or antimicrobial compounds in MBRs 6, 7. Different antimicrobial compounds have been extensively tried to control membrane biofouling such as nitrofurazone, chlorhexidine, silver salts, polymerized quaternary ammonium surfactants, anionic nanoporous hydrogels and antibacterial peptides 8. However, some antimicrobial compounds are also toxic to non-target organisms and pollute the aquatic environment. A major challenge presented by formed biofilms is that the bacteria living within biofilms have higher protection against antimicrobial compounds and are markedly more tolerant to such control treatments 9. Such complex structure of biofilms require excess use of antibiotics or synthetic antimicrobials which results in emergence of multi-antibiotic resistance among them. However, the control of biofouling does not mean the killing bacteria or limiting its growth but to block the expression of biofilm forming phenotypes. Quorum sensing, a way of bacterial population density-dependent cell to cell communication and phenotype regulation is known to associate with biofilm formation 10, 11. Thus unlike antibiotics, interrupting quorum sensing may represent a novel alternative approach to combat membrane biofouling.
Recently, interest in controlling membrane biofouling through use of quorum quenching mediated approaches has increased among scientific communities. Since quorum sensing controls a range of biological functions associated with biofilms, this approach has the potential in controlling membrane biofouling. In quorum quenching, the targets are not essential for bacterial survival and therefore are not subject to adverse effects observed like due to conventional antibiotics 12-15. It has been reported that a variety of natural compounds such as vanillin, furanones, flavonoids, curcumin etc. and few enzymes showed considerable quorum quenching activity against biofouling bacteria without interfering with its growth 16-21. Moreover, the concept of bead-entrapped quorum quenching bacteria has been introduced to MBR as a new biofouling control paradigm 22. In view of this, the present review is focused to cover all the important aspects of quorum quenching mediated approaches for control of membrane biofouling. Understanding the previous studies on quorum quenching compounds will help to design non-antibiotic biofouling control strategies and give pointers for best practices.
Quorum sensing system
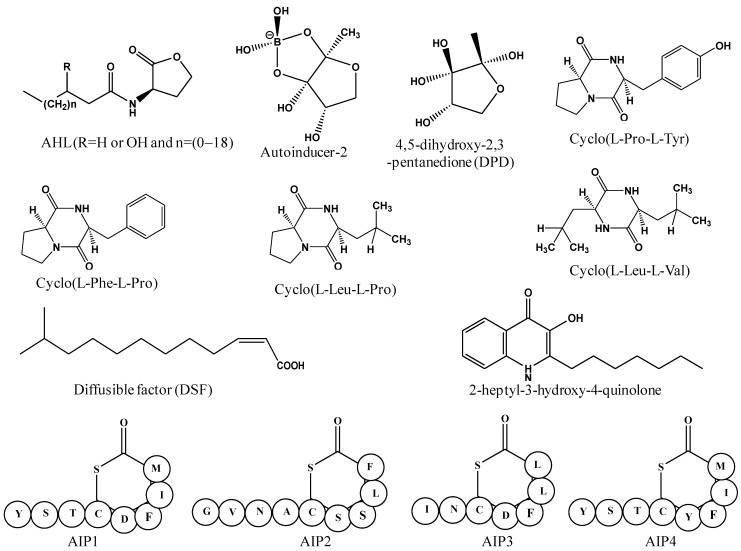
Bacteria use the language of small diffusible signaling molecules called autoinducers to communicate and assess their population densities in a process called quorum sensing. The sensing mechanism is based the synthesis, release and uptake of autoinducers in the surrounding medium, whose concentration correlates to the density of secreting bacteria in the vicinity. The basic mechanism of QS involves the interaction of autoinducer with a transcriptional regulator, either directly or through activation of a sensor kinase 23. Both the Gram-positive and Gram-negative bacteria use species-specific autoinducers for activation of quorum sensing system. A variety of quorum sensing signaling molecules functions as local sensors to communicate population densities in bacteria. These signaling molecules and their receptors have been broadly divided into three major classes: N-acyl homoserine lactones (AHLs), which vary in the length and oxidation state of the acyl side chain and produced by Gram-negative bacteria; oligopeptides or autoinducing peptides (AIP), consisting of 5-34 amino acids residues, are generally used by Gram-positive bacteria; autoinducer-2 (AI-2), a ribose derivative [4,5-dihydroxy-2,3-pentanedione] employed by both Gram-positive and Gram-negative bacteria for interspecies communication (Figure 1 and Table 1) 24, 25.
Figure 1.
Structures of bacterial quorum sensing signaling molecules, representing three major classes of autoinducers; N-acyl homoserine lactones, autoinducer-2 and autoinducing peptides 1 to 4.
Table 1.
Quorum sensing signaling molecules and phenotypes controlled in Gram-negative and Gram-positive bacteria.
| Autoinducer(s) | Producing bacteria | Phenotype(s) controlled | Ref. |
|---|---|---|---|
| Gram-negative bacteria | |||
| N-acyl homoserine lactone (AHL) | V. fischeri, P. aeruginosa, C. violaceum, Aer. hydrophila | Bioluminescence, Exopolysaccharide production, Biofilm formation, Virulence factor, Pigmentation | 30-35 |
| Autoinducer-2 (AI-2) | V. harveyi, E. coli, Y. pestis | Bioluminescence, Biofilm formation, Motility, Virulence factor | 36-39 |
| 4,5-dihydroxy-2,3-pentanedione | Sa. enteric serovar typhimurium | Virulence factor | 40, 41 |
| Cyclic dipeptides/ Diketopiperazines (DKP) (a) Cyclo(L-Pro-L-Tyr) |
P. putida WCS358, P. aeruginosa | Cross activates QS biosensors | 42, 43 |
| (b) Cyclo(L-Phe-L-Pro) | P. putida WCS358, P. aeruginosa | Cross activates QS biosensors | 42, 43 |
| (c) Cyclo(L-Leu-L-Pro) | P. putida WCS358 | Cross activates QS biosensors | 42 |
| (d) Cyclo(L-Leu-L-Val) | P. putida WCS358 | Cross activates QS biosensors | 42 |
| Quinolone (2-heptyl-3-hydroxy-4-quinolone) |
P. aeruginosa | Antibiotic production | 44-46 |
| Diffusible factor (DSF) | X. campestris | Endoglucanase production | 47 |
| Gram-positive bacteria | |||
| Autoinducing peptide (AIP1-AIP4) | Sta. aureus | Cross-signaling between strains and species, Biofilm formation, Virulence factor | 28, 29, 48, 49 |
A numbers of structurally diverse autoinducers have been identified in Gram-positive and Gram-negative bacteria. Most of them are either small (<1000 Da) organic molecules or peptides with 5-20 amino acids 26, 27. Gram-negative bacteria employ AHL, 2-heptyl-3-hydroxy-4-quinolone, fatty acid methyl esters, long-chain fatty acids and a group of inter-convertible furanones derived from DPD called AI-2. The ribose derivative AI-2 is also produced by Gram-positive bacteria, but these organisms generally prefer linear, modified or cyclic peptides AIP made by Staphylococcus sp 28, 29. The diffusible factor c-butyrolactones produced by Streptomyces sp. is structurally related to AHLs, as both classes belong to butanolides. In general, AHLs produced by Gram-negative bacteria and AIP from Gram-positive bacteria are often engaged in quorum sensing signaling and have been most intensively investigated. Many of these quorum sensing signal molecules are chemically diverse and species specific, while some of them can be recognized by inter-species communication. Such signal molecules exhibit biological properties far beyond their role in coordinating gene expression in producer strain. It is speculated that quorum sensing system allows bacteria to listen the communication signals from other bacteria and exploit this information to its own advantage.
Most of Gram-negative bacteria use AHLs to regulate quorum sensing mediated behaviors, while Gram-positive bacteria prefer linear, modified or cyclic peptides such as the AIP to control quorum sensing phenotypes. The quorum sensing systems regulate the coordination of population behavior to enhance nutrient availability, collective defense against other antagonizing organisms or community escape from adverse conditions 27. Since the discovery of quorum sensing regulation in bacteria, numerous such systems have been described. These quorum sensing systems regulates diverse functions in both Gram-negative and Gram-positive bacteria, which include biofilm formation, virulence factor, bioluminescence, motility patterns, exopolysaccharide production, antifungal or antibiotic production, endoglucanase production, pigmentation, competence, plasmid conjugal transfer, cross-signaling between strains and species etc.
AIP mediated quorum sensing in Gram-positive bacteria
Several Gram-positive bacteria are known to use modified peptides also called AIP as signaling molecules to regulate different phenotypes such as virulence (agr system in Staphylococcus sp.), bacteriocin production (pin and ssp systems in lactic acid bacteria) and competence (com system in B. subtilis) 50-52. The AIP are generated by cleavage from larger precursor peptides and subsequent modification with substitution of isoprenyl groups to form lactone and thiolactone rings 53. When the extracellular concentration of AIP becomes high, that binds to cognate membrane-bound two-component histidine kinase receptors. Which further lead into induction of receptor's kinase activity and transcription of genes in the quorum sensing regulon 54. In case of some Gram-positive bacteria, the AIP are detected by membrane-bound two component signal transduction system 55, 50; as the bacterial cell membrane is impermeable to peptides the specialized transporters are required to secrete AIP. Certain species of Gram-positive bacteria use AIP to regulate the production of virulence factors and biofilm formation 48, 49. The human pathogen Sta. aureus uses the paradigmatic Agr system to regulate adhesion and production of virulence factors 54. It is also reported that AIP-mediated quorum sensing has been used to regulate bacterial competence and conjugation in Sta. aureus and Ent. faecalis respectively 56. Thus, the evolution of AIP-mediated quorum sensing system in pathogenic bacteria could have been an effective therapeutic strategy for the control of virulence and biofilms in diseases. However, there is one report available suggesting the involvement of AI-2 mediated quorum sensing in membrane biofouling; where reduced AI-2 was positively correlated to the reduced fouling resistance of nylon membranes 57.
AHL mediated quorum sensing in Gram-negative bacteria
The best-studied quorum sensing systems in Gram-negative bacteria use LuxI-type enzymes, which produce AHLs as small diffusible signal molecules that get bind and activate members of the LuxR transcriptional activator protein family 58, 59. AHL based quorum sensing system functions through three key components: i) AHL signal molecules, ii) AHL synthase protein for synthesis of AHL signals, and iii) a regulatory protein which responds to surrounding concentration of AHL signal 60. This process initiated with the synthesis and release of AHL signals into the surrounding environment which accumulates in a cell-population-density-dependent manner. When the concentration of AHL signals reaches at higher level; the quorum sensing cells starts responding allowing them to regulate the production of secondary metabolites and control the expression of quorum sensing genes. A majority of Gram-negative bacteria regulates various phenotypes through the secretion and detection of such signaling molecules. However the efficacy of expression of quorum sensing phenotypes depends upon the presence or absence of surrounding cells. Using quorum sensing bacteria can act to express a specific set of genes responsible for variety of physiological behaviors including bioluminescence, antibiotic production, extracellular polymer production, biosurfactant synthesis, sporulation, release of virulence factors and biofilm formation (Figure 2) 61-66.
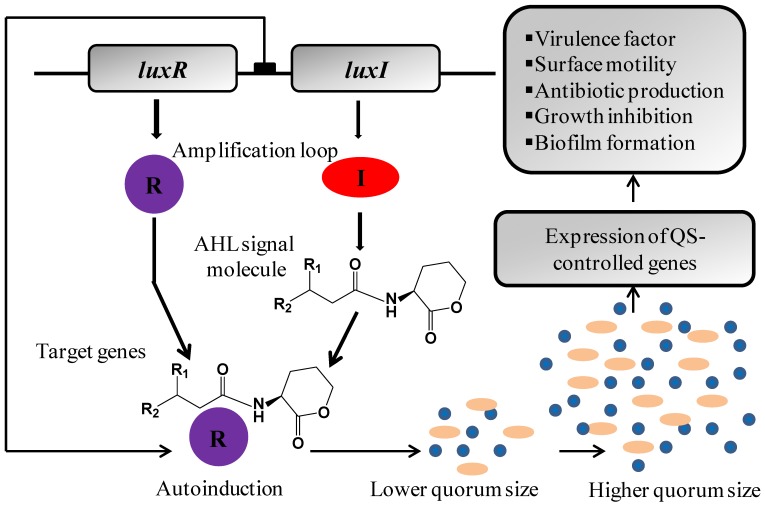
Figure 2.
The LuxR/AHL-mediated quorum sensing regulation of multiple gene expressions in Gram-negative bacteria. The 'R' and 'I' genes are homologues of the LuxR and LuxI genes in which the 'R' protein is the AHL receptor and signal transducer while I protein is AHL signal synthase. The I protein is responsible for the production of AHLs. After synthesis, AHLs get diffuse (short chain) or pumped out (long chain) of the bacterial cell into the surrounding medium prior to being taken up into nearby bacterial cells. The AHL activate R protein by direct binding to make AHL/R protein complex which rapidly increases I gene expression and hence AHL production. At a certain level of bacterial cells, the quorum sensing system becomes fully activated which leads to R-mediated expression of quorum sensing target genes.
The use of biosensor systems to detect quorum sensing signaling molecules has led to the discovery of broad range of AHLs as produced by diverse Gram-negative proteobacteria belonging to α, β, and γ subdivisions 26. The AHLs biosensors system consist of a quorum sensing-controlled promoter fused to a reporter such as lacZ or lux or gfp operon 67, 68, 34 or pigment induction e.g. violacein in C. violaceum 31. These biosensor strains have a functional R protein but lack the AHL synthase; thus the promoter activity depends on the presence of exogenous AHL 69. Use of biosensor strains revealed that wide genera of Gram-negative bacteria produce a broad range of AHL molecules ranging from C4 to C18-carbon acyl side chains and either an oxo, a hydroxy, or no substitution at the third carbon. The acyl chain varies in length, satuaration level, and oxidation state. In most cases, the acyl chain has an even number of carbon (C4-C18), although some AHLs with odd carbon (C5-C7) have been investigated 70, 71. Examples of AHLs producing bacteria includes species of Aeromonas, Acidithiobacillus, Acinetobacter, Agrobacterium, Brucella, Burkholderia, Erwinia, Enterobacter, Chromobacterium, Mesorhizobium, Pseudomonas, Ralstonia, Rhodobacter, Rhizobium, Serratia, Sinorhizobium, Vibrio and Yersinia 27. Many of these bacteria have ability to produce multiple AHLs due to the presence of more AHL synthase and thus can regulate different phenotypes.
Quorum sensing and biofouling
Quorum sensing and biofilm formations are the central and interconnected feature of bacterial social life 72-74; which enables bacteria to organize their activities at the population level and switch from acting as individual cells to concentrated multi-cellular structure in the form of biofilms 75. Biofilms are the matrix enclosed bacterial cells attaching to each other or to surfaces 76. Such complex multilayer structure of defined architecture helps bacterial communities to live in a sessile and protected environment 77, 78. The formation of biofilm is linked to a number of interacting processes beginning with secretion of signal molecules, solute diffusion, cell to cell or cell to solute interactions, EPS matrix production, colonization, surface attachment and maturation 79.
The bacterial biofilms are ubiquitous in nature and exist on all type of surfaces in water and wastewater treatment systems, which may play beneficial as well as detrimental roles. The presence of biofilms or attached bacterial cells on filtration membranes is highly prevalent and often adversely affects its efficiency. The robust biofilms are needed for treatment of wastewater by trickling filters, granular sludge and moving bed biofilm reactors 6, 80. However, deposition of biofilms and associated extracellular polysaccharides (EPS) on membrane surfaces decreases filtration flux and causes permeability loss 81, 82. The non-degraded organic matters, microbial flocs and SMP are also considered as potential foulants 83, 84, but the formation of biofilms on membrane surfaces is the major contributor in membrane biofouling and is the main obstacle restricting the development of MBR technology for advanced wastewater treatment.
The role of AHL-mediated quorum sensing in the development of specialized biofilm structure is best understood in P. aeruginosa 85. The genes and quorum sensing regulatory circuits associated with initial cell-surface interactions and biofilm maturation are also identified in some bacteria 77. Numerous links have been made between quorum sensing and biofilm formation, which includes direct demonstration of Aeromonas sp., Pseudomonas sp. and Xanthomonas sp. AHLs-mediated phenotypes important to biofilm formation in water treatment systems 85-88. The relationship between P. putida adhesions to different membrane surfaces was found to be a AHLs-mediated quorum sensing trait 89. Xia et al. 90 demonstrated that diffusion of exogenous 3-oxo-C8-HSL increased the growth rate of P. aeruginosa cells on ultra-filtration membrane biofilm and had no influence on EPS of biofilm. The quorum sensing system might enhance the growth of neighboring bacterial cells in contact with membrane surfaces into biofilm and may influence the structure and organization of biofilm. The results of the study also showed that quorum sensing system has a significant relation with protein production. A correlation between AHLs production and biofilm formation was reported among activated sludge bacterial isolates; where biosensor assay with C. violaceum 026 and A. tumefaciens A136 confirms the ability of Aeromonas, Enterobacter, Serratia, Leclercia, Pseudomonas, Klebsiella, Raoultella and Citrobacter sp. to produce AHLs and form biofilms on polystyrene surface 91.
In addition to this, several studies have linked presence of AHLs with biofilm formation and addressed the problem of membrane biofouling in MBRs 82, 89, 92. The chromatographic analysis of MBR sludge using HPLC technique has identified the presence of several AHLs molecules. Recently HPLC analysis of MBR sludge has suggested the presence of 3-oxo-C8-HSL and C8-HSL in MBR activated sludge 22. Moreover, HPLC characterization by Lade et al. 91 detected eight different AHLs v.z. C4-HSL, C6-HSL, C8-HSL, 3-oxo-C8-HSL, C10-HSL, C12-HSL, 3-oxo-C12-HSL and C14-HSL in activated sludge collected from MBR treating wastewater. Yeon et al. 93 detected C6-HSL and C8-HSL from mixed cultured biocake derived from fouled MBR treating wastewaters. These studies suggest that AHLs-mediated quorum sensing is extensively regulating biofilm formation. Though AHLs have been detected in activated sludge and biofouling bacteria, the precise role in membrane biofouling is not clear. Further investigation need to be carried out to understand the exact role of AHLs in various stages of biofouling. Still, membrane biofouling is persistent problem in MBR treating wastewaters and interfering with quorum sensing system may eliminate it and thus increase its efficiency.
Quorum quenching and biofouling control
The quorum quenching refers to a process by which autoinducer-mediated quorum sensing is interrupted 94. Quorum sensing helps bacteria to coordinate community-based behavior, but it is not essential for survival or growth. Thus, interference with quorum sensing may lead to the inhibition of desired phenotypes such as formation of biofilms. The large numbers of aquatic bacteria are Gram-negative and employ AHLs-mediated quorum sensing as their major language to coordinate population behavior 95, 10. Since quorum sensing is involved in formation of biofilms, targeting quorum sensing has offered a novel way to combat membrane biofouling without killing or inhibiting bacterial growth 96.
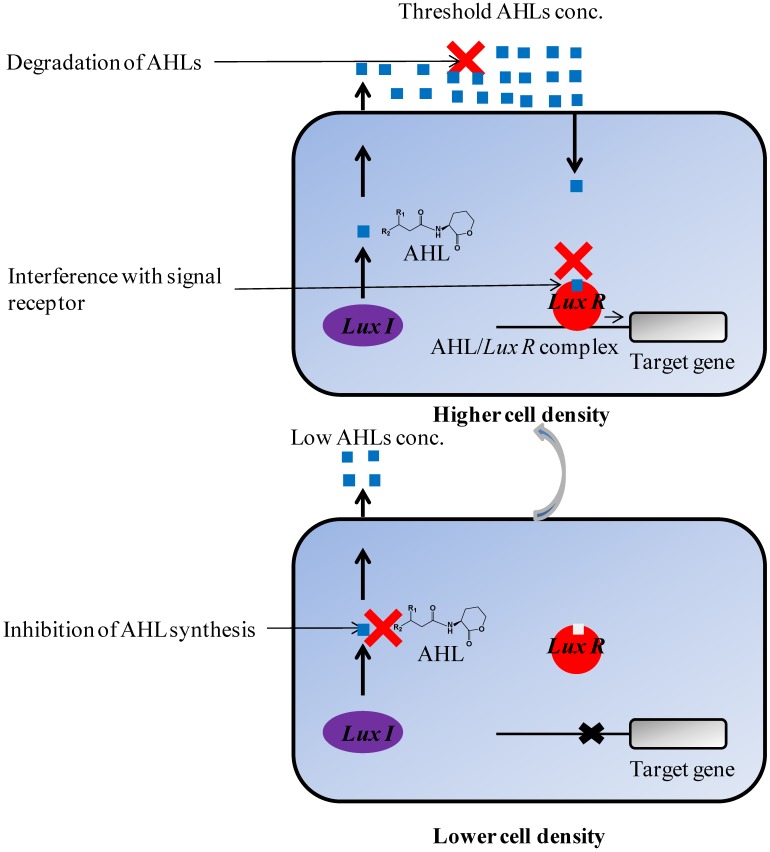
There are several quorum quenching strategies available through which the process of quorum sensing can be interrupted which includes; i) Inhibition of AHL synthesis by blocking the LuxI-type synthase proteins 97, 98 ii) Enzymatic destruction of AHLs molecules by AHL-acylase and AHL-lactonase that will prevent them from accumulating 99, 33, 100 and iii) Interference with signal receptors or blockage of formation of AHL/LuxR complex 101, 102 (Figure 3). In addition to this, quorum quenching has previously proven to be a primarily target both for quorum sensing signal synthase and sensor or response regulator proteins involved 12-15. These strategies can be applied to achieve inhibition of AHLs-mediated quorum sensing in Gram-negative and AIPs-mediated quorum sensing in Gram-positive bacteria. However, as the membrane biofouling is mainly associated with AHLs-mediated quorum sensing, our focus in this review is on inhibition of AHLs-mediated quorum sensing.
Figure 3.
Inhibition of quorum sensing in Gram-negative bacteria by various mechanisms. Three quorum quenching strategies have been used for attenuating AHL-mediated phenotypes; (i) Inhibition of AHL synthesis (ii) Degradation of AHL signal molecules (iii) Interference with signal receptor.
The traditional practices of biofouling control are based on antimicrobial compounds or antibiotics that kill or inhibit bacterial growth. However bacterial survival is essential for decomposition of contaminants and a major concern with the use of antimicrobial agent is development of multi-drug resistance among bacteria exposed 103. This leads the way for an intense search of non-antibiotic compounds that can specifically block AHL-mediated biofouling trait without interfering with growth. It is envisioned that some natural compounds and enzymes can control the AHLs-mediated biofilm formation without affecting bacterial growth and also reduce the risk of multi-drug resistance.
Natural compounds as QSI
Many natural compounds of plant origin are well known for antimicrobial activities 104, and also shown to inhibit quorum sensing while not affecting bacterial growth. The mammalian cells, plant and algae are known to secrete the novel class of such non-antibiotic quorum quenching compounds which either activate or inhibit bacterial quorum sensing 105, 106. Traditional medicinal plants are one of the most promising sources in search for natural quorum sensing inhibitory compounds. Several compounds of natural origin which can interfere with bacterial quorum sensing system and inhibit biofilm formation have been identified as secondary metabolites produced by algae, fungi, bacteria and higher plants. A major advantage of natural compounds is that it ruins the problem of bacterial resistance to conventional antibiotics, as it specifically interferes with expression of specific traits. The natural compounds are structurally similar to those of quorum sensing signal molecules and thus antagonize them and also have ability to degrade LuxR/LasR signal receptors 107, 108. A summary of the known natural quorum sensing inhibitory compounds derived from plant, fungi, algae and bacteria is provided in Table 2.
Table 2.
Natural compounds as quorum sensing inhibitors.
| Natural compound(s) | Source | QS activity | Ref. |
|---|---|---|---|
| Furanone/ 2(5H)-Furanone/ | Macroalga (Delisea pulchra) | Mimics AHL signal by occupying the binding site on putative regulatory protein which results in the disruption of QS-mediated gene regulation. Inhibit biofilm formation in Aer. hydrophila | 109, 17 |
| Repress LuxR protein dependent expression of P(luxI)-gfp(ASV) reporter fusion. Inhibit virulence factor in E. coli XL-1. | 110 | ||
| (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone. | Macroalga (Delisea pulchra) | Disrupts QS-regulated bioluminescence in V. harveyi by interacting with Hfq protein. Inhibit swarming motility and biofilm formation in E. coli | 111, 112 |
| Ajoene (1-Allyldisulfanyl-3-(prop-2-ene-1-sulfinyl)-propene) |
Garlic extract (Allium sativum) | Blocks the QS-regulated productions of rhamnolipid resulting in phagocytosis of biofilm. Targets Gac/RSM part of QS and lowers the expression of regulatory RNAs in P. aeruginosa PAO1 | 113, 114 |
| Iberin (1-Isothiocyanato-3-(methylsulfinyl)propane) |
Horseradish extract (Armoracia rusticana) | Inhibit expression of QS-regulated lasB-gfp and rhlA-gfp genes responsible for virulence factor in P. aeruginosa | 115 |
| Sulforaphane (1-Isothiocyanato-4-(methylsulfinyl)butane) |
Boroccoli | Reduce the expression of lasI-luxCDABE reporter in P. aeruginosa | 116 |
| Erucin (4-methylthiobutyl isothiocyanate) | Boroccoli | Reduce the expression of lasI-luxCDABE reporter in P. aeruginosa | 116 |
| Naringin (4'5-diOH-Flavone-7-rhgluc) |
Citrus extract | Decrease the QS mediated biofilm formation and swimming motility in Y. enterocolitica | 18 |
| Naringenin (4',5,7-Trihydroxyflavanone) |
Malagasy bark extract (Combretum albiflorum) | Reduces production of pyocyanin and elastase in P. aeruginosa PAO1. Also inhibit 3-oxo-C12-HSL and C4-HSL synthesis driven by lasl and rhll genes | 117, 118 |
| Taxifolin/ Distylin (dihydroquercetin) |
Malagasy plant extract (Combretum albiflorum) | Reduces production of pyocyanin and elastase in P. aeruginosa PAO1 | 117 |
| Morin (2',3,4',5,7-Pentahydroxyflavone) | Grapefruit (Artocarpus heterophyllus) | Inhibit LasR and RhlR dependent protease, elastase and hemolysin in P. aeruginosa PAO1 | 119, 120 |
| Patulin/ Clavacin (4-Hydroxy-4H-furo[3,2-c]pyran-2(6H)-one) |
Penicillium sp. | Targets the RhlR and LasR proteins. Down-regulates QS genes for biofilm formation and virulence in P. aeruginosa | 121 |
| Penicillic acid (3-Methoxy-5-methyl-4-oxo-2,5-hexadienoic acid) |
Penicillium sp. | Down-regulates QS genes for biofilm formation in P. aeruginosa | 121 |
| Vanillin (4-Hydroxy-3-methoxybenzaldehyde) |
Vanilla beans extract (Vanilla planifolia Andrews) | Interfere with AHL receptors. Inhibit C4-HSL, C6-HSL, C8-HSL, 3-oxo-C8-HSL. Inhibit biofilm formation in Aer. hydrophila | 122, 123, 16 |
| Agrocinopine B ( [(3S,4R,5R)-3,4,5,6-tetrahydroxy-2-oxohexyl] [(2R,3S,4S)-3,4,5-trihydroxy-1-oxopentan-2-yl] hydrogen phosphate) | Crown gall cells | Control conjugation of pTiC58 by regulating exprssion of the arc operon in A. tumefaciens | 124 |
| L-canavanine (L-α-Amino-γ-(guanidinooxy)-n-butyric acid) |
Seed exudates (Medicao sativa) | Inhibit the expression of QS-regulated phenotype exopolysaccharide II production in Si. meliloti | 125 |
| Gamma-aminobutyric acid (GABA) ( 4-Aminobutanoic acid) |
Plants (Arabidopsis sp.) | Induce the expression of attKLM operon to stimulate inactivate 3-oxo-C8-HSL by A. tumefaciens lactonase AttM | 126, 127 |
| Rosmarinic acid (R-O-(3,4-Dihydroxycinnamoyl)-3-(3,4- dihydroxyphenyl) lactic acid) | Sweet basil (Ocimum basilicum) | Inhibit protease, elastase, hemolysin production, biofilm formation and virulence factor in P. aeruginosa | 119, 108, 128 |
| Salycilic acid (2-Methyl-5-tert-butylsalicylic acid) | Plant phenolic secondary metabolite | Inhibit the expression of vir regulon in A. tumefaciens. Also stimulates AHL-lactonase expression which degrades AHLs. | 129 |
| Chlorogenic acid (3-Caffeoylquinic acid) | Plant extract (Moringa oleifera) | Inhibit QS-regulated violacein production in C. violaceum 12472 | 130 |
| Allin (2-Amino-3-[prop-2-ene-1-sulfinyl]-propionic acid) |
Garlic extract (Allium sativum) | Inhibit QS-regulated gene expression by interacting with receptors in P. aeruginosa and make biofilm sensitive to antibiotics. | 113, 131 |
| Ursolic acid (3beta-Hydroxyurs-12-en-28-oic acid) | Plant extract (Sambucus chinesis) | Inhibit biofilm formation by suppressing cystenine synthesis in E. coli | 132, 133 |
| Ellagic acid (Benzoaric acid) | Fruit extract of Terminalia chebula Retz. | Down-regulate the expression of virulence gene in P. aeruginosa PAO1. Reduces biofilm formation and swarming motility in B. cepacia | 134, 135 |
| α-Hydroxybutyric acid (2-hydroxy-butanoic acid) | Arabidopsis exudates | Induce the expression of attKLM-lacZ fusion in A. tumefaciens | 136 |
| Epigallocatechin gallate (Epigallocatechol) | Green tea (Camellia sinensis L.) | This compound has gallic acid moiety and specifically block AHL-mediated biofilm formation in Sta. aureus and B. cepacia. Inhibit transfer of conjugative R plasmid in E. coli | 135, 137-139 |
| Pyrogallol (1,2,3-Trihydroxybenzene) |
Plant extract (Punica granatum) | Inhibit AI-2 mediated bioluminescence in V. harveyi | 140, 141 |
| Cinnamon oil/ Cynnamaldheyde ( trans-Cinnamaldehyde) | Cinnamomum zeylanicum | Interfere with AI-2 based QS and decreases the DNA-binding ability of LuxR protein to reduce virulence in V. spp. Reduces LuxR-mediated transcription from the PluxI promoter which influences biofilm formation in P. aeruginosa | 142, 143 |
| Furocoumarin/ Psoralen (7H-Furo[3,2-g][1]benzopyran-7-one) |
Grapefruit juice and extract (Psoralea corylifolia L.) | The structural resemblance of furan moiety results in QS-mediated inhibition of biofilm formation in E. coli. Inhibit QS-mediated swarming motility in P. aerugionsa PAO1 | 144, 145 |
| Urolithin (3,8-Dihydroxy-benzo[c]chromen-6-one) |
Ellagitannin-rich extract from Pomegranate | Inhibit C6-HSL and 3-oxo-C6-HSL associated biofilm formation in Y. enterocolitica. Inhibit QS-mediated swarming motility in E. coli and P. aerugionsa PAO1 | 146, 147 |
| Curcumin (E,E)-1,7-bis(4-Hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione | From Curcuma longa | Down-regulates virulence factors and biofilm initiation genes in P. aerugionsa PAO1 and inhibit its phenotype expression. | 19 |
| Attenuate QS-dependent EPS production, swarming motility and biofilm formation in uropathogenic E. coli, P. aerugionsa, Pr. mirabilis and S. marcescens. | 148 | ||
| α-D-galactopyranosyl-(1→2)-glycerol (floridoside) (N), Betonicine (O), and Isethionic acid |
Red alga (Ahnfeltiopsis flabelliformis) | Inhibit C8-HSL mediated QS in A. tumefaciens NTL4 | 149 |
| Musaceae | Musaceae extract (Musa paradiciaca) | Inhibit QS-mediated elastase production and biofilm formation in P. aerugionsa PAO1 | 150 |
| Garlic | Garlic extract | Interferes with expression of QS-controlled virulence genes in P. aeruginosa | 121 |
| Piper betle | Piper betle extract | Inhibit QS-mediated biofilm formation in P. aeruginosa | 151 |
| Cuminum cyminum | Cuminum cyminum extract | Reduce LuxR dependent biofilm formation and swarming motility of P. aerugionsa | 152 |
A promising group of natural QSI is the halogenated furanones produced by the Australian red alga, Delisea pulchra which inhibit AHLs-mediated gene expression by interfering with AHL signal from its reporter protein 153, 110. Halogenated furanones are also known to inhibit quorum sensing by destabilizing and accelerating the turnover of LuxR which then impairs its ability to bind DNA and initiate transcription 154. It has now been shown that furanones inhibits AHLs as well as AI-2 based quorum sensing system as they are structural mimics of lactones and tetrahydrofuran rings of quorum sensing system 112. The marine alga Delisea pulchra produces furanones in central vesicle gland cells and secrete to the fronds to prevent bacterial colonization and thereby macro-fouling 155. Furanones are structural analogues to short-chain AHLs and appears to interact directly with LuxR-type receptors 156. Thus, they inhibit AHL-mediated quorum sensing and subsequent biofilm formation in some Gram-negative bacteria such as P. aeruginosa, V. harveyi and E. coli 122, 157-160, 112. A natural compound (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone produced by marine alga has been shown to inhibit swarming motility and biofilm formation in E. coli at concentration non-lethal to planktonic growth 112, 111.
Flavonoids are widely distributed in the plant kingdom and are known for their numerous and determinant roles in plant physiology, metabolism and development of plant-rhizobia interactions. For the human health perspective, flavonoids have shown their roles as anti-oxidant, anti-inflammatory and anticancer agents 161. In addition to these health benefits, flavonoids such as kaempferol, naringenin, quercetin and apigenein have been reported as inhibitors of AHL and AI-2 mediated pathogenic traits in P. aeruginosa PAO1, E. coli O157:H7 and V. harveyi 117, 162, 163. Quercetin and naringenin were found to inhibit quorum sensing based biofilm formation in V. harveyi BB886 and E. coli O157:H7 163. Flavanones naringenin and taxifolin derived from Malagasy plant Combretum albiflorum attenuates the production of QS-controlled virulence factors in P. aeruginosa PAO1 117. Moreover, Flavan-3-ol catechin from the bark of same plant reduce the production of quorum sensing regulated virulence factors- pyocyanin, elastase and biofilm formation in P. aeruginosa PAO1 without affecting growth 118. In addition, reduction in expression of several quorum sensing controlled genes i.e. lasI, lasR, rhlI, rhlR, lasA, lasB, phzA1 and rhlA was also reported with these compounds. Truchado et al. 18 reported the quorum sensing inhibitory effects of an orange extract enriched in O-glycosylated flavanone naringin, which diminished the levels of lactones secreted by pathogenic Y. enterocolitica and decreased quorum sensing mediated biofilm maturation without affecting bacterial growth. The naringin was also found to inhibit swimming motility and induce the transcription levels of yenR, flhDC, and fliA in Y. enterocolitica.
A sulfur-rich QSI compound ajoene have been identified from garlic extract, which can block the quorum sensing regulated production of rhamnolipids resulting in phagocytosis of biofilms. This compound also targets Gac/RSM part of quorum sensing system and lowers the expression of two small regulatory RNAs, RsmY and RsmZ in Pseudomonas aeruginosa PAO1 114. Another isothiocyanate containing sulfur-rich compound iberin extracted from horseradish has shown strong QSI activity 115. A recent study identified sulforaphane QSI from broccoli extract, which covalently bind to cys79 residue in the 3-oxo-C12-HSL binding pocket of LasR in P. aeruginosa 116, 164. An arginine analog L-Canavanine derived from seeds exudates of Medicao sativa has shown to serve as a nitrogen source for seed germination and also inhibit growth of certain bacteria and phytophagous insects 165. This compound incorporates in place of L-arginine into nascent protein chains during synthesis and results in altered protein structure and function leading to death of target bacteria 165, 125.
More recently, various plants extract including habanero (chilli), tomato, crown vetch, soybean, water lily, Daucus carota subsp. sativu, Medigo sativa, Pisum sativum seedling, Allium cepa, Allium sativum, Lycopersicum esculentu, Medicago truncatula, vanilla, Piper betle, Cuminum cyminum and some medicinal plants of southern Florida were found to possess anti quorum sensing activities 121, 156, 166, 110, 167, 151, 152, 168. A cyclic disulphur compound having strong antagonistic effect on LuxR-based quorum sensing has been identified from garlic extract 121, 169. Furocoumarin derived from grapefruit was found to inhibit quorum sensing mediated biofilm formation in E. coli and swarming motility in P. aerugionsa PAO1 144, 145. Limonoids from sour orange seeds such as isolimonic acid, ichangin and deacetyl nomilinic acid 17 β-D-glucopyranoside were found to inhibit AI-2 mediated quorum sensing in V. harveyi 170. The structural resemblance of furocoumarin and limonoids with autoinducers molecule in the furan moiety was found to be responsible for competitive quorum sensing inhibition 171, 172.
It is expected that fungi can produce QSI compounds as they have co-existed with quorum sensing bacteria from millions of years. In a recent study two QSI compounds were identified as penicilic acid and patulin produced by Pe. radicicola and Pe. coprobium respectively 121. The DNA microarray-based transcriptomics showed that penicilic acid and patulin respectively targets 60% and 45% quorum sensing genes in P. aeruginosa suggesting the RhlR and LasR quorum sensing regulators 173, 121.
Therefore natural QSI offer a potential solution to the multidrug resistance associated with traditional antibiotics or antimicrobial compounds. Several comprehensive reviews on natural compounds that antagonize quorum sensing are available 174, 161. However, further investigation is required to identify the potential QSI present in natural sources and mechanism they employ to antagonize quorum sensing. Use of natural QSI may lead to the development of novel non-antibiotic agent which will target at the inhibition of desired traits like biofilm formation or virulence factor rather than killing the bacteria.
Natural compounds for control of membrane biofouling
Natural QSI have advantages of nontoxicity, high antifouling potential and low risk of bacterial resistance development. These compounds have mostly been applied to pathogenic bacteria; however some recent studies showed that control of membrane biofouling could also be achieved by incorporation of natural QSI. An early example of the addition of natural quorum sensing compound in a CDC biofilm reactor (Center for Disease Control) was provided by Poonusamy et al. 175. The author successfully used vanillin for combating Aer. hydrophila biofilms on five different membrane surfaces. Vanillin was also reported to inhibit the short and long-chain AHL-mediated quorum sensing leading to the reduction of Aer. hydrophila biofilm on polystyrene surface 16. The Piper betle extract has been found to mitigate membrane biofouling in ultrafiltration MBR treating textile effluent 176. In another study Piper betle extract has shown the reduction in biofilm formation and EPS production caused by P. aeruginosa and bacterial consortium without raising the selective pressure for the growth of microorganisms 151. These results reveal that Piper betle extract could be used as a potential anti-quorum sensing agent for mitigation of membrane biofouling. The correlation between membrane biofouling and quorum sensing activity demonstrated that Piper betle extract could inhibit AIs production and reduce EPS and biofilm formation 177. These evidences suggest that incorporation of natural QSI on membrane surfaces and addition in MBRs could be an effective strategy for control of membrane biofouling. Such artificial quorum sensing regulatory systems might help to mimic the problem of membrane biofouling without disturbing bacterial growth. Thus, engineered membranes with natural compounds are expected to be very useful in plant scale MBR and in designing of wastewater treatment systems with economic feasibility.
Enzymes as QSI
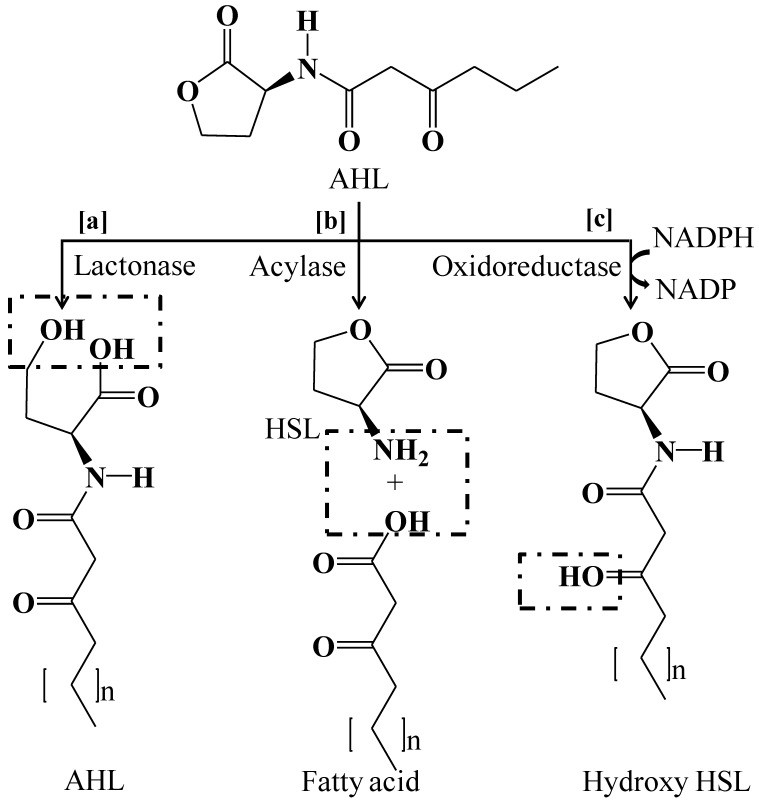
Complete degradation or inactivation of AHL signal molecules can be achieved by quorum quenching enzymes. Two major classes of enzymes that degrades AHLs signal molecules are reported, which includes lactonases that open the homoserine (HSL) ring 178, 158, 179 and acylases that cleave the acyl side chain from the HSL ring 99, 180. Another class of enzyme oxidoreductase has also been known to catalyze the oxidation or reduction of acyl side chain 181, 182 (Figure 4).
Figure 4.
Degradation mechanism of quorum sensing signal molecule N-acyl homoserine lactone by quorum quenching enzymes. (a) Lactonase open the HSL ring (b) Acylase cleaves the acyl side chain from HSL ring or hydrolyze the amide linkage (c) Oxidoreductase catalyzes the oxidation or reduction of acyl side chain.
In nature, several AHLs degrading enzymes have been reported from large number of bacteria, fungi, plants and legumes. A broad spectrum of AHLs degrading AHL-acylases are produced by Ralstonia sp. XJ12B and P. aeruginosa PAO1 99, 183. Few examples of bacterial AHL acylases reported includes AiiD from Ralstonia sp. XJ12B 99, AhlM from Streptomyces sp. 184, PvdQ and QuiP from P. aeruginosa PAO1 183, 185, 33 and AiiC from Anabaena sp. PCC7120 185. The acylases HacA and HacC produced by P. syringae B728a have shown to degrade quorum sensing signal AHLs 187. AHL-acylase produced by Streptomyces sp. has been shown to possess an ability to degrade specific AHLs having 6 or more acyl chains 184.
The other quorum quenching enzyme AHL-lactonases have been reported from various bacteria and fungi. The most promising bacteria producing AHL-lactonase are strains belonging to diverse Bacillus sp. such as B. cereus, B. subtilits and B. thuringiensis 188-191. However, B. thuringiensis present a unique property as it does not produce quorum sensing signals but has AHL-lactonase activity 192. The other bacterial lactonases includes AttM from A. tumefaciens c58 179, AiiA from Bacillus sp. 240B1 158, AiiB from A. tumefaciens 193, QIcA from Acidobacteria sp. 194 and AidH from Ochrobactrum sp. T63 195. A fungal quorum quenching enzyme gluconolactonase has been reported from As. niger IAM 2094 196. The legume alfalfa, clover, lotus, peas and yam bean shown to contains AHL degrading enzyme lactonase 197-199. A summary of the known quorum quenching enzymes is provided in Table 3.
Table 3.
Enzymes as quorum sensing inhibitors.
| Enzyme class/ Name | Source/ Producing strain | AHLs degradation | Ref. |
|---|---|---|---|
| AHL-acylase | |||
| Acylase I | Porcine (Kidney) | C4-HSL, C6-HSL, C8-HSL, 3-oxo-C10-HSL, 3-oxo-C12-HSL | 88, 200, 201 |
| C4-HSL, C6-HSL, C8-HSL | 20, 202-204 | ||
| AiiD | Ralstonia sp. XJ12B | 3-oxo-C8-HSL, 3-oxo-C10-HSL, 3-oxo-C12-HSL | 99 |
| AiiC | Anabaena sp. PCC7120 | C4-HSL ⁓ C14-HSL | 186 |
| PvdQ | Pseudomonas sp. strain PAI-A | C10-HSL, 3-oxo-C10-HSL, C12-HSL, 3-oxo-C12-HSL, C14-HSL, C16-HSL | 183, 33 |
| HacA | P. syringae strain B728a | C8-HSL, C10-HSL, C12-HSL | 187 |
| HacB | P. syringae strain B728a | C6-HSL, C8-HSL, C10-HSL, C12-HSL | 187 |
| Aac | R. solanacearum GMI1000 | C7-HSL, C8-HSL, 3-oxo-C8-HSL, C10-HSL | 205 |
| Aac | Shewanella sp. strain MIB015 | C8-HSL, C10-HSL, C12-HSL | 206 |
| AhlM | Streptomyces sp. strain M664 | C8-HSL, C10-HSL, 3-oxo-C12-HSL | 184 |
| QuiP | P. aeruginosa | C6-HSL, C8-HSL, C10-HSL, C12-HSL | 207 |
| n.d. | Pseudomonas sp. 1A1 | C6-HSL, C8-HSL, 3-oxo-C8-HSL, 3-oxo-C10-HSL, C10-HSL, 3-oxo-C12-HSL, C12-HSL | 208 |
| AHL-lactonase | |||
| Lactonase | Human (Airway epithelia) | 3-oxo-C12-HSL | 209 |
| Gluconolactonase (GL) | As. niger IAM 2094 | Lactone ring hydrolysis | 196 |
| AttM | A. tumefaciens c58 | 3-oxo-C8-HSL | 179 |
| AiiA | Bacillus sp. 240B1 | C8-HSL | 158, 210 |
| B. anthracis (Ames strain) | C6-HSL, C8-HSL, C10-HSL | 211 | |
| B. cereus and B. mycoides | C6-HSL, C8-HSL, C10-HSL | 189 | |
| B. thuringiensis | 3-oxo-C6-HSL, C6-HSL, C8-HSL | 212 | |
| AiiB | A. tumefaciens C58 | 3-oxo-C6-HSL, C6-HSL, C8-HSL, C7-HSL, 3-oxo-C8-HSL, C8-HSL | 213 |
| AiiB | A. tumefaciens | C4-HSL, 3-oxo-C6-HSL, C6-HSL, 3-oxo-C8-HSL, C8-HSL, C10-HSL | 193 |
| GKL |
Ge. kaustophilus strain HTA426 |
C6-HSL, 3-oxo-C8-HSL, C8-HSL, C10-HSL, 3-oxo-C12-HSL | 214 |
| AiiM | M. testaceum StLB037 | 3-oxo-C6-HSL, C6-HSL, 3-oxo-C8-HSL, C8-HSL, 3-oxo-C10-HSL, C10-HSL |
215 |
| MCP | My. avium subsp. paratuberculosis K-10 | C7-HSL, C8-HSL, 3-oxo-C8-HSL, C10-HSL, C12-HSL | 216 |
| PPH | My. tuberculosis | C4-HSL, 3-oxo-C8-HSL, C10-HSL | 217 |
| AidH | Ochrobactrum sp. T63 | C4-HSL, C6-HSL, 3-oxo-C6-HSL, 3-oxo-C8-HSL, C10-HSL | 195 |
| AhlS | So. silvestris StLB046 | C10-HSL | 218 |
| SsoPox | Sul. solfataricus strain P2 | 3-oxo-C8-HSL, C8-HSL, 3-oxo-C10-HSL, 3-oxo-C12-HSL | 219, 220 |
| QsdA | Rho. erythropolis W2 | 3-oxo-C6-HSL, C6-HSL, 3-oxo-C8-HSL, C8-HSL, 3-oxo-C10-HSL, C10-HSL, 3-oxo-C12-HSL, C12-HSL, 3-oxo-C14-HSL, C14-HSL |
221 |
| QIcA | Acidobacteria sp. | 3-oxo-C6-HSL, C6-HSL, C7-HSL, 3-oxo-C8-HSL, C8-HSL, 3-oxo-C10-HSL, C10-HSL | 194 |
| AhlD | Arthrobacter sp. IBN110 | C4-HSL, 3-oxo-C6-HSL, C6-HSL, C8-HSL, 3-oxo-C10-HSL, C10-HSL | 222 |
| Oxidoreductase | |||
| P450BM3 |
B. megaterium CYP102A1 |
Oxidizes; C12-HSL, 3-oxo-C12-HSL, C14-HSL, 3-oxo-C14-HSL, C16-HSL, C18-HSL, C20-HSL. | 181 |
| n.d. |
Burkholderia sp. strain GG4 |
Reduces; 3-oxo-AHL to corresponding 3-hydroxy derivatives. |
207 |
| n.d. | Rho. erythropolis W2 | Converts C8-HSL to C14-HSL to corresponding 3-hydroxy derivatives | 182 |
n.d.: Not determined.
Enzymatic control of membrane biofouling
Enzymatic inactivation of AHL molecules has recently been proved to be a promising approach for the control of membrane biofouling. The quorum quenching enzymes AHL-acylase and AHL-lactonase have shown their potential to be used as quorum quenching agent in biofouling control. Xu et al. 201 reported that formation of Aer. hydrophila biofilm on polystyrene surface would be reduced by Acylase I. In another study it was found that 5µg/ml of Acylase I dose results into 60% and 73% reduction of Aer. hydrophila and P. putida of biofilms on polystyrene surface 20. In order to overcome the loss of enzyme during operation and ensure the slow release, immobilization of Acylase I on nanofiltration membrane was carried out by Kim et al 203. The results of the laboratory-scale nanofiltration system demonstrated that newly developed acylase-immobilized membrane could inhibit quorum sensing between bacteria and biocake, thereby reducing biofouling. In addition to this, Lee et al. 223 developed an effective antifouling system by immobilizing quorum quenching acylase in magnetically separable mesoporous silica. The engineered system was found to effectively alleviate the biofilm maturation of test strain P. aeruginosa PAO1 on a membrane surface and thereby enhanced its filtration performance even under harsh conditions of high organic load and low enzyme dose. The synergetic action between enzymatic regulation of quorum sensing molecules and nanobiocatalytic enzyme stabilization has proven its high potential towards simple and effective antifouling solution in MBR. However, the exact mechanism by which the enzymatic quorum quenching can mitigate the biofilm formation in MBR is not yet fully investigated.
Yeon et al. 204 developed acylase attached magnetic particles to inhibit quorum sensing in MBR treating wastewaters. They found that immobilized enzymatic particles has reduced the biofilm formation and enhanced membrane permeability for prolonged period. A major advantage using enzymatic quorum quenching approach is that it only influences sludge characteristics and biofouling, while not impacting pollutant degradation 202. The mass production of quorum quenching enzymes by engineering gene network in bacteria whose expression is under quorum sensing system could be a futuristic approach for prolonged inhibition of membrane biofouling 224, 23. Recently, a microporous membrane encapsulated with AHL-lactonase producing recombinant E. coli has been successfully used for the control of biofouling by interspecies interference in MBR 21. In another study, a microbial-vessel containing quorum quenching bacteria encapsulated inside a porous hollow fiber membrane was found to inhibit membrane biofouling in an external submerged MBR treating synthetic wastewater 225.
Conclusions
Biofouling, a consequence of various microbial activities is a complex process in MBR treating wastewaters. Recent studies have proved evidence that quorum sensing, which was earlier known for pathogenesis, may also play key role in membrane biofouling. AHLs-based quorum sensing system associated with Gram-negative bacteria is known to have a potential role in biofilm formations. Understanding the mechanism of AHLs-based quorum sensing system in wastewater microbiology can help in targeting quorum sensing and addressing the problem of membrane biofouling. Natural compounds as QSI could act as a 'silver bullet' to solve the problem of membrane biofouling as this has great potential towards inhibiting biofilm formation without affecting bacterial growth.
Abbreviations
- Aer.
Aeromonas
- A.
Agrobacterium
- As.
Aspergillus
- B.
Bacillus
- C.
Chromobacterium
- E.
Escherichia
- Ent.
Enterococcus
- Ge.
Geobacillus
- M.
Microbacterium
- My.
Mycobacterium
- Pe.
Penicillium
- P.
Pseudomonas
- Pr.
Proteus
- R.
Ralstonia
- Rho.
Rhodococcus
- Rho.
Rhodospirillum
- S.
Serratia
- Sa.
Salmonella
- Sta.
Staphylococcus
- Si.
Sinorhizobium
- So.
Solibacillus
- Sul.
Sulfolobus
- V.
Vibrio
- X.
Xanthomonas
- Y.
Yersinia.
References
- 1.Drews A. Membrane fouling in membrane bioreactors-Characterisation, contradictions, cause and cures. J Memb Sci. 2010;363:1–28. [Google Scholar]
- 2.Judd S. 2nd ed. The MBR Book. Elsevier Ltd, UK; 2006. Principles and applications of membrane bioreactors in water and wastewater treatment. [Google Scholar]
- 3.Lesjean B, Tazi-Pain A, Thaure D. et al. Ten persistent myths and the realities of membrane bioreactor technology for municipal applications. Water Sci Technol. 2011;63:32–39. doi: 10.2166/wst.2011.005. [DOI] [PubMed] [Google Scholar]
- 4.Zheng X, Zhou Y, Chen S. et al. Survey of MBR market: Trends and perspec-tives in China. Desalination. 2010;250:609–612. [Google Scholar]
- 5.Le-Clech P. Membrane bioreactors and their uses in wastewater treatments. Appl Microbiol Biotechnol. 2010;88:1253–1260. doi: 10.1007/s00253-010-2885-8. [DOI] [PubMed] [Google Scholar]
- 6.Le-Clech P, Chen V, Fane TAG. Fouling in membrane bioreactors used in wastewater treatment. J Membr Sci. 2006;284:17–53. [Google Scholar]
- 7.Ramesh A, Lee DJ, Wang ML. et al. Biofouling in membrane bioreactor. Sep Sci Technol. 2006;41:1345–1370. [Google Scholar]
- 8.Hook AL, Chang CY, Yang J. et al. Combinatorial discovery of polymers resistant to bacterial attachment. Nat Biotech. 2012;30:868–875. doi: 10.1038/nbt.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2003;2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 10.Dobretsov S, Teplitski M, Paul V. Mini-review: quorum sensing in the marine environment and its relationship to biofouling. Biofouling. 2009;25:413–427. doi: 10.1080/08927010902853516. [DOI] [PubMed] [Google Scholar]
- 11.Sauer K, Camper AK, Ehrlich GD. et al. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams P. Quorum sensing: an emerging target for antibacterial chemo-therapy? Expert Opin Ther Targets. 2002;6:257–274. doi: 10.1517/14728222.6.3.257. [DOI] [PubMed] [Google Scholar]
- 13.Bjarnsholt T, Givskov M. Quorum sensing inhibitory drugs as next generation antimicrobials: worth the effort? Curr Infect Dis Rep. 2008;10:22–28. doi: 10.1007/s11908-008-0006-y. [DOI] [PubMed] [Google Scholar]
- 14.Lowery CA, Salzameda NT, Sawada D. et al. Medicinal chemistry as a conduit for the modulation of quorum sensing. J Med Chem. 2010;53:7467–7489. doi: 10.1021/jm901742e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uroz S, Dessaux Y, Oger P. Quorum sensing and quorum quenching: the yin and yang of bacterial communication. ChemBioChem. 2009;10:205–216. doi: 10.1002/cbic.200800521. [DOI] [PubMed] [Google Scholar]
- 16.Ponnusamy K, Paul D, Kweon JH. Inhibition of quorum sensing mechanism and Aeromonas hydrophila biofilm formation by vanillin. Environ Eng Sci. 2009;26:1359–1363. [Google Scholar]
- 17.Eberl L, Molin S, Givskov M. Surface motility of Serratia liquefaciens MG1. J Bacteriol. 1999;181:1703–1712. doi: 10.1128/jb.181.6.1703-1712.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Truchado P, Giménez-Bastida JA, Larrosa M. et al. Inhibition of quorum sensing (QS) in Yersinia enterocolitica by an orange extract rich in glycosylated flavanones. J Agric Food Chem. 2012;60:8885–8894. doi: 10.1021/jf301365a. [DOI] [PubMed] [Google Scholar]
- 19.Rudrappa T, Bais HP. Curcumin, a known phenolic from Curcuma longa, attenuates the virulence of Pseudomonas aeruginosa PAO1 in whole plant and animal pathogenicity models. J Agric Food Chem. 2008;56:1955–1962. doi: 10.1021/jf072591j. [DOI] [PubMed] [Google Scholar]
- 20.Paul D, Kim YS, Ponnusamy K. et al. Application of quorum quenching to inhibit biofilm formation. Environ Eng Sci. 2009;26:1319–1324. [Google Scholar]
- 21.Oh HS, Yeon KM, Yang CS. et al. Control of membrane biofouling in MBR for wastewater treatment by quorum quenching bacteria encapsulated in mi-croporous membrane. Environ Sci Technol. 2012;46:4877–4884. doi: 10.1021/es204312u. [DOI] [PubMed] [Google Scholar]
- 22.Kim SR, Oh HS, Jo SJ. et al. Biofouling control with bead-entrapped quorum quenching bacteria in membrane bioreactors: Physical and biological effects. Environ Sci Technol. 2013;47:836–842. doi: 10.1021/es303995s. [DOI] [PubMed] [Google Scholar]
- 23.Choudhary S, Schmidt-Dannert C. Applications of quorum sensing in biotechnology. Appl Microbiol Biotechnol. 2010;86:1267–1279. doi: 10.1007/s00253-010-2521-7. [DOI] [PubMed] [Google Scholar]
- 24.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Ann Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 25.Reading NC, Sperandio V. Quorum sensing: the many languages of bacteria. FEMS Microbiol Lett. 2006;254:1–11. doi: 10.1111/j.1574-6968.2005.00001.x. [DOI] [PubMed] [Google Scholar]
- 26.Chhabra S, Philipp B, Eberl L, et al. Extracellular communication in bacteria. In: Schulz S, editor. The chemistry of pheromones and other semiochemicals II. Vol 240: Springer Berlin Heidelberg; 2005. pp. 279–315. [Google Scholar]
- 27.Williams P. Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology. 2007;153:3923–3938. doi: 10.1099/mic.0.2007/012856-0. [DOI] [PubMed] [Google Scholar]
- 28.Yarwood JM, Bartels DJ, Volper EM. et al. Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol. 2004;186:1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.George EA, Muir TW. Molecular mechanisms of agr quorum sensing in virulent staphylococci. Chembiochem. 2007;8:847–855. doi: 10.1002/cbic.200700023. [DOI] [PubMed] [Google Scholar]
- 30.Engebrecht J, Nealson K, Silverman M. Bacterial bioluminescence: Isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983;32:773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- 31.McClean KH, Winson MK, Fish L. et al. Quorum sensing and Chromobacte-rium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 32.Karlsson T, Turkina MV, Yakymenko O. et al. The Pseudomonas eruginosa N-acylhomoserine lactone quorum sensing molecules target IQGAP1 and modulate epithelial cell migration. PLoS Pathog. 2012;8:e1002953. doi: 10.1371/journal.ppat.1002953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sio CF, Otten LG, Cool RH. et al. Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infect Immun. 2006;74:1673–1682. doi: 10.1128/IAI.74.3.1673-1682.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaw PD, Ping G, Daly SL. et al. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc Natl Acad Sci USA. 1997;94:6036–6041. doi: 10.1073/pnas.94.12.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swift S, Karlyshev AV, Fish L. et al. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J Bacteriol. 1997;179:5271–5281. doi: 10.1128/jb.179.17.5271-5281.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, Schauder S, Potier N. et al. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 37.Wood TK. Insights on Escherichia coli biofilm formation and inhibition from whole-transcriptome profiling. Environ Microbiol. 2009;11:1–15. doi: 10.1111/j.1462-2920.2008.01768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sperandio V, Torres AG, Giron JA. et al. Quorum sensing is a global regula-tory mechanism in enterohemorrhagic Escherichia coli O157:H7. J Bacteriol. 2001;183:5187–5197. doi: 10.1128/JB.183.17.5187-5197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gelhaus HC, Rozak DA, Nierman WC. et al. Exogenous Yersinia pestis quorum sensing molecules N-octanoyl-homoserine lactone and N-(3-oxooctanoyl)-homoserine lactone regulate the LcrV virulence factor. Microb Pathog. 2009;46:283–287. doi: 10.1016/j.micpath.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Miller ST, Xavier KB, Campagna SR. et al. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol Cell. 2004;15:677–687. doi: 10.1016/j.molcel.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 41.Choi J, Shin D, Ryu S. Implication of quorum sensing in Salmonella enterica serovar typhimurium virulence: The luxS gene is necessary for expression of genes in pathogenicity island 1. Infect Immun. 2007;75:4885–4890. doi: 10.1128/IAI.01942-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Degrassi G, Aguilar C, Bosco M. et al. Plant growth promoting Pseudomonas putida WCS358 produces and secretes four cyclic dipeptides: cross-talk with quorum sensing bacterial sensors. Curr Microbiol. 2002;45:250–254. doi: 10.1007/s00284-002-3704-y. [DOI] [PubMed] [Google Scholar]
- 43.Holden MTG, Chhabra SR, de Nys R. et al. Quorum sensing cross talk: isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other Gram-negative bacteria. Mol Microbiol. 1999;33:1254–1266. doi: 10.1046/j.1365-2958.1999.01577.x. [DOI] [PubMed] [Google Scholar]
- 44.Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437:422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 45.Deziel E, Lepine F, Milot S. et al. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci USA. 2004;101:1339–1344. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lepine F, Milot S, Deziel E. et al. Electrospray/mass spectrometric identification and analysis of 4-hydroxy-2-alkylquinolines (HAQs) produced by Pseudomonas aeruginosa. J Am Soc Mass Spectrom. 2004;15:862–869. doi: 10.1016/j.jasms.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 47.Wang LH, He Y, Gao Y. et al. A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol Microbiol. 2004;51:903–912. doi: 10.1046/j.1365-2958.2003.03883.x. [DOI] [PubMed] [Google Scholar]
- 48.Sturme MHJ, Kleerebezem M, Nakayama J. et al. Cell to cell communication by autoinducing peptides in gram-positive bacteria. Antonie Van Leeu-wenhoek. 2002;81:233–243. doi: 10.1023/a:1020522919555. [DOI] [PubMed] [Google Scholar]
- 49.Malone CL, Boles BR, Horswill AR. Biosynthesis of Staphylococcus aure-us autoinducing peptides by using the Synechocystis DnaB mini-intein. Appl Environ Microbiol. 2007;73:6036–6044. doi: 10.1128/AEM.00912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ji G, Beavis RC, Novick RP. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kleerebezem M, Quadri LE. Peptide pheromone-dependent regulation of antimicrobial peptide production in Gram-positive bacteria: A case of multicellular behavior. Peptides. 2001;22:1579–1596. doi: 10.1016/s0196-9781(01)00493-4. [DOI] [PubMed] [Google Scholar]
- 52.Hamoen LW, Venema G, Kuipers OP. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology. 2003;149:9–17. doi: 10.1099/mic.0.26003-0. [DOI] [PubMed] [Google Scholar]
- 53.Ansaldi M, Marolt D, Stebe T. et al. Specific activation of the Bacillus quor-um-sensing systems by isoprenylated pheromonevariants. Mol Microbiol. 2002;44:1561–1573. doi: 10.1046/j.1365-2958.2002.02977.x. [DOI] [PubMed] [Google Scholar]
- 54.Rutherford ST, Bassler BL. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harbor Perspect Med. 2012;2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Havarstein LS, Coomaraswamy G, Morrison DA. An unmodified heptade-capeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kleerebezem M, Quadri LEN, Kuipers OP. et al. Quorum sensing by peptide pheromones and two component signal transduction systems in Gram-positive bacteria. Mol Microbiol. 1997;24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 57.Xu H, Liu Y. Control and cleaning of membrane biofouling by energy un-coupling and cellular communication. Environ Sci Technol. 2011;45:595–601. doi: 10.1021/es102911m. [DOI] [PubMed] [Google Scholar]
- 58.Juhas M, Eberl L, Tummler B. Quorum sensing: the power of cooperation in the world of Pseudomonas. Environ Microbiol. 2005;7:459–471. doi: 10.1111/j.1462-2920.2005.00769.x. [DOI] [PubMed] [Google Scholar]
- 59.Duan K, Surette MG. Environmental regulation of Pseudomonas aeruginosa PAO1 Las and Rhl quorum-sensing systems. J Bacteriol. 2007;189:4287–4236. doi: 10.1128/JB.00043-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shrout JD, Nerenberg R. Monitoring bacterial twitter: does quorum sensing determine the behavior of water and wastewater treatment biofilms? Environ Sci Technol. 2012;46:1995–2005. doi: 10.1021/es203933h. [DOI] [PubMed] [Google Scholar]
- 61.Hentzer M, Givskov M, Eberl L. O'Toole MGaG, ed. Microbial Biofilms: ASM press, Washington DC; 2004. Quorum sensing in biofilms: Gossip in slime city; pp. 118–140. [Google Scholar]
- 62.Nadell CD, Xavier JB, Levin SA. et al. The evolution of quorum sensing in bacterial biofilms. PLoS Biol. 2008;6(e):14. doi: 10.1371/journal.pbio.0060014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hammer B, Bassler B. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 64.Peter SM. A network of networks: quorum-sensing gene regulation in Pseu-domonas aeruginosa. Int J Med Microbiol. 2006;296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 65.Grossman AD. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu Rev Genet. 1995;29:477–508. doi: 10.1146/annurev.ge.29.120195.002401. [DOI] [PubMed] [Google Scholar]
- 66.Parsek MR, Greenberg EP. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 67.Winson MK, Swift S, Fish L. et al. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lac-tone-mediated quorum sensing. FEMS Microbiol Lett. 1998;163:185–192. doi: 10.1111/j.1574-6968.1998.tb13044.x. [DOI] [PubMed] [Google Scholar]
- 68.Andersen JB, Heydorn A, Hentzer M. et al. gfp-based N-acyl homoser-ine-lactone sensor systems for detection of bacterial communication. Appl Environ Microbiol. 2001;67:575–585. doi: 10.1128/AEM.67.2.575-585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Kievit TR, Iglewski BH. Bacterial quorum sensing in pathogenic relation-ships. Infect. Immun. 2000;68:4839–4849. doi: 10.1128/iai.68.9.4839-4849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lithgow JK, Wilkinson A, Hardman A. et al. The regulatory locus cinRI in Rhizobium leguminosarum controls a network of quorum-sensing loci. Mol Microbiol. 2000;37:81–97. doi: 10.1046/j.1365-2958.2000.01960.x. [DOI] [PubMed] [Google Scholar]
- 71.Horng YT, Deng SC, Daykin M. et al. The LuxR family protein SpnR functions as a negative regulator of N-acylhomoserine lactone-dependent quorum sensing in Serratia marcescens. Mol Microbiol. 2002;45:1655–1671. doi: 10.1046/j.1365-2958.2002.03117.x. [DOI] [PubMed] [Google Scholar]
- 72.Bassler BL, Losick R. Bacterially speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 73.Davey ME, O'Toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 75.Kaufmann GF, Park J, Janda KD. Bacterial quorum sensing: a new target for anti-infective immunotherapy. Expert Opin Biol Ther. 2008;8:719–724. doi: 10.1517/14712598.8.6.719. [DOI] [PubMed] [Google Scholar]
- 76.Costerton JW, Lewandowski Z, Caldwell DE. et al. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–45. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 77.O'Toole G, Kaplan HD, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:81–127. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 78.Greenberg EP. Bacterial communication and group behavior. J Clin Invest. 2003;112:1288–1290. doi: 10.1172/JCI20099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yarwood JM, Schlievert PM. Quorum sensing in Staphylococcus infections. J Clinic Investig. 2003;112:1620–1625. doi: 10.1172/JCI20442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Adav SS, Lee DJ, Show KY. et al. Aerobic granular sludge: Recent advances. Biotechnol Adv. 2008;26:411–423. doi: 10.1016/j.biotechadv.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 81.Amy G. Fundamental understanding of organic matter fouling of membranes. Desalination. 2008;231:44–51. [Google Scholar]
- 82.Kim HW, Oh HS, Kim SR. et al. Microbial population dynamics and prote-omics in membrane bioreactors with enzymatic quorum quenching. Appl Microbiol Biotechnol. 2013;97:4665–4675. doi: 10.1007/s00253-012-4272-0. [DOI] [PubMed] [Google Scholar]
- 83.Bae TH, Tak TM. Interpretation of fouling characteristics of ultrafiltration membranes during the filtration of membrane bioreactror mixed liquor. J Membr Sci. 2005;264:151–160. [Google Scholar]
- 84.Defrance L, Jaffrin MY, Gupta B. et al. Contribution of various constituents of activated sludge to membrane bioreactor fouling. Bioresour Technol. 2000;73:105–112. [Google Scholar]
- 85.Davies DG, Parsek MR, Pearson JP. et al. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 86.Ya-Wen H, Lian-Hui Z. Quorum sensing and virulence regulation in Xan-thomonas campestris. FEMS Microbiol Rev. 2008;32:842–857. doi: 10.1111/j.1574-6976.2008.00120.x. [DOI] [PubMed] [Google Scholar]
- 87.Toyofuku M, Nomura N, Kuno E. et al. Influence of the Pseudomonas quinolone signal on denitrification in Pseudomonas aeruginosa. J Bacteriol. 2008;190:7947–7956. doi: 10.1128/JB.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lynch MJ, Swift S, Kirke DF. et al. The regulation of biofilm development by quorum sensing in Aeromonas hydrophila. Environ Microbiol. 2002;4:18–28. doi: 10.1046/j.1462-2920.2002.00264.x. [DOI] [PubMed] [Google Scholar]
- 89.Kim M, Lee S, Park HD. et al. Biofouling control by quorum sensing inhibition and its dependence on membrane surface. Water Sci Technol. 2012;66:1424–1430. doi: 10.2166/wst.2012.307. [DOI] [PubMed] [Google Scholar]
- 90.Xia S, Zhou L, Zhang Z. et al. Influence and mechanism of N-(3-oxooxtanoyl)-L-homoserine lactone (C8-oxo-HSL) on biofilm behaviors at the early stage. J Environ Sci. 2012;24:2035–2040. doi: 10.1016/s1001-0742(11)61060-7. [DOI] [PubMed] [Google Scholar]
- 91.Lade H, Paul D, Kweon JH. Isolation and molecular characterization of biofouling bacteria and profiling of quorum sensing signal molecules from membrane bioreactor activated sludge. Int J Mol Sci. 2014;15:2255–2273. doi: 10.3390/ijms15022255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lim SY, Kim S, Yeon KM. et al. Correlation between microbial community structure and biofouling in a laboratory scale membrane bioreactor with synthetic wastewater. Desalination. 2012;287:209–215. [Google Scholar]
- 93.Yeon KM, Cheong WS, Oh HS. et al. Quorum sensing: A new biofouling control paradigm in a membrane bioreactor for advanced wastewater treatment. Environ Sci Technol. 2009;43:380–385. doi: 10.1021/es8019275. [DOI] [PubMed] [Google Scholar]
- 94.Dong YH, Wang LH, Zhang LH. Quorum-quenching microbial infections: mechanisms and implications. Philos Trans R Soc Lond B Biol Sci. 2007;362:1201–1211. doi: 10.1098/rstb.2007.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dobretsov S, Teplitski M, Bayer M. et al. Inhibition of marine biofouling by bacterial quorum sensing inhibitors. Biofouling. 2011;27:893–905. doi: 10.1080/08927014.2011.609616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hentzer M, Givskov M, Parsek MR. Targeting quorum sensing for treatment of chronic bacterial biofilm infections. Lab Med. 2002;33:295–306. [Google Scholar]
- 97.Geske GD, O'Neill JC, Blackwell HE. Expanding dialogues: From natural autoinducers to non-natural analogues that modulate quorum sensing in Gram-negative bacteria.Chem. Soc Rev. 2008;37:1432–1447. doi: 10.1039/b703021p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Parveen N, Cornell KA. Methylthioadenosine/S-adenosylhomocysteine nucleosidase, a critical enzyme for bacterial metabolism. Mol Microbiol. 2011;79:7–20. doi: 10.1111/j.1365-2958.2010.07455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lin YH, Xu JL, Hu JY. et al. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching en-zymes. Mol Microbiol. 2003;47:849–860. doi: 10.1046/j.1365-2958.2003.03351.x. [DOI] [PubMed] [Google Scholar]
- 100.Yates EA, Philipp B, Buckley C. et al. N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect Immun. 2002;70:5635–5646. doi: 10.1128/IAI.70.10.5635-5646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Koch B, Liljefors T, Persson T. et al. The LuxR receptor: The sites of interaction with quorum-sensing signals and inhibitors. Microbiology. 2005;151:3589–3602. doi: 10.1099/mic.0.27954-0. [DOI] [PubMed] [Google Scholar]
- 102.Chen G, Swern LR, Swern DL. et al. A strategy for antagonizing quorum sensing. Mol Cell. 2011;42:199–209. doi: 10.1016/j.molcel.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vanderkooi OG, Low DE, Green K. et al. Predicting antimicrobial resistance in invasive pneumococcal infections. Clin Infect Dis. 2005;40:1288–1297. doi: 10.1086/429242. [DOI] [PubMed] [Google Scholar]
- 104.Kubo I, Masuoka N, Ha TJ. et al. Antioxidant activity of anacardic acids. Food Chem. 2006;99:555–562. [Google Scholar]
- 105.Bauer WD, Mathesius U. Plant responses to bacterial quorum sensing signals. Curr Opin Plant Biol. 2004;7:429–433. doi: 10.1016/j.pbi.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 106.Dudler R, Eberl E. Interactions between bacteria and eukaryotes via small molecules. Curr Opin Biotechnol. 2006;17:268–273. doi: 10.1016/j.copbio.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 107.Teplitski M, Mathesius U, Rambaugh KP. Perception and degradation of N-acyl homoserine lactone quorum sensing signals by mammalian and plant cells. Chem Rev. 2011;111:100–116. doi: 10.1021/cr100045m. [DOI] [PubMed] [Google Scholar]
- 108.Vattem DA, Mihalik K, Crixell SH. et al. Dietary phytochemicals as quorum sensing inhibitors. Fitoterapia. 2007;78:302–310. doi: 10.1016/j.fitote.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 109.Ponnusamy K, Paul D, Kim YS. et al. 2(5H)-Furanone: A Prospective strategy for biofouling-control in membrane biofilm bacteria by quorum sensing inhibition. Braz J Microbiol. 2010;41:227–234. doi: 10.1590/S1517-838220100001000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Manefield M, Rasmussen TB, Henzter M. et al. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology. 2002;148:1119–1127. doi: 10.1099/00221287-148-4-1119. [DOI] [PubMed] [Google Scholar]
- 111.Defoirdt T, Miyamoto CM, Wood TK. et al. The natural furanone (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone disrupts quorum sensing-regulated gene expression in Vibrio harveyi by decreasing the DNA-binding activity of the transcriptional regulator protein luxR. Environ Microbiol. 2007;9:2486–2495. doi: 10.1111/j.1462-2920.2007.01367.x. [DOI] [PubMed] [Google Scholar]
- 112.Ren D, Sims JJ, Wood TK. Inhibition of biofilm formation and swarming of Escherichia coli by (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone. Environ Microbiol. 2001;3:731–736. doi: 10.1046/j.1462-2920.2001.00249.x. [DOI] [PubMed] [Google Scholar]
- 113.Bjarnsholt T, Jensen PO, Rasmussen TB. et al. Garlic blocks quorum sensing and promotes rapid clearing of pulmonary Pseudomonas aeruginosa infec-tions. Microbiology. 2005;151:3873–3880. doi: 10.1099/mic.0.27955-0. [DOI] [PubMed] [Google Scholar]
- 114.Jakobsen TH, van Gennip M, Phipps RK. et al. Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob Agents Chemother. 2012;56:2314–2325. doi: 10.1128/AAC.05919-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jakobsen TH, Bragason SK, Phipps RK. et al. Food as a source for quorum sensing inhibitors: iberin from horseradish revealed as a quorum sensing in-hibitor of Pseudomonas aeruginosa. Appl Environ Microbiol. 2012;78:2410–2421. doi: 10.1128/AEM.05992-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ganin H, Rayo J Amara N. et al. Sulforaphane and erucin, natural isothio-cyanates from broccoli, inhibit, bacterial quorum sensing. Med Chem Com-mun. 2013;4:175–179. [Google Scholar]
- 117.Vandeputte OM, Kiendrebeogo M, Rasamiravaka T. et al. The flavanone naringenin reduces the production of quorum sensing-controlled virulence factors in Pseudomonas aeruginosa PAO1. Microbiology. 2011;157:2120–2132. doi: 10.1099/mic.0.049338-0. [DOI] [PubMed] [Google Scholar]
- 118.Vandeputte OM, Kiendrebeogo M, Rajaonson S. et al. Identification of catechin as one of the flavonoids from Combretum albiflorum bark extract that reduces the production of quorum-sensing-controlled virulence factors in Pseudomonas aeruginosa PAO1. Appl Environ Microbiol. 2010;76:243–253. doi: 10.1128/AEM.01059-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Annapoorani A, Umamageswaran V, Parameswari R. et al. Computational discovery of putative quorum sensing inhibitors against LasR and RhlR re-ceptor proteins of Pseudomonas aeruginosa. J Comput Aided Mol Des. 2012;26:1067–1077. doi: 10.1007/s10822-012-9599-1. [DOI] [PubMed] [Google Scholar]
- 120.Annapoorani A, Parameswari R, Pandian SK. et al. Methods to determine antipathogenic potential of phenolic and flavonoid compounds against uri-nary pathogen Serratia marcescens. J Microbiol Methods. 2012;91:208–211. doi: 10.1016/j.mimet.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 121.Rasmussen TB, Bjarnsholt T, Skindersoe ME. et al. Screening for quor-um-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J Bacteriol. 2005;187:1799–1814. doi: 10.1128/JB.187.5.1799-1814.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Givskov M, de Nys R, Manefield M. et al. Eukaryotic interference with homo-serine lactone mediated prokaryotic signaling. J Bacteriol. 1996;178:6618–6622. doi: 10.1128/jb.178.22.6618-6622.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Choo JH, Rukayadi Y, Hwang JK. Inhibition of bacterial quorum sensing by vanilla extract. Lett Appl Microbiol. 2006;42:637–641. doi: 10.1111/j.1472-765X.2006.01928.x. [DOI] [PubMed] [Google Scholar]
- 124.Oger P, Farrand SK. Co-evolution of the agrocinopine opines and the agro-cinopine-mediated control of TraR, the quorum-sensing activator of the Ti plasmid conjugation system. Mol Microbiol. 2001;41:1173–1185. doi: 10.1046/j.1365-2958.2001.02584.x. [DOI] [PubMed] [Google Scholar]
- 125.Keshavan ND, Chowdhary PK, Haines DC. et al. L-canavanine made by Medicao sativa interferes with quorum sensing in Sinorhizobium meliloti. J Bacteriol. 2005;187:8427–8436. doi: 10.1128/JB.187.24.8427-8436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chevrot R, Rosen R, Haudecoeur E. et al. GABA controls the level of quor-um-sensing signal in Agrobacterium tumefaciens. Proc Natl Acad Sci USA. 2006;103:7460–7464. doi: 10.1073/pnas.0600313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bouche N, Lacombe B, Fromm H. GABA signaling: a conserved and ubiqui-tous mechanism. Trends Cell Biol. 2003;13:607–610. doi: 10.1016/j.tcb.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 128.Walker TS, Bais HP, Deziel E. et al. Pseudomonas aeruginosa-plant root interactions, pathogenicity, biofilm formation, and root exudation. Plant Physiol. 2004;34:320–331. doi: 10.1104/pp.103.027888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yuan ZC, Edlind MP, Liu P. et al. The plant signal salicylic acid shuts down expression of the vir regulon and activates quormone-quenching genes in Agrobacterium. Proc Natl Acad Sci USA. 2007;104:11790–11795. doi: 10.1073/pnas.0704866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Singh BN, Singh BR, Singh RL. et al. Oxidative DNA damage protective activity, antioxidant and anti-quorum sensing potentials of Moringa oleifera. Food Chem Toxicol. 2009;47:1109–1116. doi: 10.1016/j.fct.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 131.Cady NC, McKean KA, Behnke J. et al. Inhibition of biofilm bormation, quorum sensing and infection in Pseudomonas aeruginosa by natural prod-ucts-inspired organosulfur compounds. PLoS ONE. 2012;7:e38492. doi: 10.1371/journal.pone.0038492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ren D, Zuo R, González-Barrios AF. et al. Differential gene expression for investigation of Escherichia coli biofilm inhibition by plant extract ursolic acid. Appl Environ Microbiol. 2005;71:4022–4034. doi: 10.1128/AEM.71.7.4022-4034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu J. Pharmacology of oleanolic acid and ursolic acid. J Ethanopharmacol. 1995;49:57–68. doi: 10.1016/0378-8741(95)90032-2. [DOI] [PubMed] [Google Scholar]
- 134.Sarabhai S, Sharma P, Capalash N. Ellagic acid derivatives from Terminalia chebula Retz. downregulate the expression of quorum sensing genes to at-tenuate Pseudomonas aeruginosa PAO1 virulence. PLoS One. 2013;8:e53441. doi: 10.1371/journal.pone.0053441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Huber B, Eberl L, Feucht W. et al. Influence of polyphenols on bacterial biofilm formation and quorum sensing. Z Naturforsch C. 2003;58:879–884. doi: 10.1515/znc-2003-11-1224. [DOI] [PubMed] [Google Scholar]
- 136.Chai Y, Tsai CS, Cho H. et al. Reconstitution of the biochemical activties of the AttJ repressor and the AttK, Atl and AttM catabolic enzymes of Agrobacterium tumefaciens. J Bacteriol. 2007;189:3674–3679. doi: 10.1128/JB.01274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Akiyama H, Fujii K, Yamasaki O. et al. Antibacterial action of several tannins against Staphylococcus aureus. J Antimicrob Chemother. 2001;48:487–491. doi: 10.1093/jac/48.4.487. [DOI] [PubMed] [Google Scholar]
- 138.Blanco AR, Sudano-Roccaro A, Spoto GC. et al. Epigallocatechin gallate inhibits biofilm formation by ocular Staphylococcal isolates. Antimicrob Agents Chemother. 2005;49:4339–4343. doi: 10.1128/AAC.49.10.4339-4343.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhao WH, Hu ZQ, Hara Y. et al. Inhibition by epigallocatechin gallate (EGCg) of conjugative R plasmid transfer in Escherichia coli. J Infect Chemother. 2001;7:195–197. doi: 10.1007/s101560100035. [DOI] [PubMed] [Google Scholar]
- 140.Ni N, Choudhary G, Li M. et al. Pyrogallol and its analogs can antagonize bacterial quorum sensing in Vibrio harveyi. Bioorg Med Chem Lett. 2008;18:1567–1572. doi: 10.1016/j.bmcl.2008.01.081. [DOI] [PubMed] [Google Scholar]
- 141.Sangeetha J, Vijayalakshmi K. Determination of bioactive components of ethyl acetate fraction of Punica granatum rind extract. Int J Pharm Sci Drug Res. 2011;3:116–122. [Google Scholar]
- 142.Brackman G, Defoirdt T, Miyamoto C. et al. Cinnamaldehyde and cinnamaldehyde derivatives reduce virulence in Vibrio spp. by decreasing the DNA-binding activity of the quorum sensing response regulator LuxR. BMC Microbiol. 2008;8:149–162. doi: 10.1186/1471-2180-8-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Niu C, Afre S, Gilbert ES. Sub-inhibitory concentrations of cinnamaldehyde interfere with quorum sensing. Lett Appl Microbiol. 2006;43:489–494. doi: 10.1111/j.1472-765X.2006.02001.x. [DOI] [PubMed] [Google Scholar]
- 144.Girennavar B, Cepeda ML, Soni KA. et al. Grapefruit juice and its furocoumarins inhibits autoinducer signaling and biofilm formation in bacteria. Int J Food Microbiol. 2008;125:204–208. doi: 10.1016/j.ijfoodmicro.2008.03.028. [DOI] [PubMed] [Google Scholar]
- 145.Zahin M, Hasan S, Aqil F. et al. Screening of certain medicinal plants from India for their anti-quorum sensing activity. Indian J Exp Biol. 2010;48:1219–1224. [PubMed] [Google Scholar]
- 146.Giménez-Bastida JA, Truchado P, Larrosa M. et al. Urolithins, ellagitannin metabolites produced by colon microbiota, inhibit quorum sensing in Yersinia enterocolitica: Phenotypic response and associated molecular changes. Food Chem. 2012;132:1465–1474. doi: 10.1016/j.foodchem.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 147.O'May CO, Tufenkji N. Pseudomonas aeruginosa swarming motility is blocked by cranberry proanthocyanidins and other tannin-containing materials. Appl Environ Microbiol. 2011;77:3061–3067. doi: 10.1128/AEM.02677-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Packiavathy IA, Priya S, Pandian SK. et al. Inhibition of biofilm development of uropathogens by curcumin - an anti-quorum sensing agent from Curcuma longa. Food Chem. 2014;148:453–460. doi: 10.1016/j.foodchem.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 149.Kim JS, Kim YH, Seo YW. et al. Quorum sensing inhibitors from the red alga, Ahnfeltiopsis flabelliformis. Biotechnol Bioproc Eng. 2007;12:308–311. [Google Scholar]
- 150.Musthafa KS, Ravi AV, Annapoorani A. et al. Evaluation of an-ti-quorum-sensing activity of edible plants and fruits through inhibition of the N-acyl-homoserine lactone system in Chromobacterium violaceum and Pseudomonas aeruginosa. Chemotherapy. 2010;56:333–339. doi: 10.1159/000320185. [DOI] [PubMed] [Google Scholar]
- 151.Siddiqui MF, Sakinah M, Ismail AF. et al. The anti-biofouling effect of Piper betle extract against Pseudomonas aeruginosa and bacterial consortium. Desalination. 2012;288:24–30. [Google Scholar]
- 152.Packiavathy IA, Agilandeswari P, Musthafa KS. et al. Antibiofilm and quorum sensing inhibitory potential of Cuminum cyminum and its secondary metabolite methyl eugenol against Gram negative bacterial pathogens. Food Res Int. 2012;45:85–92. [Google Scholar]
- 153.Manefield M, de Nys R, Kumar N. et al. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Mi-crobiology. 1999;145:283–291. doi: 10.1099/13500872-145-2-283. [DOI] [PubMed] [Google Scholar]
- 154.Lowery CA, Dickerson TJ, Janda KD. Interspecies and interkingdom communication mediated by bacterial quorum sensing. Chem Soc Rev. 2008;37:1337–1346. doi: 10.1039/b702781h. [DOI] [PubMed] [Google Scholar]
- 155.Maximilien R, de Nys R, Holmstrom C. et al. Chemical mediation of bacterial surface colonisation by secondary metabolites from the red alga Delisea pulchra. Aquat Microb Ecol. 1998;15:233–246. [Google Scholar]
- 156.Sanchez-Contreras M, Bauer WD, Gao M. et al. Quorum-sensing regulation in rhizobia and its role in symbiotic interactions with legumes. Philos Trans R Soc Lond B Biol Sci. 2007;362:1149–1163. doi: 10.1098/rstb.2007.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Eberhard A, Widrig CA, McBath P. et al. Analogs of the autoinducer of bioluminescence in Vibrio fischeri. Arch Microbiol. 1986;146:35–40. doi: 10.1007/BF00690155. [DOI] [PubMed] [Google Scholar]
- 158.Dong YH, Xu JL, Li XZ. et al. AiiA, an enzyme that inactivates the acylho-moserine lactone quorum-sensing signal and attenuates the virulence of Er-winia carotovora. Proc Natl Acad Sci USA. 2000;97:3526–3531. doi: 10.1073/pnas.060023897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hentzer M, Wu H, Andersen JB. et al. Attenuation of Pseudomonas aeru-ginosa virulence by quorum sensing inhibitors. EMBO J. 2003;22:3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.de Nys R, Givskov M, Kumar N. et al. Furanones. Prog Mol Subcell Biol. 2006;42:55–86. doi: 10.1007/3-540-30016-3_2. [DOI] [PubMed] [Google Scholar]
- 161.Kalia VC. Quorum sensing inhibitors: An overview. Biotechnol Adv. 2013;31:224–245. doi: 10.1016/j.biotechadv.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 162.Truchado P, Tomás-Barberán FA, Larrosa M. et al. Food phytochemicals act as quorum sensing inhibitors reducing production and/or degrading autoinducers of Yersinia enterocolitica and Erwinia carotovora. Food Control. 2012;24:78–85. [Google Scholar]
- 163.Vikram A, Jayaprakasha GK, Jesudhasan PR. et al. Suppression of bacterial cell-cell signaling, biofilm formation and type III secretion system by citrus flavonoids. J Appl Microbiol. 2010;109:515–527. doi: 10.1111/j.1365-2672.2010.04677.x. [DOI] [PubMed] [Google Scholar]
- 164.Amara N, Mashiach R, Amar D. Covalent inhibition of bacterial quorum sensing. J Am Chem Soc. 2009;131:10610–10619. doi: 10.1021/ja903292v. [DOI] [PubMed] [Google Scholar]
- 165.Rosenthal GA. L-Canavanine: a higher plant insecticidal allelochemical. Amino Acids. 2001;21:319–330. doi: 10.1007/s007260170017. [DOI] [PubMed] [Google Scholar]
- 166.Teplitski M, Robinson JB, Bauer WD. Plants secrete substances that mimic bacterial N-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol Plant Microbe Interact. 2000;13:637–648. doi: 10.1094/MPMI.2000.13.6.637. [DOI] [PubMed] [Google Scholar]
- 167.Bosgelmez-Tinaz G, Ulusoy S, Ugur A. et al. Inhibition of quorum sens-ing-regulated behaviors by Scorzonera sandrasica. Curr Microbiol. 2007;55:114–118. doi: 10.1007/s00284-006-0624-2. [DOI] [PubMed] [Google Scholar]
- 168.Adonizio AL, Downum K, Bennett BC. et al. Anti-quorum sensing activity of medicinal plants in southern Florida. J Ethnopharmacol. 2006;105:427–435. doi: 10.1016/j.jep.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 169.Persson T, Hansen TH, Rasmussen TB. et al. Rational design and synthesis of new quorum-sensing inhibitors derived from acylated homoserine lactones and natural products from garlic. Org Biomol Chem. 2005;3:253–262. doi: 10.1039/b415761c. [DOI] [PubMed] [Google Scholar]
- 170.Vikram A, Jesudhasan PR, Jayaprakasha GK. et al. Citrus limonoids interfere with Vibrio harveyi cell-cell signaling and biofilm formation by modulating the response regulator LuxO. Microbiology. 2011;157:99–110. doi: 10.1099/mic.0.041228-0. [DOI] [PubMed] [Google Scholar]
- 171.Persson T, Givskov M, Nielsen J. Quorum sensing inhibition: targeting chemical communication in Gram-negative bacteria. Curr Med Chem. 2005;12:3103–3115. doi: 10.2174/092986705774933425. [DOI] [PubMed] [Google Scholar]
- 172.Lönn-Stensrud J, Petersen FC, Benneche T. et al. Synthetic bromated furanone inhibits autoinducer-2-mediated communication and biofilm formation in oral streptococci. Oral Microbiol Immunol. 2007;22:340–346. doi: 10.1111/j.1399-302X.2007.00367.x. [DOI] [PubMed] [Google Scholar]
- 173.Rasmussen TB, Givskov M. Quorum sensing inhibitors: a bargain of effects. Microbiology. 2006;152:895–904. doi: 10.1099/mic.0.28601-0. [DOI] [PubMed] [Google Scholar]
- 174.Rasmussen TB, Givskov M. Quorum-sensing inhibitors as anti-pathogenic drugs. Int J Med Microbiol. 2006;296:149–161. doi: 10.1016/j.ijmm.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 175.Ponnusamy K, Kappachery S, Thekeettle M. et al. Anti-biofouling property of vanillin on Aeromonas hydrophila initial biofilm on various membrane sur-faces. World J Microbiol Biotechnol. 2013;29:1695–1703. doi: 10.1007/s11274-013-1332-2. [DOI] [PubMed] [Google Scholar]
- 176.Siddiqui MF, Singh L, Zularisam AW. et al. Biofouling mitigation using Piper betleextract in ultrafiltration MBR. Desalin Water Treat. 2013;51:6940–6951. [Google Scholar]
- 177.Siddiqui MF, Sakinah M, Singh L. et al. Targeting N-acyl-homoserine-lactones to mitigate membrane biofouling based on quorum sensing using a biofouling reducer. J Biotechnol. 2012;161:190–197. doi: 10.1016/j.jbiotec.2012.06.029. [DOI] [PubMed] [Google Scholar]
- 178.Dong YH, Wang LH, Xu JL. et al. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature. 2001;411:813–817. doi: 10.1038/35081101. [DOI] [PubMed] [Google Scholar]
- 179.Zhang HB, Wang LH, Zhang LH. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc Natl Acad Sci USA. 2002;99:4638–4643. doi: 10.1073/pnas.022056699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Leadbetter JR, Greenberg EP. Metabolism of acyl-homoserine lactone quor-um-sensing signals by Variovorax paradoxus. J Bacteriol. 2000;182:6921–6926. doi: 10.1128/jb.182.24.6921-6926.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chowdhary PK, Keshavan N, Nguyen HQ. et al. Bacillus megaterium CYP102A1 oxidation of acyl homoserine lactones and acyl homoserines. Biochemistry. 2007;46:14429–14437. doi: 10.1021/bi701945j. [DOI] [PubMed] [Google Scholar]
- 182.Uroz S, Chhabra SR, Cámara M. et al. N-acylhomoserine lactone quor-um-sensing molecules are modified and degraded by Rhodococcus erythropolis W2 by both amidolytic and novel oxidoreductase activities. Microbiology. 2005;151:3313–3322. doi: 10.1099/mic.0.27961-0. [DOI] [PubMed] [Google Scholar]
- 183.Huang JJ, Han JI, Zhang LH. et al. Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PAO1. Appl Environ Microbiol. 2003;69:5941–5949. doi: 10.1128/AEM.69.10.5941-5949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Park SY, Kang HO, Jang HS. et al. Identification of extracellular N-acylhomoserine lactone acylase from a Streptomyces sp. and its application to quorum quenching. Appl Environ Microbiol. 2005;71:2632–2641. doi: 10.1128/AEM.71.5.2632-2641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Huang JJ, Petersen A, Whiteley M. et al. Identification of QuiP, the product of gene PA1032, as the second acyl-homoserine lactone acylase of Pseudomonas aeruginosa PAO1. Appl Environ Microbiol. 2006;72:1190–1197. doi: 10.1128/AEM.72.2.1190-1197.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Romero M, Diggle SP, Heeb S. et al. Quorum quenching activity in Anabaena sp. PCC 7120: Identification of AiiC, a novel AHL-acylase. FEMS Microbiol Lett. 2008;280:73–80. doi: 10.1111/j.1574-6968.2007.01046.x. [DOI] [PubMed] [Google Scholar]
- 187.Shepherd RW, Lindow SE. Two dissimilar N-acyl-homoserine lactone acylases of Pseudomonas syringae influence colony and biofilm morphology. Appl Environ Microbiol. 2009;75:45–53. doi: 10.1128/AEM.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chan KG, Wong CS, Yin WF. et al. Rapid degradation of N-3-oxoacylhomoserine lactones by a Bacillus cereus isolate from Malaysian rainforest soil. Antonie Van Leeuwenhoek. 2010;98:299–305. doi: 10.1007/s10482-010-9438-0. [DOI] [PubMed] [Google Scholar]
- 189.Dong YH, Gusti AR, Zhang Q. et al. Identification of quorum-quenching N-acyl homoserine lactonases from Bacillus species. Appl Environ Microbiol. 2002;68:1754–1759. doi: 10.1128/AEM.68.4.1754-1759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Huma N, Shankar P, Kushwah J. et al. Diversity and polymorphism in AHL-lactonase gene (aiiA) of Bacillus. J Microbiol Biotechnol. 2011;21:1001–1011. doi: 10.4014/jmb.1105.05056. [DOI] [PubMed] [Google Scholar]
- 191.Liu D, Momb J, Thomas PW. et al. Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures. Biochemistry. 2008;47:7706–7714. doi: 10.1021/bi800368y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bentley SD, Chater KF, Cerdeno-Tarraga AM. et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 193.Liu D, Thomas PW, Momb J. et al. Structure and specificity of a quor-um-quenching lactonase (AiiB) from Agrobacterium tumefaciens. Biochemistry. 2007;46:11789–11799. doi: 10.1021/bi7012849. [DOI] [PubMed] [Google Scholar]
- 194.Riaz K, Elmerich C, Moreira D. et al. A metagenomic analysis of soil bacteria extends the diversity of quorum-quenching lactonases. Environ Microbi-ol. 2008;10:560–70. doi: 10.1111/j.1462-2920.2007.01475.x. [DOI] [PubMed] [Google Scholar]
- 195.Mei GY, Yan XX, Turak A. et al. AidH, an alpha/beta-hydrolase fold family member from an Ochrobactrum sp. strain, is a novel N-acylhomoserine lac-tonase. Appl Environ Microbiol. 2010;76:4933–4942. doi: 10.1128/AEM.00477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ogawa K, Nakajima-Kambe T, Nakahara T. et al. Coimmobilization of gluconolactonase with glucose oxidase for improvement in kinetic property of enzymatically induced volume collapse in ionic gels. Biomacromolecules. 2002;3:625–631. doi: 10.1021/bm025512f. [DOI] [PubMed] [Google Scholar]
- 197.Delalande L, Faure D, Raffoux A. et al. N-hexanoyl-L-homoserine lactone, a mediator of bacterial quorum-sensing regulation, exhibits plant-dependent stability and may be inactivated by germinating Lotus corniculatus seedlings. FEMS Microbiol Ecol. 2005;52:13–20. doi: 10.1016/j.femsec.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 198.Fatima Q, Zahin M, Khan MSA. et al. Modulation of quorum sensing con-trolled behavior of bacteria by growing seedling, seed and seedling extracts of leguminous plants. Ind J Microbiol. 2010;50:238–242. doi: 10.1007/s12088-010-0025-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Gotz C, Fekete A, Gebefuegi I. et al. Uptake, degradation and chiral discrimination of N-acyl-D/L-homoserine lactones by barley (Hordeum vulgare) and yam bean (Pachyrhizus erosus) plants. Anal Bioanal Chem. 2007;389:1447–1457. doi: 10.1007/s00216-007-1579-2. [DOI] [PubMed] [Google Scholar]
- 200.Sauer K, Camper AK. Characterization of phenotypic changes in Pseudomonas putida in response to surface associated growth. J Bacteriol. 2001;183:6579–6589. doi: 10.1128/JB.183.22.6579-6589.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Xu F, Byun T, Deussen HJ. et al. Degradation of N-acylhomoserine lactones, the bacterial quorum sensing molecules, by acylase. J Biotechnol. 2003;101:89–96. doi: 10.1016/s0168-1656(02)00305-x. [DOI] [PubMed] [Google Scholar]
- 202.Jiang W, Xia S, Liang J. et al. Effect of quorum quenching on the reactor performance, biofouling and biomass characteristics in membrane bioreactors. Water Res. 2013;47:187–196. doi: 10.1016/j.watres.2012.09.050. [DOI] [PubMed] [Google Scholar]
- 203.Kim JH, Choi DC, Yeon KM. et al. Enzyme-immobilized nanofiltration membrane to mitigate biofouling based on quorum quenching. Environ Sci Technol. 2011;45:1601–1607. doi: 10.1021/es103483j. [DOI] [PubMed] [Google Scholar]
- 204.Yeon KM, Lee CH, Kim J. Magnetic enzyme carrier for effective biofouling control in the membrane bioreactor based on enzymatic quorum quenching. Environ Sci Technol. 2009;43:7403–7409. doi: 10.1021/es901323k. [DOI] [PubMed] [Google Scholar]
- 205.Chen CN, Chen CJ, Liao CT. et al. A probable aculeacin a acylase from the Ralstonia solanacearum GMI1000 is N-acyl-homoserine lactone acylase with quorum-quenching activity. BMC Microbiol. 2009;9:89. doi: 10.1186/1471-2180-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Morohoshi T, Nakazawa S, Ebata A. et al. Identification and characterization of N-acylhomoserine lactone-acylase from the fish intestinal Shewanella sp. strain MIB015. Biosci Biotechnol Biochem. 2008;72:1887–1893. doi: 10.1271/bbb.80139. [DOI] [PubMed] [Google Scholar]
- 207.Chan KG, Atkinson S, Mathee K. et al. Characterization of N-acylhomoserine lactone-degrading bacteria associated with the Zingiber officinale (ginger) rhizosphere: Co-existence of quorum quenching and quorum sensing in Acinetobacter and Burkholderia. BMC Microbiol. 2011;11:51. doi: 10.1186/1471-2180-11-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Cheong WS, Lee CH, Moon YH. et al. Isolation and identification of indige-nous quorum quenching bacteria, Pesudomonas sp. 1A1, for biofouling control in MBR. Ind Eng Chem Res. 2013;52:10552–10560. [Google Scholar]
- 209.Chun CK, Ozer EA, Welsh MJ. et al. Inactivation of a Pseudomonas aeruginosa quorum-sensing signal by human airway epithelia. Proc Natl Acad Sci USA. 2004;101:3587–3590. doi: 10.1073/pnas.0308750101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wopperer J, Cardona ST, Huber B. et al. A quorum-quenching approach to investigate the conservation of quorum-sensing-regulated functions within the Burkholderia cepacia complex. Appl Environ Microbiol. 2006;72:1579–1587. doi: 10.1128/AEM.72.2.1579-1587.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ulrich RL. Quorum quenching: enzymatic disruption of N-acylhomoserine lactone-mediated bacterial communication in Burkholderia thailandensis. Appl Environ Microbiol. 2004;70:6173–6180. doi: 10.1128/AEM.70.10.6173-6180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lee SJ, Park SY, Lee JJ. et al. Genes encoding the N-acyl homoserine lac-tone-degrading enzyme are widespread in many subspecies of Bacillus thu-ringiensis. Appl Environ Microbiol. 2002;68:3919–3924. doi: 10.1128/AEM.68.8.3919-3924.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Carlier A, Uroz S, Smadja B. et al. The Ti plasmid of Agrobacterium tumefaciens harbors an attM-paralogous gene, aiiB, also encoding N-acylhomoserine lactonase activity. Appl Environ Microbiol. 2003;69:4989–4993. doi: 10.1128/AEM.69.8.4989-4993.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Chow JY, Xue B, Lee KH. et al. Directed evolution of a thermostable quor-um-quenching lactonase from the amidohydrolase superfamily. J Biol Chem. 2010;285:40911–40920. doi: 10.1074/jbc.M110.177139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wang WZ, Morohoshi T, Ikenoya M. et al. AiiM, a novel class of N-acylhomoserine lactonase from the leaf-associated bacterium Microbacte-rium testaceum. Appl Environ Microbiol. 2010;76:2524–2530. doi: 10.1128/AEM.02738-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Chow JY, Wu L, Yew WS. Directed evolution of a quorum-quenching lacto-nase from Mycobacterium avium subsp. paratuberculosis K-10 in the ami-dohydrolase superfamily. Biochemistry. 2009;48:4344–4353. doi: 10.1021/bi9004045. [DOI] [PubMed] [Google Scholar]
- 217.Afriat L, Roodveldt C, Manco G. et al. The latent promiscuity of newly identified microbial lactonases is linked to a recently diverged phosphotriesterase. Biochemistry. 2006;45:13677–13686. doi: 10.1021/bi061268r. [DOI] [PubMed] [Google Scholar]
- 218.Morohoshi T, Tominaga Y, Someya N. et al. Complete genome sequence and characterization of the N-acylhomoserine lactone-degrading gene of the po-tato leaf-associated Solibacillus silvestris. J Biosci Bioeng. 2012;113:20–25. doi: 10.1016/j.jbiosc.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 219.Merone L, Mandrich L, Rossi M. et al. A thermostable phosphotriesterase from the archaeon Sulfolobus solfataricus: Cloning, overexpression and properties. Extremophiles. 2005;9:297–305. doi: 10.1007/s00792-005-0445-4. [DOI] [PubMed] [Google Scholar]
- 220.Elias M, Dupuy J, Merone L. et al. Structural basis for natural lactonase and promiscuous phosphotriesterase activities. J Mol Biol. 2008;379:1017–1028. doi: 10.1016/j.jmb.2008.04.022. [DOI] [PubMed] [Google Scholar]
- 221.Uroz S, Oger PM, Chapelle E. et al. A Rhodococcus qsdA-encoded enzyme defines a novel class of large-spectrum quorum-quenching lactonases. Appl Environ Microbiol. 2008;74:1357–1366. doi: 10.1128/AEM.02014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Park SY, Lee SJ, Oh TK. et al. AhlD, an N-acylhomoserine lactonase in Arthrobacter sp, and predicted homologues in other bacteria. Microbiology. 2003;149:1541–1550. doi: 10.1099/mic.0.26269-0. [DOI] [PubMed] [Google Scholar]
- 223.Lee B, Yeon KM, Shim J. et al. Effective antifouling using quorum-quenching acylase stabilized in magnetically-separable mesoporous silica. Biomacro-molecules. 2014;15:1153–1159. doi: 10.1021/bm401595q. [DOI] [PubMed] [Google Scholar]
- 224.Basu S, Mehreja R, Thiberge S. et al. Spatiotemporal control of gene expression with pulse-generating networks. Proc Natl Acad Sci USA. 2004;101:6355–6360. doi: 10.1073/pnas.0307571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Jahangir D, Oh HS, Kim SR. et al. Specific location of encapsulated quorum quenching bacteria for biofouling control in an external submerged membrane bioreactor. J Memb Sci. 2012;411:130–136. [Google Scholar]